Abstract
N-Oct-3 is a neuronal transcription factor widely expressed in the developing mammalian central nervous system, and necessary to maintain neural cell differentiation. The key role of N-Oct-3 in the transcriptional regulation of a multiplicity of genes is primarily due to the structural plasticity of its so-called ‘POU’ (acronym of Pit, Oct, Unc) DNA-binding domain. We have recently reported about the unusual dual neuro-specific transcriptional regulation displayed by N-Oct-3 [Blaud,M., Vossen,C., Joseph,G., Alazard,R., Erard,M. and Nieto,L. (2004) J. Mol. Biol., 339, 1049–1058]. To elucidate the underlying molecular mechanisms, we have now made use of molecular modeling, DNA footprinting and electrophoretic mobility shift assay techniques. This combined approach has allowed us to uncover a novel mode of homodimerization adopted by the N-Oct-3 POU domain bound to the neuronal aromatic amino acids de-carboxylase and corticotropin-releasing hormone gene promoters and to demonstrate that this pattern is induced by a structural motif that we have termed ‘NORE’ (N-Oct-3 responsive element), comprising the 14 bp sequence element TNNRTAAATAATRN. In addition, we have been able to explain how the same structural motif can also induce the formation of a heterodimer in association with hepatocyte nuclear factor 3β(/Forkhead box a2). Finally, we discuss the possible role of the NORE motif in relation to neuroendocrine lung tumor formation, and in particular the development of small cell lung cancer.
INTRODUCTION
The coordinated expression of the 34 604 genes annotated in the human genome (1) is essential to the healthy development and functioning of the organism. Regulation of gene expression is ensured by a complex network of transcription factors that modulate the activity of RNA polymerase II by specifically binding to promoter sequences upstream of the starting point of transcription. Our understanding of the molecular mechanisms involved in transcriptional regulation has evolved considerably during recent years [reviewed in (2,3)], thus providing a structural framework to analyze in detail a wide spectrum of metabolic disorders, autoimmune diseases and cancers, in relation to the inappropriate functioning of transcription factors; such dysfunctions can, in general, be ascribed to mutations that modify either the expression levels or the functional structure of the factor [reviewed in (4–6)].
N-Oct-3, the human equivalent of the mouse Brn-2 (‘Brain-2’) protein, is a neuronal transcription factor widely expressed in the developing mammalian central nervous system, and necessary to maintain neural cell differentiation (7). The deletion of the Brn-2 gene results in the loss of specific neuronal lineages in the endocrine hypothalamus and subsequent loss of the posterior pituitary gland (8–10). Any imbalance in N-Oct-3 expression has critical consequences. For example, N-Oct-3 over-expression in melanocytes, cells derived from the neural crests, leads to tumorigenesis via the dysregulation of a number of genes (11–14). The fact that N-Oct-3 is such a key player in the regulation (and dysregulation) of transcription of so many genes is due to the structural plasticity of its so-called ‘POU’ (acronym of Pit, Oct, Unc) DNA-binding domain (DBD), which allows the recognition of a whole array of DNA targets.
The POU family of transcription factors is defined on the basis of a common DBD of ∼160 residues, first identified in the mammalian proteins Pit-1 and Oct-1 and the nematode factor Unc-86 [reviewed in (15)]. The POU DBD comprises two distinct, highly conserved sub-domains, termed ‘POUs’ and ‘POUh’, which contain four and three α-helices, respectively, and are connected by a linker, variable in sequence and length. All the POU domains bind specifically to the prototypic octamer ATGCAAAT. The crystallographic structure of the complex between the POU domain of the ubiquitous protein Oct-1 and the octamer has revealed that POUs interacts with the tetramer ATGC in a way similar to the phage repressors, whereas POUh interacts with the tetramer AAAT in a way similar to a homeodomain (16). Both sub-domains insert their third α-helix, the so-called ‘recognition helix’, into the DNA major groove. In addition, POUh inserts its rigid N-terminal arm into the DNA minor groove via a specific contact between a highly conserved arginine residue and the first adenine nucleotide of the AAAT sequence. Despite the robustness of the fit between a POU domain and the octamer sequence, most POU domains can also recognize various other AT-rich sequences owing to the flexibility of the linker joining the two sub-domains (17). To a certain extent, POUs and POUh recognize the respective elements of a DNA target relatively independently. These elements neither have to be contiguous nor even to belong to the same strand of the DNA (18). The extent of the adaptability of a POU domain to its target is determined by the length and sequence of the variable linker, which is specific to a given POU protein (19).
An important functional implication of the structural plasticity of the POU domain is the possibility of different patterns of homo and heterodimerization. So far, based on the crystallographic structures of various Pit-1 or Oct-1 POU/DNA complexes, two major types of POU homodimerization with palindromic or pseudo-palindromic DNA targets have been described previously (20). In previous reports, we have presented the characteristic patterns of N-Oct-3 binding to a set of neuronal promoters, including the AADC (aromatic amino acids de-carboxylase), CRH (corticotropin-releasing hormone) and aldolase C gene promoters. We have observed either an atypical homodimerization or a heterodimerization with the hepatocyte nuclear factor HNF-3β, also referred to as Forkhead box a2 (21,22). This alternative mechanism of transcriptional regulation is impossible for Oct-1, which is only able to bind as a monomer to these targets. We have shown that this differential binding pattern of Oct-1 and N-Oct-3 could be correlated with the unique capacity of a small segment of the N-Oct-3 POU linker to adopt an α-helical structure (22).
The main objective of the present study is to elucidate the molecular mechanisms underlying the unusual dual neuro-specific transcriptional regulation found for N-Oct-3. A detailed compilation of the available POU/DNA complex structures, as well as a combination of molecular modeling, DNA footprinting and electrophoretic mobility shift assay (EMSA) have enabled us to (i) uncover a new mode of homodimerization adopted by the N-Oct-3 POU domain bound to the respective neuronal AADC and CRH gene promoters, (ii) demonstrate that this pattern is induced by a novel structural motif, the NORE (N-Oct-3 responsive element) and (iii) understand how the same structural motif can also induce the formation of a heterodimer in association with HNF-3β DBD. Finally, we discuss the structure/function relationships of the NORE motif in the context of neuroendocrine tumors, and in particular small cell lung cancer (SCLC).
MATERIALS AND METHODS
Protein expression and purification
The N-Oct-3 and HNF-3β DBDs were expressed in Escherichia coli and purified as reported previously (21).
Oligonucleotides and DNA-binding assays
Twenty-four base pair oligonucleotides corresponding to the (−95/−72) and (−127/−104) fragments of the human neuronal AADC gene promoter (23) and the rat CRH gene promoter (24), respectively, and encompassing the N-Oct-3 POU homodimer binding sites (22), served as ‘wild-type’ references for a whole range of derived mutations. The double-stranded oligonucleotides were prepared and EMSA carried out as described previously (21,22).
DNase I and copper-phenanthroline footprinting
A vector plasmid (pRA103) carrying the −176 to −92 region of the rat CRH promoter (24) was constructed. A 122 bp fragment encompassing this CRH promoter sequence was then generated by PCR using flanking primers and purified on a 10% polyacrylamide gel.
The 32P-end-labeled CRH DNA fragment (5 nM concentration) was incubated at 28°C for 30 min with increasing amounts of N-Oct-3 DBD (3.2–16 nM concentration range) in 20 μl binding buffer (25 mM Tris, pH 7.5, 6% glycerol, 125 μg/ml BSA, 0.5 mM EDTA, 5 mM MgCl2, 100 mM NaCl, 2 mM DTT and 0.004% Tween-20). DNase I footprinting assays were carried out according to the standard protocols (25).
1,10-phenanthroline-copper ion (OP2-Cu) footprinting experiments were performed as described by Kuwabara and Sigman (26). The protein/DNA complexes were first separated on a 5% polyacrylamide gel and the DNA then cleaved within the gel. In both types of footprinting experiments, products of the Maxam–Gilbert chemical cleavage reaction of the identical PCR fragment served as sequence references (27).
Molecular modeling
Models were generated using the ‘Accelrys’ modules InsightII, Biopolymer, Discover, Docking and Homology (version 2000.1), run on a Silicon Graphics Fuel workstation. The structures of the N-Oct-3 POUs and POUh sub-domains were built by homology to the Oct-1 POU sub-domains [PDB accession no. 1OCT (16)] as described previously (21). Similarly, the HNF-3β DBD structure was based on that of HNF-3γ DBD [nucleic acid data bank (NDB) accession no. PDT013 (28)].
Models of the 18 bp DNA fragments from the osteopontin and immunoglobulin G (IgG) VH gene promoters were built using their respective homology with the PORE and MORE motifs of known 3D structures [PDB accession nos 1HF0 and 1E3O (20)]. Models of the 18 bp DNA fragments from the AADC and CRH gene promoters were built based on local homology with the octamer DNA and the HNF-3γ target DNA (PDB and NDB accession nos 1OCT and PDT013). The inter base pair structural parameters (rise, twist, tilt and roll) inferred from the homologous crystallographic templates were found in the ‘Double-helix/Bending Analysis’ database files (http://www.imb-jena.de) established with the ‘CURVES’ algorithm (29). These values served as input for the Biopolymer module.
The structures of the DNA-binding units were first manually docked into the sub-sites of the 18 bp DNA fragments, based on homology with the respective complex crystallographic structures cited above. The structure of the linker between the POUs and POUh sub-domains was built as described previously (21), and in all cases including the α-helical structure of the IDKIAAQ segment served as a constraint. The resulting preliminary structures were then submitted to the ‘Affinity’ program, an automatic docking refinement procedure within the Docking module which follows a Monte Carlo interaction energy minimization protocol and allows flexibility to predefined atoms of both the ligand-DBD and receptor-DNA.
RESULTS AND DISCUSSION
Structural lessons from the two prototypic PORE and MORE modes of POU domain homodimerization and their relevance to N-Oct-3
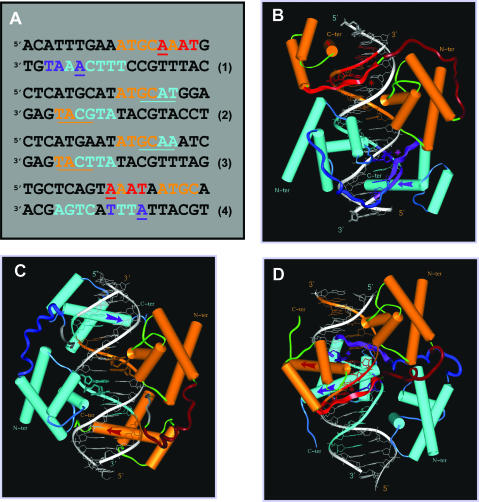
To date, the two major modes of POU domain homodimerization are generic, and are induced by two types of DNA sequence, the so-called ‘PORE’ (palindromic Oct-1 responsive elements) and ‘MORE’ (more palindromic Oct-1 responsive elements) sequences, respectively. The prototypic PORE sequence ATTTGAAATGCAAAT (Figure 1A, sequence 1) is an element of the osteopontin gene enhancer (30,31). The ability to induce a distinct type of Oct-1 POU homodimerization is maintained in the sequence ATTTGAAAGGCAAAT generated by a single T–G nucleotide transition, and the crystallographic structure of the resulting complex has been determined (20). The N-Oct-3 DBD can also homodimerize with the natural target as judged by EMSA and circular dichroism (data not shown), and we have modeled the structure of the corresponding complex by homology to that of the Oct-1 POU/PORE complex (Figure 1B). In this configuration, the relative positioning of POUs and POUh within each monomer is identical to that found in the prototypic Oct-1 POU/octamer complex.
Figure 1.
The three generic modes of N-Oct-3 POU domain homodimerization. (A) Eighteen base pair DNA sequences of: (1) the osteopontin gene enhancer fragment defining the PORE motif, (2) the prototypic MORE motif, (3) the MORE-type IgG VH gene promoter canonical fragment and (4) the AADC gene promoter fragment defining the new NORE motif. In all the cases, the first and second N-Oct-3 POU domain binding sites are coded in brown and turquoise, respectively. In the PORE (1) and NORE (4) motifs, critical nucleotides of the first and second POUh tetrameric sub-sites are coded in red and indigo respectively. In the MORE motifs (2 and 3), the first and second POUh tetrameric sub-sites are underlined in brown and turquoise, respectively, to compensate for the overlap with the POUs binding sub-sites. (B–D) Modeled structures of the homodimeric complexes between the N-Oct-3 DBD and (B) a PORE motif (sequence 1), (C) a MORE motif (sequence 3) and (D) a NORE motif (sequence 4). In all the cases, the respective display-codes for the first and second N-Oct-3 POU domains are as follows: brown- and turquoise-colored cylinders for the α-helices, red- and indigo-colored arrows for the POUh recognition helices, dark brown- and blue-colored coils for the linkers. In the PORE (B) and NORE (D) types of complexes, the first and second POUh N-terminal extensions are displayed as red- and indigo-colored ribbons, respectively, their insertion into the DNA minor groove being indicated by asterisks of the identical color.
The prototypic MORE sequence ATGCATATGCAT is a palindromic synthetic sequence (Figure 1A, sequence 2), which induces a different homodimerization pattern of the Oct-1 POU domain, as revealed by crystallography of the corresponding complex (20). In this case, the POUs of one monomer interacts with the ATGC tetramer on one strand, while its POUh counterpart interacts with the GCAT tetramer on the opposite strand. This type of homodimerization is not restricted to the Oct-1 DBD, as illustrated previously by the crystallographic structures of Pit-1 POU homodimers formed with various synthetic or natural promoter sequences (18,32). The most unexpected example of a ‘natural’ MORE sequence is no doubt provided by the previously termed ‘heptamer–octamer’ canonical sequence from the human immunoglobulin heavy chains gene promoters (IgG VH) (Figure 1A, sequence 3). The demonstration that this motif is in fact a MORE sequence (31) revealed an even more pronounced sequence degeneracy for the first and second POUh binding sites, TCAT and GCAA, respectively, in contrast with the prototypic tetramer AAAT. We have previously reported that the N-Oct-3 DBD homodimerizes cooperatively with this canonical target (21,22). N-Oct-3 DBD also binds to the prototypic MORE sequence, even more cooperatively, which is coherent with the perfect symmetry of the two binding sites (data not shown). Bearing in mind that the overall structures of the two homodimeric complexes will be similar when induced by structurally equivalent targets, we have modeled the interaction between the N-Oct-3 DBD and the canonical sequence by homology to the known structure of the Oct-1 POU/MORE complex (Figure 1C). The relative positioning of the POUs and POUh sub-domains within each monomer is clearly very different from that in the PORE mode (Figure 1B). This is owing to the relative positioning of the tetrameric binding sites, which are non-contiguous and on opposite strands in the MORE mode (Figure 1A, sequence 3), as opposed to contiguous and on the same strand in the PORE mode (Figure 1A, sequence 1).
One of the main lessons that can be drawn from the detailed analysis of the atomic structures of the different POU homodimer/DNA complexes is the paramount importance of the underlying structural symmetry of the DNA sequences involved. Thus, depending on the structural context in which it is embedded, the octamer sequence ATGCAAAT will be read by a POU domain in two totally different ways. In the PORE mode, it will be interpreted as a continuous POU binding site, while in the MORE mode, it will be segmented into the POUs binding site (ATGC) for one monomer and the overlapping POUh binding site for the other monomer (GCAA), the terminal AT being ‘mute’. The prominence of ‘structure’ over ‘sequence’ explains the significant sequence degeneracy, which is generally observed in the POU binding sites, often making it very difficult to identify a simple consensus. Another source of sequence degeneracy in the POU homodimer binding site is related to the way homodimeric ternary complexes achieve stability. An important component of stability is obviously the strength of the interaction between the two DBDs, which lessens the need for stringent contacts between all the POU sub-domains and the target DNA. Thus, in order to function as a PORE motif, a given sequence needs only display two well-conserved and symmetrically positioned POUh binding sites, since a certain amount of sequence degeneracy is allowed for the POUs binding sites. The converse is true for a MORE motif.
The N-Oct-3 DBD is thus emblematic of the adaptability of a POU domain to a variety of targets. In addition to the set of neuronal promoters with which it homodimerizes non-cooperatively (22), N-Oct-3 also forms cooperative homodimers in association with PORE sequences and with MORE sequences which include the canonical target. Even if the natural gene enhancer and promoter (osteopontin and IgG VH) described here are probably not relevant to N-Oct-3 function in vivo, they can serve as model targets to probe the structural plasticity of N-Oct-3 DBD. Indeed, we have experimental evidence that several promoter targets deregulated by N-Oct-3 in melanoma cells might be MORE sequences (R. Alazard, L. Nieto and M. Erard, unpublished data). If the N-Oct-3 DBD can adopt the same homodimerization patterns as Oct-1 DBD, the reverse is not true. Thus, the Oct-1 POU domain can only bind as a monomer on the same set of neuronal promoters which induce a non-cooperative N-Oct-3 POU homodimerization. Our objective, therefore, is to decipher the structural basis of this third pattern of N-Oct-3 DBD homodimerization and compare it with the PORE and MORE binding modes.
Similar structural elements in the neuronal AADC and CRH gene promoters induce a novel homodimerization pattern of the N-Oct-3 POU domain, the ‘NORE’ mode
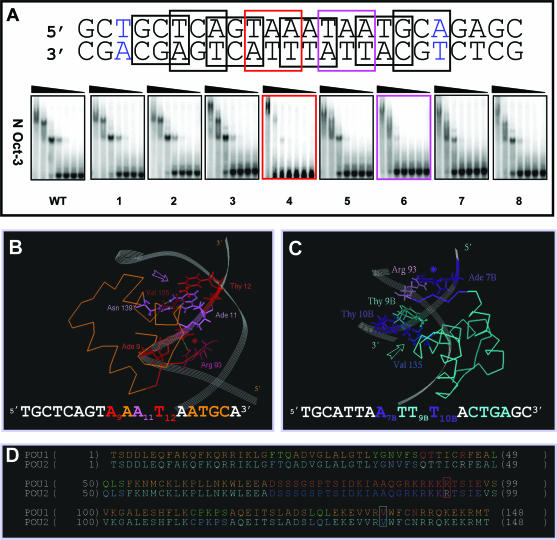
We have previously identified the binding site of the first N-Oct-3 DBD monomer to the neuronal AADC gene promoter, namely AAATAATGC, and modeled the corresponding binary complex (21). In this case, the respective targets of the POUh and POUs sub-domains are separated by a single nucleotide and in a so-called ‘switched’ order relative to the consensus octamer. This gives rise to a positioning of the first monomer (Figure 1D) which is different from that observed in the PORE or MORE complexes (Figure 1B and C). However, the principle nucleotides in contact with the POUh and POUs sub-domains are located on the same DNA strand, a feature in common with a PORE-type binary complex (Figure 1A, compare the respective upper strands in sequence 1 and 4). In order to determine the binding site for the second monomer, which is likely to be located on the opposite strand, we introduced triplet mutations into the 24 bp fragment of the AADC promoter and probed the homodimeric interaction with the N-Oct-3 DBD using EMSA (Figure 2A). Note that only the results relating to the 18 bp core fragment are presented, with nucleotides numbered accordingly (the first and last base pairs are colored in blue). Strikingly, mutations of the triplets T8A9A10 and T12A13A14 to GCC (mutants 4 and 6, respectively) are deleterious to both binding sites, with mutant 4 being the most affected. Thus, structural elements critical to both N-Oct-3 DBD binding sites, and present on both strands, overlap within the TAAATAA motif of the wild-type AADC promoter. Since the first monomer POUh sub-domain not only inserts its recognition helix into the A9A10A11T12 tetramer major groove but also tightly anchors into the minor groove via a canonical contact between the conserved Arg 93 residue in the rigid arm and the Ade 9 nt (Figure 2B), the most obvious candidate for the second POUh overlapping and symmetric binding site is the A7BT8BT9BT10B motif on the opposite strand.
Figure 2.
Structural determinants of the N-Oct-3 POU homodimerization on the AADC gene promoter (A) Mutation scanning of the 18 bp core region of the AADC fragment (the first and last base pairs are colored in blue) was carried out by selecting in turn 8 overlapping nucleotide triplets, in which A and G were substituted by C, and C and T substituted by G. The interactions between the N-Oct-3 DBD and the mutated AADC fragments were analyzed by EMSA and compared with the interaction with the non-mutated sequence (WT). The two most critical triplet mutations and their corresponding EMSA patterns have been highlighted. (B) Canonical interaction between the first monomer POUh and the optimal AAAT sub-site: anchoring into the DNA minor groove via the Arg 93/Ade 9 contact (red asterisk) and insertion of the recognition helix into the major groove via the Asn 139/Ade 11 and Val 135/Thy 12 contacts (pink arrow). (C) Interaction between the second monomer POUh and the ATTT sub-site: anchoring into the DNA minor groove via the Arg 93/Ade 7B contact (indigo asterisk) and insertion of the recognition helix into the major groove via the hydrophobic contacts between Val 135 and both Thy 9B and Thy 10B (turquoise arrow). (D) Location of Arg 93 (in red box), Val 135 (in turquoise box) and several important residues within the secondary structure elements of the first (POU1) and second (POU2) DBDs. Respective color coding is as follows: α-helices in brown and turquoise; POUh N-terminal extensions in red and indigo; linkers in dark brown and blue. POUs1 Gln 40, Thr41 and Arg 45 are colored in red, and POUs2 Thr 41 and Arg 45 in white.
Following this reasoning, we then modeled the homodimeric ternary complex according to this likely positioning of the second DBD (Figure 1A, sequence 4) and the resulting structure is displayed in Figure 1D. In addition to providing the essential contact in the DNA minor groove between the conserved Arg 93 residue from the POUh rigid arm and Ade 7B, as well as the canonical van der Waals contact in the major groove between the conserved Val 135 residue from the POUh recognition helix and the methyl group of Thy 10B, this model also predicts a second van der Waals contact between the same Val 135 and the methyl group of Thy 9B (Figure 2C). This partially compensates for the loss of the canonical double hydrogen bond in the major groove between the conserved Asn 139 of the recognition helix and Ade 11 (compare Figure 2C with 2B). In this configuration, the second POUs recognition helix is inserted into the C12BT13BG14BA15B tetramer major groove, thus generating a hydrophobic contact between the conserved Thr 41 and Thy 4 (paired with A15B), and a hydrogen bond between the Arg 45 side chain and Thy 13B (see their respective locations in the nucleotide sequence in Figure 2C and in the ‘POU2’ amino acid sequence in Figure 2D). In contrast, as previously shown (21), the first POUs inserts its recognition helix into the ATGC tetramer major groove, allowing the canonical interactions between the conserved Gln 40, Thr 41 and Arg 45 side chains and Ade 14, Thy 15 and Gua 16, respectively (see their respective location in the nucleotide sequence in Figure 2B and in the ‘POU1’ amino acid sequence in Figure 2D).
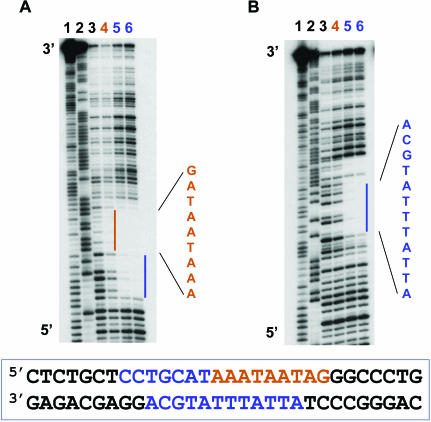
In order to determine whether this novel mode of N-Oct-3 POU homodimerization can also be elicited by the CRH promoter, we carried out DNAse I footprinting experiments. Our results show that the binding of the first monomer mainly protects the upper strand, with a clear footprint visible over the AAATAATAG sequence (Figure 3A, lane 4). This is consistent with the homology (78%) to the high-affinity binding site (AAATAATGC) on the AADC promoter (see the respective upper strands in the Figure 4A alignment). On the other hand, the binding of the second DBD is necessary to observe complete protection of the lower strand ATTATTTATGCA sequence (Figure 3B, lanes 5 and 6) and of the CCTGCAT sequence on the 5′ side of the upper strand (Figure 3A, lanes 5 and 6). To further identify the structural elements common to both promoters, we also performed a triplet mutation scanning of the 24 bp fragment of the CRH promoter. The results for triplet mutation scanning of the CRH promoter fragment were very similar to those already described for the AADC promoter (data not shown), and most importantly the conversion of the central TAA triplet to GCC virtually abolished the binding of both monomers (Figure 4B). These results again imply overlapping symmetric binding sites for the two POUh sub-domains (see Figure 4A). Moreover, the subsequent positioning of the second POUs sub-domain is entirely consistent with the total DNAse I protection observed for the lower strand ATTATTTATGCA sequence following the binding of the second monomer (Figure 3B).
Figure 3.
DNAse I footprinting analysis of N-Oct-3 POU homodimerization on the CRH gene promoter. Autoradiograms of 12% polyacrylamide denaturing gels showing the DNAse I footprints on the upper (A) and lower (B) strands of the CRH promoter fragment. Lanes 1 and 2, Maxam–Gilbert chemical sequencing references (cleavage after pyrimidine and purine residues, respectively). Lane 3, free DNA cleavage products. Lane 4, first DBD footprint (brown color coding). Lanes 5 and 6, POU homodimer footprint (blue color coding).
Figure 4.
Structural determinants of the N-Oct-3 POU homodimerization on the CRH gene promoter. (A) Alignment of the 18 bp core regions of the CRH (sequence 1) and AADC (sequence 2) promoter fragments based on the homology between the respective high-affinity binding sites for N-Oct-3 DBD. Same color coding as in Figure 1A. (B) Conversion of the TAA triplet (in boldface) of the 24 bp CRH promoter fragment (‘WT’) GCTCCTGCATAAATAATAGGGCCC to GCC (mutant ‘ΔM’) and EMSA analysis of the differential binding pattern of the N-Oct-3 DBD to the two oligonucleotides. (C and D) Modeled structure of the homodimeric complex between the N-Oct-3 DBD and the 18 bp CRH promoter fragment (sequence 1). Front views focusing on the first (C) and second (D) POU domain interactions with DNA. See the text for a detailed description. Same color coding as in Figure 1B–D. The ‘footprints’ on both strands of the first and second DBDs are shown as brown and purple-colored Connolly surfaces, respectively. (E) Same comment and color coding as in Figure 2D, except for POUs1 Gln 40, Thr41 and Arg 45 colored in yellow, and POUs2 Thr 41 and Arg 45 in red and white, respectively.
We then made use of these results as topological constraints in order to model the homodimeric complex. Figure 4C and D display a front view of the first and second POUh domains binding to their respective AAAT and ATTT tetrameric targets. Important features to note are the anchoring of the Arg 93 residues into the DNA minor groove, and the insertion of the recognition helices (indicated by red- and purple-colored arrows) into the DNA major groove via the Asn 139 and Val 135 residues for the first-bound DBD POUh and via Val 135 alone for the second-bound POUh. Again, the insertion of the first POUs recognition helix (indicated by a red triangle in Figure 4C) in the ATAG tetramer is ensured by the highly conserved Gln 40, Thr 41 and Arg 45 (see their location within the ‘POU1’ amino acid sequence in Figure 4E). As for the AADC promoter-induced complex, Thy 4 (whose location is indicated by a red asterisk in Figure 4C and D) is critical to the insertion of the second POUs recognition helix (purple triangle in Figure 4D) by allowing a hydrophobic contact with the Thr 41 methyl group. The mutation of this key nucleotide indeed abolishes the binding of the second monomer (data not shown).
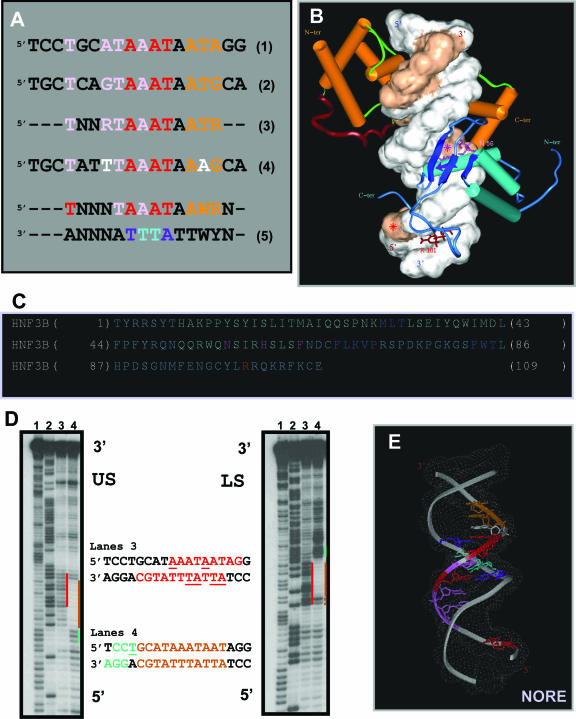
We have coined the term ‘NORE’ to designate the 14 bp sequence element TNNRTAAATAATRN (N: any nucleotide and R: purine residues), which is common to the neuronal AADC and CRH gene promoters and capable of eliciting a novel homodimerization mode exclusive to the N-Oct-3 DBD. Both the NORE and PORE motifs elicit a ‘POUh-dominant’ mode of N-Oct-3 DBD homodimerization (Figure 1D). However, in the case of the NORE mode, the two POUh binding sites are overlapping, thus creating a symmetrical ‘center’ relative to the position of the two POUs sub-domains (Figure 4C and D). This generates a unique interface, not only between the two POUs sub-domains, but also, and of equal importance, between the α-helical linker regions. In this context, we have established a strict correlation between the absence of the IDKIAAQ motif in the Oct-1 POU domain linker and its inability to form homodimers with the neuronal AADC, CRH or aldolase C gene promoters (22). Since there is an order of magnitude difference between the apparent affinity constants for N-Oct-3 DBD interacting with the two binding sites of these neuronal promoters, it is likely that the interaction energy between the two DBDs is an important component of homodimeric complex stability. Although we have already alluded to this in the case of the PORE and MORE complexes, it is only in the NORE mode that the N-Oct-3 POU linkers are in the appropriate position relative to each other to stabilize the protein/protein interface (Figure 1B–D).
NORE, a novel structural motif and a dual transcription factor binding site
Interestingly, the alignment of the homologous N-Oct-3 DBD binding sites present within the CRH and AADC promoters reveals another type of homology. The 4 nt, T4, A7, T8 and A10 of the CRH promoter are indeed homologous to T4, G7, T8 and A10, known to structurally specify the binding of HNF-3β on the AADC promoter [see the pink-coded nucleotides in the sequences 1 and 2 in Figure 5A and (21)]. In fact, for both promoters the HNF-3β binding site overlaps that of the first N-Oct-3 POU domain, defining a composite ‘POU-HNF’ binding site. By using the EMSA approach, we previously demonstrated that the N-Oct-3 and HNF-3β DBDs could simultaneously bind to the neuronal AADC, CRH and aldolase C genes promoters, despite the overlap in their binding sites (22). Furthermore, we were able to build a model of the heterodimeric complex formed on the 18 bp AADC promoter fragment (21).
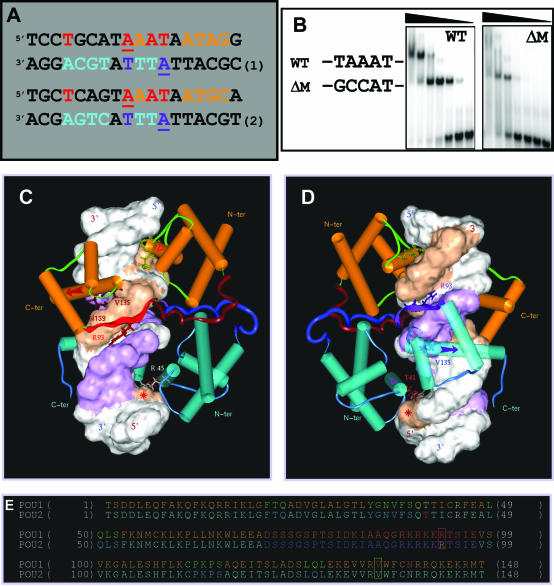
Figure 5.
Identification of the NORE as a structural motif and dual transcription factor binding site. (A) Structural homology between the HNF-3β binding sites on the CRH (sequence 1) and AADC (sequence 2) gene promoters (pink-coded nucleotides) and overlap with the N-Oct-3 high-affinity binding sites. Sequence 3, optimal NORE motif (N: any nucleotide and R: purine residues). Sequence 4, 18 bp core of the aldolase C gene promoter fragment. Sequence 5, extended consensus NORE motif and its dual binding capacity specifying N-Oct-3 DBD homo or heterodimerization (W: A or T). (B) Modeled structure of the heterodimeric complex between the N-Oct-3 and HNF-3β DBDs and the 18 bp CRH promoter. Note the two major components of HNF-3β binding and structural similarities with the second POUh binding in the homodimeric complex. See the text for a detailed description. Display-code for the DNA and N-Oct-3 DBD as in Figure 4C and D, and for the HNF-3β DBD as follows: α-helices are turquoise-colored cylinders, β-strands are dark blue ribbons, and ‘wings’ are blue coils. (C) Location of the important residues within the secondary structure elements of HNF-3β DBD (‘HNF3B’); color coding as in (B). (D) Autoradiograms of 12% polyacrylamide denaturing gels showing the OP2-Cu cleavage products of the upper (‘US’) and lower (‘LS’) strands of the CRH promoter fragment. Lanes 1 and 2, Maxam–Gilbert chemical sequencing references (cleavage after pyrimidine and purine residues, respectively). Lanes 3, free DNA; cleavage enhancement sites highlighted in red. Lanes 4, DNA in the 1/1 high-affinity complex with the N-Oct-3 DBD; footprint and flanking cleavage enhancement sites highlighted in brown and green, respectively. (E) Model of the bound-form of the NORE motif [same color coding as in (A), sequence 5].
We now present a model of the ternary complex formed by the two DBDs and the 18 bp CRH promoter fragment (Figure 5B). While the N-Oct-3 DBD occupies the high-affinity POU binding site as in the case of the homodimer, the two major components of HNF-3β DBD binding are (i) the insertion of its third helix (indicated by a purple-colored arrow) into the DNA major groove—mainly via the Asn 56/Ade 10 double hydrogen bond contact and (ii) the interaction in the minor groove between the HNF-3β C-terminal ‘wing’ Arg 101 and Thy 4 (Figure 5C, see the location of amino acid residues). Our modeling reveals striking global similarities between the binding of the HNF-3β DBD to the DNA in the heterodimeric complex and the binding of the second N-Oct-3 POUh in the homodimeric complex (compare Figure 5B with Figure 4D). In particular, it is worth underlining the critical importance of Thy 4 (marked by a red asterisk) in both contexts. Interestingly, the relative interchangeability of the N-Oct-3 and HNF-3β DBDs with respect to the formation of a structural ternary complex is in agreement with competitive-binding EMSA experiments and in addition the DNA bending is the same in both ternary complexes as shown by circular permutation assays (data not shown).
Thus, not only can the NORE motif TNNRTAAATAATRN (Figure 5A, sequence 3) elicit a novel type of N-Oct-3 DBD homodimerization, but it can also induce heterodimerization with the HNF-3β DBD. As is the case for the PORE and MORE motifs, the underlying structure of the NORE motif must play a fundamental role in its function. We have used copper bis-phenanthroline as a probe to monitor the structure of the CRH promoter fragment, either free or in complex with a single molecule of N-Oct-3 DBD. Beyond its primary role as a chemical agent for footprinting, copper bis-phenanthroline has proved to be a powerful reporter for identifying local DNA structure (33). Upon intercalation of its two phenanthroline rings into the DNA minor groove, this drug cleaves both DNA strands on either side with an efficiency which is sequence-dependent (34). As shown in Figure 5D (lanes 3), the free DNA is preferentially cleaved at two sites, the AAATAATAG sequence in the upper strand and the ATTATTTATGCA sequence in the lower strand, with hot spots at A9 and A13 and A4BT5B and A7BT8B, respectively, with numbering referring to the 18 bp core fragment. Significantly, these hypersensitive sites map to the identical region as seen for the footprint created by the binding of a single N-Oct-3 DBD monomer as shown in lanes 4. This clearly implies that it is the specific DNA minor groove structure of the NORE motif core sequence TAAATAAT that directs the binding of the first DBD. A striking consequence of this binding is the enhancement of nucleotide cleavage at C2C3T4, and in particular an increased accessibility to the critical Thy 4, thereby facilitating binding of either the POUs of a second N-Oct-3 DBD or the second wing of HNF-3β DBD.
Interestingly, the neuronal aldolase C gene promoter stimulates the same pattern of N-Oct-3 binding as the AADC and CRH promoters [(22) and C. Vossen, G. Joseph, L. Nieto and M. Erard, unpublished data] although the nucleotides in positions 4 and 12 of the aldolase C NORE motif are a thymine and an adenine residue, respectively, instead of purine and thymine residues (Figure 5A, see the white-coded nucleotides in the sequence 4). In the context of N-Oct-3 POU homodimer formation, both nucleotide changes fall within the POUs binding sites, which are unlikely to affect POUh-driven homodimerization in the NORE mode. In the case of heterodimer formation, the purine in position 4 stacks with the Phe residue of the HNF-3β DBD recognition helix and it can probably be replaced by a thymine nucleotide. This again emphasizes that the most critical mutations affecting function are the ones which occur in the TAAATAA core sequence of the NORE motif. In conclusion, the structural reading of the extended consensus 14-residue NORE motif and the predicted 3D structure of its bound form are displayed in Figure 5A (sequence 5) and Figure 5E, respectively.
Potential role of the NORE motif in SCLC dysregulation
Based on its properties, the NORE motif can clearly be qualified as a so-called ‘composite element’ (CE). This notion was first introduced by Diamond et al. (35), who showed that the glucocorticoid response element comprised a glucocorticoid receptor binding site adjacent to an AP-1 binding site. Since then, the concept has gained importance, leading to the establishment of the TRANSCompel dedicated database, which now contains several hundreds of CEs (36). As defined by Makeev et al. (37), a CE corresponds to a pair of adjacent, or even overlapping, binding sites, involved in the formation of specific ternary complexes. It defines a minimal functional unit in which both protein–DNA and protein–protein interactions contribute to a specific pattern of gene transcriptional regulation, and identical CEs are expected to perform related functions in genes. Thus, the regulatory regions of the FGF-4, UTF1 and Sox-2 genes, all involved in the maintenance of the pluripotent state of embryonic stem cells, share a CE which specifies the co-binding of the Oct-3/4 and Sox-2 transcription factors to adjacent sites (38–40). Regarding the AADC, CRH and aldolase C gene neuronal promoters, we can ask what is the potential function/dysfunction specified by the NORE motif as a CE?
‘HNF-3β’ has recently been renamed ‘Foxa2’, thus referring rather to the presence of the highly conserved winged-helix DBD than to any particular cellular localization. Deregulation of human HNF-3(/Fox) genes leads to a whole range of diseases, including congenital disorders, diabetes mellitus and carcinogenesis (41). As reviewed by Costa et al. (42), mouse HNF-3β is first expressed shortly after the onset of gastrulation in the foregut and visceral endoderm, the notochordal mesoderm and the neurotube floorplate, and then during the formation of the definitive endoderm. The adult mouse expression pattern of HNF-3β includes epithelial cells of the lung, liver, thyroid gland, pancreas, stomach and intestine (42). Expression profiling techniques have revealed that human HNF-3β can regulate complex networks of target genes, such as those involved in insulin secretion (43) or in lung epithelial cell maturation (44).
Most importantly, HNF-3β(/Foxa2) expression has very recently been detected in the whole spectrum of human neuroendocrine lung tumors, while it is absent from normal pulmonary neuroendocrine (PNE) cells (45). Among the neuroendocrine lung tumors expressing HNF-3β, SCLC is currently the object of extensive research [reviewed in (46–48)]. Classic SCLC cell lines are characterized by the presence of several neuroendocrine markers, such as chromogranin A and AADC, and very high amounts of AADC mRNA have been detected by RT–PCR (49). Derived from neuroectodermal cells, SCLC expresses N-Oct-3, as well as a number of neuropeptides and their receptors (50). Thus, elevated CRH levels have been observed in SCLC (51), with a subsequent increase in cyclic AMP levels, arachidonic acid release and lung cancer cell growth (52). Finally, the ectopic expression of aldolase C in neuroendocrine tumors, including SCLC, has been reported (53). In the light of these different observations, we propose that the NORE motif, shared by the AADC, CRH and aldolase C gene promoters, becomes functional as a composite N-Oct-3/HNF-3β binding site, following the appearance of HNF-3β expression, characteristic of SCLC dysregulation. In contrast, in the normal PNE cells, N-Oct-3 would exist as a homodimer bound to the NORE motif, leading to a basal level of expression of the AADC, CRH and aldolase C genes. An example where the N-Oct-3 homodimer has a reduced capacity to mediate transcriptional activation has been reported previously (54).
CONCLUSION
The capacity to successfully predict transcription factor binding sites within promoters and potential synergies between adjacent sites and bound factors is a major objective in the post-genomic era (55,56). An essential prerequisite for this is without doubt the need for a better understanding of the complex mechanisms involved in transcriptional regulation. As thoroughly reviewed by Johnson and Jameson (4), critical parameters include the homo and heterodimerization of transcription factor DBDs. Well-known examples are the nuclear receptors and the basic leucine zipper (‘b-Zip’) proteins (57–59), and the palindromic [or more often pseudo-palindromic (60)] characteristics of their respective binding sites. In this context, our study emphasizes the necessity (i) to employ a structural as opposed to a purely linear sequence-based reading of the nucleic regulatory sequences [reviewed in (61)] and (ii) to take into account the often sophisticated ‘combinatorial code’ used to achieve cell-specific gene expression (4).
Owing to its remarkable plasticity, the N-Oct-3 POU domain can adopt three different configurations and corresponding homodimerization patterns, with the POUs and POUh sub-domains acting as sensors for the distinct structures which characterize the respective PORE, MORE and NORE DNA motifs. We have experimental evidence that, in solution, the N-Oct-3 DBD is monomeric and without any preferential configuration (unpublished data). This implies that it is the pre-existing DNA conformation that dictates the mode of interaction with N-Oct-3 by providing a particular distribution of potential POUs and POUh tetrameric binding sites. Indeed, a number of recent articles have reassessed the so-called ‘indirect’ (i.e. structure-based) recognition mechanism as an important determinant of specificity (62–64).
The NORE motif, present in a set of neuronal promoters, elicits a particular type of POU domain homodimerization or heterodimerization with HNF-3β, which is exclusive to the neuronal transcription factor N-Oct-3. Interestingly, it is the joint action of a structural DNA motif and a unique α-helical structure adopted by part of the N-Oct-3 POU linker that provides the basis for this alternative neuro-specific transcriptional regulation. This process possibly involved in SCLC might also be relevant to other types of neuroendocrine tumors, for which HNF-3β expression clearly needs to be investigated.
Acknowledgments
We thank David Barker for careful reading of the manuscript. This work was supported by a research grant from the Région Midi-Pyrénées (AO No. 03001137). Funding to pay the Open Access publication charges for this article was provided by Centre National de la Recherche Scientifique.
REFERENCES
- 1.Veeramachaneni V., Makalowski W., Galdzicki M., Sood R., Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren A.J. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 2002;12:107–114. doi: 10.1016/s0959-440x(02)00296-8. [DOI] [PubMed] [Google Scholar]
- 3.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nature Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson W., Jameson J.L. Transcriptional control of gene expression. In: Jameson J.L., editor. Principles of Molecular Medicine. Towota, NJ: Humana Press Inc.; 1998. [Google Scholar]
- 5.Latchman D.S. Transcription factors as potential targets for therapeutic drugs. Curr. Pharm. Biotechnol. 2000;1:57–61. doi: 10.2174/1389201003379022. [DOI] [PubMed] [Google Scholar]
- 6.Darnell J.E., Jr Transcription factors as targets for cancer therapy. Nature Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 7.Fujii H., Hamada H. A CNS-specific POU transcription factor, Brn-2, is required for establishing mammalian neural cell lineages. Neuron. 1993;11:1197–1206. doi: 10.1016/0896-6273(93)90231-f. [DOI] [PubMed] [Google Scholar]
- 8.Nakai S., Kawano H., Yudate T., Nishi M., Kuno J., Nagata A., Jishage K., Hamada H., Fujii H., Kawamura K., et al. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 9.Schonemann M.D., Ryan A.K., McEvilly R.J., O'Connell S.M., Arias C.A., Kalla K.A., Li P., Sawchenko P.E., Rosenfeld M.G. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 10.Andersen B., Rosenfeld M.G. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 2001;22:2–35. doi: 10.1210/edrv.22.1.0421. [DOI] [PubMed] [Google Scholar]
- 11.Eisen T., Easty D.J., Bennett D.C., Goding C.R. The POU domain transcription factor Brn-2: elevated expression in malignant melanoma and regulation of melanocyte-specific gene expression. Oncogene. 1995;11:2157–2164. [PubMed] [Google Scholar]
- 12.Eisen T.G. The control of gene expression in melanocytes and melanomas. Melanoma Res. 1996;6:277–284. doi: 10.1097/00008390-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Thomson J.A., Murphy K., Baker E., Sutherland G.R., Parsons P.G., Sturm R.A., Thomson F. The brn-2 gene regulates the melanocytic phenotype and tumorigenic potential of human melanoma cells. Oncogene. 1995;11:691–700. [PubMed] [Google Scholar]
- 14.Goodall J., Wellbrock C., Dexter T.J., Roberts K., Marais R., Goding C.R. The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol. Cell. Biol. 2004;24:2923–2931. doi: 10.1128/MCB.24.7.2923-2931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latchman D.S. POU family transcription factors in the nervous system. J. Cell. Physiol. 1999;179:126–133. doi: 10.1002/(SICI)1097-4652(199905)179:2<126::AID-JCP2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Klemm J.D., Rould M.A., Aurora R., Herr W., Pabo C.O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 17.Herr W., Cleary M.A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson E.M., Li P., Leon-del-Rio A., Rosenfeld M.G., Aggarwal A.K. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 19.van Leeuwen H.C., Strating M.J., Rensen M., de Laat W., van der Vliet P.C. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 1997;16:2043–2053. doi: 10.1093/emboj/16.8.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remenyi A., Tomilin A., Pohl E., Lins K., Philippsen A., Reinbold R., Scholer H.R., Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 21.Millevoi S., Thion L., Joseph G., Vossen C., Ghisolfi-Nieto L., Erard M. Atypical binding of the neuronal POU protein N-Oct3 to noncanonical DNA targets. Implications for heterodimerization with HNF-3 beta. Eur. J. Biochem. 2001;268:781–791. doi: 10.1046/j.1432-1327.2001.01934.x. [DOI] [PubMed] [Google Scholar]
- 22.Blaud M., Vossen C., Joseph G., Alazard R., Erard M., Nieto L. Characteristic patterns of N Oct-3 binding to a set of neuronal promoters. J. Mol. Biol. 2004;339:1049–1058. doi: 10.1016/j.jmb.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 23.Le Van Thai A., Coste E., Allen J.M., Palmiter R.D., Weber M.J. Identification of a neuron-specific promoter of human aromatic L-amino acid decarboxylase gene. Brain Res. Mol. Brain Res. 1993;17:227–238. doi: 10.1016/0169-328x(93)90006-b. [DOI] [PubMed] [Google Scholar]
- 24.Thompson R.C., Seasholtz A.F., Herbert E. Rat corticotropin-releasing hormone gene: sequence and tissue-specific expression. Mol. Endocrinol. 1987;1:363–370. doi: 10.1210/mend-1-5-363. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel F., Brent R., Kingston R., Moore D., Seidman J., Struhl K. Current Protocols in Molecular Biology. Canada: John Wiley and Sons; 1993. [Google Scholar]
- 26.Kuwabara M.D., Sigman D.S. Footprinting DNA–protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase–lac promoter complexes. Biochemistry. 1987;26:7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- 27.Maxam A.M., Gilbert W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 29.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 30.Botquin V., Hess H., Fuhrmann G., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomilin A., Remenyi A., Lins K., Bak H., Leidel S., Vriend G., Wilmanns M., Scholer H.R. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell. 2000;103:853–864. doi: 10.1016/s0092-8674(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 32.Scully K.M., Jacobson E.M., Jepsen K., Lunyak V., Viadiu H., Carriere C., Rose D.W., Hooshmand F., Aggarwal A.K., Rosenfeld M.G. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 33.Spassky A., Sigman D.S. Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry. 1985;24:8050–8056. doi: 10.1021/bi00348a032. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer F., Rimsky S., Spassky A. DNA-stacking interactions determine the sequence specificity of the deoxyribonuclease activity of 1,10-phenanthroline-copper ion. J. Mol. Biol. 1996;260:523–539. doi: 10.1006/jmbi.1996.0419. [DOI] [PubMed] [Google Scholar]
- 35.Diamond M.I., Miner J.N., Yoshinaga S.K., Yamamoto K.R. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 36.Kel-Margoulis O.V., Kel A.E., Reuter I., Deineko I.V., Wingender E. TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 2002;30:332–334. doi: 10.1093/nar/30.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makeev V.J., Lifanov A.P., Nazina A.G., Papatsenko D.A. Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res. 2003;31:6016–6026. doi: 10.1093/nar/gkg799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrosetti D.C., Basilico C., Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein–protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimoto M., Fukushima A., Okuda A., Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh M. Human FOX gene family (Review) Int. J. Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 42.Costa R.H., Kalinichenko V.V., Lim L. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- 43.Lantz K.A., Vatamaniuk M.Z., Brestelli J.E., Friedman J.R., Matschinsky F.M., Kaestner K.H. Foxa2 regulates multiple pathways of insulin secretion. J. Clin. Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan H., Xu Y., Ikegami M., Stahlman M.T., Kaestner K.H., Ang S.L., Whitsett J.A. Foxa2 is required for transition to air breathing at birth. Proc. Natl Acad. Sci. USA. 2004;101:14449–14454. doi: 10.1073/pnas.0404424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khoor A., Stahlman M.T., Johnson J.M., Olson S.J., Whitsett J.A. Forkhead box A2 transcription factor is expressed in all types of neuroendocrine lung tumors. Hum. Pathol. 2004;35:560–564. doi: 10.1016/j.humpath.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Stupp R., Monnerat C., Turrisi A.T., III, Perry M.C., Leyvraz S. Small cell lung cancer: state of the art and future perspectives. Lung Cancer. 2004;45:105–117. doi: 10.1016/j.lungcan.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Rossi A., Maione P., Colantuoni G., Guerriero C., Gridelli C. The role of new targeted therapies in small-cell lung cancer. Crit. Rev. Oncol. Hematol. 2004;51:45–53. doi: 10.1016/j.critrevonc.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Muller-Hagen G., Beinert T., Sommer A. Aspects of lung cancer gene expression profiling. Curr. Opin. Drug Discov. Devel. 2004;7:290–303. [PubMed] [Google Scholar]
- 49.Vachtenheim J., Novotna H. Expression of the aromatic L-amino acid decarboxylase mRNA in human tumour cell lines of neuroendocrine and neuroectodermal origin. Eur. J. Cancer. 1997;33:2411–2417. doi: 10.1016/s0959-8049(97)00302-x. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber E., Himmelmann A., Malipiero U., Tobler A., Stahel R., Fontana A. Human small cell lung cancer expresses the octamer DNA-binding and nervous system-specific transcription factor N-Oct 3 (brain-2) Cancer Res. 1992;52:6121–6124. [PubMed] [Google Scholar]
- 51.Auchus R.J., Mastorakos G., Friedman T.C., Chrousos G.P. Corticotropin-releasing hormone production by a small cell carcinoma in a patient with ACTH-dependent Cushing's syndrome. J. Endocrinol. Invest. 1994;17:447–452. doi: 10.1007/BF03347737. [DOI] [PubMed] [Google Scholar]
- 52.Moody T.W., Zia F., Venugopal R., Korman L.Y., Goldstein A.L., Fagarasan M. Corticotropin-releasing factor stimulates cyclic AMP, arachidonic acid release, and growth of lung cancer cells. Peptides. 1994;15:281–285. doi: 10.1016/0196-9781(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 53.Inagaki H., Eimoto T., Haimoto H., Hosoda S., Kato K. Aldolase C in neuroendocrine tumors: an immunohistochemical study. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1993;64:297–302. doi: 10.1007/BF02915126. [DOI] [PubMed] [Google Scholar]
- 54.Rhee J.M., Gruber C.A., Brodie T.B., Trieu M., Turner E.E. Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem. 1998;273:34196–34205. doi: 10.1074/jbc.273.51.34196. [DOI] [PubMed] [Google Scholar]
- 55.Hannenhalli S., Levy S. Predicting transcription factor synergism. Nucleic Acids Res. 2002;30:4278–4284. doi: 10.1093/nar/gkf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wasserman W.W., Krivan W. In silico identification of metazoan transcriptional regulatory regions. Naturwissenschaften. 2003;90:156–166. doi: 10.1007/s00114-003-0409-4. [DOI] [PubMed] [Google Scholar]
- 57.Khorasanizadeh S., Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 58.Newman J.R., Keating A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 59.Garvie C.W., Wolberger C. Recognition of specific DNA sequences. Mol. Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 60.Selvaraj S., Kono H., Sarai A. Specificity of protein–DNA recognition revealed by structure-based potentials: symmetric/asymmetric and cognate/non-cognate binding. J. Mol. Biol. 2002;322:907–915. doi: 10.1016/s0022-2836(02)00846-x. [DOI] [PubMed] [Google Scholar]
- 61.Hoglund A., Kohlbacher O. From sequence to structure and back again: approaches for predicting protein–DNA binding. Proteome Sci. 2004;2:3. doi: 10.1186/1477-5956-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steffen N.R., Murphy S.D., Tolleri L., Hatfield G.W., Lathrop R.H. DNA sequence and structure: direct and indirect recognition in protein–DNA binding. Bioinformatics. 2002;18(Suppl. 1):S22–S30. doi: 10.1093/bioinformatics/18.suppl_1.s22. [DOI] [PubMed] [Google Scholar]
- 63.Paillard G., Lavery R. Analyzing protein–DNA recognition mechanisms. Structure (Camb.) 2004;12:113–122. doi: 10.1016/j.str.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 64.Dragan A.I., Read C.M., Makeyeva E.N., Milgotina E.I., Churchill M.E., Crane-Robinson C., Privalov P.L. DNA Binding and Bending by HMG Boxes: Energetic Determinants of Specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]