Abstract
The retinoblastoma tumor suppressor (RB) is functionally inactivated in the majority of cancers and is a critical mediator of DNA damage checkpoints. Despite the critical importance of RB function in tumor suppression, the coordinate impact of RB loss on the response to environmental and therapeutic sources of damage has remained largely unexplored. Here, we utilized a conditional knockout system to ablate RB in adult fibroblasts. This model system enabled us to investigate the temporal role of RB loss on cell cycle checkpoints and DNA damage repair following ultraviolet (UV) and ionizing radiation (IR) damage. We demonstrate that RB loss compromises rapid cell cycle arrest following UV and IR exposure in adult primary cells. Detailed kinetic analysis of the checkpoint response revealed that disruption of the checkpoint is concomitant with RB target gene deregulation, and is not simply a manifestation of chronic RB loss. RB loss had a differential effect upon repair of the major DNA lesions induced by IR and UV. Whereas RB did not affect resolution of DNA double-strand breaks, RB-deficient cells exhibited accelerated repair of pyrimidine pyrimidone photoproducts (6-4 PP). In parallel, this repair was coupled with enhanced expression of specific factors and the behavior of proliferating cell nuclear antigen (PCNA) recruitment to replication and repair foci. Thus, RB loss and target gene deregulation hastens the repair of specific lesions distinct from its ubiquitous role in checkpoint abrogation.
INTRODUCTION
Cells have evolved complex mechanisms of genome surveillance and DNA repair to maintain genetic stability in the face of bombardment by exogenous insult (1–3). Cell cycle checkpoint pathways are examples of evolutionarily conserved responses to DNA damage (4). Following recognition of DNA lesions, such as those induced by ultraviolet radiation (UV) and ionizing radiation (IR), cell cycle checkpoints are elicited to limit the propagation of deleterious mutations to daughter cells. Several checkpoint proteins play essential roles in the maintenance of appropriate DNA damage response. A critical mediator of cell cycle control involved in the DNA damage checkpoint is the retinoblastoma tumor suppressor protein (RB). During early G1 phase of the cell cycle, hypophosphorylated RB is active and binds to members of the E2F transcription factor family to antagonize their function. The RB–E2F complex forms on the promoters of a multitude of E2F target genes to repress transcription. E2F is known to regulate many downstream targets that are involved in cell cycle progression (e.g. cyclin A, cyclin E, cdc2 and cdk2) and DNA replication [e.g. proliferating cell nuclear antigen (PCNA), mini-chromosome maintenance-7 (MCM-7), topoisomerase IIα, thymidine kinase] (5,6). Due to the requisite nature of these target genes, RB-mediated transcriptional repression inhibits progression into S-phase. Control of RB binding to E2Fs is exerted in mid-G1 by the activation of cdk4/cyclin D1 and cdk2/cyclin E, which phosphorylate and inactivate RB thereby allowing S-phase entry (7–9). DNA damage has the general influence of activating RB by promoting dephosphorylation. Following DNA damage, the presence of RB is required for cell cycle inhibition (10–13). This response has typically been assessed using mouse embryonic fibroblasts, wherein RB is believed to facilitate arrest by transcriptional repression of key targets. However, prior studies have been limited to analysis of the effect of chronic RB loss, rather than the acute inactivation evident in cancer.
It has been reported that RB function is impaired in the majority of cancers as the activities of several disparate mechanisms result in its functional inactivation (14–18). Presumably, RB loss contributes to genetic instability by allowing cells to evade cell cycle regulation and facilitating DNA damage checkpoint bypass. Consistent with this idea, it has been shown that RB suppresses the development of aneuploidy following damage (19). While RB is implicated in gross chromosome instability, its effect on DNA repair remains unexplored. However, a role for RB in repair has recently been suggested by the finding that several RB/E2F regulated genes are involved in the repair of UV and IR damage (20–24). Therefore, it can be envisioned that RB loss and downstream target deregulation could have distinct effects upon the cellular response to genotoxic insult, including both checkpoint deregulation and aberrant repair.
To probe these responses, we investigated the role of RB in UV and IR damage signaling, checkpoint activation and lesion repair in adult primary cells containing acute RB loss. Here, we report that RB function is critical for induction of a rapid cell cycle checkpoint in response to these agents. Additionally, we find that the DNA damage checkpoint bypass is concomitant with RB deletion and downstream target deregulation. Abrogation of the DNA damage checkpoint was associated with accelerated pyrimidine pyrimidone photoproduct (6-4 PP) repair and rapid engagement of DNA damage repair factors. Taken together, our data demonstrate that RB loss facilitates abrogation of transient cell cycle arrest following environmentally and therapeutically relevant doses of UV and IR, while contributing specifically to the acceleration of UV lesion repair.
MATERIALS AND METHODS
Isolation of primary RbloxP/loxP murine adult fibroblasts
Floxed Rb mice (RbloxP/loxP) of mixed 129/FVBN background (25), at least five weeks of age, were sacrificed by CO2 anesthetization followed by cervical dislocation. Fibroblasts were isolated from the peritoneal fascia by excision, mincing of the peritoneum and constant agitation for 40 min at 37°C in 0.2 mg/ml collagenase (Type I, Sigma) supplemented with 100 U Dnase I (Roche). The dissociated tissue was washed with PBS and subsequently incubated for 20 min at 37°C in 0.25% trypsin (Gibco) with constant agitation. After two PBS washes, the isolated cells were plated in tissue culture dishes.
Cell culture, recombinant adenoviral infections
RbloxP/loxP murine adult fibroblasts (MAFs) were subcultured in DMEM containing 10% fetal bovine serum supplemented with 100 U/ml penicillin/streptomycin and 2 mM L-glutamine at 37°C in air containing 5% CO2. In this study, all primary cells were between passages 2 and 4. Replication defective recombinant adenovirus expressing green fluorescent protein (Ad-GFP) or GFP in addition to Cre recombinase (Ad-GFP-Cre) were obtained from G. Leone (Department of Molecular Genetics, Ohio State University). The conditional RB knockout in primary RbloxP/loxP MAFs were attained by infecting cells with adenovirus at approximately 2 × 107 virus particles per dish to achieve an infection efficiency of 90–95% as determined by GFP fluorescence. Cells were cultured for at least four days post-adenoviral infection prior to use while the passage number and length of time post-infection remained consistent throughout all experiments unless otherwise stated.
Immunoblotting
Cells infected with Ad-GFP or Ad-GFP-Cre were harvested by trypsinization and lysed in RIPA buffer. Equal amounts of protein, as determined by Bio-RAD DC assay, were resolved by SDS–PAGE. Specific proteins were detected by standard immunoblotting procedures using the following primary antibodies: (Santa Cruz, 1:500 dilution) PCNA (pc10), Cyclin E (HE12), Cyclin A (C-19), MCM-7 (141.2), Cyclin B1 (GNS1), Lamin B (sc-6217), anti-RB (G3-245, Becton Dickson, 1:100 dilution), total p53 Ab-3 (Oncogene OP29, 1:250 dilution) and phospho-p53 ser-18 (Cell Signaling 9284S, 1:500 dilution).
RT–PCR analysis of recombination
RT–PCR analysis was performed to verify adenoviral-Cre-mediated recombination in primary MAFs. Total RNA was extracted using Trizol (Gibco) and cDNA was synthesized from 1 μg of RNA with the SuperScript RT–PCR system (Gibco) according to the manufacturer's protocol. cDNAs were amplified using PCR and the following primers: (sense) 5′-CCTTGAACCTGCTTGTCCTC-3′ and (antisense) 5′-GAAGGCGTGCACAGAGTGTA-3′. PCR conditions consisted of initial denaturation for 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 52°C and 1 min at 72°C, followed by a final extension for 5 min at 72°C. Ten microliters of PCR product was run on a 2% agarose gel and visualized by ethidium bromide staining.
DNA damage, PCNA extraction and immunofluorescence
Primary MAFs infected with Ad-GFP or Ad-GFP-Cre were seeded on coverslips in 6-well dishes and allowed to adhere. Cells were treated at room temperature either with ionizing radiation through exposure to 137Cs (dose rate: 0.67 Gy/min) in tissue culture media or with ultraviolet irradiation (UVC) (low pressure mercury lamp; Mineralight lamp model UVG-11; UVP, Inc. San Gabriel, CA) following removal of DMEM and washing the cells twice with PBS. Treated cells were labeled with BrdU (Amersham Pharmacia Biotech) to detect DNA synthesis, then washed, fixed in 3.7% formaldehyde, and processed to detect and quantitate BrdU incorporation by immunofluorescence and cell scoring as previously described (26). All BrdU results are expressed as a percentage of untreated control cells set to 100%. PCNA extraction and immunofluorescence was performed as previously described (27) using the monoclonal pc10 PCNA antibody (Santa Cruz). γH2AX immunofluorescence was performed as previously described (28) using an anti-phospho-H2AX ser-139 mouse primary antibody (Upstate Biotechnology). CPD and 6-4 photoproduct (6-4 PP) staining was performed as described in Wang et al. (29) using antibodies generously contributed by Dr Tsukasa Matsunaga (Kanazawa University, Japan). Relative staining intensities of γH2AX, CPD lesions and 6-4 PP were quantified by capturing images of equal exposure using microscopy and performing densitomery using Metamorph. All data are from 10 nuclei captured on random fields.
Immunoassay for repair of CPD and 6-4 PP
CPD and 6-4 PP present in cellular DNA were detected and quantified by slot blot. Total genomic DNA was extracted using the DNEasy tissue kit (Qiagen) according to manufacturer's instructions. DNA was quantified through spectrophotometry and gel electrophoresis prior to denaturation through boiling and sonication. Increasing concentrations of DNA were loaded for slot blot transfer using a vacuum blotter and hybridization onto a Nytran membrane. The membrane was blocked in 10% milk/1× saline-tween and probed with either 1:1000 primary monoclonal antibody specific for CPD or 6-4 PP DNA lesions (T. Matsunaga). Filters were then processed according to standard western blot protocol and lesion abundance was quantified using densitometry following the subtraction of background. To control for equal loading, experiments were done in triplicate and percent reduction was calculated between equal DNA concentrations from different time points within the same cell type.
RESULTS
Acute downregulation of RB protein abrogates the DNA damage checkpoint response to UV and IR
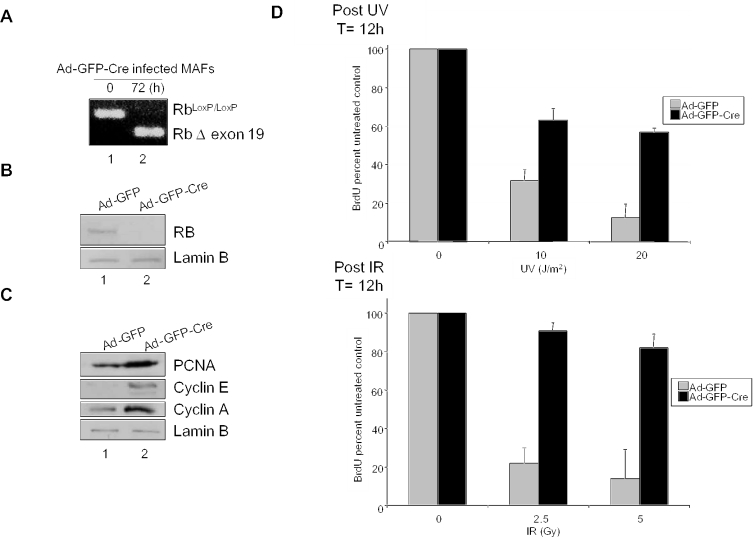
Studies of RB function have historically utilized mouse embryo fibroblasts (MEFs) harboring loss of RB throughout development or extensively cultured tumor lines. There is a caveat in these models, in that RB loss is compensated by RB related pocket proteins (i.e. p107 and p130) during development (30,31). In contrast, RB is acutely lost in the majority of cancer cases (14–18). Thus, we utilized an knockout system involving mice harboring a conditional Rb allele in which loxP sites flank Rb exon 19 (RbloxP/loxP mice) (25). Through adenoviral expression of Cre recombinase, acute RB loss can be achieved in genetically stable MAFs. To examine the action of RB in cells, MAFs were initially infected with recombinant adenoviruses expressing both GFP and Cre recombinase (Ad-GFP-Cre) or GFP alone (Ad-GFP) as a control. Efficient infection of cells was evident following 16–24 h, as >90% of Ad-GFP-Cre infected cells demonstrated high levels of GFP fluorescence (data not shown). Confirmation of Cre-mediated recombination was performed by RT–PCR analysis using primers in exons 18 and 20 of the Rb gene. RNA was prepared from uninfected MAFs or those infected with Ad-GFP-Cre at 72 h post-infection. RT–PCR analysis revealed loss of Rb RNA and accumulation of the Δexon19 transcript in the infected cells relative to the control (Figure 1A). Immunoblotting with anti-RB monoclonal antibody revealed that the Cre-mediated recombination resulted in acute downregulation of RB protein in Ad-GFP-Cre infected MAFs (Figure 1B).
Figure 1.
Acute downregulation of RB protein abrogates the DNA damage checkpoint response to UV and IR. (A) Asynchronously proliferating primary RbloxP/loxP MAFs were infected with Ad-GFP or Ad-GFP-Cre adenovirus. RNA was isolated at 0 and 72 h post-infection and RT–PCR was performed employing primers specific for regions flanking the loxP sites in the murine RB gene. The appearance of the smaller transcript post-infection indicates recombination at the floxed RB locus. (B) MAFs infected with adenoviruses encoding either GFP (lane 1) or GFP-Cre (lane 2) were harvested five days post-infection in RIPA buffer. Equal amounts of protein were separated by SDS–PAGE and immunoblotted with an anti-RB monoclonal antibody. Lysates were immunoblotted with the polyclonal Lamin B antibody to control for equal loading. (C) MAFs infected with Ad-GFP (lane 1) or Ad-GFP-Cre (lane 2) were harvested five days post-infection and equal concentrations of each protein were separated by electrophoresis. The effect of acute RB loss on downstream target expression was analyzed by immunoblotting for PCNA, cyclin E and cyclin A. Lamin B serves as a loading control. (D) Top panel: asynchronously proliferating primary RbloxP/loxP MAFs infected with either Ad-GFP control or Ad-GFP-Cre adenoviruses were irradiated with 0, 10 or 20 J/m2 UV. Treated cells were cultured for 12 h and labeled with BrdU for the final 2 h prior to harvest. The proliferative fraction of treated cells was determined with respect to untreated control through immunofluorescence using an anti-BrdU antibody. Bottom panel: Ad-GFP and Ad-GFP-Cre infected MAFs were exposed to 0, 2.5 or 5 Gy gamma irradiation and were cultured for 12 h while in the presence of BrdU for the final 2 h prior to harvest. Immunofluorescence for BrdU was performed to determine the percent of cells that progressed through S-phase during the labeling period.
To delineate the consequence of conditional RB ablation on the RB/E2F signaling axis, MAFs infected with either Ad-GFP or Ad-GFP-Cre were harvested five days post-infection and levels of specific RB target proteins were analyzed by immunoblot. Relative to control (Figure 1C, lane 1), the Ad-GFP-Cre infected MAFs exhibited increased levels of proteins downstream of RB signaling including, PCNA, cyclin E and cyclin A (lane 2). No changes were detected in lamin B protein levels, which served as a loading control. Therefore, RB deletion in primary adult cells results in target gene deregulation.
To evaluate the role of RB in the DNA damage response of adult fibroblasts, asynchronously proliferating Ad-GFP or Ad-GFP-Cre infected MAFs were exposed to 0, 10 or 20 J/m2 UV and subsequently cultured for 10 h to elicit the checkpoint response. Cells were pulse labeled with bromodeoxyuridine (BrdU) for 2 h and the replicative fraction of treated cells was determined by immunofluorescent detection of BrdU incorporation. MAFs containing functional RB exhibited a dose-dependent cell cycle inhibition (relative to untreated control), whereas cells lacking RB exhibited minimal responses at each dose (Figure 1D, top panel). Similar results were evident when the response to therapeutic doses of IR was investigated in the same manner. Following exposure to 0, 2.5 or 5 Gy IR, Ad-GFP infected MAFs exhibited a robust dose-dependent inhibition of cell cycle in which BrdU incorporation was reduced by greater than 75%, while Ad-GFP-Cre infected cells were largely unaffected (Figure 1D, bottom panel). Taken together, these data demonstrate that acute deletion of RB in primary adult cells results in abrogation of the DNA damage checkpoint response to both IR and UV irradiation.
RB loss compromises the rapid checkpoint response to IR/UV
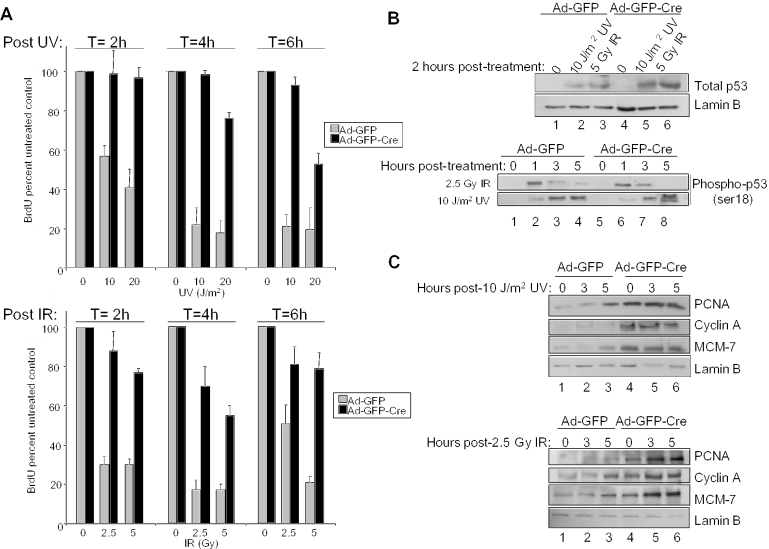
Traditionally, RB has been characterized as participating in checkpoint responses with delayed kinetics. In part, this is due to the use of chemotherapeutic agents wherein the induction of DNA damage is delayed due to drug action (32–34). Thus, one of the advantages of studying the cellular response to UV or IR is the immediate induction of DNA damage. To understand the kinetics of cell cycle attenuation following exposure to UV and IR, Ad-GFP and Ad-GFP-Cre infected MAFs were exposed to either 0, 10 or 20 J/m2 UV or 0, 2.5 or 5 Gy IR and cultured. Following damage, cells were pulsed with BrdU for the final 2 h in culture prior to harvesting at 2, 4 and 6 h. The DNA damage checkpoint was evident as early as 2 h following exposure to either UV or IR as determined by immunofluorescent detection of BrdU incorporation (Figure 2A). Additionally, this response was maintained for at least 6 h following damage. Surprisingly, this rapid response to UV and IR damage was compromised in RB-deficient cells, as Ad-GFP-Cre infected MAFs largely bypassed cell cycle inhibition at each time point. These data indicate that RB loss is sufficient to bypass the rapid checkpoint response to DNA damage.
Figure 2.
Acute RB loss compromises the rapid checkpoint response to UV and IR. (A) Top panel: Ad-GFP and Ad-GFP-Cre infected MAFs were exposed to 0, 10 or 20 J/m2 UV and cultured for 2, 4 or 6 h in the presence of BrdU for the final 2 h. Immunofluorescence was utilized to detect BrdU labeled nuclei, which were then scored and represented as percent untreated control. Bottom panel: the aforementioned experiment was repeated following treatment with 0, 2.5 or 5 Gy IR. (B) To assess the role of RB in rapid DNA damage signaling, asynchronously growing MAFs infected with either Ad-GFP or Ad-GFP-Cre were exposed to 2.5 Gy IR or 10 J/m2 UV, and harvested at various time points in RIPA buffer. Equal amounts of protein were separated by electrophoresis and immunoblotting for total p53 (top panel) and phosphorylated p53 (ser-18) (bottom panel) (C) To analyze the role of RB in DNA damage signaling to downstream targets, cells were treated and harvested as in (B) and immunoblotting for PCNA, cyclin A and MCM-7 (top and bottom panels) was performed. Lamin B serves as a control for equal loading.
Since it is postulated that RB signals via repression of target genes, we investigated the rapid action of DNA damage on RB target genes. In response to IR or UV damage, ser-18 of p53 (homologous to human ser-15) is known to be rapidly phosphorylated by ATM family kinases (35). Therefore, as a control for upstream signaling, immunoblotting for total p53 and phospho-p53 ser-18 was performed. To analyze rapid DNA damage signaling, MAFs infected with either Ad-GFP or Ad-GFP-Cre were treated with 10 J/m2 UV or 2.5 Gy IR and harvested 2 h following treatment and analyzed for total p53 expression by immunoblot (Figure 2B, top panel). Lamin B serves as a control for equal loading. As expected, both cultures exhibited similar inductions of p53 following either UV or IR damage. To further probe the induction kinetics of p53, phospho-p53 expression was analyzed at 0, 1, 3 and 5 h post-treatment (Figure 2B, bottom panel). Not surprisingly, both cultures exhibited relatively equal kinetics of phospho-p53 induction (10,13). However, phospho-p53 induction was slightly faster in response to IR than UV and its response to UV persisted longer than that from IR exposure. To subsequently characterize expression levels of downstream RB targets, Ad-GFP and Ad-GFP-Cre infected MAFs were analyzed at 0, 3 and 5 h following 10 J/m2 UV (Figure 2C, top panel) or 2.5 Gy IR (Figure 2C, bottom panel) exposure. Interestingly, expression levels of PCNA, MCM-7 and cyclin A remained relatively constant during the rapid response to UV and IR indicating that RB action did not affect levels of these downstream targets during checkpoint induction. Equal loading was verified by lamin B immunoblot. These data argue that the rapid function of RB in cell cycle arrest following DNA damage does not apparently involve attenuation of target genes.
RB loss is directly coupled with target gene deregulation to promote abrogation of the DNA damage checkpoint response
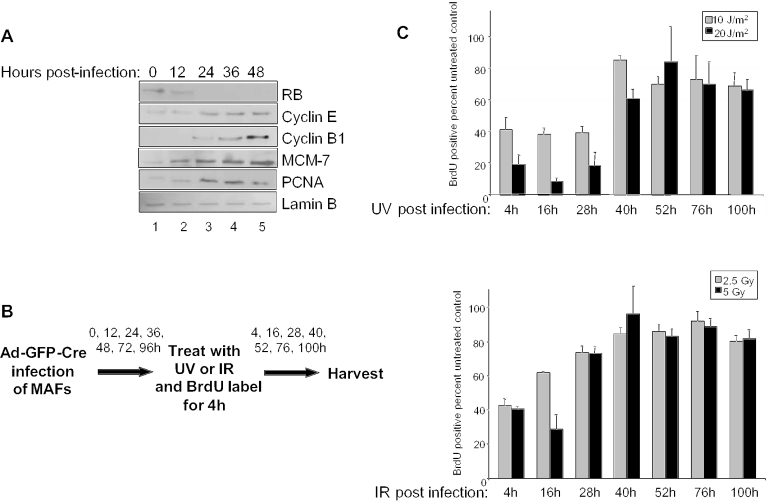
Although it is understood that RB function is necessary for proper regulation of downstream targets and the DNA damage checkpoint response, the kinetic ordering of these events has not been established. Specifically, the data shown above suggests that while RB may not actively cause the checkpoint, RB loss could enable checkpoint bypass through the accumulation of RB target gene products. To determine whether loss of RB protein or target gene deregulation is more closely coupled to loss of checkpoint function, we examined the discrete kinetics of this pathway. Asynchronous MAFs were infected (T = 0 h) and harvested for immunoblot every 12 h for 48 h. Immunoblot analysis revealed complete loss of RB protein by 24 h post-Ad-GFP-Cre infection (Figure 3A). Analysis of RB target genes showed that the expression of several downstream targets including, MCM-7, PCNA, cyclin B1 and cyclin E, all became deregulated concurrent with RB loss.
Figure 3.
Kinetics of RB loss is concomitant with target gene deregulation to promote abrogation of the DNA damage checkpoint response. (A) Asynchronously proliferating primary MAFs were infected with Ad-GFP-Cre and harvested every 12 h for a period of 48 h. Equal protein concentrations from each lysate were separated by electrophoresis and immunoblotted for the expression of RB, cyclin E, cyclin B1, MCM-7 and PCNA. Immunoblot for lamin B was used to control for equal loading. (B) This schematic illustrates the experimental approach used to determine whether acute RB loss or target gene deregulation more directly influence the kinetics of DNA damage checkpoint bypass. Asynchronously proliferating MAFs were infected with Ad-GFP-Cre and irradiated at various timepoints. Following irradiation, cells were cultured with BrdU for 4 h prior to harvesting at their indicated times post-infection. Immunofluorescence was performed to determine the proliferative fraction of treated cells as compared with the untreated controls. (C) As illustrated in (B), MAFs were infected with Ad-GFP-Cre and the temporal cell cycle response to 0, 10 or 20 J/m2 UV during the RB knockout was examined and represented as a percent of BrdU positive untreated controls (top panel). Cells were treated with 0, 2.5 or 5 Gy IR and analyzed for BrdU incorporation as described in (B) (bottom panel).
Since disruption of checkpoint function could simply be a manifestation of chronic RB loss, we examined the nature of checkpoint function throughout the RB knockout time course. In parallel with the previously outlined experiments, Ad-GFP-Cre infected MAFs were treated with UV or IR every 12 h post-infection for 96 h. Following damage, the cells were pulsed with BrdU for 4 h and harvested for checkpoint analysis (Figure 3B). Scoring of the populations of DNA damaged cells throughout the time course of RB knockout revealed that DNA damage checkpoint function remained intact in response to 0, 10 or 20 J/m2 UV irradiation through 28 h post-infection. However, by 40 h post-infection the checkpoint response became impaired (Figure 3C, top panel), concurrent with maximal deregulation of target genes following RB protein loss (Figure 3A). Similarly, the equivalent experimental setup was employed to investigate checkpoint function in response to IR damage signaling. Abrogation of proper checkpoint function occurred slightly more rapidly in response to 0, 2.5 or 5 Gy IR, such that cells were able to incorporate BrdU in the presence of DNA damage by 28 h post-infection (Figure 3C, bottom panel). Together, these data reveal that the kinetics of RB loss are concomitant with target gene deregulation and impaired DNA damage checkpoint response. Therefore, the closely coupled dynamics of these events suggest that RB functional inactivation coincident with downstream target deregulation is required for the loss of proper cell cycle arrest in response to DNA damage.
Acute RB loss differentially influences UV- and IR-induced DNA lesion removal
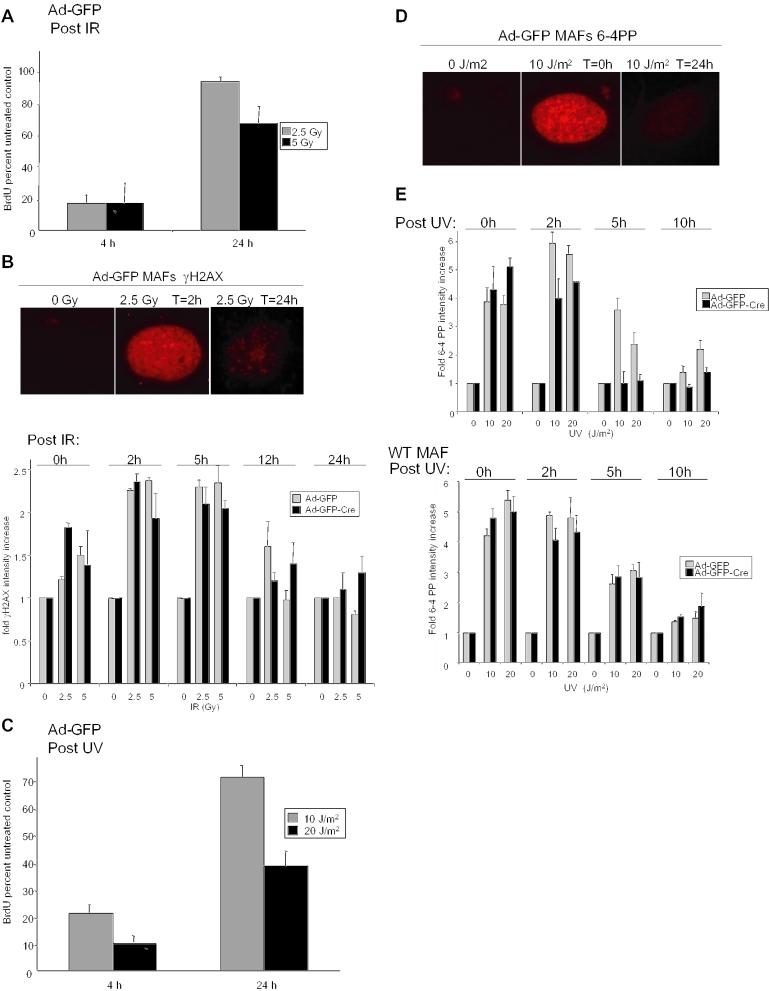
Checkpoint responses are by definition reversible, presumably due to DNA damage repair (36). Consistent with this notion, Ad-GFP infected cells were able to recover from the UV- and IR-induced checkpoints and re-enter the cell cycle. As before, following 0, 2.5 or 5 Gy IR, Ad-GFP infected cells were labeled with BrdU for the final 4 h prior to harvest at 4 or 24 h post-damage. The results demonstrated a significant increase in BrdU incorporation compared with untreated controls (set to 100%) in RB-proficient MAFs 24 h following damage, suggesting that the doses of IR used in these experiments are repairable (Figure 4A). Since IR directly elicits DNA double-strand breaks, we next investigated the influence of RB loss upon the accumulation and relative repair of these lesions. Here, we employed immunofluorescence with antibodies recognizing γH2AX, an efficient measure of double-strand break accumulation, to demonstrate the extent to which these lesions are repaired in the 24 h following damage (Figure 4B, top panel). Interestingly, RB loss had no significant effects upon γH2AX foci intensity among images of the cell population taken at equal exposures following IR damage. Both Ad-GFP and Ad-GFP-Cre infected MAFs displayed a similar increase in staining intensity during the first 5 h post-damage and a similar kinetic decrease in intensity from 5 to 24 h (Figure 4B, bottom panel).
Figure 4.
Differential effects of acute RB loss on UV- and IR-induced DNA lesion removal. (A) Ad-GFP or Ad-GFP-Cre infected MAFs were exposed to 0, 2.5 or 5 Gy IR and cultured for 4 or 24 h in the presence of BrdU for the final 4 h prior to harvest. Immunofluorescent detection was employed and BrdU positive cells were scored and represented as percent untreated control. (B) Asynchronously growing MAFs infected with either Ad-GFP or Ad-GFP-Cre were exposed to 0 or 2.5 Gy IR. At 0, 2, 5, 12 and 24 h after cell irradiation, samples were fixed and analyzed for γH2AX foci formation by immunofluorescence using an anti-γH2AX monoclonal antibody (top panel). The relative abundance of γH2AX foci was determined through quantification of staining intensity in images taken at equal exposures using Metamorph software (bottom panel). These data are represented graphically as the relative increase in γH2AX intensity. (C) Adenovirally infected MAFs were treated with 0, 10, 20 J/m2 UV and cultured for 4 or 24 h in the presence of BrdU for the final 4 h. Scoring of BrdU immunofluorescence revealed the percent of treated cells that were BrdU positive with respect to untreated controls. (D) Ad-GFP and Ad-GFP-Cre infected MAFs were treated with 0 or 10 J/m2 UV and harvested at 0 and 24 h post-treatment for 6-4 PP immunofluorescence. Images of equal exposure were taken. (E) MAFs from (D) were treated with 0, 10 or 20 J/m2 UV and cultured for 0, 2, 5 or 10 h post-UV. Cells were then harvested and immunofluorescence for 6-4 PP was performed. Microscopic images of equal exposures were obtained and the relative abundance of 6-4 PPs were quantified using Metamorph software. The data are represented graphically as fold 6-4 PP intensity increase (top panel). Wild-type MAF control cells lacking loxP sites were cultured and infected with Ad-GFP or Ad-GFP-Cre. Five days post-infection, wild-type cells were treated with UV and analyzed for 6-4 PP repair as in top panel (bottom panel).
In order to determine the corresponding impact of UV repair on the cell cycle, the ability of the RB-proficient cells to recover from the checkpoint and re-enter cell cycle was investigated. Ad-GFP infected MAFs were treated with 0, 10, 20 J/m2 UV and propagated in culture while being pulse labeled with BrdU prior to harvest at 4 and 24 h. Detection of BrdU incorporation revealed the ability of RB-proficient cells to significantly recover from the UV-induced DNA damage checkpoint and resume cell cycle progression. The percentage of BrdU positive cells significantly increased from 4 to 24 h following each dose of UV damage as compared with untreated controls (set to 100%) (Figure 4C). To monitor the induction of UV lesions in the single-cell, the abundance of 6-4 PPs was examined by immunofluorescence immediately following damage (T = 0 h) and after 24 h of recovery (T = 24 h) (Figure 4D). These lesions are clearly induced in MAFs by 10 J/m2 UV at T = 0 h and are largely resolved by 24 h. To more closely examine the kinetics of 6-4 PP repair, Ad-GFP and Ad-GFP-Cre infected cells were treated with 0, 10 or 20 J/m2 UV and harvested for 6-4 PP analysis at 0, 2, 5 and 24 h following damage. The average pixel intensities of images taken at equal exposures were compared using Metamorph software and displayed graphically to reveal that RB-deficient cells exhibit a kinetic difference in the loss of 6-4 PP staining intensities, as compared with RB-proficient cells (Figure 4E, top panel). To ensure that acceleration of 6-4 PP resolution was not due to infection with Ad-GFP-Cre, wild-type MAF control cells retaining Rb were cultured and infected with Ad-GFP or Ad-GFP-Cre as before. Wild-type cells were treated with UV and monitored for 6-4 PP repair as in the top panel of Figure 4E, to reveal that it is indeed RB loss which enhances 6-4 PP repair rather than Ad-GFP-Cre infection (Figure 4E, bottom panel). Together, these data indicate that while RB loss plays no apparent role in the repair of IR lesions (e.g. DNA double-strand breaks), the loss of RB accelerates the repair of UV-induced 6-4 PPs.
Acute RB loss accelerates UV-induced DNA damage repair
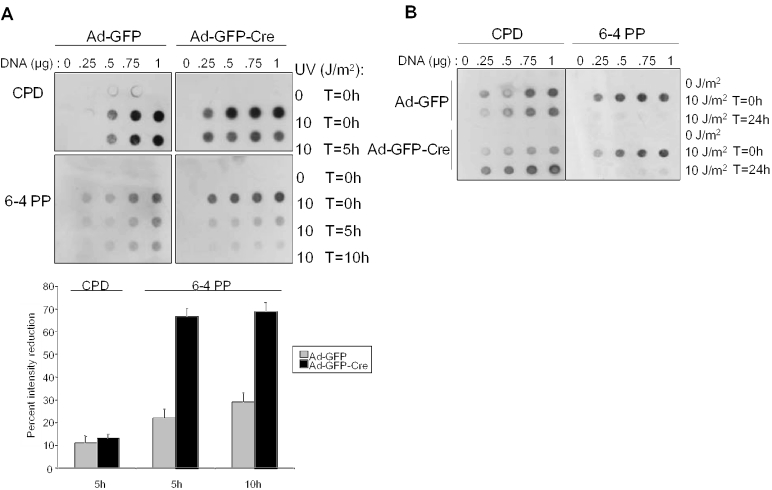
Since the role of RB in UV-induced damage repair has been largely unexplored, we dissected the consequence of RB loss on cyclobutane pyrimidine dimmers (CPD) and 6-4 PP repair kinetics in greater detail using a more quantitative analysis. Ad-GFP and Ad-GFP-Cre infected MAFs exposed to 0 or 10 J/m2 UV were harvested at 0, 5 and 10 h post-UV treatment and lysates were used to purify genomic DNA. Increasing amounts of DNA were spotted onto Nytran membranes and immunoblotted for the abundance of both CPD and 6-4 PP lesions. In confirmation with other studies, we reveal that MAFs are compromised for CPD repair (37–40) and RB had no apparent effect upon lesion repair during the time course examined (Figure 5A, top panel). However, the murine system has competent 6-4 PP repair pathways (41). Clearly, 6-4 PP repair is functional in both RB-proficient and RB-deficient MAFs. However, Ad-GFP-Cre infected MAFs exhibited a greater reduction in 6-4 PP lesions (∼68%) as compared with cells infected with Ad-GFP control (∼25%) at 5 and 10 h post-treatment (Figure 5A, bottom panel), indicating that RB loss contributes to the increased kinetics of UV damage-induced lesion repair. Despite the early differential in repair kinetics, both RB-proficient and -deficient MAFs demonstrated largely complete repair of 6-4 PPs induced by 10 J/m2 UV by 24 h (Figure 5B). Thus, RB loss accelerates 6-4 PP repair kinetics.
Figure 5.
Acute RB loss accelerates UV-induced DNA damage repair. (A) To examine the role of RB in the kinetics of UV-induced CPD and 6-4 PP lesion repair, Ad-GFP and Ad-GFP-Cre infected MAFs were treated with 0 or 10 J/m2 UV and harvested for genomic DNA purification at 0, 5 and 10 h. Increasing concentrations of denatured genomic DNA were vacuum blotted onto a membrane and probed with anti-CPD and anti-6-4 PP antibodies (top panel). The lesion intensities in the top panel were quantified using Metmorph and plotted as percent reduction in lesion intensity from cells treated with 10 J/m2 UV and harvested at T = 0 to those harvested at the indicated timepoint (bottom panel). (B) As performed in (A), cells were treated with UV and cultured prior to harvesting at 0 and 24 h. Dot blot analysis was performed as previously described to confirm previous reports of impaired repair of CPD lesions in murine cells and to reveal the effect of acute RB loss on acceleration of 6-4 PP repair.
RB modifies repair factor dynamics
There are two possible explanations for the differential 6-4 PP repair kinetics among RB-proficient and -deficient cells. First, RB has recently been implicated in the negative regulation of a wide array of repair factors (e.g. PCNA, RAD50, RAD51, MLH1, MSH2 and FEN1) (21–24), thus it is possible that the elevated levels of these factors in RB-deficient cells may allow for accelerated repair of DNA damage. Second, loss of RB-dependent DNA damage checkpoint function and resulting ongoing replication in RB-deficient cells may more quickly initiate repair factor engagement with the DNA lesion, thereby enhancing repair.
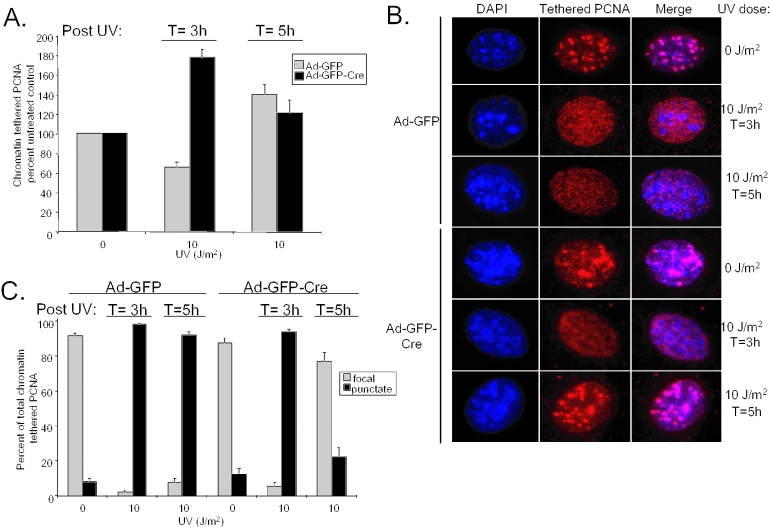
In order to examine these possibilities, we investigated the role of RB loss on the kinetics of PCNA engagement with chromatin following UV damage. PCNA is an interesting repair factor, not only because it is regulated by RB, but also because it performs dual activities upon recruitment to chromatin in DNA replication and DNA repair (42–44). Therefore, to dissect the significance of PCNA involvement in UV damage repair, chromatin extractions were performed on Ad-GFP and Ad-GFP-Cre infected MAFs at 0, 3 and 5 h following UV treatment. Next, PCNA immunofluorescence was used to determine the percent of cells in the population that exhibited chromatin tethered PCNA with respect to unextracted controls. RB-proficient cell populations exhibited a slight decrease in PCNA chromatin tethering from 0 to 3 h followed by a marked increase by 5 h following damage (Figure 6A). In contrast, PCNA engagement with chromatin was accelerated in the RB-deficient populations, peaking at 3 h and becoming similar to Ad-GFP infected controls by 5 h post-damage. In confirmation with previous studies, these results reveal an increase in PCNA tethering following UV exposure, indicating the involvement of PCNA in nucleotide excision repair. In addition, this involvement in repair is accelerated in RB-deficient cells. These experiments also revealed differential PCNA chromatin tethering patterns within the nuclei of cells following UV damage, which indicated that a shift in PCNA function was occurring during UV damage repair (Figure 6B). Approximately 90% of tethered PCNA staining appeared in large foci in undamaged cells, indicative of replication foci (Figure 6C). However, 3 h after exposure to 10 J/m2 UV, nearly 100% of both RB-proficient and -deficient populations of cells exhibited diffuse small punctate patterns of PCNA tethering within the nucleus. The global nature of this PCNA staining pattern suggests that PCNA function has shifted to repair foci at this time point. Strikingly, 5 h post-UV damage, nearly 100% of Ad-GFP infected cells continue to exhibit diffuse punctate PCNA chromatin tethering while more than 75% of Ad-GFP-Cre infected cells demonstrate localized focal patterning, as observed in the undamaged state. These data indicate that the RB-deficient cells are able to initiate UV-induced damage repair with enhanced kinetics and that in these cells the PCNA distribution more rapidly shifted its association with repair foci to replication foci following damage.
Figure 6.
RB modifies repair factor dynamics following DNA damage. (A) Ad-GFP and Ad-GFP-Cre infected MAFs treated with 0 or 10 J/m2 UV were harvested for chromatin extraction at 3 and 5 h post-treatment. Immunofluorescence for PCNA abundance was performed on extracted cells in parallel with unextracted controls. PCNA positive cells in each sample were counted and set as a percent of their respective unextracted control. These data were graphed as percent chromatin tethered PCNA of the unirradiated control. (B) Images of cells from (A) were taken at equal exposures to display the differential PCNA chromatin tethering patterns throughout the UV damage repair time course. (C) The PCNA tethering patterns of the irradiated chromatin extracted populations of MAFs from (A) were analyzed as displaying discrete ‘foci’ colocalized with heterochromatin, indicating PCNA is involved in replication, or diffuse ‘punctate’ staining throughout the nucleus, suggesting PCNA is engaged in global repair. The chromatin tethered PCNA positive cells were scored with respect to their staining pattern and represented as a percent of the total chromatin tethered PCNA positive cells.
DISCUSSION
Consistent with previous studies in MEFs, here we observed impairment of the DNA damage checkpoint and deregulated cell cycle progression following IR and UV damage in adult primary cells harboring acute RB loss (Figure 1) (10,26). Several models have been proposed which aim to describe how RB could be functioning to inhibit cell cycle progression following DNA damage. One model places RB in direct contact with replication machinery to inhibit replication (45–52), while another suggests that RB inhibits replication indirectly, via repression of downstream targets (31). Kinetic analysis of the DNA damage response in MAFs allowed us to probe the nature of RB function in checkpoint activation. Our data indicates that downstream targets such as PCNA, cyclin A and MCM-7 are not repressed by RB during the induction of the rapid cell cycle checkpoint (Figure 2). This data a priori supports the first model, wherein RB acts directly to inhibit replication and arrest cell cycle following recognition of DNA damage. However, it is equally possible that the vast target gene deregulation, which occurs concomitant with RB loss, facilitates checkpoint bypass. In an attempt to differentiate these possibilities, we closely examined the kinetics of RB deletion and DNA damage response in adult cells. Results presented indicate that RB loss is intimately coupled with target deregulation, together facilitating checkpoint abrogation (Figure 3). Thus, either model of RB function in checkpoint induction could be appropriate, in that RB loss prevents its direct action in replication inhibition while concurrently disrupting its function in control of downstream transcriptional targets.
The ability of cells to recognize damaged DNA and elicit cell cycle checkpoints following genotoxic insult depends upon complex signaling pathways. Although many of the downstream pathway components in the G1/S checkpoint are involved in signaling from both UV and IR induced lesions, many of the initial upstream components vary. In the case of IR-induced DNA double-strand break signaling, ATM kinase activity is immediately stimulated to phosphorylate a number of downstream effectors including histone H2AX, p53 and chk2 (35,53–57). Similarly, ATM and the rad 3 related (ATR) protein senses UV damage and participates in signal transduction via phosphorylation of many of the same effectors as ATM, such as chk2 and p53 (58). Our studies reveal that rapid phosphorylation of H2AX and p53 following IR and UV (p53 only) are not compromised by acute RB loss, despite the observed impairment of DNA damage checkpoint function (Figure 2). As such, these results are consistent with literature that places RB downstream of H2AX and p53 phosphorylation in the DNA damage signaling and repair pathway.
As the function of DNA damage signaling effectors were unaffected by RB loss, we sought to understand the consequence of the differential DNA damage checkpoint function on the cellular response to DNA damage. Because control cells exhibiting a functional checkpoint were able to resume cell cycle progression 24 h following damage, we assumed that these cells must have been able to repair a significant portion of the DNA lesions. Thus, we sought to monitor the actual induction and removal of these lesions imparted by IR and UV damage. Although monitoring the reduction in γH2AX to analyze DNA double-strand break repair following IR damage is rather qualitative, our studies revealed that RB did not play a dramatic role in IR-induced damage repair (Figure 4). This may suggest that participants in the DNA double-strand break repair pathway are upstream of the RB/E2F axis or that those repair factors deregulated via RB loss are not rate-limiting for repair. Next, we explored the role of RB in repair of UV-induced 6-4 PPs. Our investigations revealed that RB loss significantly accelerated removal of 6-4 PPs during the first 10 h following exposure to UV. However, by 24 h post-damage, the RB-proficient cells exhibited equal levels of lesion removal with the RB-deficient cells (Figures 4 and 5). This is the first evidence indicating that RB inactivation modifies DNA damage repair. The underlying mechanisms for this change could be multiple. First, recent evidence suggests that RB regulates the expression of several DNA damage repair factors involved in UV damage repair processes including: FEN1, XPC, RPA2-3, RFC4 and PCNA (20–24). As we found that the rate of repair of UV-induced damage was modified by RB loss, we investigated its effect upon the function of an RB-regulated UV damage repair factor, PCNA. We presume that high basal levels of proteins such as PCNA enable the RB-deficient cells to complete repair processes more quickly, as we found that PCNA protein expression rapidly shifted from diffuse punctate patterning (indicative of its function in repair) to focal expression (evident in normal replication) (Figure 6). Therefore, the observed UV repair factor dynamics are consistent with the kinetics of 6-4 PP lesion removal for each cell type. Second, the loss of cell cycle arrest following damage may initiate faster repair, modifying the conventional view of checkpoint function being necessary for efficient lesion resolution. Traditionally, DNA damage checkpoints are viewed as providing necessary time for recruitment of repair or apoptotic factors. However, due to the fact that RB-deficient cells demonstrate elevated basal levels of a variety of repair factors, recruitment time may inevitably be shortened, thereby eliminating the necessity for cell cycle arrest.
In summary, the data presented indicate that RB participates in the response to UV and IR damage signaling in a differential manner. Although RB loss abrogates the checkpoint in response to both forms of DNA damage, the consequence of this loss in each instance differs. Following IR, the loss of checkpoint function did not affect lesion repair. This suggests that RB-deficient cells were replicating in the presence of DNA damage for a prolonged period of time, possibly inducing secondary lesions which contribute to genomic instability and activation of apoptotic pathways. However, following UV, RB-deficient cells rapidly repaired 6-4 PPs, potentially limiting the accumulation of detrimental replication-mediated lesions. Although the consequences of checkpoint bypass have not been fully elucidated, there exist several possible influences of the observed rapid repair of UV lesions in RB-deficient cells. First, accelerated 6-4 PP repair kinetics may facilitate secondary lesion development, which would explain the clinical observations that RB-deficient tumor cells are more sensitive to death upon challenge (59,60) and that hereditary retinoblastoma survivors are at an increased risk for melanoma (61,62). Second, loss of cell cycle arrest coupled with rapid 6-4 PP lesion repair may prevent proper activation of apoptotic cascades in cells harboring other UV-induced lesions (e.g. CPD), facilitating the propagation of mutations. Because all cells are assaulted by damaging environmental signals and a high proportion of RB-deficient cancers are treated with DNA damaging therapy, understanding the effect of RB inactivation on the response to DNA damage will enhance our perceptions of tumorigenesis and cancer therapeutics. Our results provide the framework for understanding the critical role of RB in the DNA damage response. These data indicate that RB is required for the rapid induction of cell cycle arrest following recognition of both UV and IR damage in adult primary cells. Additionally, RB loss in these cells is closely coupled with target gene deregulation and contributes to abrogation of checkpoint function. Lastly, we demonstrate that although damage signaling remains unaffected, RB loss accelerates UV lesion repair and modifies repair factor dynamics.
Acknowledgments
We are grateful to Drs Karen Knudsen and Christopher Mayhew for their helpful comments on the manuscript and all members of the Knudsen laboratories for insightful discussions. We would like to thank Drs David Johnson and John Powers for sharing ideas and protocols. This work was supported by NIH CA106412 to E.S.K. The NIEHS Core grant E30-ES-06096 provided infrastructure support for this study. E.E.B. is supported by DOD BCRP Grant BC030315 and the Albert J. Ryan Foundation. Funding to pay the Open Access publication charges for this article was provided by NIH CA106412.
REFERENCES
- 1.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C. Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem. Sci. 1995;20:381. doi: 10.1016/s0968-0004(00)89082-9. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell L.H., Weinert T.A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 5.Chellappan S.P., Hiebert S., Mudryj M., Horowitz J.M., Nevins J.R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 6.Stevaux O., Dyson N.J. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 7.Sherr C.J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 8.Harbour J.W., Luo R.X., Dei Santi A., Postigo A.A., Dean D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 9.Bartek J., Bartkova J., Lukas J. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 1997;237:1–6. doi: 10.1006/excr.1997.3776. [DOI] [PubMed] [Google Scholar]
- 10.Harrington E.A., Bruce J.L., Harlow E., Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl Acad. Sci. USA. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen E.S., Buckmaster C., Chen T.T., Feramisco J.R., Wang J.Y. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrera R.E., Sah V.P., Williams B.O., Makela T.P., Weinberg R.A., Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayhew C.N., Perkin L.M., Zhang X., Sage J., Jacks T., Knudsen E.S. Discrete signaling pathways participate in RB-dependent responses to chemotherapeutic agents. Oncogene. 2004;23:4107–4120. doi: 10.1038/sj.onc.1207503. [DOI] [PubMed] [Google Scholar]
- 14.Yamasaki L. Role of the RB tumor suppressor in cancer. Cancer Treat Res. 2003;115:209–239. doi: 10.1007/0-306-48158-8_9. [DOI] [PubMed] [Google Scholar]
- 15.Harbour J.W., Lai S.L., Whang-Peng J., Gazdar A.F., Minna J.D., Kaye F.J. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241:353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W.H., Bookstein R., Lee E.Y. Studies on the human retinoblastoma susceptibility gene. J. Cell. Biochem. 1988;38:213–227. doi: 10.1002/jcb.240380309. [DOI] [PubMed] [Google Scholar]
- 17.Knudson A., Jr. Genetics of tumors of the head and neck. Arch. Otolaryngol. Head Neck Surg. 1993;119:735–737. doi: 10.1001/archotol.1993.01880190031006. [DOI] [PubMed] [Google Scholar]
- 18.Bookstein R. Tumor suppressor genes in prostatic oncogenesis. J. Cell. Biochem. Suppl. 1994;19:217–223. [PubMed] [Google Scholar]
- 19.Lan Z., Sever-Chroneos Z., Strobeck M.W., Park C.H., Baskaran R., Edelmann W., Leone G., Knudsen E.S. DNA damage invokes mismatch repair-dependent cyclin D1 attenuation and retinoblastoma signaling pathways to inhibit CDK2. J. Biol. Chem. 2002;277:8372–8381. doi: 10.1074/jbc.M108906200. [DOI] [PubMed] [Google Scholar]
- 20.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 21.Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J.R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polager S., Kalma Y., Berkovich E., Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 23.Markey M.P., Angus S.P., Strobeck M.W., Williams S.L., Gunawardena R.W., Aronow B.J., Knudsen E.S. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- 24.Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nature Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 25.Marino S., Vooijs M., van Der Gulden H., Jonkers J., Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen K.E., Booth D., Naderi S., Sever-Chroneos Z., Fribourg A.F., Hunton I.C., Feramisco J.R., Wang J.Y., Knudsen E.S. RB-dependent S-phase response to DNA damage. Mol. Cell. Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sever-Chroneos Z., Angus S.P., Fribourg A.F., Wan H., Todorov I., Knudsen K.E., Knudsen E.S. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol. Cell. Biol. 2001;21:4032–4045. doi: 10.1128/MCB.21.12.4032-4045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q.E., Zhu Q., Wani M.A., Wani G., Chen J., Wani A.A. Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Repair (Amst.) 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 30.Sage J., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 31.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 32.Siddik Z.H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 33.Van Triest B., Pinedo H.M., Giaccone G., Peters G.J. Downstream molecular determinants of response to 5-fluorouracil and antifolate thymidylate synthase inhibitors. Ann. Oncol. 2000;11:385–391. doi: 10.1023/a:1008351221345. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence T.S., Blackstock A.W., McGinn C. The mechanism of action of radiosensitization of conventional chemotherapeutic agents. Semin. Radiat. Oncol. 2003;13:13–21. doi: 10.1053/srao.2003.50002. [DOI] [PubMed] [Google Scholar]
- 35.Chao C., Saito S., Kang J., Anderson C.W., Appella E., Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartek J., Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 37.Hwang B.J., Toering S., Francke U., Chu G. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohr V.A., Smith C.A., Okumoto D.S., Hanawalt P.C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 39.van der Horst G.T., van Steeg H., Berg R.J., van Gool A.J., de Wit J., Weeda G., Morreau H., Beems R.B., van Kreijl C.F., de Gruijl F.R., et al. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 40.Schul W., Jans J., Rijksen Y.M., Klemann K.H., Eker A.P., de Wit J., Nikaido O., Nakajima S., Yasui A., Hoeijmakers J.H., et al. Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J. 2002;21:4719–4729. doi: 10.1093/emboj/cdf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell D.L. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 42.Rieger K.E., Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 2004;32:4786–4803. doi: 10.1093/nar/gkh783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paunesku T., Mittal S., Protic M., Oryhon J., Korolev S.V., Joachimiak A., Woloschak G.E. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int. J. Radiat. Biol. 2001;77:1007–1021. doi: 10.1080/09553000110069335. [DOI] [PubMed] [Google Scholar]
- 44.Cleaver J.E., Karplus K., Kashani-Sabet M., Limoli C.L. Nucleotide excision repair ‘a legacy of creativity’. Mutat. Res. 2001;485:23–36. doi: 10.1016/s0921-8777(00)00073-2. [DOI] [PubMed] [Google Scholar]
- 45.Takemura M., Kitagawa T., Izuta S., Wasa J., Takai A., Akiyama T., Yoshida S. Phosphorylated retinoblastoma protein stimulates DNA polymerase alpha. Oncogene. 1997;15:2483–2492. doi: 10.1038/sj.onc.1201431. [DOI] [PubMed] [Google Scholar]
- 46.Sterner J.M., Dew-Knight S., Musahl C., Kornbluth S., Horowitz J.M. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 1998;18:2748–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pennaneach V., Salles-Passador I., Munshi A., Brickner H., Regazzoni K., Dick F., Dyson N., Chen T.T., Wang J.Y., Fotedar R., et al. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell. 2001;7:715–727. doi: 10.1016/s1097-2765(01)00217-9. [DOI] [PubMed] [Google Scholar]
- 48.Gladden A.B., Diehl J.A. The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB.MCM7 binding. J. Biol. Chem. 2003;278:9754–9760. doi: 10.1074/jbc.M212088200. [DOI] [PubMed] [Google Scholar]
- 49.Kennedy B.K., Barbie D.A., Classon M., Dyson N., Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dimitrova D.S., Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell. Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 51.Bosco G., Du W., Orr-Weaver T.L. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nature Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- 52.Harbour J.W., Dean D.C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 53.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 54.Khanna K.K., Keating K.E., Kozlov S., Scott S., Gatei M., Hobson K., Taya Y., Gabrielli B., Chan D., Lees-Miller S.P., et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nature Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 55.Siliciano J.D., Canman C.E., Taya Y., Sakaguchi K., Appella E., Kastan M.B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirao A., Kong Y.Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 57.Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Lobrich M., Jeggo P.A. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 58.Motoyama N., Naka K. DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev. 2004;14:11–16. doi: 10.1016/j.gde.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Nahle Z., Polakoff J., Davuluri R.V., McCurrach M.E., Jacobson M.D., Narita M., Zhang M.Q., Lazebnik Y., Bar-Sagi D., Lowe S.W. Direct coupling of the cell cycle and cell death machinery by E2F. Nature Cell. Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 60.Samuelson A.V., Lowe S.W. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl Acad. Sci. USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moll A.C., Imhof S.M., Bouter L.M., Tan K.E. Second primary tumors in patients with retinoblastoma. A review of the literature. Ophthalmic Genet. 1997;18:27–34. doi: 10.3109/13816819709057880. [DOI] [PubMed] [Google Scholar]
- 62.Fletcher O., Easton D., Anderson K., Gilham C., Jay M., Peto J. Lifetime risks of common cancers among retinoblastoma survivors. J. Natl Cancer Inst. 2004;96:357–363. doi: 10.1093/jnci/djh058. [DOI] [PubMed] [Google Scholar]