Abstract
Adherent-invasive Escherichia coli strain LF82 recovered from a chronic lesion of a patient with Crohn's disease is able to invade cultured intestinal epithelial cells. Three mutants with impaired ability to invade epithelial cells had the Tn5phoA transposon inserted in the yfgL gene encoding the YfgL lipoprotein. A yfgL- negative isogenic mutant showed a marked decrease both in its ability to invade Intestine-407 cells and in the amount of the outer membrane proteins OmpA and OmpC in the culture supernatant, as shown by analysis of the culture supernatant protein contents by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry. Transcomplementation of the LF82-ΔyfgL isogenic mutant with the cloned yfgL gene restored invasion ability and outer membrane protein release in the culture supernatant. The outer membrane proteins in the culture supernatant of strain LF82 resulted from the formation of vesicles. This was shown by Western blot analysis of periplasmic and outer membrane fraction markers typically found in outer membrane vesicles and by transmission electron microscopic analysis of ultracentrifuged cell-free LF82 supernatant pellets, indicating the presence of vesicles with a bilayered structure surrounding a central electron-dense core. Thus, deletion of the yfgL gene in strain LF82 resulted in a decreased ability to invade intestinal epithelial cells and a decreased release of outer membrane vesicles.
Crohn's disease (CD) is a human inflammatory bowel disease characterized by chronic transmural, segmental, and granulomatous inflammation of the intestine (17). CD exhibits features that might be the result of a microbial process in the gut. Various studies have addressed the hypothesis that pathogenic bacteria contribute to the pathogenesis of inflammatory bowel disease (9, 28, 31, 37, 38). Of the bacteria that could play a role in the pathogenesis of CD, pathogenic Escherichia coli strains have been incriminated. E. coli bacteria are abnormally predominant (between 50 and 100% of the total number of aerobes and anaerobes) in early and chronic ileal lesions of CD, and most E. coli strains isolated from the ileal mucosa of CD patients adhere to intestinal epithelial cells (14). In addition to their ability to adhere, E. coli strains isolated from 36% of CD patients are able to invade intestinal epithelial cells (13) and belong to a new pathogenic group of E. coli, adherent-invasive E. coli (AIEC) (8). AIEC strains have none of the virulence factors of invasive bacteria known to be involved in gastrointestinal infections by pathogenic strains of E. coli, Salmonella spp., and Shigella flexneri (8).
The invasive ability of E. coli strain LF82, the AIEC reference strain, was studied. Electron microscopic examination of LF82-infected intestinal epithelial cells revealed a macropinocytosis-like process of entry dependent on actin microfilaments and microtubule recruitment and characterized by the elongation of membrane extensions, which surround the bacteria at the site of contact between entering bacteria and epithelial cells (8). Type 1 pilus-mediated adherence plays an essential role in the invasive ability of strain LF82 by inducing membrane extensions (7). However, type 1 pili have to be expressed in the genetic background of strain LF82 to promote bacterial uptake, since their expression in E. coli strain K-12 is not sufficient to confer invasiveness. Flagella also play a direct role in the adhesion and invasion processes of AIEC strain LF82 via motility and an indirect role in the interaction between bacteria and epithelial cells by down-regulating the expression of type 1 pili (3). In addition, the lipoprotein NlpI, which is probably located in the inner membrane, is thought to operate in a regulatory pathway involved in the synthesis of flagella, type 1 pili, and other virulence factors yet to be identified (4).
Other genetic determinants involved in the invasion process of AIEC strain LF82 were identified by analyzing an LF82 Tn5phoA insertion mutant library (7). Of 16 mutants with decreased ability to invade epithelial cells compared to the wild-type strain LF82, 11 mutants had a transposon inserted in various genes of the type 1 pilus operon, one mutant had a transposon inserted in the yibP gene, one mutant had a transposon inserted in the nlpI gene, and three mutants had a transposon inserted in the yfgL gene (b2512) of E. coli strain MG1655 (GenBank accession no AE000337). The yfgL gene encodes the YfgL lipoprotein, which is involved in the synthesis and/or degradation process of peptidoglycan and in the susceptibility of E. coli strain K-12 to killing by glycolipid derivatives of vancomycin (18).
The aim of this study was to characterize the role of the YfgL lipoprotein in the invasive process of CD-associated adherent-invasive E. coli strain LF82. We show here that yfgL is required for the invasion ability of adherent-invasive E. coli strain LF82, regardless of type 1 pilus and flagellum expression, but in correlation with outer membrane vesicle (OMV) release.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cell line.
E. coli strain LF82 was isolated from a chronic ileal lesion of a patient with CD and belongs to E. coli serotype O83:H1. It adhered to and strongly invaded Intestine-407, HEp-2, and Caco-2 cells (8). E. coli strains JM109 and C600 were used as host strains for cloning experiments. The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth or on LB agar plates (Institut Pasteur Production) overnight at 37°C. Antibiotics were added to media at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 25 μg/ml. Intestine-407 cells (derived from human intestinal embryonic jejunum and ileum) were purchased from Flow Laboratories and cultured as previously described (8).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| LF82 | E. coli isolated from an ileal biopsy of a CD patient | 14 |
| 2E4 | Tn5phoA insertion into yfgL of strain LF82 | 7 |
| OA8 | Tn5phoA insertion into yfgL of strain LF82 | 7 |
| 51F8 | Tn5phoA insertion into yfgL of strain LF82 | 7 |
| 52D11 | Tn5phoA insertion into fimA of strain LF82 | 7 |
| LF82-ΔyfgL | LF82 isogenic mutant with the yfgL gene deleted | This study |
| MC4100 | E. coli K-12 | Laboratory stock |
| MC4100-ΔyfgL | MC4100 isogenic mutant with the yfgL gene deleted | This study |
| E12860/0 | Enteroinvasive E. coli strain | 16 |
| E12860/0-ΔyfgL | Enteroinvasive E. coli isogenic mutant with the yfgL gene deleted | This study |
| Plasmids | ||
| pHSG575 | E. coli cloning vector, chloramphenicol resistant | 39 |
| pKOBEG | pBAD cloning vector harboring λ phage redγβα operon, chloramphenicol resistant | 12 |
| pBAD33 | E. coli cloning vector, chloramphenicol resistant | 20 |
| pPBI01 | pHSG575 harboring the 11.2-kb SalI fragment with the entire fim operon of E. coli K-12 strain J96 | 7 |
| pPBI07 | pBAD33 harboring the 1.2-kb XbaI-PstI fragment with the entire yfgL gene of strain LF82 | This study |
Invasion assay.
Bacterial invasion of human intestinal epithelial cells was performed by the gentamicin protection assay as described previously (8). Briefly, monolayers were seeded in 24-well tissue culture plates (Polylabo, Strasbourg, France) with 4 × 105 cells per well and incubated for 20 h. Monolayers were then infected at a multiplicity of infection (MOI) of 10 bacteria per cell in 1 ml of the cell culture medium without antibiotics and with heat-inactivated fetal calf serum (FCS) (BioWhittaker, Cambrex BioSciences, Verviers, Belgium). After a 3-h incubation period at 37°C, fresh cell culture medium containing 100 μg of gentamicin (Sigma-Aldrich, St. Louis, Mo.) per ml was added for 1 h to kill extracellular bacteria. Monolayers were washed three times in phosphate-buffered saline (PBS) (pH 7.2) and lysed with 1% Triton X-100 (Sigma-Aldrich) in deionized water. Samples were diluted and plated onto Mueller-Hinton agar plates to determine the number of CFU corresponding to the total number of intracellular bacteria. For coinfection experiments, both E. coli LF82-ΔyfgL isogenic mutant and wild-type LF82 strain or E. coli K-12 strain MC4100 were added to the cell monolayer at an equivalent MOI of 10. The number of intracellular LF82-ΔyfgL isogenic mutant bacteria was determined on Mueller-Hinton agar plates containing kanamycin. For pretreatment of the intestinal epithelial cells with OMVs, monolayers were incubated for 1 h in cell culture medium with heat-inactivated FCS in the presence of OMVs prepared as described below from 100 ml of the supernatant of a bacterial culture grown overnight in LB broth. Monolayers were then washed two times in PBS and infected in 1 ml of the cell culture medium at an equivalent MOI of 10.
Colony immunoblotting.
After overnight incubation at 37°C in LB broth without shaking, 1 ml of bacterial culture was harvested by centrifugation and resuspended in 100 μl of PBS. A 5-μl sample was spotted onto a nitrocellulose membrane (Amersham International, Buckinghamshire, England), and the membrane was treated as previously described (3).
Yeast cell aggregation assay.
Commercial baker's yeast (Saccharomyces cerevisiae) was suspended in PBS (5 mg [dry weight] per ml). Bacterial strains were grown overnight at 37°C without agitation on LB broth, washed, and resuspended in PBS at an optical density at 620 nm of 0.4. Equal volumes of yeast cell suspension and bacterial suspension were mixed on a glass slide. Aggregation was monitored visually, and the titer was recorded as the last dilution of bacteria giving a positive aggregation reaction.
Motility assay.
Bacterial strains were grown overnight at 37°C without agitation in LB broth, and 2-μl portions of the culture were inoculated into the center of 0.3% LB agar plates. The plates were incubated at 37°C for 18 h, and motility was assessed qualitatively by examining the circular swimming motion formed by the growing motile bacterial cells.
Transmission electron microscopy (TEM). (i) Negative staining.
Bacteria were grown overnight in LB broth without shaking, fixed, and negatively stained with 1% ammonium molybdate on carbon-Formvar copper grids (Electron Microscopy Sciences, Hatfield, Pa.).
Monolayers of Intestine-407 cells were infected at a MOI of 100 bacteria per cell in 2 ml of cell culture medium without antibiotics and with heat-inactivated FCS. After a 5-h incubation period at 37°C, monolayers were washed five times in PBS. Thin sections of Intestine-407 cells were prepared as previously described (8).
OMV preparations in 10 mM Tris-HCl (pH 8.0)-150 mM NaCl were placed on carbon-Formvar copper grids (Electron Microscopy Sciences). The grids were then stained with 1% ammonium molybdate.
(ii) Immunolabelling. Gold immunolabelling was performed by the method of Levine et al. (29). A drop of OMV preparation in 10 mM Tris-HCl (pH 8.0)-150 mM NaCl was placed on carbon-Formvar copper grids (Electron Microscopy Sciences). The grids were placed face down on a suitable dilution of antiserum raised against E. coli lipopolysaccharide O83 for 15 min. After three washes, the grids were placed on a drop of gold-labeled anti-rabbit serum (BB International, Cardiff, United Kingdom) for 15 min. After thorough washing, the grids were negatively stained with phosphotungstic acid (pH 6.0).
Construction of ΔyfgL isogenic mutants.
Isogenic mutants with the yfgL gene deleted were generated with a PCR product by the method of Chaveroche et al. (12). Briefly, the strategy was to replace a chromosomal sequence with a selectable antibiotic resistance gene (kanamycin) generated by PCR. This PCR product was generated by using primers with 50-nucleotide extensions that are homologous to regions adjacent to the yfgL gene and template E. coli strain carrying a kanamycin resistance gene (Table 2). In addition, E. coli strains were transformed with pKOBEG, a plasmid encoding Red proteins from phage λ under the control of a promoter induced by l-arabinose. These proteins protect linear DNA from degradation in bacteria. The plasmid was maintained in bacteria at 30°C with 25 μg of chloramphenicol per ml and was inactivated at 42°C.
TABLE 2.
Oligonucleotides used for PCR experiments
| Primer | Oligonucleotide sequence (5′ → 3′) | PCR product size (bp) | Use |
|---|---|---|---|
| yfgL1 | ATGATGCAGATGAAAATTAATAATTTGTCCATCTGA | 1,148 | ΔyfgL isogenic mutant construction |
| GAGGGACCCGAAAGCCACGTTGTGTCTCAA | |||
| yfgL2 | CGGCCCCTGTCCAGGAGCCGTTTTCAAAGTGAACG | ||
| ACAGAGACGAGCGCTGAGGTCTGCCTCGTG | |||
| A2GBL-3 | AAAGCCACGTTGTGTCTCAA | 957 | Kanamycin resistance cassette amplification |
| B2GBLnp5 | TTAGAAAAACTCATCGAGCA | ||
| yfgL3 | GTAGTGCATGGGAAGCAGGC | 1,378 | Isogenic mutant verification |
| yfgL4 | TAAATCATCAGACAACGCACGC | ||
| yfgL5 | CGTGGAGCACTTCCGTTGG | 941 | Isogenic mutant verification |
| yfgL6 | GCTGGGCAACGAAACGACC | ||
| yfgLXbaI | CCCGGGTCTAGATCCATCTGAGAGGGACCCGATGC | 1,263 | Cloning of yfgL |
| yfgLPstl | CTGCAGCGGCCCCTGTCCAGGAGCCG |
E. coli strains carrying pKOBEG were grown at 30°C with 1 mM l-arabinose to induce Red protein expression. When the optical density at 620 nm reached 0.5, the bacterial culture was incubated for 10 min at 42°C in order to inactivate the plasmid. Bacteria were washed three times with 10% glycerol, and PCR products were electroporated. ΔyfgL isogenic mutants were then selected on LB agar containing 50 μg of kanamycin per ml. The replacement of the yfgL gene by the kanamycin resistance cassette in ΔyfgL isogenic mutants was confirmed by PCR.
Transcomplementation assay of E. coli LF82-ΔyfgL isogenic mutant.
The yfgL gene was amplified by PCR (Perkin-Elmer thermal cycler) from E. coli LF82 genomic DNA (1 to 10 ng) using 2.5 U of Pfu DNA polymerase (Promega) and primers yfgLXbaI and yfgLPstI (Table 2) in Pfu DNA polymerase buffer containing 200 μM of each deoxynucleoside triphosphate. The amplified DNA was purified using NucleoSpin extract kit (Macherey-Nagel, Düren, Germany), digested with XbaI and PstI, and ligated to XbaI-PstI-digested arabinose-inducible expression vector pBAD33 (Table 1). In the resulting recombinant vector, named pPBI07, yfgL is expressed under the control of the ara promoter. This construct was used to transform the LF82-ΔyfgL isogenic mutant.
DNA manipulations and PCR experiments.
Standard DNA procedures were performed as described elsewhere (36). DNA to be amplified was released from bacteria by boiling. Bacteria were harvested from 1-ml samples of a culture grown overnight in LB broth, resuspended in 150 μl of sterile water, and incubated at 100°C for 15 min. The cell debris was pelleted by centrifugation, and 5-μl samples of the lysate were used in the PCRs. All PCR primer sequences are listed in Table 2.
Cell fractionation and OMV preparation.
Bacteria were grown without shaking at 37°C in LB broth. For whole-cell lysates, cells were pelleted and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (2% SDS, 50 mM Tris-HCl [pH 6.8], 12.5% glycerol, 400 mM β-mercaptoethanol, 0.01% bromophenol blue). For outer membrane preparations, cells were recovered and treated as previously described (35). For supernatants, 50-ml samples of culture were centrifuged at 20,000 × g, and the supernatants were collected and centrifuged further to remove cells and cell debris. Supernatants were filtered through a 0.20-μm-pore-size filter (Millipore, Billerica, Mass.). Proteins were precipitated on ice with 10% (wt/vol) trichloroacetic acid (Sigma-Aldrich) for 1 h. Proteins were collected by centrifugation, and pellets were washed twice with acetone. Proteins were resuspended in SDS-PAGE loading buffer. OMVs were isolated essentially by the method of Wai et al. (42). The culture supernatants were filtered, and OMVs were collected by ultracentrifugation at 150,000 × g for 3 h at 4°C. OMV pellets were resuspended in 10 mM Tris-HCl (pH 8.0)-150 mM NaCl.
SDS-PAGE and Western blot analysis.
Samples were resuspended in SDS-PAGE loading buffer and heated for 5 min at 95°C, and equivalent amounts of protein extract were separated by SDS-PAGE (12% polyacrylamide). Coomassie brilliant blue staining was performed as described elsewhere (36). Western immunoblotting was performed by the procedure of Towbin et al. (40). Proteins were electroblotted onto nitrocellulose membranes (Amersham International), and the membranes were immunoblotted for type 1 pili (rabbit antiserum raised against purified type 1 pilus preparations, diluted 1:1,000), Lep (rabbit anti-Lep, 1:1,000), MalE (rabbit anti-MalE, 1:1,000), OmpA (rabbit anti-OmpA, 1:1,000), OmpC/F (rabbit anti-OmpC/F, 1:1,000), and RNA polymerase alpha subunit (rabbit anti-alpha, 1:10,000). Immunoreactants were detected using horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (1:10,000), enhanced chemiluminescence reagents (Amersham International), and autoradiography.
MALDI-TOF MS analysis and protein identification.
After Coomassie brilliant blue staining, bands of interest were excised from the gel and washed for 1 h with 0.1 ml of a solution of 0.1 M ammonium bicarbonate and 50% acetonitrile and then for 15 min with 0.1 ml of 100% acetonitrile. The remaining solvent was removed by drying the gel pieces for 10 min in a vacuum centrifuge. Proteins were digested overnight at 37°C with 400 ng of trypsin (Promega). The peptide mixture was extracted from the gel by a solution of 50% acetonitrile and 0.05% trifluoroacetic acid, desalted, and concentrated using C18ZipTips (Millipore) according to the manufacturer's instructions. Typically, samples were eluted in 3 μl of 3 volumes of acetonitrile and 2 volumes of 0.1% trifluoroacetic acid and mixed 1:1 with a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% (vol/vol) acetonitrile-0.3% trifluoroacetic acid. Samples were applied to a target matrix-assisted laser desorption ionization (MALDI) plate and dried before analysis in the mass spectrometer. Mass spectrometry (MS) was performed using a PerSeptive Biosystems Voyager DE-Pro peptide mass spectrometer. Mass fingerprinting spectra were obtained with about 200 laser shots and calibrated using trypsin autolysis peaks as internal standards. Peptide mass fingerprint data from MALDI-time of flight (TOF) MS were compared with protein databases using Profound (Proteometrics) and Mascot search engine (33).
Statistical analysis.
For analysis of the significance of differences in invasion levels, Student's t test was used. All experiments were repeated at least three times. A P value less than or equal to 0.05 was considered statistically significant.
RESULTS
Phenotypes of E. coli strain LF82 mutants with the yfgL gene disrupted by Tn5phoA.
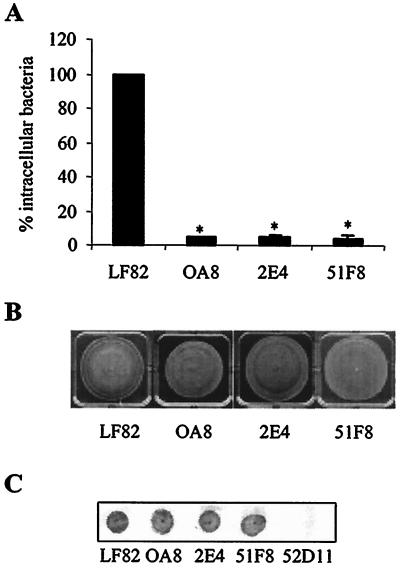
A quantitative invasion assay using intestinal epithelial cells (Intestine-407 cells) was used to measure the invasion levels of mutants with the Tn5phoA transposon inserted into the yfgL gene. The ability of three mutants, strains OA8, 2E4, and 51F8, to invade Intestine-407 cells was significantly decreased (P < 0.001) and was only 4.5, 5.2, and 4.1%, respectively, of that of strain LF82 (Fig. 1A). Since type 1 pili, flagella, and motility are known to play key roles in the invasion process of strain LF82 (3, 7), we examined the corresponding phenotypes of these mutants. On swim agar plate motility assays, the mutants were motile (Fig. 1B). Colony immunoblotting assay using polyclonal antibodies to type 1 pili showed that they expressed type 1 pili (Fig. 1C). These findings indicated that the decrease in the ability of mutants with the yfgL gene disrupted by Tn5phoA to invade intestinal epithelial cells was not caused by lack of type 1 pili or motility.
FIG. 1.
Invasion ability, motility, and type 1 pilus expression of E. coli mutants with the Tn5phoA transposon inserted in the yfgL gene. A. Intracellular bacteria in human intestinal epithelial cells (Intestine-407 cells) were quantified after a 3-h infection period, followed by gentamicin treatment for an additional hour. Results are expressed as the percentage of intracellular bacteria relative to that of E. coli strain LF82, which is set at 100%. The invasion level of the wild-type strain LF82 expressed as a mean percentage of the original inoculum was 0.95% ± 0.10%. Each value is the mean ± standard error of the mean (error bar) of at least four separate experiments. Values that were significantly different from the value for wild-type strain LF82 (P < 0.001) are indicated by an asterisk. B. Motility assay on 0.3% agar after 18 h at 37°C. Motility was visualized as a halo of radial diffusion of bacteria around the primary inoculum. C. Expression of type 1 pili, analyzed by colony immunoblotting using rabbit antiserum raised against purified type 1 pili. Mutant 52D11 with an insertion of Tn5phoA within the fimA gene was used as a negative control for pilus type 1 expression (7).
Phenotype of the E. coli LF82-ΔyfgL isogenic mutant.
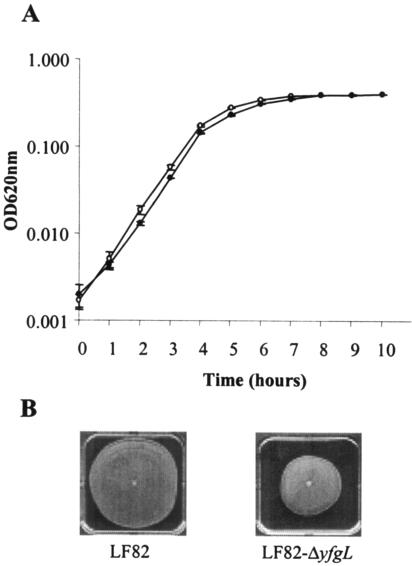
An LF82 isogenic mutant with the yfgL gene deleted was constructed as described in Materials and Methods. It has been suggested that the YfgL lipoprotein is involved in the regulation of lytic transglycosylases (autolysins) and may interfere with bacterial growth and/or bacterial viability (18). Therefore, we compared the growth rate of the LF82-ΔyfgL mutant with that of the wild-type strain LF82. Growth analysis was performed at 37°C without shaking in LB broth. As shown in Fig. 2A, the growth curves of strain LF82 and LF82-ΔyfgL isogenic mutant were similar.
FIG. 2.
Phenotype of the E. coli LF82-ΔyfgL isogenic mutant. A. Time course of bacterial growth in LB broth of AIEC LF82 wild-type strain (open circles) and LF82-ΔyfgL isogenic mutant (black circles). The initial inoculum was 4 × 106 bacteria per ml. Results are means ± standard errors of the means (error bars) of six separate experiments. OD620nm, optical density at 620 nm. B. Motility assay on 0.3% agar after 18 h at 37°C. Motility was visualized as a halo of radial diffusion of bacteria around the primary inoculum.
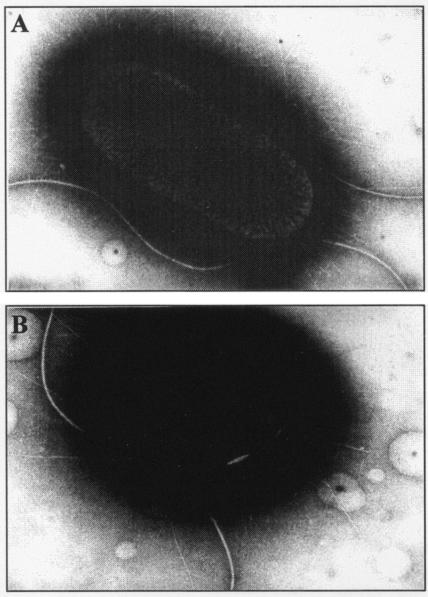
Motility assays indicated that the E. coli LF82-ΔyfgL isogenic mutant was less motile than the wild-type strain (Fig. 2B). To analyze whether the difference in swim size between LF82 and LF82-ΔyfgL isogenic mutant was dependent on flagellation, we performed TEM analysis. As shown in Fig. 3, the levels of flagella expressed by the LF82-ΔyfgL isogenic mutant were similar to those expressed by the wild-type strain. The difference observed in motility between the wild-type strain LF82 and the LF82-ΔyfgL isogenic mutant could involve factors other than the number of flagella, such as rotary speed or chemotactic ability. As E. coli mutants with transposon insertions were not affected in motility ability and since these mutants were PhoA positive, it is possible that YfgL domain fusions with alkaline phosphatase activity located outside the inner membrane are able to allow the expression of fully functional flagella.
FIG. 3.
Transmission electron micrograph of negatively stained E. coli LF82 bacteria (A) and LF82-ΔyfgL isogenic mutant (B). Magnifications: ×22,000 (A) and ×27,000 (B).
Yeast cell aggregation assays performed to analyze type 1 pilus expression indicated that the E. coli LF82-ΔyfgL isogenic mutant synthesized fourfold-lower titers of type 1 pili than the wild-type strain LF82 did (Table 3). The decrease in the number of surface-exposed type 1 pili in the LF82-ΔyfgL isogenic mutant was confirmed by TEM analysis, as shown in Fig. 3.
TABLE 3.
Enhanced expression of type 1 pili in E. coli LF82-ΔyfgL isogenic mutant does not modify invasion level
| Strain | Yeast aggregation (titer)a | Invasion level (%)b |
|---|---|---|
| LF82 | 1/8 | 100.00 ± 0.00 |
| LF82-ΔyfgL | 1/2 | 8.31 ± 0.69 |
| LF82-ΔyfgL/pPBI01 | 1/8 | 9.64 ± 0.44 |
Yeast cell aggregation was monitored visually, and the titer was recorded as the last dilution giving a positive aggregation reaction.
Percentage of intracellular bacteria relative to that obtained for AIEC strain LF82, which was set at 100%. Each value is the mean ± standard error of the mean of at least four separate experiments.
Role of yfgL gene in E. coli LF82 invasion.
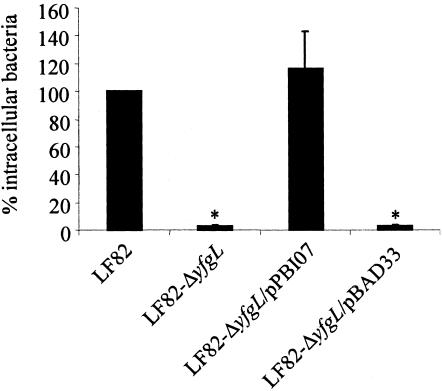
Quantitative invasion assays showed that the LF82-ΔyfgL isogenic mutant had consistently reduced ability to invade Intestine-407 cells, with 2.9% ± 0.7% residual invasion level compared to that of wild-type strain LF82, which was set at 100% (Fig. 4). Complementation experiments were performed using the PCR product of the yfgL gene from strain LF82 cloned into arabinose-induced pBAD33 expression vector. The resulting recombinant vector, named pPBI07, was electroporated into the LF82-ΔyfgL isogenic mutant. The invasion level of LF82-ΔyfgL isogenic mutant transformed with plasmid pPBI07 harboring cloned yfgL gene was restored to 116.0% ± 27.5% of that of strain LF82. Transformation with vector pBAD33 alone did not modify the invasion level of the LF82-ΔyfgL isogenic mutant. Therefore, the yfgL gene affects the ability of strain LF82 to invade intestinal epithelial cells.
FIG. 4.
Restoration of the invasion defect of the E. coli LF82-ΔyfgL isogenic mutant after transcomplementation with the plasmid pPBI07 harboring the entire LF82 yfgL gene. Invasion of Intestine-407 cells was determined after a 3-h infection period, followed by gentamicin treatment for an additional hour. Results are expressed as the percentage of intracellular bacteria relative to that obtained for wild-type strain LF82, which was set at 100%. The invasion level of the wild-type strain LF82 expressed as the mean percentage of the original inoculum was 1.31% ± 0.13%. Each value is the mean ± standard error of the mean (error bar) of at least three separate experiments. Values that were significantly different from the value for wild-type strain LF82 (P < 0.001) are indicated by an asterisk.
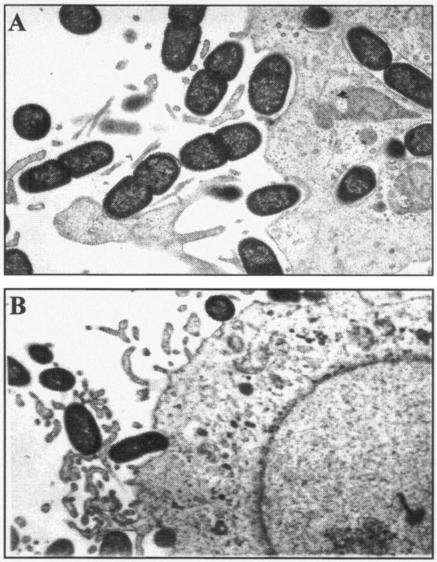
The interaction of the E. coli LF82-ΔyfgL isogenic mutant with intestinal epithelial cells was analyzed by TEM on cell monolayer sections after a 5-h infection period. LF82-infected Intestine-407 cells had numerous bacteria interacting with the host cell surface, elongated membrane extensions at the site of intimate contact between the entering bacteria and the epithelial cells, and several bacteria internalized within endocytic vacuoles (Fig. 5A). When epithelial cell monolayers were infected with LF82-ΔyfgL isogenic mutant, bacteria interacting with the intestinal epithelial cells induced morphological modifications of the host cell membranes similar to those observed with the wild-type strain LF82. However, no intracellular bacteria were found within the intestinal epithelial cells (Fig. 5B). This confirmed the decrease in invasion observed by quantitative invasion assays.
FIG. 5.
Transmission electron micrographs of Intestine-407 cells infected with E. coli strain LF82 (A) and LF82-ΔyfgL isogenic mutant (B). Strain LF82 adhered to Intestine-407 cells and induced the elongation of membrane extensions upon contact with the eukaryotic cell membranes. Numerous bacteria were found inside Intestine-407 cells. In contrast, no intracellular bacteria were observed with the LF82-ΔyfgL isogenic mutant, even if adhering bacteria induced membrane elongations. Magnification,×6,000.
The decreased ability of E. coli LF82-ΔyfgL isogenic mutant to invade intestinal epithelial cells could be related to the observed decreased expression of type 1 pili. Transformation of the LF82-ΔyfgL isogenic mutant was performed with plasmid pPBI01 harboring the cloned fim operon to induce overexpression of type 1 pili. The yeast cell aggregation assay showed that the levels of type 1 pili expressed in the LF82-ΔyfgL isogenic mutant harboring the plasmid pPBI01 and in the wild-type strain LF82 were similar (Table 3). However, the invasion level of LF82-ΔyfgL isogenic mutant harboring the plasmid pPBI01 was not increased, indicating that the yfgL gene is necessary for the full ability of strain LF82 to invade intestinal epithelial cells, regardless of type 1 pili. This prompted us to analyze secreted bacterial components involved in the invasive process of the AIEC strain LF82 that are absent in the LF82-ΔyfgL isogenic mutant.
Analysis of proteins released into the culture supernatant of E. coli strain LF82.
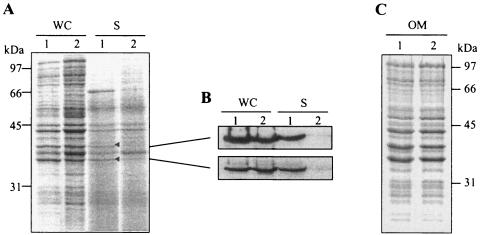
Proteins secreted from the wild-type strain LF82 and the LF82-ΔyfgL isogenic mutant were compared after collection of the supernatant, protein precipitation with trichloroacetic acid, and analysis by SDS-PAGE. In the supernatant of a culture of strain LF82 grown to early stationary phase, six prominently stained bands were observed in the range of 36 to 66 kDa, with apparent molecular masses of 36, 37, 39, 44, 45, and 66 kDa (Fig. 6A). The banding pattern of the LF82-ΔyfgL isogenic mutant differed from that of the wild-type strain. Two bands at 36 and 66 kDa were less intense than in the wild-type strain, and one band with an apparent molecular mass of 39 kDa was barely detected. As the 66-kDa protein was in the molecular mass range of flagellin subunits, we performed Western blot analysis with antibodies raised against flagellin H1. It revealed that this 66-kDa protein was the major flagellin subunit FliC (data not shown). The presence of flagellin FliC in the supernatant is mainly due to broken flagellar filaments or flagellar filaments detached from the surface of the bacteria and does not represent a quantitative measurement of FliC secretion. Moreover, as stated above, TEM analysis showed that the levels of flagella expressed by the LF82-ΔyfgL isogenic mutant were similar to those expressed by the wild-type strain. Therefore, we did not further investigate the decreased amount of FliC in the LF82-ΔyfgL isogenic mutant culture supernatant.
FIG. 6.
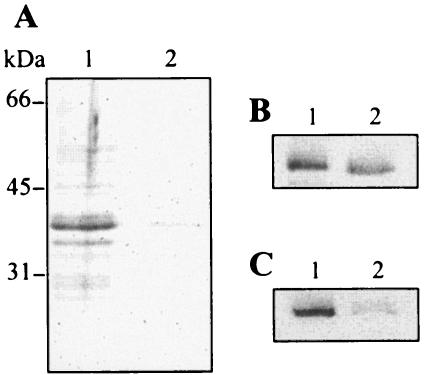
Release of proteins into the E. coli LF82 culture supernatant. Bacterial cells were grown without shaking at 37°C in LB broth to early stationary phase. A. Whole-cell lysates (WC) and secreted proteins in culture supernatants (S) of strain LF82 and LF82-ΔyfgL isogenic mutant were separated by SDS-PAGE and stained with Coomassie blue. The positions of secreted proteins in the culture supernatant of strain LF82 identified by MS are marked by a small black triangle. B. Western immunoblot analysis of OmpC/F (top) and OmpA (bottom) outer membrane proteins in whole-cell lysates (WC) and culture supernatants (S). The positions of OmpC/F and OmpA protein bands detected by antibodies correlate with the protein bands analyzed by MS. C. Outer membrane preparations (OM) of strain LF82 and LF82-ΔyfgL isogenic mutant were separated by SDS-PAGE and stained with Coomassie blue. The positions of molecular mass markers (in kilodaltons) are shown at the sides of the gels. Lanes: 1, strain LF82; 2, LF82-ΔyfgL isogenic mutant.
To analyze the 36- and 39-kDa proteins that were expressed differently by E. coli strain LF82 (wild type) and the LF82-ΔyfgL isogenic mutant, we performed MALDI-TOF MS. These two bands were unequivocally identified as outer membrane proteins using Profound (Proteometrics) and Mascot search engine. The 36-kDa protein was OmpA, and the 39-kDa protein was OmpC. These findings were confirmed by Western blot analysis using specific antibodies raised against OmpA and OmpC/F (Fig. 6B). Thus, while both OmpA and OmpC were accumulated in the supernatant of strain LF82, the LF82-ΔyfgL isogenic mutant had a defect for this accumulation. However, this defect was not seen in the whole-cell fraction of LF82-ΔyfgL mutant compared to that of strain LF82 (Fig. 6B). Similarly, strain LF82 and LF82-ΔyfgL isogenic mutant showed an overall equivalent pattern of protein accumulation in their outer membrane fractions (Fig. 6C). These observations indicate that even though strain LF82 and the mutant exported similar amounts of OmpA and OmpC to their outer membrane, their presence was decreased in the culture supernatant of the LF82-ΔyfgL isogenic mutant.
OMV formation by E. coli strain LF82 and LF82-ΔyfgL isogenic mutant.
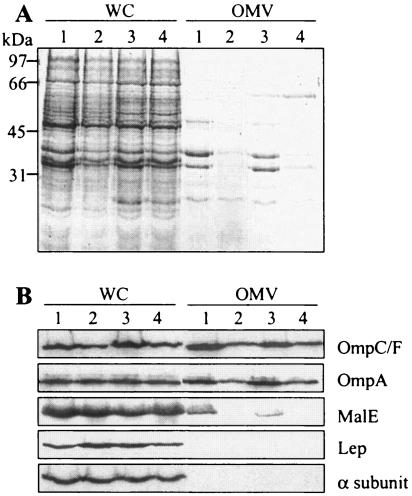
The presence of a large amount of outer membrane proteins in the culture supernatant of strain LF82 indicated the putative formation of OMVs discharged from the surface of the cell during bacterial growth. Using the protocol of Wai et al. (42) to concentrate vesicles, analysis of protein content of the culture supernatant of strain LF82 by SDS-PAGE showed five prominently stained bands with apparent molecular masses of 27, 30, 36, 39, and 48 kDa (Fig. 7A). MALDI-TOF MS indicated that the 36- and 39-kDa major bands were OmpA and OmpC. The three other bands at 27, 30, and 48 kDa were identified as the MltA-interacting protein A (MipA protein), the nucleoside-specific channel-forming protein Tsx, and a putative capsid protein of bacteriophage HK620, respectively. The overall protein contents of samples from the LF82-ΔyfgL mutant and from the LF82-ΔyfgL mutant transformed with pBAD33 were markedly lower than that of the wild-type strain (Fig. 7A). Transcomplementation of the LF82-ΔyfgL isogenic mutant with the cloned yfgL gene led to restoration of the proteins released in the culture supernatant to a level similar to that of strain LF82.
FIG. 7.
Analysis of protein content of OMVs released by E. coli strain LF82 and LF82-ΔyfgL isogenic mutant. Bacterial cells were grown overnight without shaking at 37°C in LB broth. A. Whole-cell lysates (WC) and OMV preparations were separated by SDS-PAGE and stained with Coomassie blue. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. B. Western immunoblot analysis of the OmpC/F and OmpA outer membrane proteins, the MalE periplasmic protein, the Lep inner membrane protein, and the α subunit of RNA polymerase cytoplasmic protein in whole-cell lysates (WC) and OMV preparations. Lanes: 1, strain LF82; 2, LF82-ΔyfgL isogenic mutant; 3, LF82-ΔyfgL isogenic mutant transcomplemented with pPBI07 harboring the entire yfgL gene; 4, LF82-ΔyfgL isogenic mutant transformed with the pBAD33 vector alone.
The presence of OMVs was confirmed by analysis of different subcellular fraction markers of OMV contents. As shown in Fig. 7B, the E. coli strain LF82 vesicle preparation was derived from the outer membrane, as shown by the presence of OmpA and OmpC, and specifically contained the maltose-binding protein MalE, a marker for periplasmic proteins. Vesicles were not contaminated by cytoplasmic or inner membrane markers, as shown by the Western blot analysis using anti-RNA polymerase alpha subunit or anti-leader peptidase (Lep) antisera, respectively. Western blot analysis revealed that in the LF82-ΔyfgL isogenic mutant vesicle preparation, the outer membrane proteins OmpA and OmpC were less abundant than in the vesicle preparation of the wild-type strain and that the periplasmic protein MalE was not detected. Analysis of vesicle preparation content of LF82-ΔyfgL isogenic mutant transcomplemented with the cloned yfgL gene revealed outer membrane and periplasmic protein patterns similar to those of wild-type strain LF82. All these results indicated that LF82 produced OMVs and that LF82-ΔyfgL mutant was impaired in the amount or composition of OMVs released, as the periplasmic MalE protein was absent or not detected in OMVs prepared from LF82-ΔyfgL mutant culture supernatant. The LF82-ΔyfgL mutant OMV preparation was concentrated 10-fold, and Western blot analysis using MalE antibodies indicated that the MalE protein, in addition to outer membrane proteins, was actually present in the resulting OMV preparation (data not shown). Therefore, the results of all the experiments taken together indicated that the absence of yfgL gene induced a decrease in the amount of OMVs released by the bacteria but did not affect the composition of the OMVs, at least for OmpA, OmpC, and MalE proteins.
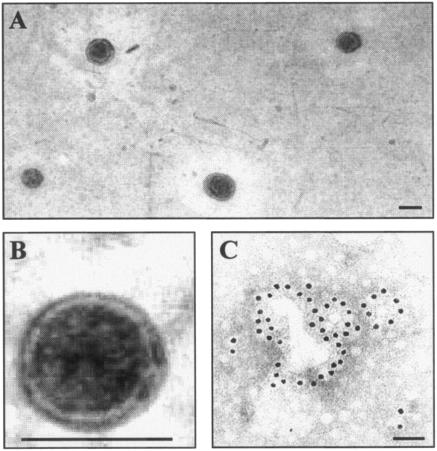
The presence of OMVs in culture supernatant of AIEC strain LF82 was confirmed by TEM analysis performed on ultracentrifuged cell-free supernatant pellets negatively stained with ammonium molybdate. Vesicles with a diameter of 50 to 200 nm were observed (Fig. 8A). Higher-resolution micrographs showed that they had a spherical and bilayered structure surrounding a central electron-dense core (Fig. 8B). Gold immunolabelling using O83 lipopolysaccharide-specific antibodies showed homogeneous distribution of gold particles, labeling round structures with a diameter of 50 to 200 nm, confirming the presence of OMVs in the culture supernatant of strain LF82 (Fig. 8C).
FIG. 8.
Transmission electron micrographs of negatively stained OMVs isolated from the culture supernatant of E. coli strain LF82 (A and B) and immunogold labeling of OMV preparation (C) using antibodies raised against E. coli lipopolysaccharide O83. Bars, 100 nm.
Effect of yfgL deletion in E. coli K-12.
E. coli K-12 strain MC4100 is known to release OMVs (25). To determine whether the yfgL gene was involved in the production of OMVs by E. coli K-12, we constructed a yfgL isogenic mutant of E. coli K-12 strain MC4100. As shown in Fig. 9A, the amount of OMVs released by the bacteria was lower in the isogenic mutant culture supernatant than that released in the wild-type strain culture supernatant. Western blot analysis revealed that in MC4100-ΔyfgL isogenic mutant vesicle preparation, the outer membrane protein OmpA and the periplasmic protein MalE were less abundant than in the wild-type strain vesicle preparation (Fig. 9B and C). Therefore, the yfgL gene product had a similar effect on OMV production in E. coli K-12 strain MC4100 and AIEC strain LF82.
FIG. 9.
Analysis of protein content of OMVs released by E. coli strain MC4100 and MC4100-ΔyfgL isogenic mutant. Bacterial cells were grown overnight without shaking at 37°C in LB broth. A. OMV preparations were separated by SDS-PAGE and stained with Coomassie blue. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. B and C. Western immunoblot analysis of OmpA (B) and MalE (C) in OMV preparation. Lanes: 1, strain MC4100; 2, MC4100-ΔyfgL isogenic mutant. Compared to Fig. 7, the E. coli K-12 OMV preparations were concentrated threefold.
Effects of OMVs on the invasion of the E. coli LF82-ΔyfgL isogenic mutant.
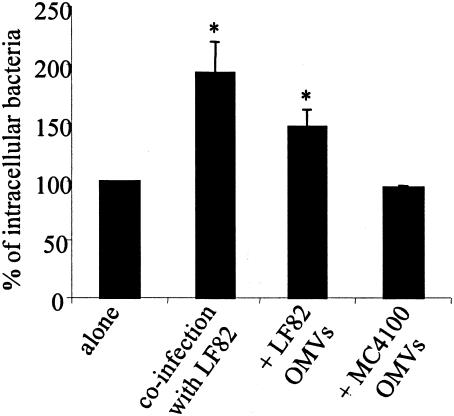
The ability of the LF82-ΔyfgL isogenic mutant to invade intestinal epithelial cells was analyzed in coinfection experiments together with the wild-type strain LF82. The invasion level of the LF82-ΔyfgL isogenic mutant was increased in coinfection experiments with LF82, reaching 193.0% ± 27.0% compared to the LF82-ΔyfgL isogenic mutant in monoinfection assay, which is set at 100% (Fig. 10). The invasion level of LF82-ΔyfgL isogenic mutant was also increased when the intestinal epithelial cells were pretreated with LF82 OMVs, reaching 142.2% ± 14.9% (Fig. 10). Interestingly, when the intestinal epithelial cells were pretreated with E. coli K-12 strain MC4100 OMVs, no increase in the invasion level of the LF82-ΔyfgL isogenic mutant was observed. In addition, in order to test whether the yfgL gene product is needed for invasion of other invasive bacteria, we engineered an yfgL isogenic mutant of the enteroinvasive E. coli strain E12860/0. The mean invasion level of the wild-type enteroinvasive E. coli strain E12860/0 was 17.32% ± 8.07% of the original inoculum and the mean invasion level of the enteroinvasive E. coli-ΔyfgL isogenic mutant was only 0.23% ± 0.14%, indicating that the yfgL gene product plays a role in the invasiveness of not only AIEC strain LF82 but also enteroinvasive E. coli strain E12860/0.
FIG. 10.
Effect of coinfection with wild-type E. coli strain LF82 or pretreatment of intestinal epithelial cells with LF82 OMVs on the invasive level of LF82-ΔyfgL isogenic mutant. For coinfection experiments, invasion of Intestine-407 cells by LF82-ΔyfgL isogenic mutant was determined after a 3-h infection period, followed by gentamicin treatment for an additional hour and determination of the number of bacteria on Mueller-Hinton agar plates containing kanamycin. For experiments in the presence of LF82 OMVs, Intestine-407 cells were pretreated with OMVs for 1 h and then infected by bacteria. Results are expressed as the percentage of intracellular bacteria relative to that obtained for LF82-ΔyfgL isogenic mutant, which was set at 100%. The invasion level of LF82-ΔyfgL isogenic mutant expressed as the mean percentage of the original inoculum was 0.15% ± 0.11%. Each value is the mean ± standard error of the mean (error bar) of at least three separate experiments. Values that were significantly different from the value for the LF82-ΔyfgL isogenic mutant alone (P < 0.001) are indicated by an asterisk.
DISCUSSION
The aim of the present study was to investigate the role of YfgL in CD-associated adherent-invasive E. coli strain LF82, since mutants with the Tn5phoA transposon inserted in the yfgL gene showed a large decrease in their ability to invade intestinal epithelial cells. An isogenic mutant with the yfgL gene deleted was engineered. The LF82-ΔyfgL mutant showed a 34-fold decrease in its invasion level. Its motility was moderately reduced, and it synthesized fewer type 1 pili than the wild-type strain LF82. Transcomplementation of the LF82-ΔyfgL mutant with the cloned yfgL gene fully restored the wild-type phenotype. To investigate whether the decrease in type 1 pilus expression could be responsible for the decrease in invasion ability of the yfgL mutant, we forced expression of type 1 pili in this mutant by transforming the LF82-ΔyfgL isogenic mutant with plasmid pPBI01 harboring the cloned fim operon. This fully restored type 1 pilus expression to a level similar to that of the wild-type strain. However, the defect in invasion was still observed. Similarly, the moderate attenuation of motility observed for the LF82-ΔyfgL isogenic mutant could not have been responsible for its invasion defect, since a flagellum-negative mutant of strain LF82 with overexpression of type 1 pili showed only a twofold decrease in its ability to invade intestinal epithelial cells (3).
Defects in released proteins from the E. coli LF82-ΔyfgL isogenic mutant were observed by analyzing the culture supernatant protein contents. SDS-PAGE analysis in parallel with MALDI-TOF MS of concentrated cell-free culture supernatants unequivocally indicated that the amounts of two prominent proteins present in wild-type strain LF82 supernatant were much lower in LF82-ΔyfgL isogenic mutant supernatant. They were identified as the outer membrane proteins OmpA and OmpC. The integral outer membrane proteins in the culture supernatant of strain LF82 resulted from the formation of vesicles, as confirmed by analysis of periplasmic and outer membrane fraction markers typically found in the composition of OMVs (6). Moreover, TEM analysis of ultracentrifuged cell-free LF82 supernatant pellets showed the presence of vesicles with a diameter of 50 to 200 nm which had a bilayered structure surrounding a central electron-dense core. Thus, deletion of the yfgL gene affected the release of the outer membrane proteins OmpA and OmpC in the culture supernatant by decreasing release of OMVs. We did not observe any difference in the composition of the OMVs released by the wild-type strain and the LF82-ΔyfgL isogenic mutant. However, our analysis was restricted to OmpA, OmpC, and MalE, and we cannot rule out the possibility that other bacterial proteins, including some virulence factor(s), were present in LF82 OMVs and absent in LF82-ΔyfgL OMVs.
The shedding of OMVs during the growth of gram-negative bacteria is a common phenomenon (6). OMVs arise from the surfaces of gram-negative bacteria and consist of outer membrane and entrapped periplasmic components. Two different mechanisms for the generation of OMVs have been proposed; one proposal is that OMVs are formed when the outer membrane expands faster than the underlying peptidoglycan layer (43) and another proposal is that the formation of OMVs is linked to turgor pressure of the cell envelope during bacterial growth (44). In the latter, the cell wall is excised and released from the peptidoglycan by lytic transglycosylases, and the accumulation of released muramyl peptides in the periplasmic space creates turgor pressure on the outer membrane and triggers OMV blebbing. The presence of MipA which forms a complex between the murein polymerase PBP1B and the lytic transglycosylase MltA (41) in the LF82 OMV preparation could indicate that LF82 OMVs contain peptidoglycan fragments.
There are at least two possible explanations of the origin of the defect in OMV production when the yfgL gene was deleted in E. coli strain LF82. A yfgL mutant could produce greater amounts of peptidoglycan than the wild-type strain as already shown for the E. coli K-12 strain MC4100 (18). To test this hypothesis, chloramphenicol was added to the bacterial culture to cause an accumulation of cell wall peptidoglycan (32). After treatment with chloramphenicol, a decrease in OMV release by wild-type strain LF82 was observed (data not shown), indicating that an accumulation of peptidoglycan could be involved in the AIEC LF82 OMV formation. However, another hypothesis needs to be considered, since it has been suggested that the yfgL gene product up-regulates lytic transglycosylases and is therefore involved in peptidoglycan turnover (18). Deletion of the yfgL gene in the E. coli K-12 strain MC4100 also affected OMV formation. It is likely that in the absence of the yfgL gene product in AIEC strain LF82 or in E. coli K-12, there is a down-regulation of lytic transglycosylases and consequently a decreased accumulation in the periplasm of muramyl peptides leading to less turgor pressure on the outer membrane and hence decreased OMV blebbing.
Mutations that affect the formation of vesicles in E. coli negatively or positively have already been reported. The histone-like protein HNS acting as a global regulator was shown to negatively affect the amount of vesicles in enterotoxigenic E. coli strains (21). Defects in proteins linking the outer membrane to the peptidoglycan layer or involved in a structural network between the inner and outer membranes and the peptidoglycan layer result in the shedding of large amounts of OMVs. Mutations in the major lipoprotein (Lpp) gene or in the tol-pal genes in E. coli K-12 strain modified the amount of OMVs released (5, 11). The level of vesicle formation increases when the tolA, tolQ, and tolR genes are mutated and decreases when the tolB and pal genes are mutated. Interestingly, TolB carries a six-blade beta-propeller domain (1, 10), and predictive structure analysis indicates that YfgL may have a similar domain (ExPASy [http://www.expasy.org]).
The yfgL gene product is required for invasion of other invasive bacteria. Indeed, we show in the present study that the deletion of the yfgL gene in the reference enteroinvasive E. coli strain E12860/0 led to a strong decrease in its invasion level. In addition, while the present article was being reviewed, Amy et al. reported that deletion of the yfgL gene in Salmonella enterica serovar Enteritidis induced a decrease in its ability to invade intestinal epithelial cells and a decreased secretion of bacterial effectors by the type III secretion system (TTSS) (2). The role of yfgL in enteroinvasive E. coli and S. enterica serovar Enteritidis invasion could be indirectly related to a defect in invasin secretion by TTSS. It has been shown in S. enterica serovar Typhimurium that in the absence of protein-peptidoglycan interactions due to structural modifications of the peptidoglycan, the assembly of the TTSS needle complex is abolished, leading to a reduction in protein secretion and in bacterial invasion of cultured eukaryotic cells (34). Therefore, at least in invasive bacteria, the yfgL gene product interfering with peptidoglycan synthesis and/or degradation does overall play a crucial role in virulence. It can modulate the secretion of bacterial compounds essential for the bacteria to invade epithelial cells by acting either on the assembly of the TTSS or on OMV release.
OMVs are a potential vehicle for intercellular transport of virulence factors by various gram-negative bacteria. Through their interaction with eukaryotic cells, they can deliver vesicle components and virulence factors. Interestingly, we observed that pretreatment of intestinal epithelial cells with E. coli LF82 OMVs increased the invasion level of the LF82-ΔyfgL isogenic mutant as well as coincubation of LF82-ΔyfgL isogenic mutant and wild-type strain LF82. In addition, the effect of LF82 OMVs was specific, since no increase in the invasion level of LF82-ΔyfgL isogenic mutant was observed when cells were pretreated with OMVs prepared from E. coli K-12 MC4100 culture supernatant, suggesting that LF82 OMVs can contain bacterial effectors not present in K-12 OMVs. Besides, transport of virulence factors via OMVs was reported in enterohemorrhagic E. coli strains with Shiga-like toxin, in Helicobacter pylori with VacA, in enterotoxigenic E. coli strains with heat-labile enterotoxin, and in Actinobacillus actinomycetemcomitans with leukotoxin (15, 19, 21-24, 26, 27, 30). Interestingly, the secretion of some virulence factors via OMVs is necessary for their full maturation, as recently reported for the activation of the pore-forming cytotoxin ClyA before its delivery to epithelial cells (42). Moreover, the outer membrane adhesin and invasin Ail from Yersinia enterocolitica was shown to be transported and delivered to epithelial cells via OMVs when expressed heterologously in E. coli (25).
In conclusion, our results show that the yfgL gene product plays a key role in the virulence of AIEC strain LF82, since the engineered LF82-ΔyfgL isogenic mutant had a strong decreased ability to invade intestinal epithelial cells which is concomitant with a decrease in OMV release. It is tempting to speculate that the OMVs released by AIEC strain LF82 contain virulence factors that contribute to the invasion process.
Acknowledgments
This study was supported in part by a grant from the Ministère de la Recherche et de la Technologie (PRFMMIP 2000) and by grants from the Association F. Aupetit (AFA) and Institut de Recherche des Maladies de l'Appareil Digestif (IRMAD, Laboratoire Astra France).
We thank Michael Donnenberg (Division of Infectious Diseases, University of Maryland School of Medicine, Baltimore) for enteroinvasive E. coli strain E12860/0, Karen Krogfelt and the World Health Organization International Escherichia coli Centre (Copenhagen, Denmark) for E. coli type 1 pilus antibodies, Roland Lloubès (CNRS UPR 9027, Institut de Biologie Structurale et Microbiologie, Marseille, France) for OmpA and OmpC/F antibodies, Jean Michel Betton (Unité de Repliement et Modélisation des Protéines, Institut Pasteur, Paris, France) for MalE antibodies, Annie Kolb (Unité des Régulations Transcriptionnelles, Département de Microbiologie Fondamentale et Médicale, Institut Pasteur, Paris, France) for α subunit of RNA polymerase antibodies, Jan-Willer de Gier (Department of Biochemistry and Biophysics, Arrhenius Laboratories, Stockholm University, Stockholm, Sweden) for Lep antibodies, and Lothar Beutin for O83 antibodies (Division of Emerging Bacterial Pathogens, Department of Biological Safety, Robert Koch Institut, Berlin, Germany). We also thank Annie Fraisse and Monique Oron (Département de Microscopie Electronique, Faculté de Médecine, Université d'Auvergne, Clermont-Ferrand, France) for technical assistance with electron microscopy and Christophe Chambon (Plateforme Protéomique, Station de Recherche sur la Viande, INRA, Theix, France) for help with MALDI-TOF MS analysis.
REFERENCES
- 1.Abergel C., E. Bouveret, J. M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Benedetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Struct. Fold Des. 7:1291-1300. [DOI] [PubMed]
- 2.Amy, M., P. Velge, D. Senocq, E. Bottreau, F. Mompart, and I. Virlogeux-Payant. 2004. Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res. Microbiol. 155:543-552. [DOI] [PubMed] [Google Scholar]
- 3.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781-794. [DOI] [PubMed] [Google Scholar]
- 4.Barnich, N., M. A. Bringer, L. Claret, and A. Darfeuille-Michaud. 2004. Involvement of lipoprotein NlpI in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 72:2484-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubes. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272-1284. [DOI] [PubMed] [Google Scholar]
- 8.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke, D. A., and A. T. Axon. 1988. Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhoea. BMJ 297:102-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure Fold. Des. 8:57-66. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [Online.] http://nar.oupjournals.org/cgi/content/full/28/22/e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darfeuille-Michaud, A., J. Boudeau, P. Bulois, C. Neut, A. L. Glasser, N. Barnich, M. A. Bringer, A. Swidsinski, L. Beaugerie, and J. F. Colombel. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412-421. [DOI] [PubMed] [Google Scholar]
- 14.Darfeuille-Michaud, A., C. Neut, N. Barnich, E. Lederman, P. Di Martino, P. Desreumaux, L. Gambiez, B. Joly, A. Cortot, and J. F. Colombel. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405-1413. [DOI] [PubMed] [Google Scholar]
- 15.Demuth, D. R., D. James, Y. Kowashi, and S. Kato. 2003. Interaction of Actinobacillus actinomycetemcomitans outer membrane vesicles with HL60 cells does not require leukotoxin. Cell. Microbiol. 5:111-121. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg, M. S., A. Donohue-Rolfe, and G. T. Keusch. 1990. A comparison of HEp-2 cell invasion by enteropathogenic and enteroinvasive Escherichia coli. FEMS Microbiol. Lett. 57:83-86. [DOI] [PubMed] [Google Scholar]
- 17.Duchmann, R., H. Lochs, and W. Kruis. 1999. Crohn disease, ulcerative colitis. When bacteria attack the intestinal wall. MMW Fortschr. Med. 141:48-51. [In German.] [PubMed] [Google Scholar]
- 18.Eggert, U. S., N. Ruiz, B. V. Falcone, A. A. Branstrom, R. C. Goldman, T. J. Silhavy, and D. Kahne. 2001. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294:361-364. [DOI] [PubMed] [Google Scholar]
- 19.Fiocca, R., V. Necchi, P. Sommi, V. Ricci, J. Telford, T. L. Cover, and E. Solcia. 1999. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188:220-226. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horstman, A. L., and M. J. Kuehn. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538-32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadurugamuwa, J. L., and T. J. Beveridge. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615-621. [DOI] [PubMed] [Google Scholar]
- 23.Kadurugamuwa, J. L., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, S., Y. Kowashi, and D. R. Demuth. 2002. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 32:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Kesty, N. C., and M. J. Kuehn. 2004. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesty, N. C., K. M. Mason, M. Reedy, S. E. Miller, and M. J. Kuehn. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolling, G. L., and K. R. Matthews. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamps, L. W., K. T. Madhusudhan, J. M. Havens, J. K. Greenson, M. P. Bronner, M. C. Chiles, P. J. Dean, and M. A. Scott. 2003. Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn's disease. Am. J. Surg. Pathol. 27:220-227. [DOI] [PubMed] [Google Scholar]
- 29.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, and R. Patnaik. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Z., A. J. Clarke, and T. J. Beveridge. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180:5478-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Y., H. J. van Kruiningen, A. B. West, R. W. Cartun, A. Cortot, and J. F. Colombel. 1995. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 108:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattingly, S. J., L. Daneo-Moore, and G. D. Shockman. 1977. Factors regulating cell wall thickening and intracellular iodophilic polysaccharide storage in Streptococcus mutans. Infect. Immun. 16:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 34.Pucciarelli, M. G., and F. Garcia-del Portillo. 2003. Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48:573-585. [DOI] [PubMed] [Google Scholar]
- 35.Pugsley, A. P., and C. A. Schnaitman. 1979. Factors affecting the electrophoretic mobility of the major outer membrane proteins of Escherichia coli in polyacrylamide gels. Biochim. Biophys. Acta 581:163-178. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sartor, R. B., H. C. Rath, S. N. Lichtman, and E. A. van Tol. 1996. Animal models of intestinal and joint inflammation. Baillieres Clin. Rheumatol. 10:55-76. [DOI] [PubMed] [Google Scholar]
- 38.Schultsz, C., M. Moussa, R. van Ketel, G. N. Tytgat, and J. Dankert. 1997. Frequency of pathogenic and enteroadherent Escherichia coli in patients with inflammatory bowel disease and controls. J. Clin. Pathol. 50:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 40.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer, W., M. von Rechenberg, and J. V. Holtje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 42.Wai, S. N., B. Lindmark, T. Soderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 43.Wensink, J., and B. Witholt. 1981. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 116:331-335. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, L., R. Srisatjaluk, D. E. Justus, and R. J. Doyle. 1998. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol. Lett. 163:223-228. [DOI] [PubMed] [Google Scholar]