Abstract
Acetate kinase catalyzes the reversible magnesium-dependent synthesis of acetyl phosphate by transfer of the ATP γ-phosphoryl group to acetate. Inspection of the crystal structure of the Methanosarcina thermophila enzyme containing only ADP revealed a solvent-accessible hydrophobic pocket formed by residues Val93, Leu122, Phe179, and Pro232 in the active site cleft, which identified a potential acetate binding site. The hypothesis that this was a binding site was further supported by alignment of all acetate kinase sequences available from databases, which showed strict conservation of all four residues, and the recent crystal structure of the M. thermophila enzyme with acetate bound in this pocket. Replacement of each residue in the pocket produced variants with Km values for acetate that were 7- to 26-fold greater than that of the wild type, and perturbations of this binding pocket also altered the specificity for longer-chain carboxylic acids and acetyl phosphate. The kinetic analyses of variants combined with structural modeling indicated that the pocket has roles in binding the methyl group of acetate, influencing substrate specificity, and orienting the carboxyl group. The kinetic analyses also indicated that binding of acetyl phosphate is more dependent on interactions of the phosphate group with an unidentified residue than on interactions between the methyl group and the hydrophobic pocket. The analyses also indicated that Phe179 is essential for catalysis, possibly for domain closure. Alignments of acetate kinase, propionate kinase, and butyrate kinase sequences obtained from databases suggested that these enzymes have similar catalytic mechanisms and carboxylic acid substrate binding sites.
Acetate kinase catalyzes the reversible magnesium-dependent phosphorylation of acetate with ATP (equation 1) and is very important for the energy-yielding metabolism of anaerobic microbes.
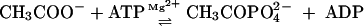 |
(1) |
In most fermentative anaerobes this enzyme is responsible for production of a major portion of the ATP (reverse of equation 1). Acetate kinase functions in the energy-yielding pathway for conversion of the methyl group of acetate to methane (equation 2) in Methanosarcina species.
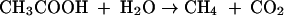 |
(2) |
The first reaction in this pathway is the formation of acetyl phosphate catalyzed by acetate kinase (equation 1). The phosphoryl group of acetyl phosphate is then displaced by coenzyme A (CoA), producing acetyl-CoA and orthophosphate, which is catalyzed by phosphotransacetylase (12, 36). Acetyl-CoA is subsequently cleaved to a methyl group, a carbonyl group, and CoA via the multisubunit carbon monoxide dehydrogenase/acetyl-CoA synthase (12). The methyl group is reduced to methane with electrons that originate from oxidation of the acetyl-CoA carbonyl group to CO2 (12). The proton gradient generated by a membrane-bound electron transport chain is utilized to drive ATP synthesis (12).
The reaction catalyzed by acetate kinase was initially described in 1944 by Lipmann (24). Following the first purification in 1954 from Escherichia coli (31), the enzyme was the subject of several investigations, yet questions concerning the catalytic mechanism and particularly the substrate binding sites have remained unanswered. Investigations with the E. coli enzyme have led to two proposals for the catalytic mechanism: direct in-line transfer of the γ-phosphoryl group of ATP to acetate (6, 34) and a covalent triple-displacement mechanism involving two phosphoenzyme intermediates (35). Although the phosphorylated E. coli acetate kinase is able to transfer the phosphoryl group to acetate (39, 40), it is not kinetically competent (6). Furthermore, the phosphoenzyme has been shown to phosphorylate enzyme I of the bacterial phosphotransferase system (13) and CheY (11), a member of the flagellar motor cascade; thus, the phosphoenzyme may function only in sugar transport and chemotaxis rather than play an essential role in the catalytic mechanism of equation 1. Finally, the acetate kinase of Methanosarcina thermophila has been shown to be inhibited by a putative transition state analogue, ADP-AlF3-acetate, in which the AlF3 is proposed to mimic the meta-phosphate in a direct phosphoryl transfer mechanism (26). The E. coli and M. thermophila acetate kinases exhibit 44% sequence identity, suggesting that they utilize similar catalytic mechanisms (32).
The first crystal structure reported for acetate kinase was that of the M. thermophila enzyme containing ADP bound in the active site cleft proximal to a solvent-accessible pocket of hydrophobic residues proposed to accept the methyl group of acetate or acetyl phosphate (8). A recent crystal structure of the M. thermophila acetate kinase containing acetate, ADP, and the transition state analog AlF3 further supported the hypothesis that there is a direct in-line mechanism and showed the methyl group of acetate located in the previously proposed binding pocket (14). Kinetic analyses of active site replacement variants also supported the hypothesis that there is a direct in-line mechanism for the M. thermophila enzyme and suggested roles for conserved arginine, histidine, and glutamate residues (16, 26, 27, 32, 33). Here we report kinetic analyses of variants designed to characterize the proposed hydrophobic binding pocket which indicated that the pocket has a role in both acetate binding and specificity. Finally, our results also suggested that the conserved Phe179 residue has a role in catalysis by contributing to domain movement.
MATERIALS AND METHODS
Materials.
Chemicals were purchased from Sigma Chemical, VWR Scientific Products, or Fisher Scientific. Oligonucleotides for DNA sequencing and site-directed mutagenesis were purchased from Integrated DNA Technologies (Coralville, Iowa). ATP solutions were adjusted to pH 7.0 with sodium hydroxide, and concentrations were determined by utilizing the extension coefficient. All ATP solutions were equimolar with magnesium chloride. Acetate, propionate, and butyrate stock solutions were adjusted to pH 7.0 with sodium hydroxide. Acetyl phosphate was made fresh daily.
Analysis of acetate, propionate, and butyrate kinase sequences.
The nonredundant protein and nucleotide sequence databases at the National Center for Biotechnology Information were searched for acetate, propionate, and butyrate kinase sequences by using the BLAST network server and the BLASTp and tBLASTn programs (3, 4). Sequences were aligned with ClustalX (38) by using a Gonnet PAM 250 weight matrix with a gap opening penalty of 10.0 and a gap extension penalty of 0.05.
Site-directed mutagenesis.
Mutagenesis was performed by the oligonucleotide-directed in vitro mutagenesis method (21) with a QuikChange mutagenesis kit (Stratagene). Plasmid pML703 (23), a derivative of the expression vector pT7-7 (37) containing the M. thermophila ack gene, was the target for mutagenesis with the primers listed in Table 1. Mutations were verified by dye termination cycle sequencing by using an ABI PRISM 377 DNA sequencer (Applied Biosystems) at the Nucleic Acid Facility at Pennsylvania State University.
TABLE 1.
Mutagenic oligonucleotide primers
| Variant | Sequencea |
|---|---|
| Val93 Ala | 5′ GTC GGA CAC AGA GTT GCG CAT GGT GGA GAG 3′ |
| 5′ CTC TCC ACC ATG CGC AAC TCT GTG TCC GAC 3′ | |
| Val93 Gly | 5′ GTC GGA CAC AGA GTT GGG CAT GGT GGA GAG 3′ |
| 5′ CTC TCC ACC ATG CCC AAC TCT GTG TCC GAC 3′ | |
| Leu122 Ala | 5′ TTT GAA CTG GCA CCC GCG CAC AAC CCT CCA 3′ |
| 5′ TGG AGG GTT GTG CGC GGG TGC CAG TTC AAA 3′ | |
| Phe179 Ala | 5′ GTC AGG AAA TAC GGT GCG CAC GGC ACA TCC 3′ |
| 5′ GGA TGT GCC GTG CGC ACC GTA TTT CCA GAC 3′ | |
| Pro232 Ala | 5′ AGC ATG GGC TTC ACA GCG CTT GAA GGG CTT 3′ |
| 5′ AAG CCC TTC AAG CGC TGT GAA GCC CAT GCT 3′ |
For each variant, the forward and reverse primer sequences are given. The mutation is underlined.
Heterologous production and purification of acetate kinases.
The wild-type and variant acetate kinases were overproduced in E. coli BL21(DE3) [F− dcm ompT hsdS (rB− mB−) gal λ(DE3)] and were purified as described previously (23). Protein purity was examined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (22), and protein concentrations were determined by the Bradford method (7) by using the Bio-Rad dye reagent with bovine serum albumin as the standard.
Molecular masses.
The native molecular masses of the wild-type and variant enzymes were determined by gel filtration chromatography by using a Superose 12 gel filtration column (Amersham Pharmacia Biotech) calibrated with blue dextran (2,000 kDa), urease (trimer, 272 kDa; hexamer, 545 kDa), bovine serum albumin (monomer, 66 kDa; dimer, 132 kDa), chicken egg albumin (45 kDa), bovine erythrocyte carbonic anhydrase (29 kDa), and bovine milk α-lactalbumin (14.2 kDa). The column was preequilibrated with 50 mM potassium phosphate (pH 6.8) containing 150 mM KCl and was developed at a flow rate of 0.4 ml/min.
Kinetic parameters of wild-type and variant acetate kinases.
The hydroxamate assay, an adaptation of the methods of Lipmann and Rose et al. (1, 24, 31), detects acetyl phosphate formation from acetate and ATP and was used to measure the initial acetate kinase activities and kinetic parameters in the forward (ADP/acetyl phosphate-producing) direction. By utilizing the pyruvate kinase (PK)-lactate dehydrogenase (LDH) coupled assay system (1), hydroxylamine was found to be an inhibitor of acetate kinase activity (14); therefore, kinetic parameters of wild-type and variant acetate kinases were determined by utilizing the PK-LDH coupled assay system. Briefly, each assay solution contained 60 mM HEPES (pH 7.0), 5 mM MgCl2, 16.7 U of PK, 36 U of LDH, 3 mM phosphoenolpyruvate, and 0.2 mM NADH along with a fixed concentration of substrate (200 mM acetate or 1 to 2 mM equimolar ATP-MgCl2) as appropriate. The wild-type or variant enzyme concentrations ranged from 0.5 to 50 μg/ml, depending upon the specific activity. Changes in absorbance at 340 nm were monitored for 1 to 5 min at 1-s intervals with a Beckman DU640 spectrophotometer. The Km and kcat values were determined by nonlinear regression data analysis fit to the Michaelis-Menten equation by using the Kaleidagraph program (Synergy Software, Reading, Pa.). The kinetic parameters reported below are averages of at least three independent trials, with kcat values independent of the variable substrate.
To determine kinetic parameters of the ATP-producing reaction, the previously described hexokinase-glucose 6-phosphatase enzyme-linked assay was used (1). Briefly, the assay mixtures contained 100 mM Tris (pH 7.4), 0.2 mM dithiothreitol, 10 mM MgCl2, 4.4 mM glucose, 1 mM NADP, 10 U of hexokinase (yeast), 10 U of glucose-6-phosphate dehydrogenase (yeast), 5 mM ADP, and different acetyl phosphate concentrations. Kinetic constants were determined by using nonlinear regression to fit data with the Kaleidagraph program (Synergy Software).
Kinetic parameters with the alternative substrates propionate and butyrate.
The abilities of the wild-type and variant acetate kinases to utilize the substrates propionate and butyrate in the propionyl or butyryl phosphate reaction direction were determined by utilizing the PK-LDH coupled assay system described above, with the ATP-MgCl2 concentration kept at 2 mM and various concentrations of propionate or butyrate as appropriate for the enzyme of interest.
Cavity volume determination and modeling of propionate and butyrate in the active site pocket.
Models of Val93Ala and Val93Gly variant enzymes were generated by utilizing O (17), and GROMOS96 energy minimization was performed (41). The volumes of the cavities in the wild-type, Val93Ala, and Val93Gly enzymes were determined by utilizing VOIDOO (19) with a 1.4-Å rolling sphere probe and a starting point of 32.0, 44.0, 55.8 (x, y, z). PDB, topology, parameter, and connectivity files for acetate, propionate, and butyrate were obtained from the HICUP website (18). Acetate, propionate, and butyrate were positioned in the cavities located in the wild-type, Val93Ala, and Val93Gly active sites, respectively, and GROMOS96 energy minimization was performed for each preparation (41).
RESULTS
Putative acetate binding pocket.
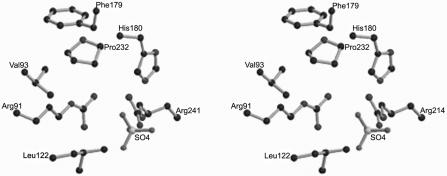
Located in the M. thermophila acetate kinase active site cleft are the residues Val93, Phe179, and Pro232, which form a hydrophobic pocket (Fig. 1) predicted to bind the methyl group of acetate (8, 14). Sequence alignments (data not shown) revealed that Val93, Phe179, and Pro232 of the M. thermophila enzyme are strictly conserved in all other acetate kinases obtained from the databases, implying that these residues have important roles, possibly in substrate binding. Although not previously proposed to be a member of the hydrophobic binding pocket (8), Leu122 was found to be highly conserved among all acetate kinase sequences obtained from the databases (data not shown) and to be located in the vicinity of the putative pocket (Fig. 1); thus, Leu122 is a potential member of the pocket.
FIG. 1.
Stereo view of the proposed acetate binding site in the acetate kinase from M. thermophila. The figure was generated with previously published coordinates (9) by using MOLSCRIPT (20) and Raster3D (25). All residues are labeled at the corresponding α carbons.
It was postulated previously that propionate kinase and butyrate kinase have hydrophobic pockets similar to that of acetate kinase that bind their respective substrates (8); thus, propionate kinase and butyrate kinase sequences obtained from the databases were aligned with the M. thermophila acetate kinase sequence (Fig. 2). The catalytically essential residues Arg91, Asp148, His180, Arg241, and Glu384 of the M. thermophila acetate kinase (1, 16, 23, 32, 33) were found to be strictly conserved in propionate kinase and butyrate kinase (Fig. 2), suggesting that the propionate kinase and butyrate kinase active site structures and catalytic mechanisms are similar to those of acetate kinase. Although hydrophobic residues were found in the propionate kinase and butyrate kinase sequences at positions equivalent to the positions of the acetate kinase hydrophobic pocket residues (Val93, Phe179, and Pro232), the conservation was less strict than that for the catalytically essential residues of acetate kinase. The less bulky residues alanine and serine were found in the propionate kinase sequences, and a glycine was found in all the butyrate kinase sequences at positions equivalent to the position of Val93 in acetate kinase (Fig. 2). The residues in the propionate kinase and butyrate kinase sequences at positions equivalent to the position of acetate kinase Phe179 were found to be strictly conserved except for the Enterococcus faecalis butyrate kinase sequence, in which the equivalent residue was glycine. Although leucine was identified in all the propionate kinase sequences at the position equivalent to the position of Leu122 in acetate kinase, alanine was found in the equivalent position in all butyrate kinase sequences (Fig. 2). Finally, the acetate kinase Pro232 residue was found to be strictly conserved at the equivalent position in the propionate kinases; however, glycine was identified at the position equivalent to the position of Pro232 in all the butyrate kinase sequences.
FIG. 2.
Alignment of acetate kinase, propionate kinase, and butyrate kinase sequences. The deduced amino acid sequences of the enzymes were aligned by using ClustalX (38). Residues essential for acetate kinase activity (number signs) and for the hydrophobic pocket (percent signs) are indicated above the alignment. Identical (asterisks) and similar (colons or periods) residues in the sequences are indicated below the alignment. The values in parentheses are the levels of identity followed by the levels of similarity (expressed as percentages) to the acetate kinase from M. thermophila. Abbreviations: MT AK, M. thermophila acetate kinase (accession number gi:584720); LC PK, Lactobacillus collinoides propionate kinase (gi:29335735); SE PK, S. enterica subsp. enterica serovar Typhimurium propionate kinase (gi:5069465); EC PK, E. coli propionate kinase (gi:1176151); CB BK1, Clostridium acetobutylicum butyrate kinase I (gi:20137334); CP BK, Clostridium perfringens butyrate kinase (gi:4239872); CB BK2, C. acetobutylicum butyrate kinase II (gi:20137415); TT BK, Thermoanaerobacter tengcongensis butyrate kinase (gi:20517209); EF BK, Enterococcus faecalis butyrate kinase (gi:20137247).
Purification of wild-type and variant acetate kinases.
The residues Val93, Leu122, Phe179, and Pro232 were subjected to site-specific replacement to test the hypothesis that they are involved in acetate binding. The wild-type and variant enzymes were overexpressed in E. coli and purified to apparent homogeneity, as judged by SDS-PAGE (data not shown). The yields of the purified variants were similar to that of the wild-type enzyme, and the subunit molecular masses of the variants were indistinguishable from that of the wild type, as determined by SDS-PAGE (data not shown). Native gel filtration chromatography indicated that the variants were dimeric, like the wild-type enzyme (data not shown). These results indicated that the purified variants were not compromised by major structural changes. A Phe179Leu variant was produced at high levels in E. coli based on the SDS-PAGE analysis; however, no acetate kinase activity was detected in the cell extracts. This variant also displayed markedly different chromatographic properties than the wild type during purification attempts, suggesting that there was improper folding.
Kinetic parameters of wild-type and variant acetate kinases.
The kinetic constants for the wild-type and variant enzymes assayed in the direction of acetyl phosphate synthesis are shown in Table 2. All variants showed less than twofold decreases in the Km for ATP relative to the wild-type enzyme, further supporting the hypothesis that the overall structure of the active site was not affected in the variant enzymes. However, all of the variants showed increases in the Km for acetate relative to the wild type; these increases ranged from 7-fold for the Phe179Ala variant to 26-fold for the Val93Gly variant. The increases in the Km for acetate for the variants indicate that these residues contribute to acetate affinity. The kcat values decreased 8-fold or less relative to the wild type for all variants, except for a 480-fold decrease for Phe179Ala. The kinetic parameters for acetyl phosphate (Table 3) were considerably less affected than the kinetic parameters for acetate by replacements in the hydrophobic pocket (Table 2). The values for the Km for acetyl phosphate ranged from 1.5-fold (Leu122Ala) to 3.5-fold (Pro232Ala) greater than the wild-type value, with the exception of Phe179Ala, which showed a 2.3-fold decrease in the Km. The moderate changes in the Km suggest that factors other than the putative hydrophobic pocket residues contribute to acetyl phosphate affinity. While kcat changed less than 2- to 3-fold for the majority of variants, the Phe179Ala variant showed 233- and 479-fold decreases in kcat for the acetate- and acetyl phosphate-forming directions, respectively, suggesting that this residue may have a catalytic role. While substantial increases in the Km for acetate were observed for all the variants relative to the wild type (Table 2), there was relatively little change in the Km values for propionate and butyrate (Table 4). In contrast, the kcat values for the Val93Ala and Val93Gly variants with propionate and butyrate increased relative to the wild type, while the kcat values for the same variants decreased with acetate (Table 2), indicating that the relationship between pocket size and substrate size correlates with the rate of catalysis. While a Km value is only an approximation of an enzyme's affinity for a substrate, in the absence of true dissociation constants, it can provide useful information.
TABLE 2.
Kinetic parameters of wild-type and variant acetate kinases assayed in the direction of acetyl phosphate synthesis
| Enzyme | kcat (s−1) | Acetate
|
ATP
|
||
|---|---|---|---|---|---|
| Km (mM) | kcat/Km (s−1 mM−1) | Km (μM) | kcat/Km (s−1 μM−1) | ||
| Wild type | 1,055 ± 57 | 1.5 ± 0.16 | 703 ± 84 | 71.3 ± 7.0 | 14.8 ± 1.7 |
| Val93 Ala | 844 ± 34 | 21.4 ± 1.8 | 39 ± 2.0 | 49.9 ± 3.3 | 16.9 ± 1.3 |
| Val93 Gly | 347 ± 21 | 39.5 ± 1.7 | 8.8 ± 0.7 | 68.7 ± 1.9 | 5.1 ± 0.3 |
| Leu122 Ala | 150 ± 12 | 19.5 ± 1.8 | 7.7 ± 0.9 | 57.6 ± 2.7 | 2.6 ± 0.24 |
| Phe179 Ala | 2.2 ± 0.3 | 10.4 ± 1.1 | 0.21 ± 0.04 | 50.5 ± 0.6 | 0.043 ± 0.006 |
| Pro232 Ala | 132 ± 2 | 32.0 ± 2.6 | 4.1 ± 0.3 | 54.2 ± 1.5 | 2.4 ± 0.08 |
TABLE 3.
Kinetic parameters for wild-type and variant acetate kinases assayed in the direction of ATP synthesis
| Enzyme | Acetyl phosphate
|
||
|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | |
| Wild type | 2,680 ± 45 | 0.34 ± 0.01 | 7,882 ± 267 |
| Val93 Ala | 3,869 ± 85 | 0.74 ± 0.03 | 5,228 ± 241 |
| Val93 Gly | 2,572 ± 44 | 0.71 ± 0.04 | 3,623 ± 213 |
| Leu122 Ala | 1,039 ± 5 | 0.50 ± 0.01 | 2,078 ± 43 |
| Phe179 Ala | 12 ± 1 | 0.15 ± 0.01 | 80 ± 9 |
| Pro232 Ala | 2,396 ± 7 | 1.19 ± 0.02 | 2,013 ± 34 |
TABLE 4.
Kinetic parameters for wild-type and variant acetate kinases assayed in the direction of propionyl phosphate and butyryl phosphate synthesis
| Enzyme | Propionate
|
Butyrate
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | |
| Wild type | 218 ± 15 | 14.4 ± 1.4 | 24 ± 5.4 | 0.18 ± 0.016 | 39 ± 13 | 0.005 ± 0.002 |
| Val93 Ala | 1,029 ± 45 | 6.2 ± 0.9 | 165 ± 25 | 42.4 ± 9.0 | 33.4 ± 3.5 | 1.26 ± 0.3 |
| Val93 Gly | 840 ± 3 | 25.0 ± 0.6 | 33.6 ± 0.8 | 294 ± 21 | 63 ± 6.0 | 4.6 ± 0.3 |
| Leu122 Ala | 6.7 ± 0.3 | 10.7 ± 1.4 | 0.63 ± 0.09 | NDa | ND | ND |
| Phe179 Ala | 0.37 ± 0.01 | 11 ± 0.8 | 0.033 ± 0.003 | ND | ND | ND |
| Pro232 Ala | 8.5 ± 0.3 | 46 ± 3 | 0.19 ± 0.01 | ND | ND | ND |
ND, activity was below the detection limit (change in absorbance at 340 nm of <0.0001 U/min).
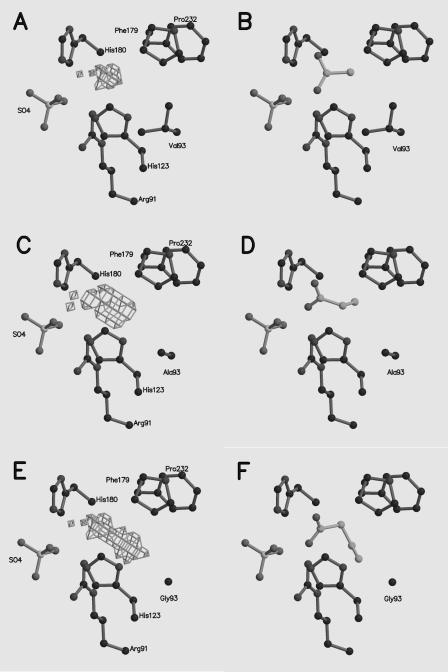
Modeling of wild-type and variant acetate kinases.
With the improved catalytic efficiency of the Val93Ala and Val93Gly variants with propionate and butyrate relative to the wild type, molecular models of the variant acetate kinases were constructed to determine if the hydrophobic cavities could accommodate substrates in support of the kinetic results. Masks of the cavities determined for the wild type, the Val93Ala variant, and the Val93Gly variant are shown in Fig. 3A, C, and E. The volumes determined for the wild type, Val93Ala variant, and Val93Gly variant hydrophobic cavities were 35, 43, and 61 Å3, respectively, indicating that there was an increase in the pocket size as the size of the position 93 side chain decreased, which is consistent with the kinetic results. Propionate was successfully modeled in the cavity of the wild-type structure, whereas butyrate was not. These results are consistent with substantial activity of the wild-type enzyme with propionate and minimal activity with butyrate. Both propionate and butyrate were successfully modeled in the cavity of the Val93Ala variant, which is consistent with the ability of this variant to utilize both substrates. Figure 3B, D, and F shows the results for modeling with the substrate yielding the greatest kcat for each enzyme. The results show the aliphatic carbons accommodated by the hydrophobic cavities of the respective enzymes with the carboxyl group approximately 3.5 Å from Arg91 and oriented toward the sulfate ion in the modeled structures.
FIG. 3.
Solvent-accessible cavities in wild-type acetate kinase and the modeled Val93Ala and Val93Gly variants. (A, C, and E) Wild-type acetate kinase and the Val93Ala and Val93Gly variants, respectively, with the active site cavity volume masks. (B, D, and F) Acetate, propionate, and butyrate modeled into wild-type acetate kinase and the Val93Ala and Val93Gly variants, respectively. All diagrams were generated by using MOLSCRIPT (20) and Raster3D (25).
DISCUSSION
The acetate kinase variants reported here all exhibited severalfold increases in the Km for acetate, supporting the hypothesized roles for the hydrophobic pocket residues Val93, Leu122, Phe179, and Pro232 as important contributors to acetate binding. The progressive increase in Km for acetate with a decrease in the side chain volume at position 93 supports the hypothesis that there is a decrease in hydrophobic interactions with acetate as the pocket size increases. Furthermore, the overall hydrophobicity of the pocket is decreased in the variants. Attempts to measure the Kd values of the variants for acetate and acetyl phosphate have been unsuccessful so far due to the upper solubility limit of acetate kinase (20 μM) relative to the Km values of the variants for acetate and acetyl phosphate, which were as high as 39.5 and 1.19 mM, respectively. The recently reported crystal structure of acetate kinase from M. thermophila containing acetate shows the methyl group located in the hydrophobic pocket (14). The kinetic results reported here are consistent with the crystal structure and further suggest that acetate is in a catalytically competent location in the structure.
The results obtained from modeling acetate in the previously published wild-type acetate kinase structure were consistent with the kinetic results, suggesting that the hydrophobic pocket has a role in binding the methyl group of acetate. The model also showed the carboxyl group of acetate pointing toward the sulfate ion which is the proposed site for the γ phosphate of ATP (8). Furthermore, the carboxyl group of acetate in the model is poised to interact with the guanidinium cation of Arg91 previously proposed to facilitate the binding of acetate (14, 33). The modeling results are consistent with the recently determined crystal structure of M. thermophila acetate kinase containing ADP, acetate, and the transition state analog AlF3 (14). In this structure, acetate is bound with the methyl group in the pocket and the carboxyl group pointing toward AlF3, a proposed transition state analog of the γ phosphate of ATP, supporting the hypothesis that there is a direct in-line mechanism. The kinetic and modeling results presented here are consistent with this mechanism.
In addition to an essential role for the hydrophobic pocket in binding acetate, the results suggest that the size of the pocket is a determinant of substrate specificity and also is important for catalysis. As the side chain volume decreased in the position 93 variants, the kcat increased for propionate and butyrate relative to the wild type, in accord with the modeling results. Notably, the kcat of the Val93Gly variant with butyrate was 1,600-fold greater than that for the wild type. Furthermore, the kcat for the Val93Ala variant with propionate was equivalent to that for the wild-type enzyme with acetate, although the catalytic efficiency of the former was lower due to the slightly larger Km for propionate. As kinetic studies have not been performed with any propionate kinase, the Km values and catalytic efficiencies cannot be compared. These results suggest that while hydrophobic interactions appear to be the main determinant of affinity for acetate, specificity for acetate is through limiting the size of the hydrophobic pocket, determined in large part by Val93 located in the floor of the pocket. These results also suggest that the side chain size at position 93 is important for catalysis in the direction of acetyl phosphate synthesis, most likely due to positioning of the carboxyl group of acetate in proximity to the γ-phosphate of ATP for nucleophilic attack. However, the kcat values for the Leu122Ala, Phe179Ala, and Pro232Ala variants with acetate and propionate were substantially lower than the values for the position 93 variants, and no activity was detected with butyrate. These results suggest that the Leu122, Phe179, and Pro232 residues have greater impact than Val93 on substrate positioning and catalysis. The wild-type M. thermophila acetate kinase had substantial propionate kinase activity, which may have physiological relevance. It was recently reported that pduW encodes propionate kinase (29) in Salmonella enterica and that acetate kinase is able to function in propionate metabolism in the absence of pduW.
If the affinity of acetate kinase for acetyl phosphate occurs through the same hydrophobic interactions as the affinity for acetate, then similar changes would be expected in the Km values for acetate and propionate for the variants. However, the increases in the Km for acetyl phosphate were severalfold less than the increases in the Km for acetate for all of the variants, suggesting that the affinity of acetyl phosphate is mediated to a greater extent by interaction with the phosphoryl group than by hydrophobic interactions with the methyl group. This interpretation is further supported by the fivefold-lower Km for acetyl phosphate than for acetate for the wild-type enzyme. Except for the Phe179Ala variant, the kcat for the variants changed less in the direction of ATP synthesis than in the direction of acetyl phosphate synthesis, which is consistent with less influence for the hydrophobic pocket on positioning acetyl phosphate for optimum catalysis. Although Arg91 was hypothesized to interact with the phosphoryl group of acetyl phosphate based upon the crystal structure complexed with ADP (8), the kinetic analysis results for variants are inconsistent with this role for Arg91 (26, 33; unpublished data); thus, the residue interacting with acetyl phosphate remains unknown. Neither propionyl phosphate nor butyryl phosphate is commercially available; thus, the effect of the pocket size on the specificity for the phosphorylated substrates has not been tested.
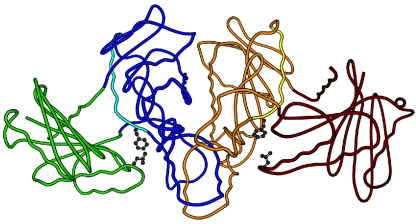
Members of the ASKHA superfamily are known to undergo domain closure which is required for catalysis (15, 28, 30). The catalytically essential Phe179 residue, which is conserved in all three kinases (Fig. 2), is located at the N terminus of helix α3, which is at the bridge between domains I and II (8) (Fig. 4). The ASKHA family members glycerol kinase (10, 30), hexokinase (2), and phosphoglycerate kinase (5) have a conserved glycine residue in their equivalent helices (α3 in glycerol kinase and hexokinase, α7 in phosphoglycerate kinase) that is postulated to be part of the hinge at which the catalytically essential domain closure occurs (2, 5, 10, 30). In glycerol kinase and phosphoglycerate kinase a phenylalanine immediately follows this hinge glycine, and this motif is also present in the acetate kinase and propionate kinase sequences (Fig. 2). Therefore, this Gly-Phe pair may identify a previously unrecognized conserved motif required for domain motion in the ASKHA superfamily. Phe179 likely participates in the closure of domain II down onto domain I by creating a greasy hydrophobic patch that allows the sliding shear action required for movement (10, 15). The Leu122Ala variant also lies at the domain I-domain II interface (Fig. 4), implying that it contributes to the proposed hydrophobic patch. Indeed, the Leu122Ala variant showed a reduction in kcat relative to the wild type in the direction of ATP synthesis, which is consistent with a role in addition to the proposed positioning of the carboxyl group of acetate for optimal catalysis in the direction of acetyl phosphate synthesis.
FIG. 4.
Overall structure of acetate kinase, highlighting Phe179 and Leu122 in the hinge region. Monomer A is blue and green (domain I, green; domain II, blue), monomer B is orange and red (domain I, red; domain II, orange), and the α3 helices are cyan (monomer A) and yellow (monomer B). Phe179 and Leu122 are shown in ball-and-stick form. The figure was generated by using MOLSCRIPT (20) and Raster3D (25).
Comparisons of acetate kinase, propionate kinase, and butyrate kinase sequences resulted in identification of conserved residues that are consistent with a catalytic mechanism for propionate kinase and butyrate kinase similar to that of acetate kinase. This conservation with acetate kinase was found to extend to hydrophobic pocket residues of propionate kinase and butyrate kinase, with exceptions that are consistent with the hypothesized role for the pockets in binding the alkyl groups of the respective carboxylic acid substrates (8). Exceptions to conservation with the acetate kinase pocket residues were found for Val93 in the floor of the pocket, where the equivalent position contained alanine or serine in propionate kinase and glycine in butyrate kinase. This replacement with progressively smaller residues in the propionate kinases and butyrate kinases supports the role proposed for the hydrophobic pockets in view of kinetic results obtained with the acetate kinase Val93 variants, in which replacement with progressively smaller residues resulted in increased kcat values for propionate and butyrate relative to the wild type. The modeling results further support the hypothesis that substitution at the position equivalent to Val93 allows accommodation of propionate and butyrate, strengthening the proposed role of the hydrophobic pockets in propionate kinase and butyrate kinase. However, the relatively higher Km for butyrate and lower kcat with butyrate for both acetate kinase Val93 variants suggest that the butyrate kinase active site architecture may deviate from the architecture of propionate kinase and acetate kinase.
Conclusions.
Kinetic analyses of acetate kinase variants have resulted in identification of residues that form a hydrophobic pocket and are important for binding acetate. The results further suggest that the size of the pocket is an important determinant of substrate specificity and support the hypothesis that the acetate identified in the recently reported crystal structure (14) is located in the catalytically competent position. Furthermore, the results reported here and the recently reported crystal structure both support a direct in-line mechanism. Finally, we identified a previously unrecognized catalytically essential residue, Phe179, and hypothesize that this residue has a role in domain closure during catalysis.
Acknowledgments
This work was supported by NIH grant GM44661 to J.G.F.
REFERENCES
- 1.Aceti, D. J., and J. G. Ferry. 1988. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. Evidence for regulation of synthesis. J. Biol. Chem. 263:15444-15448. [PubMed] [Google Scholar]
- 2.Aleshin, A. E., C. Zeng, G. P. Bourenkov, H. D. Bartunik, H. J. Fromm, and R. B. Honzatko. 1998. The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate. Structure 6:39-50. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipmann. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbach, G., R. Huber, M. Grattinger, K. Zaiss, H. Schurig, R. Jaenicke, and U. Jacob. 1997. Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability. Structure 5:1475-1483. [DOI] [PubMed] [Google Scholar]
- 6.Blattler, W. A., and J. R. Knowles. 1979. Stereochemical course of phosphokinases. The use of adenosine [gamma-(S)-16O,17O,18O]triphosphate and the mechanistic consequences for the reactions catalyzed by glycerol kinase, hexokinase, pyruvate kinase, and acetate kinase. Biochemistry 18:3927-3933. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Buss, K. A., D. R. Cooper, C. Ingram-Smith, J. G. Ferry, D. A. Sanders, and M. S. Hasson. 2001. Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 183:680-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buss, K. A., C. Ingram-Smith, J. G. Ferry, D. A. Sanders, and M. S. Hasson. 1997. Crystallization of acetate kinase from Methanosarcina thermophila and prediction of its fold. Protein Sci. 6:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bystrom, C. E., D. W. Pettigrew, B. P. Branchaud, P. O'Brien, and S. J. Remington. 1999. Crystal structures of Escherichia coli glycerol kinase variant S58→W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion. Biochemistry 38:3508-3518. [DOI] [PubMed] [Google Scholar]
- 11.Dailey, F. E., and H. C. Berg. 1993. Change in direction of flagellar rotation in Escherichia coli mediated by acetate kinase. J. Bacteriol. 175:3236-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferry, J. G. 1997. Enzymology of the fermentation of acetate to methane by Methanosarcina thermophila. Biofactors 6:25-35. [DOI] [PubMed] [Google Scholar]
- 13.Fox, D. K., N. D. Meadow, and S. Roseman. 1986. Phosphate transfer between acetate kinase and enzyme I of the bacterial phosphotransferase system. J. Biol. Chem. 261:13498-13503. [PubMed] [Google Scholar]
- 14.Gorrell, A., S. H. Lawrence, and J. G. Ferry. Structural and kinetic analyses of arginine residues in the active-site of the acetate kinase from Methanosarcina thermophila. J. Biol. Chem., in press. [DOI] [PubMed]
- 15.Hurley, J. H. 1996. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 25:137-162. [DOI] [PubMed] [Google Scholar]
- 16.Ingram-Smith, C., R. D. Barber, and J. G. Ferry. 2000. The role of histidines in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 275:33765-33770. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Kleywegt, G. J., and T. A. Jones. 1998. Databases in protein crystallography. Acta Crystallogr. Sect. D 54:1119-1131. [DOI] [PubMed] [Google Scholar]
- 19.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. Sect. D 50:178-185. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 21.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Latimer, M. T., and J. G. Ferry. 1993. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J. Bacteriol. 175:6822-6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipmann, F. 1944. Enzymatic synthesis of acetyl phosphate. J. Biol. Chem. 155:55-70. [Google Scholar]
- 25.Merritt, E., and D. Bacon. 1997. Raster3D photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 26.Miles, R. D., A. Gorrell, and J. G. Ferry. 2002. Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 277:22547-22552. [DOI] [PubMed] [Google Scholar]
- 27.Miles, R. D., P. P. Iyer, and J. G. Ferry. 2001. Site-directed mutational analysis of active site residues in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 276:45059-45064. [DOI] [PubMed] [Google Scholar]
- 28.Page, R., U. Lindberg, and C. E. Schutt. 1998. Domain motions in actin. J. Mol. Biol. 280:463-474. [DOI] [PubMed] [Google Scholar]
- 29.Palacios, S., V. J. Starai, and J. C. Escalante-Semerena. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettigrew, D. W., G. B. Smith, K. P. Thomas, and D. C. Dodds. 1998. Conserved active site aspartates and domain-domain interactions in regulatory properties of the sugar kinase superfamily. Arch. Biochem. Biophys. 349:236-245. [DOI] [PubMed] [Google Scholar]
- 31.Rose, I. A., M. Grunberg-Manago, S. R. Korey, and S. Ochoa. 1954. Enzymatic phosphorylation of acetate. J. Biol. Chem. 211:737-756. [PubMed] [Google Scholar]
- 32.Singh-Wissmann, K., C. Ingram-Smith, R. D. Miles, and J. G. Ferry. 1998. Identification of essential glutamates in the acetate kinase from Methanosarcina thermophila. J. Bacteriol. 180:1129-1134. (Erratum, J. Bacteriol. 180:3018.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh-Wissmann, K., R. D. Miles, C. Ingram-Smith, and J. G. Ferry. 2000. Identification of essential arginines in the acetate kinase from Methanosarcina thermophila. Biochemistry 39:3671-3677. [DOI] [PubMed] [Google Scholar]
- 34.Skarstedt, M. T., and E. Silverstein. 1976. Escherichia coli acetate kinase mechanism studied by net initial rate, equilibrium, and independent isotopic exchange kinetics. J. Biol. Chem. 251:6775-6783. [PubMed] [Google Scholar]
- 35.Spector, L. B. 1980. Acetate kinase: a triple-displacement enzyme. Proc. Natl. Acad. Sci. USA 77:2626-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadtman, E. (ed.). 1955. Phosphotransacetylase from Clostridium kluyveri, vol. 1. Academic Press, New York, N.Y.
- 37.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todhunter, J. A., and D. L. Purich. 1974. Evidence for the formation of a gamma-phosphorylated glutamyl residue in the Escherichia coli acetate kinase reaction. Biochem. Biophys. Res. Commun. 60:273-280. [DOI] [PubMed] [Google Scholar]
- 40.Todhunter, J. A., K. B. Reichel, and D. L. Purich. 1976. A kinetically important phosphoryl-enzyme intermediary in the intrinsic purine nucleoside-5′-diphosphokinase activity of Escherichia coli acetate kinase. Arch. Biochem. Biophys. 174:120-128. [DOI] [PubMed] [Google Scholar]
- 41.van Gunsteren, W., X. Daura, and A. Mark. 1998. GROMOS force field. Encycl. Comput. Chem. 2:1211-1216. [Google Scholar]