Abstract
Cohesive ends of 16-3, a temperate phage of Rhizobium meliloti 41, have been identified as 10-base-long, 3′-protruding complementary G/C-rich sequences. terS and terL encode the two subunits of 16-3 terminase. Significant homologies were detected among the terminase subunits of phage 16-3 and other phages from various ecosystems.
The terminase enzyme is part of a large nucleoprotein complex that packages viral DNA into the capsid (4, 16). It has been well studied in the case of phage λ, but quite a few other members have been identified in different phages and studied in detail (6). Studying the packaging reaction of new phages provides the opportunity to identify alternative mechanisms and can extend our understanding of the packaging process and the formation and functioning of nucleoprotein complexes in general. Studies of similar functions in diverse systems are also required to understand the evolution of complex biological machines.
Phage 16-3 is a temperate phage of Rhizobium meliloti 41. The genetic and physical maps of the phage have been established previously (9, 11, 13, 22) and summarized in reference 7. Genes, proteins, and chromosomal sites for several functions of the phage have been studied in more detail. These include (i) the main repressor protein, C, required for establishing and maintaining lysogeny, and the operator regions where the C protein binds (8, 10, 25); (ii) the components of the site-specific recombination system (5, 12, 23, 29, 30); (iii) the immX regulatory region conferring immunity against homoimmune phages (7); and (iv) the recently identified h gene encoding the tail fiber protein (26).
Identification of the cohesive ends of phage 16-3
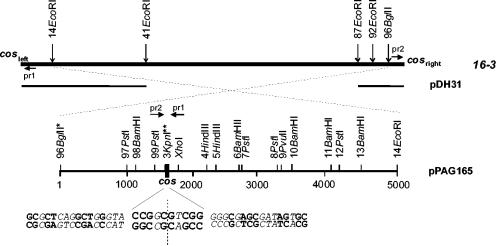
The plasmids used during the course of the study are listed in Table 1. The 96BglII-14EcoRI fragment (Fig. 1) (numbering was based on the map of restriction sites in reference 13) of phage 16-3 was subcloned from cosmid pDH31 (11), which contains the region where the covalently ligated ends of the phage chromosome are located. The resulting plasmid, pPAG165, was used to determine the nucleotide sequence of the entire fragment. Nucleotide sequence determination was performed by the dideoxy chain termination method (28) with the fmol DNA cycle sequencing system (Promega). Primer 1, 5′-CACGGCTTCGGCGGCGCTGTC-3′, and primer 2, 5′-GGCAAGAAGGTCGTGACCTATG-3′, were designed close to the expected ends, and the isolated 92EcoRI-cosright and cosleft-14EcoRI fragments from phage DNA were used as templates for a pair of sequencing reactions to distinguish between 5′ and 3′ overhangs (Fig. 2). A 10-bp sequence region existing in pPAG165 was absent from the sequences of both end-fragments, hence we concluded that the chromosome of phage 16-3 ended in 10-base-long, 3′-protruding, single-stranded, complementary sequences (5′-…CCGGCGTCGG-3′ and 3′-GGCCGCAGCC…-5′). The cos sequence has high G/C content and shows dyad symmetry. The symmetry can be recognized in patches within the duplex DNA flanking the single-stranded ends (Fig. 1).
TABLE 1.
Plasmids used in the studies
| Plasmid | Relevant features | Reference |
|---|---|---|
| pRK2013 | Helper in conjugative transfer of pBBR1MCS-5 derivative plasmids | 15 |
| pCU101 | Helper in conjugative transfer of plasmid with pCU1 mob | 33 |
| pBBR1MCS-5 | Derivative of the broad-host-range cloning vector pBBR1MCS; Gmr; replicate in Escherichia coli and R. meliloti | 19 |
| pDH31 | Cosmid clone, contains the 25-kb 87EcoRI-41EcoRI region of phage 16-3 | 11 |
| pCU999 | Source of kanamycin cassette | 24 |
| pCU996Ω | Source of spectinomycin cassette (Ω) | 2 |
| pSEM155 | pSC101 replicon, pCU1 mob, attP, Kmr | 14 |
| pPAG160 | attP was deleted from pSEM155, and the Km cassette was replaced by the spectinomycin cassette, derived from pCU996Ω | This work |
| pPAG165 | 96BglII-14EcoRI fragment (nt 1-5006) cloned in pPAG160 | This work |
| pPAG168 | pPAG165 derivative, Km cassette replaced codons 34-81 in ORF525 | This work |
| pPAG169 | pPAG165 derivative, Km cassette inserted at codon 59 in ORF154 | This work |
| pPAG185 | pPAG165 derivative, Km cassette inserted at codon 56 in ORF171 | This work |
| pPAG201 | Similar to pPAG165 but contains the 6-kb 94BamHI-14EcoRI region and the Km cassette replaced codons 60-75 in ORF249 | This work |
| pCS473 | pBBR1MCS-5 containing the region from nt 1650-2466 including ORF154 | This work |
| pCS474 | pBBR1MCS-5 containing from nt 1823-3984 including ORF525 | This work |
| pCS475 | pBBR1MCS-5 containing from nt 1-932 including ORF249 | This work |
FIG. 1.
Physical map of phage 16-3 surrounding the cos region. The genome of phage 16-3 is indicated by the top bar. Only the phage content of the cosmid clone pDH31 and plasmid pPAG165 are shown. Numbered restriction sites were published previously (11). *, indicates that the BglII site was lost in pPAG165 because of cloning; **, indicates that the KpnI site, previously placed for the left end-fragment, was found on the right end-fragment. The sequence of the cos site (nt 1597 to 1606) and its flanking regions are shown on the bottom, letters in italics indicate positions with a mismatch within the palindrome, the dotted line indicates the axis of dyad symmetry. Location of primers (pr1 and pr2) used for sequence determination of the ends of the phage are also shown.
FIG. 2.
Principle used to distinguish between 5′ (A) and 3′ (B) overhangs of the cohesive ends. The sequence of the cohesive ends would appear in both end-fragment sequences if the phage has 5′ overhangs, while it would appear in neither sequence in the case of 3′ protrusion. End-fragment sequences were compared to the sequence of the covalently closed cos site carried by pPAG165.
Identification of genes around the cos region essential for phage production
Since the coding sequence of a DNA binding protein and its target site are often found in close proximity in prokaryotic systems, we expected the terminase genes to be located in the vicinity of cos. ORF171 and ORF249 on the right end and ORF154 and ORF525 on the left end of the phage genome were identified as candidate genes and tested for terminase function.
To investigate the roles of the different open reading frames (ORFs), kanamycin (Km) cassette insertional mutagenesis was applied. We used a modified version of our previously developed procedure (7) which utilized homologous recombination for inserting a Km cassette, derived from pCU999 (24), into the genome of R. meliloti lysogenic for 16-3cti3 prophage. The 1.2-kb Km cassette was usually inserted at desired locations, but in some cases, due to difficulties in cloning or maintenance of the clone, small deletions were generated in the target sequence simultaneously with the insertion of the cassette. The deletion end points for the relevant plasmids are recorded in Table 1. Proper insertion of the Km cassette was detected in each case by PCRs.
Plasmids pPAG201, pPAG185, pPAG169, and pPAG168 were constructed and contained the kanamycin cassette in ORF249, ORF171, ORF154, and ORF525, respectively. The plasmids were introduced into the lysogenic strain R. meliloti 41(16-3cti3) by triparental mating. Growth conditions, media, and conditions for triparental mating have been described previously (29). Since these plasmids were not able to replicate in R. meliloti, Km-resistant colonies indicated homologous recombination between the plasmid and the prophage either with one crossover (resulted in the integration of the entire plasmid into the prophage) or with two crossovers (resulting in the insertion of the Km cassette into the targeted ORF of the prophage). Selection for Km-resistant but streptomycin-sensitive (antibiotic resistance of the vector part of the plasmids) colonies resulted in lysogenic R. meliloti strains with prophages 16-3cti3Km-249, 16-3cti3Km-171, 16-3cti3Km-154, and 16-3cti3Km-525. That is, the prophages contained the Km gene in ORF249, ORF171, ORF154, and ORF525, respectively. Primers complementary to the Km gene and to the different ORFs were used in PCR tests to confirm the structures of the insertion alleles (data not shown).
Due to the thermoinducible mutation (ti3) in the c gene (required to maintain the lysogenic state) of the prophages, shifting the temperature from 28 to 37°C resulted in prophage excision. Overnight cultures of lysogenic R. meliloti strains were diluted 40-fold in fresh yeast-tryptone broth (YTB) medium and shaken at 28°C until the optical density at 600 nm reached 0.3 to 0.4. The cells were then resuspended in fresh YTB medium to an optical density of 600 nm of 0.3. Prophages were induced by incubating the samples at 37°C for 30 min. To detect phage yield upon prophage induction, cultures were further shaken at 28°C for 4 h and then chloroform was added to kill surviving bacteria. Phage titration of the cell lysates on the R. meliloti 41 lawn indicated that, in the case of 16-3cti3Km-171, normal phage burst occurred (1,85 × 1010 PFU/ml; induction of 16-3cti3 prophage resulted in 2,6 × 1010 PFU/ml). No phage productions (<102) were detected after induction of prophages 16-3cti3Km-249, 16-3cti3Km-154, and 16-3cti3Km-525. The results indicated that cistrons, represented by ORF249, ORF154, and ORF525, are vital for phage growth.
Phage productions were fully regained at the wild-type level when pCS475, pCS473, and pCS474 were introduced into lysogenic strains R. meliloti 41(16-3cti3Km-249), R. meliloti 41(16-3cti3Km-154), and R. meliloti 41(16-3cti3Km-525), respectively. Phage titers were determined on both R. meliloti 41 containing the appropriate plasmid (pCS475, pCS473, or pCS474) and R. meliloti 41 without a plasmid. Complementation was indicated by high titers that varied between 0.2 × 1010 to 2.9 × 1010 PFU/ml in the lawn of bacteria with plasmids, while recombination (i.e., the wild-type region from the plasmid replaced the Km cassette in the phage genome) was indicated by low titers between 7.5 × 104 to 1.0 × 106 PFU/ml in the lawn of R. meliloti without plasmids.
Since the function of all three cistrons could be reestablished by complementation, we conclude that the insertion of the Km resistance gene did not disrupt any cis-acting control element of the cognate genes, and that in each case, the loss of function was due to the inactivation of the gene product itself.
Functional tests for identification of the terminase genes of phage 16-3
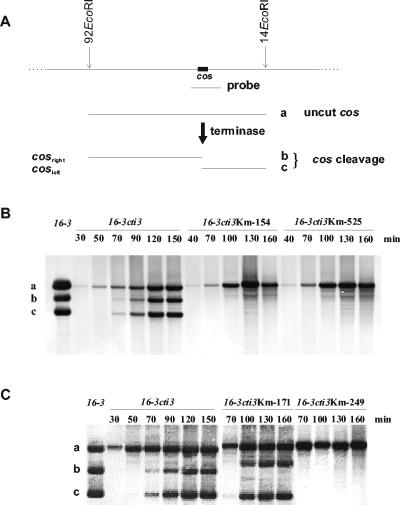
If ORF249, ORF154, and ORF525 encoded the subunits of a terminase enzyme, their inactivation would result in uncut cos sites and failure in packaging of the phage DNA. Southern hybridization (27, 32) was performed to test the hypothesis. Samples were collected at various times following heat induction of the prophages. Total bacterial DNA was prepared by a method described previously (3). DNA (300 ng) from each sample, digested overnight with EcoRI, was separated on a 0.8% agarose gel. Before loading, samples were kept at 80°C for 15 min to separate unligated cohesive ends. Filters, obtained after capillary transfer of DNA from the gels, were probed by the 1,600-bp-long 98BamHI-6BamHI fragment (nucleotides [nt] 1138 to 2766) overlapping the cos site (440-bp and 1,160-bp overlaps at the right and left ends, respectively) (Fig. 3A). DNA was labeled by using a DIG-Chem-Link labeling and detection set (Roche). Hybridizations and detections were carried out by using a DIG nucleic acid detection kit (Roche). In these conditions, three bands could be detected in samples derived from the control R. meliloti 41(16-3cti3). The top band was the 92EcoRI-14EcoRI fragment which represented the covalently closed cos sites derived from the 16-3 concatemer DNAs, while the other two bands (92EcoRI-cosright and cosleft-14EcoRI) derived from cutting of the cos site by active terminase (Fig. 3B). Southern hybridization of samples obtained from prophage induction in R. meliloti 41(16-3cti3Km-249), R. meliloti 41(16-3cti3Km-154), and R. meliloti 41(16-3cti3Km-525) showed only covalently closed cos sites, indicative of the malfunction of terminase activity. Southern analyses of samples derived from induction of prophages, containing the kanamycin cassette inserted into ORF171 [i.e., R. meliloti 41(16-3cti3Km-171)] indicated wild-type terminase activity since fragments of 92EcoRI-cosright and cosleft-14EcoRI appeared (Fig. 3C).
FIG. 3.
Functional test for identification of the terminase genes. Outline of detecting cos cleavage by Southern hybridization (A). The probe overlapped the cos site, and hybridized to three fragments, derived from the EcoRI-digested concatemeric phage DNA. The uncut cos fragment (a) was indicated by the intact 92EcoRI-14EcoRI fragment (8,317 bp), while cos cleavage resulted in two more fragments, the right end-fragment (b) (92EcoRI-cosright; 4,915 bp) and left end-fragment (c) (cosleft-14EcoRI; 3,402 bp). Detection of cos cleavage following prophage induction of lysogenic R. meliloti 41 containing phage 16-3cti3Km-154 and 16-3cti3Km-525 (B) and 16-3cti3Km-171 and 16-3cti3Km-249 (C). Lane 16-3 contained purified phage DNA digested with EcoRI, and samples from 16-3cti3 induction served as a positive control. Elapsed time following prophage induction is indicated in minutes. (Note that fragments a and b, derived from the Km mutant phage DNAs, migrated slower than the appropriate control fragments, since they contained the 1.2-kb Km cassette.)
Bioinformatic analyses of the ORFs
Putative ORFs were searched against the available databases (nonredundant protein database) by using BLAST (Basic Local Alignment Search Tool) available at GenomeNet (Bioinformatics Center, Kyoto University) (1) in order to establish amino acid sequence homologies. Multiple alignment of sequences were produced with the ClustalW program with the following parameters: gap open penalty = 10, gap extension penalty = 0.05, BLOSUM weight matrix (17, 34).
ORF249 indicates the longest open reading frame, although the use of alternative start codons (to ATG) would result in shorter products (ORF215 and ORF197) in the same frame. Insertion of the Km cassette at codon position 60 (counting according to ORF249) disrupts even the shortest possible ORF. To mark the gene we prefer to use ORF249, since homology with homing endonuclease sequences starts with its first codon (data not shown). The N-terminal 72 residues of ORF249 showed significant homology to homing endonucleases (32 out of 72 residues [44%] were identical with both MobE homing endonuclease of bacteriophage Aeh1 and a probable mobile endonuclease E of phage T4), while the rest of the proteins did not show any relationship (data not shown). Determination of the role of the protein encoded by ORF249 requires further studies. If the protein acts as an endonuclease, it could have a direct role in DNA cleavage at the cos site, alone or complexed with other terminase subunits as a part of the packaging machinery. It has been shown that lambda phages with an amber mutation in any head gene or in FI, the gene encoding the accessory packaging protein gpFI, results in the loss of cos cutting and phage production (21, 31). The product of ORF249 could therefore be an accessory packaging protein.
Homology of ORF171 to other proteins was based on its 62 C-terminal residues. The closest relatives were holin proteins of different phages (identity, 32 out of 62 residues [51%]; similarity, 62%) (data not shown). Although ORF171 did not seem to be essential for cos cleavage and phage production, there are also alternative translational start sites resulting in ORF165 or ORF76. Insertion of a Km cassette at codon position 56 (counting according to ORF171) ruled out the role of ORF171 and ORF165 but not ORF76. Homologies found with other known sequences (holin proteins of different phages) fall into the region of ORF76. We have tested a mutant prophage in which the Km cassette replaced almost the entire ORF76 (1 to 63 codons deleted). The mutation resulted in no cos cutting and no phage production (data not shown). Since phage production could not be complemented by ORF76 supplemented in trans, it is likely that the changes disrupted a cis element involved in cos cutting and/or in phage production.
ORF154 showed high homology only to the small terminase subunit of phage HK022 and HK97 (Fig. 4A) (identity, 52 out of 154 residues [34%]; similarity, 53%) and no reasonable homology was found to other proteins in the database. Alignments of ORF525 with its closest relatives are shown on Fig. 4B. The highest homology was found to the terminase large subunit of phages HK97 and HK022 (identity, 275 out of 525 residues [52%]; similarity, 66%). Residues of the putative functional motifs (adenine binding, Walker A and B, and motif III) of the ATPase domains (20) of the HK97 and 16-3 large terminase subunits are completely identical. Significant homology was also found to putative terminase proteins encoded by regions of phage origin in Bacillus cereus and Streptococcus pyogenes genomes. Homology found with large subunits of other terminases, identified in different phages and bacterial genomes, remained below 20% identity and 30% similarity.
FIG. 4.
Alignment of ORF154 (A) and ORF525 (B) with the highest-scoring protein sequences in the databases. Under the protein sequences, asterisks represent identical residues, while colons and periods indicate amino acids with similar characteristic (more and less, respectively) residues at a given position.
In databanks we have found genes and proteins of various phages which showed extensive homologies with ORF154 and ORF525, and from the alignment we concluded that these ORFs encode the subunits of the terminase enzyme of phage 16-3. The highest homologies were found with the terminase of phage HK97 and HK022. Indeed these phages also have 10-bp-long, 3′-protruding cohesive ends (18). However, the sequence of the cohesive ends of phage 16-3 and HK phages are not similar. We renamed ORF154 and ORF525 as genes terS and terL, respectively.
Nucleotide sequence accession number
The sequence of the 5,006-bp-long region was deposited in GenBank (accession no. AJ557020).
Acknowledgments
We thank Csilla Sánta Török, Magdolna Tóth Péli, and Kornélia Szóráth Gál for excellent technical assistance and Dhruba Chattoraj for discussion and helpful comments on the manuscript.
This work was supported by grants from the Hungarian Scientific Research Fund (OTKA) (T 032205 and T 032255); from the National Research and Development Program (NKFP) (OM 0028/2001 and OM 278/2001); and from the Hungarian Academy of Sciences (MTA/TKI/AKT-F 1999-2001 and MTA/TKI/AKT-F 2003-2006).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S., B. T. Luck, H. Y. Kim, and V. N. Iyer. 1992. Three clustered origins of replication in a promiscuous-plasmid replicon and their differential use in a PolA+ strain and a ΔPolA strain of Escherichia coli K-12. J. Bacteriol. 174:8139-8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, M. L., L. W. Enquist, and T. J. Silhavy (ed.). 1982. Advanced bacterial genetics, p. 132. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 4.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 5.Blaha, B., S. Semsey, S. Ferenczi, Z. Csiszovszki, P. P. Papp, and L. Orosz. 2004. A proline tRNA(CGG) gene encompassing the attachment site of temperate phage 16-3 is functional and convertible to suppressor tRNA. Mol. Microbiol. 54:742-754. [DOI] [PubMed] [Google Scholar]
- 6.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csiszovszki, Z., Z. Buzas, S. Semsey, T. Ponyi, P. P. Papp, and L. Orosz. 2003. immX immunity region of rhizobium phage 16-3: two overlapping cistrons of repressor function. J. Bacteriol. 185:4382-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallmann, G., F. Marincs, P. Papp, M. Gaszner, and L. Orosz. 1991. The isolated N-terminal DNA binding domain of the c repressor of bacteriophage 16-3 is functional in DNA binding in vivo and in vitro. Mol. Gen. Genet. 227:106-112. [DOI] [PubMed] [Google Scholar]
- 9.Dallmann, G., L. Orosz, and B. Sain. 1979. Restriction mapping of DNA of temperate Rhizobium meliloti phage 16-3: comparison of genetic and physical maps indicates a long, genetically silent chromosomal arm. Mol. Gen. Genet. 176:439-448. [DOI] [PubMed] [Google Scholar]
- 10.Dallmann, G., P. Papp, and L. Orosz. 1987. Related repressor specificity of unrelated phages. Nature 330:398-401. [Google Scholar]
- 11.Dorgai, L., F. Olasz, M. Berenyi, G. Dallmann, A. Pay, and L. Orosz. 1981. Orientation of the genetic and physical map of Rhizobium meliloti temperate phage 16-3. Mol. Gen. Genet. 182:321-325. [Google Scholar]
- 12.Dorgai, L., I. Papp, P. Papp, M. Kalman, and L. Orosz. 1993. Nucleotide sequences of the sites involved in the integration of phage 16-3 of Rhizobium meliloti 41. Nucleic Acids Res. 21:1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorgai, L., G. Polner, E. Jonas, N. Garamszegi, Z. Ascher, A. Pay, G. Dallmann, and L. Orosz. 1983. The detailed physical map of the temperate phage 16-3 of Rhizobium meliloti 41. Mol. Gen. Genet. 191:430-433. [DOI] [PubMed] [Google Scholar]
- 14.Ferenczi, S., A. Ganyu, B. Blaha, S. Semsey, T. Nagy, Z. Csiszovszki, L. Orosz, and P. P. Papp. 2004. Integrative plasmid vector for constructing single-copy reporter systems to study gene regulation in Rhizobium meliloti and related species. Plasmid 52:57-62. [DOI] [PubMed] [Google Scholar]
- 15.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 17.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 18.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 19.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, I. I. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, M. S., S. Matsuzaki, S. Imai, and V. B. Rao. 2002. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30:4009-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murialdo, H., and W. L. Fife. 1987. Synthesis of a trans-acting inhibitor of DNA maturation by prohead mutants of phage lambda. Genetics 115:3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orosz, L., Z. Svab, A. Kondorosi, and T. Sik. 1973. Genetic studies on rhizobiophage 16-3. I. Genes and functions on the chromosome. Mol. Gen. Genet. 27:341-350. [PubMed] [Google Scholar]
- 23.Papp, I., L. Dorgai, P. Papp, E. Jonas, F. Olasz, and L. Orosz. 1993. The bacterial attachment site of the temperate Rhizobium phage 16-3 overlaps the 3′ end of a putative proline tRNA gene. Mol. Gen. Genet. 240:258-264. [DOI] [PubMed] [Google Scholar]
- 24.Papp, P. P., and V. N. Iyer. 1995. Determination of the binding sites of RepA, a replication initiator protein of the basic replicon of the IncN group plasmid pCU1. J. Mol. Biol. 246:595-608. [DOI] [PubMed] [Google Scholar]
- 25.Papp, P. P., T. Nagy, S. Ferenczi, P. Elo, Z. Csiszovszki, Z. Buzas, A. Patthy, and L. Orosz. 2002. Binding sites of different geometries for the 16-3 phage repressor. Proc. Natl. Acad. Sci. USA 9:8790-8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putnoky, P., V. Deak, K. Bekasi, A. Palvolgyi, A. Maasz, Z. Palagyi, G. Hoffmann, and I. Kerepesi. 2004. H protein of bacteriophage 16-3 and RkpM protein of Sinorhizobium meliloti 41 are involved in phage adsorption. J. Bacteriol. 186:1591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Sanger, F. S., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semsey, S., B. Blaha, K. Koles, L. Orosz, and P. P. Papp. 2002. Site-specific integrative elements of rhizobiophage 16-3 can integrate into proline tRNA (CGG) genes in different bacterial genera. J. Bacteriol. 184:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semsey, S., I. Papp, Z. Buzas, A. Patthy, L. Orosz, and P. P. Papp. 1999. Identification of site-specific recombination genes int and xis of the Rhizo-bium temperate phage 16-3. J. Bacteriol. 181:4185-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sippy, J., and M. Feiss. 2004. Initial cos cleavage of bacteriophage λ concatemers requires proheads and gpFI in vivo. Mol. Microbiol. 52:501-513. [DOI] [PubMed] [Google Scholar]
- 32.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 33.Thatte, V., D. E. Bradley, and V. N. Iyer. 1985. N conjugative transfer system of plasmid pCU1. J. Bacteriol. 163:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]