Abstract
The expression of scaffoldin-anchoring genes and one of the major processive endoglucanases (CelS) from the cellulosome of Clostridium thermocellum has been shown to be dependent on the growth rate. For the present work, we studied the gene regulation of selected cellulosomal endoglucanases and a major xylanase in order to examine the previously observed substrate-linked alterations in cellulosome composition. For this purpose, the transcript levels of genes encoding endoglucanases CelB, CelG, and CelD and the family 10 xylanase XynC were determined in batch cultures, grown on either cellobiose or cellulose, and in carbon-limited continuous cultures at different dilution rates. Under all conditions tested, the transcript levels of celB and celG were at least 10-fold higher than that of celD. Like the major processive endoglucanase CelS, the transcript levels of these endoglucanase genes were also dependent on the growth rate. Thus, at a rate of 0.04 h−1, the levels of celB, celG, and celD were threefold higher than those obtained in cultures grown at maximal rates (0.35 h−1) on cellobiose. In contrast, no clear correlation was observed between the transcript level of xynC and the growth rate—the levels remained relatively high, fluctuating between 30 and 50 transcripts per cell. The results suggest that the regulation of C. thermocellum endoglucanases is similar to that of the processive endoglucanase celS but differs from that of a major cellulosomal xylanase in that expression of the latter enzyme is independent of the growth rate.
The cellulosome complex of the thermophilic anaerobe Clostridium thermocellum is a highly active extracellular cellulolytic system (5, 8, 14, 15, 18, 28, 29, 31, 33, 38, 39). Individual cellulosomes consist of up to nine catalytic subunits attached to a noncatalytic scaffolding protein, or scaffoldin (1, 3, 12, 13, 20, 21, 37), originally termed CipA (cellulosome-integrating protein A). To date, >20 cellulosomal enzymes are known to be produced by C. thermocellum (5, 6, 38, 39, 40). These catalytic components include mostly endoglucanases, exoglucanases, xylanases, and other hemicellulases.
Evidence from various sources has indicated that the expression of cellulosomal components from different cellulosome-producing bacteria is generally regulated by growth conditions rather than being constitutive. These changes in cellulosome composition have been shown to influence the activity of the entire complex (4, 9-11, 16, 19, 22-24, 26, 29, 35, 36, 41). Cellulosomes prepared from cells grown on cellulose exhibited higher activities on crystalline cellulose than cellulosomes prepared from cells grown on cellobiose (16). The increase in cellulosome activity was found to be accompanied by an increase in the transcript level of the major cellulosomal processive endoglucanase, CelS (Cel48A). More insights into the factors that influence the transcript level of celS were obtained from studies involving continuous cultures operated at different dilution rates. With this approach, it was determined that the expression level of celS increased with decreasing growth rates. The influence of the growth rate was not exclusively limited to the transcription of the celS gene, as transcription of the scaffoldin gene, cipA, and of other cipA-related genes was also affected (17).
In order to study the alteration of the expression patterns of cellulosomal genes due to changes in physiological conditions, we determined their transcript levels. Among the genes selected as targets for this study, we chose two that encode major cellulosomal endoglucanases, CelB (Cel5A) and CelG (Cel5D), from glycoside hydrolase family 5 (7, 32), and another that encodes a family 9 endoglucanase, CelD (Cel9A) (27). In addition, the regulation of expression of the family 10 xylanase XynC (25) was also investigated in order to compare whether gene regulation of a hemicellulase subunit is similar to that of the endoglucanases. Our results established that the patterns of transcription are similar for the various endoglucanase genes studied in this work, which are, in turn, similar to that observed for previously studied cellulosomal genes, i.e., the major processive endoglucanase celS and the primary scaffoldin cipA (16, 17). In contrast, the expression of xynC represented a distinctive regulation pattern.
MATERIALS AND METHODS
Organism, substrates, and culture conditions.
C. thermocellum YS was originally isolated from soil samples obtained at the hot springs of Yellowstone National Park (2, 29, 30). Cells were grown in batch culture at 60°C in Duran anaerobic bottles (Schott Corporation, Mainz, Germany) in a medium containing the following reagents (per liter): 0.65 g of K2HPO3 · 3H2O, 0.5 g of KH2PO4, 1.3 g of (NH4)SO4, 42 g of morpholinepropanesulfonic acid (MOPS), 5 g of yeast extract, 1 g of cysteine, 0.5 g of MgCl2, and 2 mg of resazurin. The medium included 1.0% of the desired carbon source, either cellobiose from Acros Organics (Geel, Belgium) or microcrystalline cellulose (Avicel), obtained from Teva-Pharmaceutical Industries (Kfar Sava, Israel). Before the turbidity was measured, cellulose-grown cultures were vortexed vigorously and centrifuged at 100 × g for 1 min to remove the insoluble substrate. Continuous cultures were performed in a BIOFLO 3000 fermentor (New Brunswick Scientific, Edison, N.J.) in a 1.5-liter working volume at 60°C. The pH was maintained at 7.2 by the automatic addition of 5 N NaOH. Agitation was kept constant at 100 rpm. For the maintenance of anaerobic conditions, the headspace of the bioreactor contained 99.99% CO2 for initial growth and then changed to 99.99% N2 while the continuous process started. Continuous cultures were operated under conditions of cellobiose limitation (2 g/liter), whereby C. thermocellum adapted to different growth rates. The residual cellobiose concentration at the steady state was assessed by a reducing sugar assay using the dinitrosalicylic acid reagent (34). The cellobiose concentration correlated, as expected, to the dilution rate. Typical values were 0.22, 0.17, and 0.14 g/liter for dilution rates of 0.23, 0.19, and 0.15 h−1, respectively. Note that C. thermocellum YS cannot utilize yeast extract as a carbon source. The establishment of steady-state conditions was assumed when the culture had been growing for a period of at least three generations and the cell density (monitored spectrophotometrically) had remained unchanged for at least one generation.
RNA extraction.
Precipitated cells, derived from 10 ml of culture medium, were washed twice with 10 mM Tris-HCl, pH 7.5, and the pellets were snap-frozen in liquid nitrogen and stored at −80°C. The total RNA was extracted from cells by use of an RNeasy kit (QIAGEN GmbH, Hilden, Germany), with minor modifications as described earlier (16). The RNA concentration was estimated by measuring the A260, and the resultant RNA preparations were stored in aliquots at −80°C.
RPA.
RNase protection assays (RPAs) were performed by use of an RPAII kit (Ambion Inc., Austin, Tex.). Different amounts of RNA were hybridized to 32P-labeled antisense probes, and the protected RNAs were placed directly in a scintillation counter for quantification. For visualization, the protected RNAs were separated in 5% polyacrylamide gels containing 7 M urea and visualized by use of a phosphorimager system. Probes were labeled by use of a Maxiscript in vitro transcription kit (Ambion) and T7 RNA polymerase. The appropriate fragments were cloned into pGEM (Promega, Madison, Wis.). The different probes included the following DNA fragments (positions are relative to the initial ATG): celG, from −77 to +283; celB, from −101 to +119; celD, from −88 to +499; celF, from −211 to +338; and xynC, from −174 to +419.
Determination of amount of RNA per cell.
The amount of RNA was established for each of the cultures wherein the transcript levels of the genes were determined. A pellet derived from 30 ml of culture was washed with 10 mM Tris-Cl, pH 7.6, and resuspended in 10 ml of 10% cold trichloroacetic acid (TCA). The suspension was kept on ice for 30 min. Supernatant fluids were discarded after 10 min of centrifugation at 10,000 × g. The cells were then resuspended in 5% TCA and centrifuged, and the pellet was dissolved gently in 1.5 ml of 0.1 N NaOH by use of a sealed Pasteur pipette. The samples were incubated overnight at 37°C to allow for the complete hydrolysis of RNAs. The solution was neutralized with 1 ml of 10% TCA, incubated for 15 min on ice, and then centrifuged. For determination of the RNA concentration in the sample, 1 volume of appropriately diluted supernatant fluid was mixed with 1 volume of orcinol reagent (1% orcinol dissolved in 0.1% FeCl3 in concentrated HCl), and the solution was boiled for 15 min. After the tubes were cooled under running tap water, 2 volumes of distilled water was added. The absorbance was recorded at two wavelengths, namely, 600 nm (background) and 660 nm (background plus green complex). For determination of the RNA or nucleotide concentration, a deoxyadenosine solution at 50 μg/ml was used as a standard. Since the results were obtained as purine riboside equivalents of RNA, the values were doubled to obtain nucleotide equivalents (purines plus pyrimidines) and multiplied by the average nucleotide molecular weight. For determination of the amount of total RNA per cell, cells were counted under a microscope by use of a Petroff-Hausser counter chamber. The number of transcripts of each gene per cell was then calculated from the resultant values of counts per minute per microgram of total RNA, the specific activity of the probe, and the average amount of total RNA determined per cell.
RESULTS
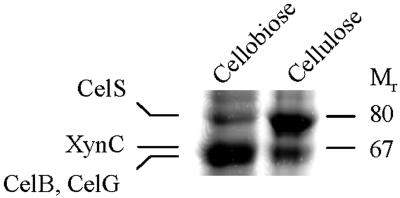
In previous studies of C. thermocellum (4, 16), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses revealed that the compositions of cellulosome subunits derived from cellulose- versus cellobiose-grown stationary-phase cultures differed greatly. The most prominent difference was a reduction in the family 48 CelS band in cellobiose-grown cells compared to cellulose-grown cells (Fig. 1). The regulation of celS was investigated in detail and was described in an earlier work (16). Another major difference revealed by SDS-PAGE analysis was an enhancement of a 67-kDa band upon growth on cellobiose (4). An analysis of the N-terminal amino acid sequence of this 67-kDa band indicated that it actually represented three separate cellulosomal subunits, namely, the two endoglucanases CelB and CelG and the xylanase XynC (16). For further study of the factors responsible for the regulation of these proteins, cultures of C. thermocellum were grown under more defined growth conditions.
FIG. 1.
SDS-PAGE analysis of cellulosome preparations purified from cellobiose- and cellulose-grown cells. Cellulosomes, derived from the late stationary phase of C. thermocellum grown either on microcrystalline cellulose or on cellobiose, were isolated from the culture medium by affinity chromatography with cellulose. The preparations were separated by SDS-6% PAGE. The cellulosomal subunits XynC, CelB, and CelG were identified previously from polyvinylidene difluoride blots of the gels by amino acid sequence analysis (16).
Due to the difficulty in distinguishing among these cellulosomal subunits by SDS-PAGE, an alternative strategy was required for estimating the level of each enzyme. Another problem with studying the regulation of cellulosomal genes at the protein level is that the cellulosome complex is present in both cell-free and cell-bound forms. In this context, most studies have focused on the cell-free fraction, due to the relatively facile methods for its isolation, and the cell-bound fraction (e.g., from exponential-phase cells) has often been ignored. For the circumvention of such problems, the expression level of each gene was determined at the transcriptional level by RPAs. RPA was the method of choice because of its capacity to provide quantitative results whereby the expression levels of genes can be obtained as numbers of transcripts per cell.
Effect of carbon source on transcript levels of selected cellulosomal genes.
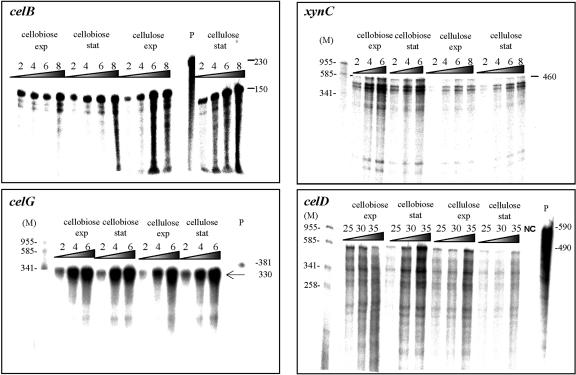
As mentioned above, our initial results indicated that the overall combined level of expression of three cellulosomal enzymes (the endoglucanases CelB and CelG and the xylanase XynC, represented by a common 67-kDa band) was higher in late-stationary-phase cellobiose-grown cells than in cellulose-grown cells. In order to determine the effect of the carbon source on the expression of these genes, we assessed the mRNA level of each in cultures grown on the relevant substrate. Cells were harvested during the exponential and late-exponential phases, when culture turbidities (A660) reached 0.85 and 1.8, respectively, for cellobiose-grown cells, and 0.65 and 2.0, respectively, for cellulose-grown cells. An RPA was used to assess the level of mRNA for each gene. To ensure that the amount of the probe in the hybridization reaction was not limiting, we hybridized different amounts of the isolated RNAs with each 32P-labeled antisense probe. After digestion with RNases, the RPA product was precipitated, the supernatant fluids were removed, and the radioactivity was assessed in a scintillation counter. The protected product was separated in a polyacrylamide gel for visualization by a phosphorimager system (Fig. 2). A linear correlation was obtained in all cases examined between the amount of total RNA used in the assay and the radioactivity of the protected product. The calculated number of transcripts of each gene is presented in Table 1.
FIG. 2.
Transcript levels of endoglucanase genes celB, celG, and celD and the xylanase gene xynC. RPAs were performed with a relevant 32P-labeled antisense probe as described earlier (16), using RNAs from exponential (exp)- or stationary (stat)-phase cultures grown on either cellobiose or crystalline cellulose, as designated. The different amount (in micrograms) of RNA used to hybridize with the probe is indicated at the top of each lane. Autoradiographs of the respective RPA products were visualized with a phosphorimager system. Negative control lanes (NC) contained RNAs from yeast. Lanes M, size markers (in base pairs). The full-length probe is shown in lanes P. Values at the right indicate the size of the undigested full-length antisense probe and the estimated sizes of the protected products, in bases.
TABLE 1.
Transcript levels of selected endoglucanase genes and a xylanase gene in C. thermocellum batch cultures grown on either cellobiose or cellulose
| Gene | No. of transcripts per cell on indicated substratea
|
|||
|---|---|---|---|---|
| Cellobiose
|
Crystalline cellulose
|
|||
| Exponential phase | Late exponential phase | Exponential phase | Late exponential phase | |
| celB | 6 | 8 | 11 | 12 |
| celG | 22 | 22 | 20 | 24 |
| celD | 0.7 | 0.9 | 1 | 0.8 |
| celF | 1 | 1 | 2 | 1 |
| xynC | 54 | 80 | 28 | 30 |
Values represent the averages ± 15% standard error of at least three determinations of mRNA levels.
The results indicated that among the three 67-kDa-band-related genes, only the transcript level of xynC was higher in cellobiose-grown cells than in cellulose-grown cells. During exponential growth on cellobiose, the expression of xynC was 54 transcripts per cell, and the value increased to 80 upon entering the stationary phase of growth (Fig. 2). The xynC mRNA level in cellulose-grown cells was only about 30 transcripts per cell. In contrast to the case for xynC, the transcript level of celG was only slightly affected by the carbon source and was about 20 transcripts per cell during exponential growth on both substrates. The transcript level of the endoglucanase gene celB was twice as high in cellulose-grown cells but was still two- to threefold lower than that obtained for celG (6 to 12 versus 20 transcripts per cell). These two endoglucanase genes were unaffected by the phase of growth, since no increase was observed in the transcript level when cells entered stationary phase. Nevertheless, the overall combined transcript level of the two endoglucanases, together with that of the xylanase, during the late exponential phase of cellobiose-grown cells proved to be higher than that observed for cellulose-grown cells in the corresponding phase of growth (110 compared to 66 transcripts per cell). This finding is in accord with the SDS-PAGE results described above, wherein the intensity of the 67-kDa band in cellobiose-grown cells was higher than that observed in cellulose-grown cells. These results suggest that this difference mainly reflects an increase in the expression of the xylanase gene xynC upon entry into the stationary phase of growth. To compare the transcription patterns of celB and celG to those of other cellulosomal endoglucanases, we also determined the expression levels of two different family 9 genes, celD and celF. The regulation of the last two genes was previously characterized (35), and thus they were used as a reference for the present work. The transcript levels of celD and celF were previously shown to undergo an increase at the end of the exponential phase of growth, while during the early phase the levels of both genes were undetectable. In agreement with these earlier findings, the transcript levels obtained for both genes in the present work were only about one transcript per cell under all conditions studied (Table 1). The low transcript levels of these genes were consistent with the fact that it was not possible to detect these proteins by SDS-PAGE and N-terminal sequencing of candidate bands.
One possible factor that has been shown to play a role in the expression of the endoglucanases studied here is the rate of cell growth, as demonstrated previously for other cellulosome-related genes (16, 17). The difference in maximal growth rates of cellobiose- versus cellulose-grown cultures probably resulted from the difference in the immediate availability of the two substrates. The growth rates achieved on cellobiose and cellulose were 0.35 and 0.23 h−1 (i.e., doubling times of 2 and 3 h), respectively. In this context, it seems that the growth rate either has a contrary effect on the transcription of xynC or that the transcription of this gene is influenced by another factor(s). To address these issues, we used chemostat cultures.
Transcript levels of selected cellulosomal genes during growth in continuous culture with limited cellobiose.
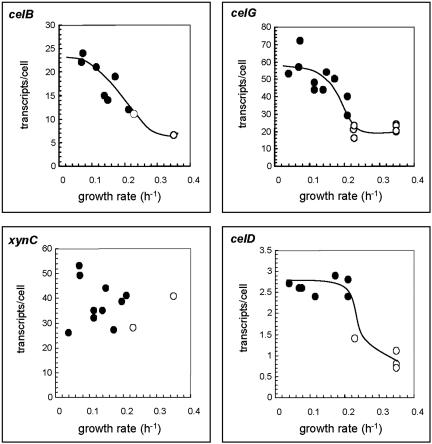
Chemostat studies were conducted to investigate the influence of the growth rate (i.e., the cellobiose concentration) on the transcription of the desired cellulosomal genes of C. thermocellum. This system allowed the study of lower growth rates than those achieved with cellulose-grown batch cultures. Moreover, the chemostat enabled us to study the regulation of the desired genes under defined steady-state conditions. Continuous cultures were operated with limiting concentrations of cellobiose. The ability to control the growth rate is achieved by controlling the flow rate of the medium (dilution rate) introduced into the fermentor. The dilution rates established ranged from 0.21 to 0.04 h−1, and the resultant cell turbidities (A660) were within the range of 0.5 to 0.7.
As shown in Fig. 3, the levels of transcripts obtained for the endoglucanase genes celB and celG were only slightly higher at a dilution rate of 0.21 h−1 (13 and 29 transcripts per cell, respectively) than those achieved in cultures grown on cellulose at a growth rate of 0.23 h−1(11 and 24 transcripts per cell, respectively). In contrast, the transcript level of celD increased up to threefold when the rate of growth decreased from 0.23 to 0.21 h−1. A further decrease in the dilution rate to 0.04 h−1 failed to influence the transcript level of celD, and the level stabilized at about two to three transcripts per cell. In the cases of celB and celG, a decrease in the dilution rate was accompanied by a gradual increase in the transcript level. Thus, at a dilution rate of 0.04 h−1, the transcript levels of these genes were two- to threefold higher than those obtained in cellulose-grown cells (25 and 55 transcripts per cell for celB and celG, respectively). Interestingly, no clear trend was observed for the transcript level of xynC upon altering the dilution rate. The level of xynC mRNA fluctuated from 30 to 55 transcripts per cell, irrespective of the growth rate.
FIG. 3.
Transcript levels of the indicated cellulosomal genes as a function of the growth rate. The amounts of the respective mRNAs were determined by RPAs and converted to the numbers of transcripts per cell, based on the average measured amount of total RNA in a single cell (0.18 pg/cell). Values represent the averages of several (at least three) measurements, with accuracies of ±25%. Continuous cultures (filled circles) were operated with carbon (cellobiose) limitation at dilution rates from 0.04 to 0.21 h−1. Batch cultures (open circles) were grown to exponential phase on either cellulose or cellobiose at rates of 0.23 and 0.35 h−1, respectively. Note the differences in the scales of the different graphs.
DISCUSSION
The most important conclusion to be drawn from the results of the present study is that there appears to be a common trend in the regulation of the different cellulosomal genes. In this context, the expression of celG and celB was found to be growth rate dependent, as observed earlier for the genes encoding the major processive endoglucanase and the primary scaffoldin (celS and cipA, respectively). As observed in our earlier studies on the expression of cellulosomal components (16, 17), upon the transition from conditions of rapid exponential growth to very low growth rates, the transcript levels of celB, celG, and celD increased about threefold. In contrast, transcription of the xylanase gene xynC appeared to follow a different pattern of regulation, and its expression remained at a relatively high level, irrespective of the growth rate.
In general, the influence of the growth phase on the expression of the genes studied in this work was relatively minor, although the transcript level of xynC exhibited a defined increase during stationary-phase growth on cellobiose. Interestingly, this finding is in contrast to the response observed earlier for the transcript levels of celS and cipA, both of which decreased upon entry into the stationary phase of growth (16, 17). The observed difference in the transcript levels of xynC between the stationary- and exponential-growth phases may in fact denote a physiological function essential to the fundamental lifestyle of the bacterial cell. In this context, C. thermocellum specializes in the hydrolysis and utilization of cellulose and its degradation products alone. Although it readily hydrolyzes xylan and other hemicelluloses, it cannot assimilate their components.
In the native ecosystem, the plant cell wall is structured such that the cellulose microfibrils are embedded in a colloidal matrix of lignin and hemicellulose. To commence effective biodegradation of the growth-relevant cellulose substrate, the cells must first penetrate the hemicellulose barrier. Therefore, relatively high levels of xylanases, such as XynC, would serve as a primary means for gaining access to the main substrate. In the next stage, the cells attach to and begin to degrade the cellulosic substrate. Under these conditions, the growth rate is relatively low and the production of cellulases would be stimulated in order to support further growth. The growth rate would then increase as a function of the accumulation of soluble degradation products; further production of cellulases would be less critical, and their transcript levels would decrease, as supported by the results of this work.
To date, more than two dozen C. thermocellum enzymes have been sequenced by conventional means, the majority of which appear to be dockerin-containing cellulosomal enzymes. About half are endoglucanases and/or processive cellulases, and the other half are xylanases and other hemicellulases. Moreover, the C. thermocellum draft genome sequence is now available at the Joint Genome Institute website (http://www.jgi.doe.gov). The genome sequence has ensured the identification of additional enzymes and other cellulosome-related genes. It will thus be interesting to determine in future research whether the trends observed in the present work reflect a more general feature of this bacterium, particularly with respect to the regulation of cellulases versus hemicellulases. In this context, Doi and colleagues (23, 24) have recently examined the regulation of expression of cellulosomal enzymes in a mesophilic cellulosome-producing bacterium, Clostridium cellulovorans, and the coordinated expression of cellulases and hemicellulases was demonstrated.
Acknowledgments
This research was supported by the Israel Science Foundation (grants 394/03, 771/01, and 446/01) and by a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. Additional support was provided by the Otto Meyerhof Center for Biotechnology, The Technion, established by the Minerva Foundation (Munich, Germany).
REFERENCES
- 1.Ali, B. R., M. P. Romaniec, G. P. Hazlewood, and R. B. Freedman. 1995. Characterization of the subunits in an apparently homogeneous subpopulation of Clostridium thermocellum cellulosomes. Enzyme Microbiol. Technol. 17:705-711. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, E. A., L. J. W. Shimon, R. Lamed, and Y. Shoham. 1998. Cellulosomes: structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. Cellulose-decomposing prokaryotes and their enzyme systems. .In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y. [Online.] http://link.springer.de/link/service/books/10125/index.htm.
- 7.Béguin, P., P. Cornet, and J. Millet. 1983. Identification of the endoglucanase encoded by the celB gene of Clostridium thermocellum. Biochimie 65:495-500. [DOI] [PubMed] [Google Scholar]
- 8.Béguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 9.Bhat, K. M., P. W. Goodenough, E. Owen, and T. M. Wood. 1993. Cellobiose: a true inducer of cellulosome in different strains of Clostridium thermocellum. FEMS Microbiol. Lett. 111:73-78. [Google Scholar]
- 10.Bhat, S., P. W. Goodenough, E. Owen, B. Brooker, R. Stenning, and M. K. Bhat. 1995. Evaluation of the cellulase system produced by three strains of Clostridium thermocellum on cellobiose and Avicel. Biochem. Soc. Trans. 23:587S. [DOI] [PubMed] [Google Scholar]
- 11.Bhat, S., J. F. Kennedy, P. W. Goodenough, E. Owen, and M. K. Bhat. 1997. Effect of d-glucono-1,4-lactone on the production of CMCase, pNPCase and true cellulase by Clostridium thermocellum. Carbohydr. Polymers 34:95-99. [Google Scholar]
- 12.Ciruela, A., H. J. Gilbert, B. R. S. Ali, and G. P. Hazlewood. 1998. Synergistic interaction of the cellulosome integrating protein (CipA) from Clostridium thermocellum with a cellulosomal endoglucanase. FEBS Lett. 422:221-224. [DOI] [PubMed] [Google Scholar]
- 13.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 15.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J. Bacteriol. 185:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felix, C. R., and L. G. Ljungdahl. 1993. The cellulosome—the exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 19.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerngross, U. T., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 22.Halliwell, G., T. M. Philips, and N. Halliwell. 1995. Microcrystalline forms of cellulose as substrates for strains of Clostridium thermocellum and cellulase formation. Proc. Biochem. 30:243-250. [Google Scholar]
- 23.Han, S. O., H. Y. Cho, H. Yukawa, M. Inui, and R. H. Doi. 2004. Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J. Bacteriol. 186:4218-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 185:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi, H., K. I. Takagi, M. Fukumura, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1997. Sequence of xynC and properties of XynC, a major component of the Clostridium thermocellum cellulosome. J. Bacteriol. 179:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, E. A., F. Bouchot, and A. L. Demain. 1985. Regulation of cellulase formation in Clostridium thermocellum. J. Gen. Microbiol. 131:223-232. [Google Scholar]
- 27.Joliff, G., P. Béguin, and J.-P. Aubert. 1986. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 14:8605-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamed, R., and E. A. Bayer. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1-46. [Google Scholar]
- 29.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microbiol. Technol. 7:37-41. [Google Scholar]
- 30.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamed, R., E. Setter, R. Kenig, and E. A. Bayer. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163-181. [Google Scholar]
- 32.Lemaire, M., and P. Béguin. 1993. Nucleotide sequence of the celG gene of Clostridium thermocellum and characterization of its product, endoglucanase CelG. J. Bacteriol. 175:3353-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, G. L. R., W. E. Blum, and A. L. Burton. 1960. Measurements of carboxymethylcellulase activity. Anal. Biochem. 2:127-132. [Google Scholar]
- 35.Mishra, S., P. Béguin, and J. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nochur, S. V., A. L. Demain, and M. F. Roberts. 1990. True cellulase production by Clostridium thermocellum grown on different carbon sources. FEMS Microbiol. Lett. 71:199-204. [Google Scholar]
- 37.Poole, D. M., E. Morag, R. Lamed, E. A. Bayer, G. P. Hazlewood, and H. J. Gilbert. 1992. Identification of the cellulose binding domain of the cellulosome subunit S1 from Clostridium thermocellum. FEMS Microbiol. Lett. 99:181-186. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 39.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W. K., K. Kruus, and J. H. D. Wu. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weimer, P. J., and J. G. Zeikus. 1977. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence of Methanobacterium thermoautotrophicum. Appl. Environ. Microbiol. 33:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]