Abstract
The binding of SeqA protein to hemi-methylated GATC sequences (hemi-sites) regulates chromosome initiation and the segregation of replicated chromosome in Escherichia coli. We have used atomic force microscopy to examine the architecture of SeqA and the mode of binding of one molecule of SeqA to a pair of hemi-sites in aqueous solution. SeqA has a bipartite structure composed of a large and a small lobe. Upon binding of a SeqA molecule to a pair of hemi-sites, the larger lobe becomes visibly separated into two DNA binding domains, each of which binds to one hemi-site. The two DNA binding domains are held together by association between the two multimerization domains that make up the smaller lobe. The binding of each DNA binding domain to a hemi-site leads to bending of the bound DNA inwards toward the bound protein. In this way, SeqA adopts a dimeric configuration when bound to a pair of hemi-sites. Mutational analysis of the multimerization domain indicates that, in addition to multimerization of SeqA polypeptides, this domain contributes to the ability of SeqA to bind to a pair of hemi-sites and to its cooperative behavior.
INTRODUCTION
DNA methylation plays key roles in epigenetic regulation of chromatin assembly, gene expression, development and differentiation in mammals (1–3). In Escherichia coli, the adenine residues of GATC sequences on both strands are methylated at the 6-amino group by Dam methyltransferase (4,5). Replication of the chromosome generates hemi-methylated GATC sequences, since the newly synthesized strand is transiently unmethylated. SeqA protein preferentially binds to hemi-sites and sequesters them from methylation by Dam, especially at the origin of chromosomal replication (oriC), so preventing over-initiation of chromosomal replication (6–10). SeqA foci appear to be formed in the hemi-methylated region behind the replication fork and track the replication fork (11–15). SeqA function is required for nucleoid organization and proper segregation of replicated chromosomes (7,11,16,17), and these functions require the interaction of SeqA with topoisomerase IV, an enzyme that relaxes negatively and positively supercoiled DNA and decatenates replicated chromosomes (18). Also, SeqA appears to be involved in maintaining the negative superhelicity of DNA (16,17).
One molecule of SeqA, as a functional unit, binds to two hemi-sites that can be separated by up to 31 bp, and induces bending of the bound DNA (12,19–21). The binding affinity and extent of bending are maximal when the two hemi-sites are present on the same side (or phase) of the DNA helix. In addition, two SeqA molecules can interact cooperatively when the pairs of hemi-sites to which they are bound are separated by ≤30 bp, and as a result their capacity to interact with another molecule bound at distal hemi-sites is increased (20,21). This interaction between the bound proteins triggers further aggregation of free SeqA proteins onto the bound proteins, leading to the formation of large aggregates that can be visualized as foci in vivo (11–13).
Gel filtration, sedimentation and chemical cross-linking experiments have suggested that SeqA behaves as a homotetramer of 21 kDa polypeptides that aggregates reversibly in a concentration-dependent manner enhanced by the presence of DNA (22). Trypsin digestion of SeqA protein has revealed that the N-terminal fragment (residues 1–50) is responsible for multimerization and aggregation (21,23,24). The structure of co-crystals of a C-terminal fragment (amino acid residues 51–181) and DNA containing a hemi-site has permitted localization of the contact sites of SeqA with the hemi-site. It appears that the narrow major groove and distorted backbone structure of a hemi-site contributes to its recognition by SeqA (25).
Although the structure of co-crystals of the C-terminal fragment and DNA containing a hemi-site has been resolved, the structure of the intact SeqA molecule remains unclear. The properties of the latter differ from those of the C-terminal fragment in the following respects: SeqA behaves as a multimer, but the fragment as a monomer (22,23); in the presence of a large excess of competitor DNA, SeqA forms stable complexes with hemi-methylated DNA, whereas the fragment does not (21); and finally, even in the absence of competitor DNA, formation of the fragment–DNA complex requires a much greater concentration of the participating fragment than in the case of the SeqA–DNA complex (23,26,27). These discrepancies led us to determine the structures of SeqA and of SeqA–DNA complexes by atomic force microscopy (AFM) under physiologically relevant conditions. We have also shown, by mutational analysis, that the N-terminal region contributes to the binding of SeqA to a pair of hemi-sites and to its cooperative binding to multiple hemi-sites.
MATERIALS AND METHODS
Materials
Sources were as follows: restriction enzymes and cloning enzymes, Promega; [γ-32P]ATP (5000 Ci/mmol) and poly(dI–dC), Amersham Biosciences; T4 polynucleotide kinase, New England Biolabs; Pyrobest DNA polymerase and T4 DNA ligase, Takara; QIAEX II gel extraction kit, Qiagen; site-directed mutagenesis kit, Stratagene; and unmethylated and methylated synthetic oligonucleotides, Genotech. Unless otherwise indicated, additional reagents were from Sigma.
Proteins
The mutant forms of seqA were constructed by site-directed mutagenesis as previously described (22). Wild-type SeqA and its mutants were expressed and purified from W3SQT [pBAD18-seqA], as previously described (21).
Crude fractions of W3SQT expressing mutant proteins were prepared according to Lee et al. with minor modifications. E.coli W3SQT cells harboring the indicated mutations in pBAD18-seqA were grown in LB medium to an optical density at 600 nm of 0.4, and L-(+)-arabinose was added to a final concentration of 0.1%. Three hours later, the cells were harvested, suspended in Buffer A [25 mM Tris (pH 7.6), 1 mM EDTA, 10% glycerol and 1 mM DTT] and lysed, and partially purified fraction II was prepared as described (22). Wild-type and SeqA(T18G) proteins were further purified by heparin agarose chromatography; fraction II containing SeqA protein was diluted 25-fold with Buffer A containing 100 mM KCl and immediately loaded onto a heparin agarose column equilibrated with the same buffer. The protein was eluted with a linear salt gradient up to 1 M KCl. Both wild-type and SeqA(T18G) protein eluted around 350 mM KCl.
DNAs containing hemi-sites
The sequences of the DNA fragments used are given in Table 1. The DNA fragments containing four hemi-methylated sites with variable spacing between the second and third hemi-sites were prepared from plasmids described previously (20).
Table 1.
Nucleotide sequences of hemi-methylated DNA
| Spacing (bp) | Figures | DNA sequences |
|---|---|---|
| Synthetic 75 bp DNAs containing two hemi-sites | ||
| 11a | Figure 2C | 5′-ctgggtattaaaaagaagatctatttatgatcagttctgttctgttctgttctcttattaggctcgcactgccct-3′ |
| 5′-agggcagtgcgagcctaataagagaacagaacagaacagaactgatcataaatagatcttctttttaatacccag-3′ | ||
| 21a | Figure 2A and Figure 4A | 5′-ctgggtattaaaaagaagatctatttatgttcagttctgatctgttctgttctcttattaggctcgcactgccct-3′ |
| 5′-agggcagtgcgagcctaataagagaacagaacagatcagaactgaacataaatagatcttctttttaatacccag-3′ | ||
| 31a | Figure 2D | 5′-ctgggtattaaaaagaagatctatttatgttcagttctgttctgttctgatctcttattaggctcgcactgccct-3′ |
| 5′-agggcagtgcgagcctaataagagatcagaacagaacagaactgaacataaatagatcttctttttaatacccag-3′ | ||
| 16a | Figure 2E | 5′-ctgggtattaaaaagaagatctatttatgttcagatctgttctgttctgttctcttattaggctcgcactgccct-3′ |
| 5′-agggcagtgcgagcctaataagagaacagaacagaacagatctgaacataaatagatcttctttttaatacccag-3′ | ||
| 16a | Figure 3A | 5′-ctgggtattaaaaagaagatctatttatttagagatctgttctattgtgatctcttattaggatcgcactgccct-3′ |
| 5′-agggcagtgcgatcctaataagagatcacaatagaacagatctctaaataaatagatcttctttttaatacccag-3′ | ||
| DNAs containing four hemi-sites | ||
| 16b | Figure 3B and Figure 4B | 5′-agctccaccgcggtggcggccgctctagaactagtggatccgactgtgatccatggagctagcgatctcttattacgatccaattgtaccaagctactggaattcgatatcaagcttatcgataccgtcgacctcgagggggggcccggtac-3′ |
| 5′-cgggccccccctcgaggtcgacggtatcgataagcttgatatcgaattccagtagcttggtacaattggatcgtaa taagagatcgctagctccatggatcacagtcggatccactagttctagagcggccgccaccgcggtgg-3′ | ||
| 19b | Figure 3C | 5′-gcggtggcggccgctctagaactagtggatccgactgtgatccatggccgagctagcgatctcttattacgatccaattgtaccaagctactggaattcgatatcaagcttatcgataccgtcgacctcgagggggggcccggtac-3′ |
| 5′-cgggccccccctcgaggtcgacggtatcgataagcttgatatcaattccagtagcttggtacaattggatcgtaataagagatcgctagctcggccatggatcacagtcggatccactagttctagagcggccgccacc-3′ | ||
| 32b | Figure 4C | 5′-ctagaactagtggatccgactgtgatccatggggaaacagctatgaggagctagcgatctcttattacgatccaattgtaccaagctactggaattcgatatcaagcttatcgataccgtcgacctcgagggggggcccggtac-3′ |
| 5′-cgggccccccctcgaggtcgacggtatcgataagcttgatatcaattccagtagcttggtacaattggatcgtaataagagatcgctagctagctcctcatagctgtttccccatggatcacagtcggatccactagtt-3′ | ||
| 41b | Figure 3D | 5′-ctagaactagtggatccgactgtgatccatggggaaacagctatgaggatgagtacgcgctagcgatctcttattacgatccaattgtaccaagctactggaattcgatatcaagcttatcgataccgtcgacctcgagggggggcc-3′ |
| 5′-ccccctcgaggtcgacggtatcgataagcttgatatcaattccagtagcttggtacaattggatcgtaataagagatcgctagcgcgtactcatcctcatagctgtttccccatggatcacagtcggatccactagtt-3′ | ||
The GATC sequences containing N6-methyladenine are underlined. The spacing is given as the number of nucleotides from one modified adenine residue to the next. The top and bottom strands of synthetic 75 bp DNAs were annealed as previously described (20). DNAs containing four hemi-sites were prepared from fully methylated and unmethylated plasmids harboring each fragment, as previously described.
aThe spacing between two hemi-sites.
bThe spacing between the second and the third hemi-sites.
Atomic force microscopy
Bioscope AFM images were obtained with a Nanoscope IIIa controller (Digital Instruments, Santa Barbara, CA) using the tapping mode in an aqueous environment. The cantilevers had oxide-sharpened silicon nitride tips (Digital Instruments) with a nominal spring constant of 0.32 N/m. The resonance frequency in the aqueous environment was set at 8.5 ± 0.3 kHz, and the microscope was operated at drive amplitudes between 50 and 500 kHz. AFM images (512 × 512 pixels) were collected at a scan speed of 0.8–2.1 Hz, and image analyses were performed with Nanoscope version 4.32 software (Digital Instruments).
Gel shift assays
Unless otherwise indicated, the 20 μl reaction mixtures contained 10 mM Tris–HCl (pH 7.6), 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 μg of poly(dI–dC), 10% glycerol, 5 μg of BSA, and ∼2 fmol of hemi-methylated DNA together with the specified amount of SeqA protein. The mixtures were incubated for 15 min at 32°C, and the subsequent steps have been described (9).
RESULTS
Binding of SeqA to hemi-sites induces conformational changes in both SeqA and DNA
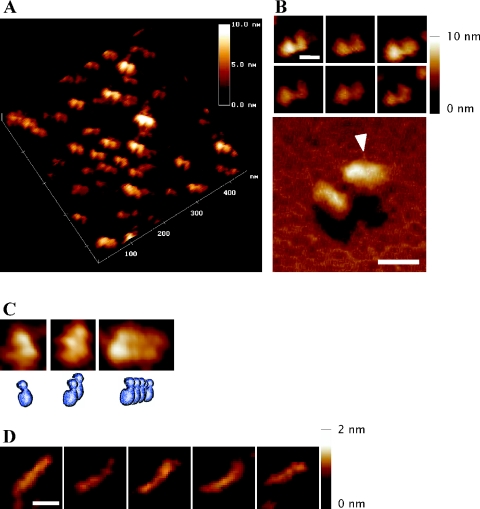
AFM has been used to study biological processes such as protein dynamics and DNA–protein interactions in aqueous media (28–30). We used AFM in tapping mode to examine the structure of SeqA in a buffer solution in the absence of DNA (Figure 1). In the tapping mode, the oscillating tip only touches the sample at the end of its downward movement, thereby reducing the contact time and the frictional forces. Compared with the contact mode of AFM, the tapping mode causes less damage to samples in liquid media. Most of the images of SeqA bound to the mica surface were uniformly of bipartite structures of similar size, with a large lobe (or domain) linked to a smaller one, and there appeared to be a groove in the large lobe (arrow in Figure 1B). SeqA aggregates in a reversible, concentration-dependent manner (22). To observe these aggregates, we increased the SeqA concentration on the mica 4-fold (Figure 1C). This resulted in oligomeric structures in which two or more bipartite structures were stacked repetitively in the same direction. Even at lower SeqA concentrations than that used in Figure 1A, no forms smaller than bipartite structures were observed. These results indicate that the bipartite structure is the functional unit of SeqA.
Figure 1.
Atomic force microscopy of SeqA and 75 bp hemi-methylated DNA. (A) An aliquot of 10 μl of purified SeqA (50 μg/ml in buffer A containing 0.1 M KCl and no glycerol) was deposited on freshly cleaved mica and incubated for 30 min under ambient humidity to adsorb to the mica surface. Surface scanning of a 450 × 450 nm field was performed in height and amplitude mode. (B) Representative images of SeqA in computerized three dimensions (3D) (upper panel), and an enlarged 3D view (lower panel). The arrowhead points to the groove in the large lobe of SeqA. (C) Representative images of multimeric forms of SeqA molecules, with schematic pictures. A 10 μl of purified SeqA (200 μg/ml in buffer A containing 0.1 M KCl and no glycerol) was deposited on freshly cleaved mica and the surface was scanned as in (A). (D) A 75 bp DNA containing two hemi-sites with a spacing of 21 bp (Table 1) was deposited on a Ni-treated mica surface, and images were recorded in the tapping mode in aqueous solution. The horizontal scale bars represent 20 nm, and the upright scale bars, height of the particles from baseline to the top.
The AFM image of 75 bp DNA containing two hemi-sites was linear with an average contour length of 29.8 ± 2.5 nm (Figure 1D). Although the limit of resolution caused by the tip radius (<20 nm) did not allow accurate measurements, the estimated length is in fairly good agreement with the expected value of 25.5 nm for B-DNA (0.34 nm/bp) and with previous measurements by AFM (31,32).
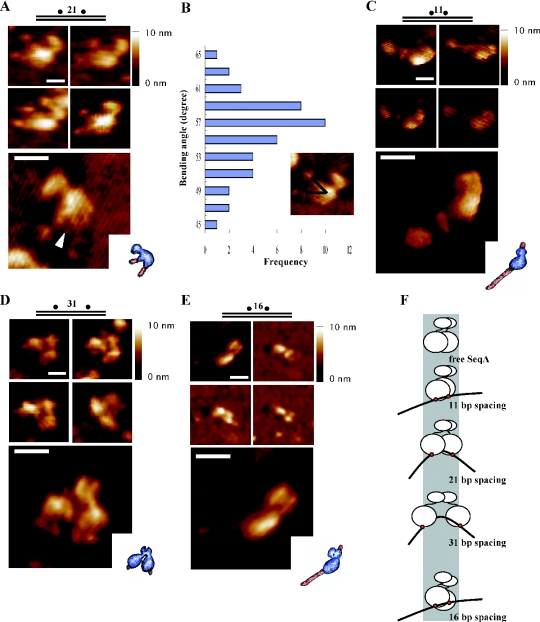
In SeqA complexes with 75 bp DNA containing two hemi-sites separated by 21 bp, the large lobe appeared to be bound to the DNA, linked to the smaller lobe (Figure 2A). The grooves in the complexes were wider than in free SeqA (Figure 1B) and the unbound regions of the bound DNA looked like a pair of a ‘duck's web feet’ connected to two ‘legs’ separated by the groove in the large lobe. The distance between the two ends of the bound DNA was 20.4 ± 1.9 nm, compared with 29.8 ± 2.5 nm for the free DNA, pointing to a conformational change, a bending, of the bound DNA. The most frequent bending angle of the bound DNA was 55–59° (Figure 2B), and the DNA was bent inwards towards the bound protein. The binding of one molecule of SeqA to a pair of hemi-sites and the bending of the bound DNA agree with previous conclusions from gel-shift experiments (20–22). SeqA complexes with DNA containing one hemi-site were barely detectable by AFM.
Figure 2.
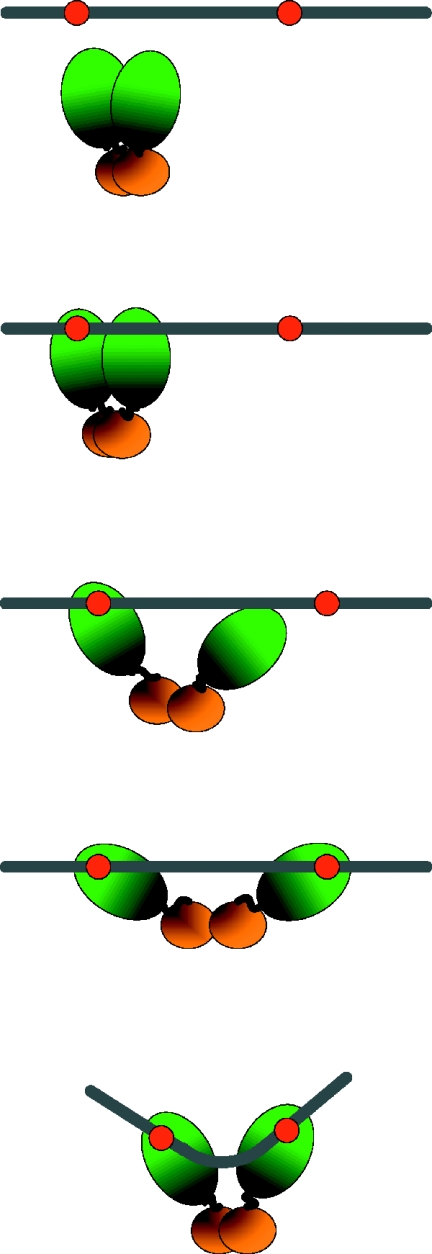
Dimeric configuration of SeqA bound to a pair of hemi-sites. To observe the complexes of SeqA with hemi-methylated DNA, 10 μl of a reaction mixture in buffer A containing SeqA (0.2–20 ng), the indicated DNA (50 fmol) and no glycerol was incubated for 15 min at 32°C, and the mixture was directly deposited on freshly cleaved mica. After further incubation for 30 min at room temperature, the specimen was gently washed with buffer A, fixed with 0.5% glutaraldehyde for 2–3 min, and washed several times again with buffer A. Images were recorded in the tapping mode in aqueous solution. (A) Representative images of complexes formed by SeqA and the 21 bp spacing DNA (upper panel), and a computerized 3D view of the complexes (lower panel) with a schematic picture (inner box). The arrowhead points to the portion of SeqA opened up as a result of binding to the DNA. (B) Relative frequency of the DNA bending angles in the complexes of SeqA with 21 bp spacing DNA. The degree of DNA bending was measured by drawing lines from the ends of the DNA strand to the apparent bending point of the bound DNA. (C) Representative images of complexes formed by SeqA and the 11 bp spacing DNA, and a computerized 3D view of the complexes. (D) Representative images of complexes formed by SeqA and 31 bp spacing DNA, and a computerized 3D view of the complexes. (E) Representative images of complexes formed by SeqA and 16 bp spacing DNA, and a computerized 3D view of the complexes. All the horizontal scale bars represent 20 nm, and the upright scale bars represent height of the particles from the baseline to the top. (F) Cartoon of the complexes formed between SeqA and a pair of hemi-sites spaced increasing distances apart.
Dimeric configuration of SeqA protein
DNA with an 11 bp spacing between the two hemi-sites resulted in SeqA complexes that differed from the 21 bp spacing DNA–SeqA complexes (Figure 2C). The location of two hemi-sites towards one end of the 75 bp DNA (nucleotide numbers of 44–47 and 55–58) permitted the unbound region to be distinguished from the bound region and the bound SeqA appeared to be more compact than when bound to the 21 bp spacing DNA.
When the spacing between the two hemi-sites was increased to 31 bp (Figure 2D), the large lobe of SeqA that was bound to the 31 bp spacing DNA was split into two domains that were bound to different points on the DNA. The increase in spacing resulted in a less compacted structure of the bound SeqA as depicted in Figure 2F. The similar mobility of the SeqA complexes formed with DNAs with different spacing of the two hemi-sites (20) eliminates the possibility that two separate molecules of SeqA bind to the DNA when there is 31 bp spacing. Instead, this result shows that the large lobe of each SeqA molecule actually consists of two DNA binding domains each of which binds to a hemi-site; the small lobe was also split with a cleft between the two parts.
When two hemi-sites were separated by 16 bp they faced in opposite directions on the helix. SeqA bound to 16 bp spacing DNA had a structure different from that bound to 11 or 21 bp spacing DNA (Figure 2E): the large lobe was lengthened and the small lobe split in two. SeqA has the highest affinity for DNA with the 21 bp spacing followed by the 11, 31 and 16 bp spacing (20), indicating that it achieves the most favorable structure with the 21 bp spacing DNA.
Both SeqA and its N-terminal fragment (amino acids 1–50) eluted as a tetramer in gel-filtration studies (22,24). Chemical cross-linking experiments showed that pairs of monomers of SeqA are closely associated. Hence, we have proposed that the SeqA molecule is composed of two dimers (20–22). A recent crystal structure revealed that the N-terminal fragment is a dimer that oligomerizes to form a filament (24). These observations agree with our finding that SeqA has a bipartite structure and oligomerizes on one direction (Figure 1). Taken together these results suggest that SeqA is a dimer of two 21 kDa polypeptides, not a tetramer. The tetrameric behavior of SeqA protein observed in previous experiments may have been a consequence of the extended, non-globular structure of SeqA.
The AFM structures of the SeqA bound to DNA, especially those obtained with the 31 bp spacing DNA, thus reveal that the large lobe actually contains two DNA binding regions and the small lobe two multimerization sites. The latter become associated and serve as an anchor that supports the dimeric configuration of SeqA. Amino acids 71–181 region of SeqA form a hemi-site binding domain and the 51–70 region forms a flexible hinge region (23,27). The N-terminal fragment containing amino acid residues 1–50 is responsible for multimerization. We also propose that the DNA binding and multimerization domains in the AFM structures correspond to the C-terminal region (amino acids 71–181) and N-terminal region (amino acids 1–50), respectively, with the two domains connected by a flexible hinge region (amino acids 51–70).
Thr-18 in the multimerization domain is necessary for cooperative binding of SeqA to hemi-methylated DNA
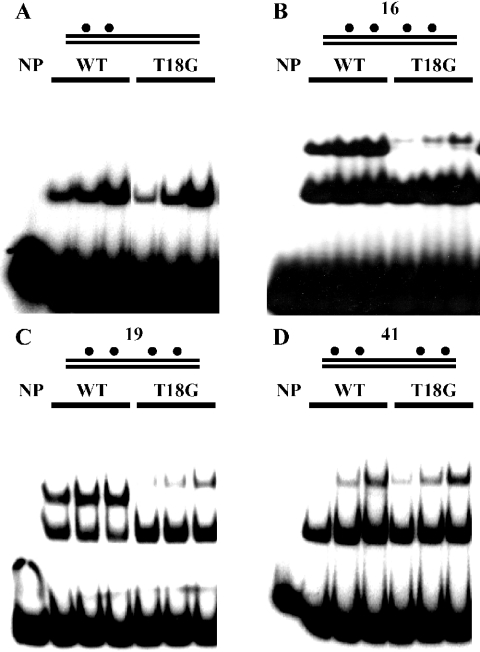
We have identified the amino acids participating in the interaction between SeqA molecules by bacterial two-hybrid analysis with randomly mutagenized seqA (22). One of the mutated proteins defective in the interaction had Thr-18 replaced by Gly (data not shown). We have analyzed the binding of SeqA(T18G) to hemi-sites (Figure 3). Because one SeqA molecule binds to a pair of hemi-sites, both wild-type SeqA and SeqA(T18G) formed a complex with DNA containing two hemi-sites (Figure 3A), and their affinity for the DNA was similar. Binding of the wild-type and mutant proteins to DNAs harboring four hemi-sites with 16 or 19 bp spacing between the pairs of hemi-sites generated an additional slow-mobility complex formed by the binding of two SeqA molecules (20,21) (Figure 3B and C). However SeqA(T18G) formed much less of this complex and the latter migrated more slowly. We have shown that two SeqA molecules bind cooperatively to DNA with ≤30 bp spacing between pairs of hemi-sites, but independently to DNA with >30 bp spacing (20). It is this cooperative binding that is more efficient with wild-type SeqA than with SeqA(T18G) and that generates a faster moving slow-mobility complex. On the other hand, with the DNA with pairs of hemi-sites spaced 41 bp apart, the wild-type and mutant proteins formed similar amounts of the slow-mobility complexes and these migrated with the same speed (Figure 3D), and the absence of cooperative binding of SeqA to the 41 bp spaced DNA reduced formation of the slow-mobility complex. Evidently, SeqA requires Thr-18 in the multimerization domain for cooperative binding to hemi-methylated GATC sequences.
Figure 3.
The Thr-18 of SeqA contributes to cooperative interaction between SeqA molecules. The binding behavior of purified wild-type SeqA and SeqA(T18G) was analyzed by gel shift experiments: 10, 20 and 40 ng of wild-type SeqA (designated as WT) and SeqA(T18G) were incubated with P32-labeled DNA containing a pair of hemi-sites, which were separated by 16 bp (A), and two pairs of hemi-sites in which the pairs were separated by 16 bp (B), 19 bp (C) and 41 bp (D). Complexes of SeqA and DNA were separated on 5% polyacrylamide gels, dried and visualized by autoradiography.
Glu-5, Asp-7 and Glu-9 of the multimerization domain contribute to proper binding of SeqA to a pair of hemi-sites
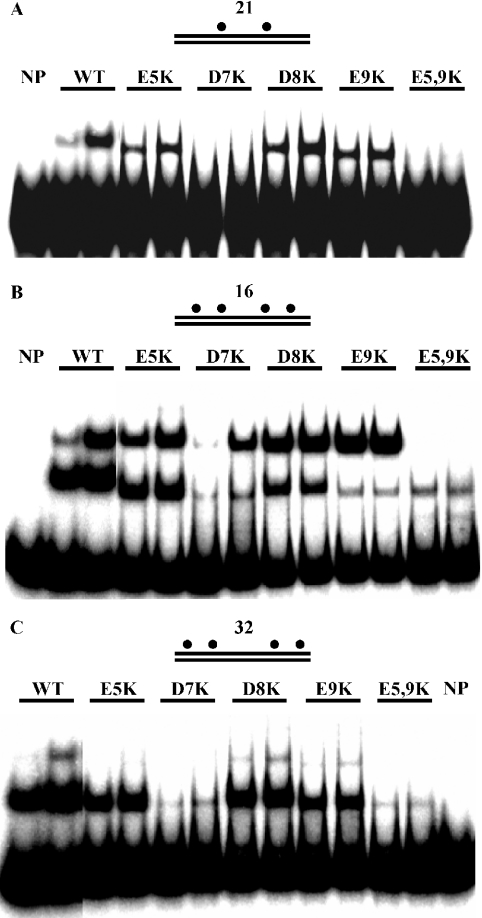
The N-terminus of SeqA has abundant negatively charged amino acids: Glu-5, Asp-7, Asp-8 and Glu-9 (7). We substituted each of these amino acids with lysine (K), and examined the binding of the resulting proteins to hemi-methylated DNA (Figure 4). While SeqA(E5K), (D8K) and (E9K) bound to DNA containing a pair of hemi-sites with similar affinity to wild-type protein, SeqA(D7K) and SeqA(E5, 9K) lost their binding ability (Figure 4A). When we tested cooperative binding of the mutant proteins to DNA containing two pairs of hemi-sites with 16 bp spacing (Figure 4B), SeqA(D7K) formed less of the complexes than the other proteins and favored the slow-mobility complex over the fast-mobility complex, and SeqA(E9K) also formed more of the slow-mobility complex than the fast-mobility complex (20). SeqA(E5,9K) did not form stable complexes with this DNA on its own, but was able to bind cooperatively when mixed with wild-type or other mutant SeqA proteins (20). When the spacing between the two hemi-sites was increased to 32 bp, SeqA(D7K) no longer formed a slow-mobility complex (Figure 4C). These results indicate that cooperative interaction between SeqA(D7K) molecules permits SeqA(D7K) to form slow-mobility complexes with 16 bp spacing DNA.
Figure 4.
N-terminal SeqA mutant proteins are defective in binding to a pair of hemi-sites, but interact cooperatively. Fractions containing mutant SeqA proteins were incubated with DNA containing a pair of hemi-sites separated by 21 bp (A), with DNA containing four hemi-sites with a 16 bp spacing between the pairs of hemi-sites (B) and DNA containing four hemi-sites with 32 bp spacing (C). In each experiment, 33 and 100 ng of wild-type SeqA (designated as WT) and SeqA(E5K), (D7K), (E8K), (E9K) and (E5,9K) partially purified fractions were added to the reaction. The amount of SeqA protein in each case was similar in western blot (data not shown).
It thus appears that SeqA(D7K) and SeqA(E5,9K) possess a defect in forming a complex with DNA containing two hemi-sites, but that cooperative interaction between the mutant proteins themselves (Figure 4B) or with other proteins (20) permits them to form a complex with the DNA containing four hemi-sites. This suggests that the mutant proteins recognize pairs of hemi-sites, but cannot form stable complexes with them. Hence, we conclude that Glu-5, Asp-7 and Glu-9 are needed for proper binding of SeqA to a pair of hemi-sites.
DISCUSSION
The binding of a SeqA molecule to a pair of hemi-sites separated by up to three helical turns (31 bp) (13,21) of the DNA suggests that the conformation of SeqA is flexible. The free SeqA proteins observed by AFM were homogeneous in appearance with a groove of constant width in the large lobes (Figure 1). No free molecules with split DNA binding domains were observed, such as the complexes with 31 bp spacing DNA (Figure 2). These observations exclude the possibility that, prior to DNA binding, the free SeqA molecules are a mixture of conformations with the two DNA binding domains at different distances from one another and that different molecules in the population bind to differently spaced hemi-sites. Instead we suggest that, since the C-terminal domains of SeqA are able on their own to bind to a hemi-site with a low affinity (23,26,27), the transient binding of one of the two DNA binding domains to a hemi-site causes a conformational change of the molecule that allows the other DNA binding domain to make contact with an unbound hemi-site, as depicted in Figure 5.
Figure 5.
Binding of a SeqA to a pair of hemi-methylated GATC sequences. The binding of one of the DNA binding domains of a SeqA molecule to a hemi-site leads to conformational change of the molecule that allows the other DNA binding domain to bind to an adjacent hemi-site within 32 bp. Binding is stabilized by bending the bound DNA.
The extended conformation of SeqA bound to two hemi-sites on a linear DNA molecule is stabilized by bending the region between the two hemi-sites. Increasing the spacing between two hemi-sites to three helical turns results in greater separation of the DNA binding domains of the SeqA (Figure 2). Presumably, it is for this reason that SeqA has a lower affinity for 31 bp spacing DNA than for 21 bp spacing (20) and is not able to bind to DNA with hemi-sites spacing of more than 31 bp (12). These results suggest either that its two DNA binding domains cannot reach two hemi-sites separated by more than three helical turns, or that SeqA has limited space into which to fit the bent DNA, or both. The distortion of SeqA that accompanies its binding to hemi-sites on opposite sides of the DNA helix accounts for its preference for hemi-sites on the same side (Figure 2E).
A SeqA molecule bound to a pair of hemi-sites can interact cooperatively with another SeqA molecule bound to an adjacent pair of hemi-sites (20,21). The cooperative interaction between the bound molecules enhances their capacity for interaction and permits them to interact with another SeqA molecule bound to distal hemi-sites. This step-wise interaction is followed by aggregation of free SeqA proteins onto the bound proteins. Thus, SeqA possesses three distinct and consecutive biochemical properties: binding to a pair of hemi-sites, cooperative binding to two or more pairs of sites and aggregate formation.
We have shown that Glu-5, Asp-7 and Glu-9 are required for proper binding of SeqA to a pair of hemi-sites (Figure 4), and that Thr-18 is necessary for cooperativity (Figure 3). The N-terminal fragment (amino acids 1–50) is responsible for aggregation of SeqA proteins onto long hemi-methylated DNA (21) as well as the multimerization of individual SeqA polypeptides to form a multimer (23). Thus, the multimerization domain influences many aspects of the biochemical behavior of SeqA protein: multimerization of SeqA polypeptides, proper binding of the DNA binding domain to a pair of hemi-sites, cooperative binding to multiple hemi-sites and aggregation onto hemi-methylated DNA.
The GATC sequences that become transiently hemi-methylated upon DNA replication are distributed with variable spacing between the sequences in E.coli genome (12). The flexible dimeric configuration of SeqA and its cooperative and aggregation properties permit it to interact with the diversely spaced hemi-sites. In this way, SeqA can perform its roles in DNA replication: the regulation of chromosome initiation, formation of foci, maintenance of negative superhelicity and segregation of the replicated DNA.
Acknowledgments
This work was supported by a grant from the 21C Frontier Microbial Genomics and Applications Center Program and from Systems Biology, Ministry of Science and Technology, Republic of Korea. K.P.K. and C.B.H. were supported by a grant from the Crop Functional Genomics Center Program (CG1434), Ministry of Science and Technology, Republic of Korea. S.K., J.S.H. and K.Y.L. were supported by a BK21 Research Fellowship from the Ministry of Education, Republic of Korea. Funding to pay the Open Access publication charges for this article was provided by the BK program from the Ministry of Education, Republic of Korea.
REFERENCES
- 1.Sarraf S.A., Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 2.Wade P.A. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- 3.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 4.Geier G.E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J. Biol. Chem. 1979;254:1408–1413. [PubMed] [Google Scholar]
- 5.Campbell J.L., Kleckner N. E.coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 6.Messer W., Weigel C. Initiation of chromosome replication. In: Neidhardt F.C., Curtiss R. III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996. pp. 1580–1582. [Google Scholar]
- 7.Lu M., Campbell J.L., Boye E., Kleckner N. SeqA: a negative modulator of replication initiation in E.coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 8.Slater S., Wold S., Lu M., Boye E., Skarstad K., Kleckner N. E.coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 9.Kang S., Lee H., Han J.S., Hwang D.S. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J. Biol. Chem. 1999;274:11463–11468. doi: 10.1074/jbc.274.17.11463. [DOI] [PubMed] [Google Scholar]
- 10.Boye E., Stokke T., Kleckner N., Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraga S., Ichinose C., Niki H., Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA–protein complexes in E.coli. Mol. Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 12.Brendler T., Sawitzke J., Sergueev K., Austin S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 2000;19:6249–6258. doi: 10.1093/emboj/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onogi T., Niki H., Yamazoe M., Hiraga S. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol. Microbiol. 1999;31:1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga S., Ichinose C., Onogi T., Niki H., Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 15.Bach T., Krekling M.A., Skarstad K. Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J. 2003;22:315–323. doi: 10.1093/emboj/cdg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitao T., Nordstrom K., Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- 17.Weitao T., Nordstrom K., Dasgupta S. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 2000;1:494–499. doi: 10.1093/embo-reports/kvd106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang S., Han J.S., Park J.H., Skarstad K., Hwang D.S. SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 2003;278:48779–48785. doi: 10.1074/jbc.M308843200. [DOI] [PubMed] [Google Scholar]
- 19.Brendler T., Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J.S., Kang S., Kim S.H., Ko M.J., Hwang D.S. Binding of SeqA protein to hemi-methylated GATC sequences enhances their interaction and aggregation properties. J. Biol. Chem. 2004;279:30236–30243. doi: 10.1074/jbc.M402612200. [DOI] [PubMed] [Google Scholar]
- 21.Han J.S., Kang S., Lee H., Kim H.K., Hwang D.S. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J. Biol. Chem. 2003;278:34983–34989. doi: 10.1074/jbc.M304923200. [DOI] [PubMed] [Google Scholar]
- 22.Lee H., Kang S., Bae S.H., Choi B.S., Hwang D.S. SeqA protein aggregation is necessary for SeqA function. J. Biol. Chem. 2001;276:34600–34606. doi: 10.1074/jbc.M101339200. [DOI] [PubMed] [Google Scholar]
- 23.Guarné A., Zhao Q., Ghirlando R., Yang W. Insights into negative modulation of E.coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nature Struct. Biol. 2002;9:839–843. doi: 10.1038/nsb857. [DOI] [PubMed] [Google Scholar]
- 24.Guarné A., Brendler T., Zhao Q., Ghirlando R., Austin S., Yang W. Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. EMBO J. 2005 doi: 10.1038/sj.emboj.7600634. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae S.H., Cheong H.K., Cheong C., Kang S., Hwang D.S., Choi B.S. Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition. J. Biol. Chem. 2003;278:45987–45993. doi: 10.1074/jbc.M306038200. [DOI] [PubMed] [Google Scholar]
- 26.Fujikawa N., Kurumizaka H., Yamazoe M., Hiraga S., Yokoyama S. Identification of functional domains of the Escherichia coli SeqA protein. Biochem. Biophys. Res. Commun. 2003;300:699–705. doi: 10.1016/s0006-291x(02)02891-7. [DOI] [PubMed] [Google Scholar]
- 27.Fujikawa N., Kurumizaka H., Nureki O., Tanaka Y., Yamazoe M., Hiraga S., Yokoyama S. Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein. Nucleic Acids Res. 2004;32:82–92. doi: 10.1093/nar/gkh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viani M.B., Pietrasanta L.I., Thompson J.B., Chand A., Gebeshuber I.C., Kindt J.H., Richter M., Hansma H.G., Hansma P.K. Probing protein–protein interactions in real time. Nature Struct. Biol. 2000;7:644–647. doi: 10.1038/77936. [DOI] [PubMed] [Google Scholar]
- 29.Engel A., Schoenenberger C.A., Müller D.J. High resolution imaging of native biological sample surfaces using scanning probe microscopy. Curr. Opin. Struct. Biol. 1997;7:279–284. doi: 10.1016/s0959-440x(97)80037-1. [DOI] [PubMed] [Google Scholar]
- 30.Möller C., Allen M., Elings V., Engel A., Müller D.J. Tapping-mode atomic force microscopy produces faithful high-resolution images of protein surfaces. Biophys. J. 1999;77:1150–1158. doi: 10.1016/S0006-3495(99)76966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivetti C., Guthold M., Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietrasanta L.I., Thrower D., Hsieh W., Rao S., Stemmann O., Lechner J., Carbon J., Hansma H. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl Acad. Sci. USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]