Abstract
Universal DNA base analogs having photocleavable properties would be of great interest for development of new nucleic acid fragmentation tools. The photocleavable 7-nitroindole 2′-deoxyribonucleoside d(7-Ni) was previously shown to furnish a highly efficient approach to photochemically trigger DNA backbone cleavage at preselected position when inserted in a DNA fragment. In the present report, we examine its potential use as universal DNA nucleoside, by analogy with the 5-nitroindole analog that is generally considered as universal base. The d(7-Ni) phosphoramidite was incorporated into oligonucleotides. Hybridization properties of resulting 11mer duplexes indicated a behavior close to that of the 5-nitroindole analog. Enzymatic recognition by Klenow fragment exonuclease-free using 40mers containing the unnatural bases as templates indicated notably a decrease of the polymerase activity with preferential incorporation of dAMP opposite both the 7-Ni and 5-Ni bases. Incorporation of the d(7-Ni) triphosphate was also studied indicating absence of significant differences between the incorporation kinetics opposite each natural base in the template. All the hybridization and enzymatic data indicate that 7-nitroindole can be considered as a cleavable base analog, although not strictly fulfilling, like the 5-nitro isomer, all properties required for a universal base.
INTRODUCTION
Design of modified nucleosides or triphosphates able to substitute for natural nucleic acids components and possessing photochemically cleavable properties is of high interest in the development of new nucleic acids fragmentation tools. DNA fragmentation is frequently required for nucleic acids analysis (sequencing, structural analysis) and a number of cleavage techniques exist, each presenting limitations. Restriction enzymes have been used quite frequently in many applications, however their size and sequence do not allow systematic use. Artificial compounds able to cleave DNA have been developed. These chemical nucleases including notably metallic complexes (1–5) or photoactive derivatives (6) necessitate addition of reagents and consequently removal of byproducts. Direct DNA fragmentation via photochemical processes is an attractive approach allowing both spatial and temporal control and offering the advantage of being fully reagent-free, permitting DNA fragmentation in cell or during PCR reactions. However, only a limited number of photocleavable DNA ‘building blocs’ have been developed. Most consist of nucleoside analogs chemically inserted in DNA fragments and are based on the photochemical properties of the o-nitrobenzyl group (7–13).
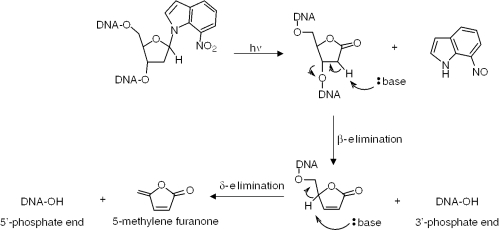
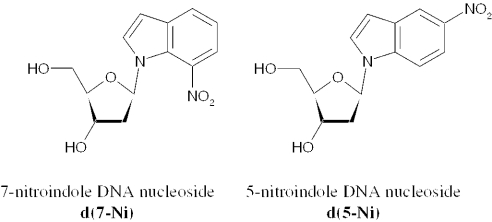
We reported the synthesis (14), structure (15) and photochemical properties (16) of the 7-nitroindole nucleoside d(7-Ni). Illumination of oligonucleotides containing d(7-Ni) triggers a radical process in which the excited nitro group induces an intramolecular H-1′ abstraction leading quantitatively to the highly labile deoxyribonolactone moiety accompanied with loss of the 7-nitrosoindole fragment. Subsequent mild basic or thermal treatment leads to cleavage of the DNA strand (Figure 1). The reaction is total, rapid and sequence independent. This photoactive 7-nitroindole heterocycle is indeed an analog of the 4-, 5- and 6-nitroindoles which have been investigated as ‘universal’ bases in oligonucleotides, among which the authors concluded that the 5-nitro isomer was superior giving highest duplex stability and behaving indiscriminately towards each of the four natural bases in duplex melting experiments (17). ‘Universal base analogs’ have indeed attracted much attention in recent years due to their intrinsic fundamental interest and to their potential applications as biochemical tools (use as primers for PCR and sequencing or as substrates for enzymes as triphosphates) (18). Apart from the afore-mentioned 5-nitroindole considered as being among the best base analogs such compounds as nitropyrroles (17–19), 8-aza-7-deazapurine-2,6-diamine (20,21) or hydrophobic structures containing notably the isocarbostyryl ring have been developed (22–27). It was thus of interest to investigate the photocleavable 7-nitroindole as a potential ‘universal’ base and compare it to its 5-nitroindole isomer (Figure 2). This compound could thus possess the two complementary properties of being universal and photocleavable, an approach that has been elegantly pioneered by Pirrung in developing the DNA base analog nitropiperonyl 2′-deoxyriboside (28). In the present paper, we report on the base-pairing properties of d(7-Ni) in oligonucleotide duplexes and compare it to the 5-nitroindole isomer. We also describe enzymatic experiments to study the recognition of this unnatural nucleoside incorporated in oligonucleotides by the DNA polymerase Klenow fragment exonuclease-free as well as the synthesis and incorporation of the d(7-Ni) triphosphate by the polymerase.
Figure 1.
Irradiation of DNA containing the 7-nitroindole nucleoside results in the formation of the alkali labile deoxyribonolactone lesion followed by backbone cleavage.
Figure 2.
Structure of the deoxynucleosides derived from 7-nitroindole [d(7-Ni)] and 5-nitroindole [d(5-Ni)].
MATERIALS AND METHODS
Materials
All reagents were of the highest quality and commercially available. Oligonucleotides were labeled using [γ-32P]ATP (specific activity 3000 Ci/mmol) (NEM™ Life Science) and T4 polynucleotide kinase purchased from MBI fermentas. The Klenow fragment of Escherichia coli DNA polymerase I deficient in 3′→5′ proofreading exonuclease activity (KF exo−) was obtained from MBI fermentas. Nucleotide triphosphates (dNTPs) were purchased from Pharmacia Biotech. Reagents used for automated synthesis were obtained from PE Biosystems.
Oligonucleotide synthesis
Conventional or modified [d(7-Ni) or d(5-Ni)] oligonucleotides were synthesized using standard solid-phase β-cyanoethylphosphoramidite method on a Expedite DNA synthesizer (Perseptive Biosystem 8900). The nitroindole nucleoside phosphoramidite [d(7-Ni)] was prepared as previously reported (14,16). The 40 nt long oligonucleotides were used as template in enzymatic reactions. Melting experiments and CD were conducted on shorter oligonucleotides (11mer). During the automated synthesis, the standard cycles for 1 μmol DNA synthesis were modified since double amounts of phosphoramidites were used for coupling and after this step also double amount of capping reagent was added. For incorporation of the nitroindole nucleosides [d(7-Ni) or d(5-Ni)], standard coupling time was prolonged by 10 min and quantity of capping reagent multiplied by 5. After automated synthesis, the oligomers for CD and melting experiments keeping the DMT group in 5′ terminal were cleaved from the support by concentrated aqueous ammonia for 1 h, deprotected by heating at 55°C for 24 h. Finally, oligomers were purified by high-performance liquid chromatography (HPLC) on a reversed-phase nucleosil C-18 column (Macherey-Nagel 10 × 250 mm) with a linear gradient of acetonitrile (5–35% in 20 min) in 20 mM ammonium acetate solution (pH 7). Oligomers for enzymatic reaction were detritylated on column, cleaved from support then deprotected as described previously. These oligomers were purified by electrophoresis on a 20% polyacrylamide gel in the presence of 7 M urea. The concentration of oligodeoxynucleotides was determined by UV. Oligonucleotide sequences used for experiments have been reported in Table 1.
Table 1.
Oligonucleotide sequences
| Number | Sequence (5′→3′) |
|---|---|
| 1 | CGCAC 7-Ni CACGC |
| 2 | CGCAC 5-Ni CACGC |
| 3 | GCGTGAGTGCG |
| 4 | GCGTGCGTGCG |
| 5 | GCGTGGGTGCG |
| 6 | GCGTGTGTGCG |
| 7 | CGCACACACGC |
| 8 | CGCACGCACGC |
| 9 | GCGTG 7-Ni GTGCG |
| 10 | CATGCTGATGAATTCAGGCCT 7-Ni GAGGAATCGCTGGTACCG |
| 11 | CATGCTGATGAATTCAGGCCT 5-Ni GAGGAATCGCTGGTACCG |
| 12 | CGGTACCAGCGATTCCTC |
| 13 | CATGCTGATGAATTCAGGCCTTGAGGAATCGCTGGTACCG |
| 14 | CATGCTGATGAATTCAGGCCTCGAGGAATCGCTGGTACCG |
| 15 | CATGCTGATGAATTCAGGCCTGGAGGAATCGCTGGTACCG |
| 16 | CATGCTGATGAATTCAGGCCTAGAGGAATCGCTGGTACCG |
| 17 | CATGCTGATGAATTCAGGCCA 7-Ni GAGGAATCGCTGGTACCG |
| 18 | CATGCTGATGAATTCAGGCCC 7-Ni GAGGAATCGCTGGTACCG |
| 19 | CATGCTGATGAATTCAGGCCG 7-Ni GAGGAATCGCTGGTACCG |
| 20 | CATGCTGATGAATTCAGGCCATGAGGAATCGCTGGTACCG |
| 21 | CATGCTGATGAATTCAGGCCCTGAGGAATCGCTGGTACCG |
| 22 | CATGCTGATGAATTCAGGCCGTGAGGAATCGCTGGTACCG |
Melting temperature experiments
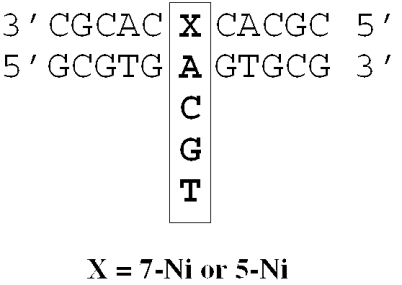
Experiments were performed on 11 bp DNA duplexes containing d(7-Ni) or d(5-Ni) paired with dG, dC, dA or dT (Figure 3). Equimolar solutions (3 μM) of unmodified or d(7-Ni) modified oligonucleotides were mixed with their complementary strands in buffer consisting of 10 mM sodium phosphate/1 mM EDTA/100 mM NaCl adjusted to pH 7. UV absorption spectra (at 260 nm) and melting experiments were recorded using a CARY 400 Scan UV-Visible spectrophotometer (Varian) equipped with a Peltier thermoelectric Cary temperature controller (Varian). Before each melting experiment, samples were heated at 80°C for 5 min and cooled down slowly to ensure that the oligonucleotides were in the duplex state. The absorbance was monitored at 260 nm from 5 to 80°C at a heating rate of 0.3°C per min. Data were recorded every 1 min. Melting temperatures were determined from the derivative method using the ‘Cary Win UV-bio’ (version 2) application software. Error in the determination of Tm has been estimated as ±0.6°C (average of three independent measurements).
Figure 3.
DNA-modified duplexes used for melting temperature studies. The four natural bases were located successively opposite the unnatural base X (X = 7-Ni or 5-Ni). The four variants of unnatural base pair have been indicated in box region.
Synthesis
1-[3′-O-Acetyl-2′-deoxy-β-D-erythro-pentofuranosyl]-7-nitroindole (3)
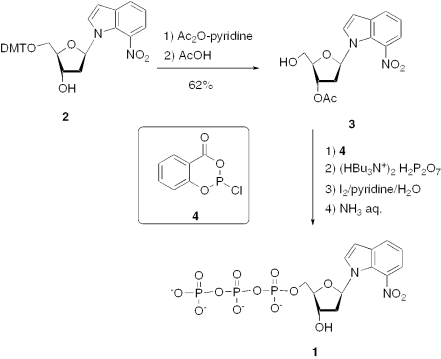
To a solution of the compound 2 (650 mg, 1.12 mmol) in 4 ml of pyridine, acetic anhydride (230 μl, 2.24 mmol) was added (see Figure 4). The mixture was stirred at room temperature overnight. The solvents were removed under reduced pressure. The crude intermediate product was purified by chromatography on silica gel (elution with AcOEt/cyclohexane 40/60 to 50/50). The resulting product was then dissolved in 4.5 ml of aqueous acetic acid (80%) and stirred for 90 min at room temperature. After evaporation of solvents under reduced pressure, the residual oil was purified on silica gel (elution with AcOEt/cyclohexane 40/60 to 60/40) to afford 3 (yellow oil, 222 mg, 62%). Rf = 0.14 (AcOEt/cyclohexane 40/60). 1H NMR (300 MHz, CDCl3): δ = 7.84 (d, J = 7.8 Hz, 1 H), 7.81 (d, 1 H, J = 7.9 Hz), 7.59 (d, J = 3.6 Hz, 1 H), 7.15 (t, J = 7.9 Hz, 1 H), 6.68 (d, J = 3.4 Hz, 1 H), 6.36 (t, J = 6.4 Hz, 1 H), 5.29 (m, 1 H), 4.04 (m, 1 H), 3.72 (m, 2 H), 2.66 (m, 2 H), 2.16 (br. s, 1 H), 2.11 (s, 3 H). 13C NMR (75 MHz, CDCl3): δ = 171.0, 137.2, 133.6, 127.3 (2C), 126.6, 120.3, 119.5, 104.6, 87.0, 84.4, 74.2, 62.6, 38.7, 21.1. MS (DCI, NH3/isobutane): 337.7 [M+NH3]+. Anal. Calcd for C15H16N2O6 (320.3): C 55.47, H 5.12, N 8.62. Found: C 55.69, H 5.21, N 8.44.
Figure 4.
Synthesis of the unnatural triphosphate [d(7-Ni)TP].
5′-Triphosphate of 1-[2′-deoxy-β-D-erythro-pentofuranosyl]-7-nitroindole (1)
The 3′-acetyl nucleoside 2 (50 mg, 150 μmol) was dehydrated by coevaporation with dry pyridine (twice) and then dissolved under argon in dry pyridine (150 μl) and dry dioxane (450 μl). To this solution, 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one (200 μl of 1 M solution freshly prepared in dry dioxane, 200 μmol) was added and stirred at room temperature under argon for 20 min. Then tributylammonium pyrophosphate (480 μl of 0.5 M solution in dry DMF, 240 μmol) and tributylamine (195 μl, 0.8 mmol) were added simultaneously and stirred for 30 min. The reaction mixture was then treated with iodine (3 ml of 1% solution in aqueous pyridine solution: pyridine/water = 98/2 v/v, 240 μmol) for 20 min and then the excess of iodine was decomposed by addition of sodium hydrogen sulfite aqueous solution (5%, 1.8 ml). After stirring for 10 min, solvents were removed in vacuo, then the residue was dissolved in 15 ml of water and 30 ml of concentrate ammoniac aqueous solution. The solution was stirred for 1 h at room temperature. The aqueous solution was washed three times with dichloromethane (50 ml) and concentrated in vacuo. The crude product was purified by reverse-phase silica gel chromatography (LiChroprep RP 18 silica gel, 40–63 μm, elution with 0–100% MeOH/H2O) to afford 15 mg (20%) of 1. Rf = 0.62 (propan-1-ol/H2O/NH3 11/2/7). 1H NMR (300 MHz, D2O): δ = 8.01 (d, J = 7.8 Hz, 1 H), 7.89 (d, J = 8.0 Hz, 1 H), 7.76 (d, J = 3.6 Hz, 1 H), 7.24 (t, J = 7.9 Hz, 1 H), 6.85 (d, J = 3.4 Hz, 1 H), 6.44 (t, J = 6.5 Hz, 1 H), 4.68 (m, 1 H), 4.13 (m, 1 H), 4.05 (m, 2 H), 2.70 (m, 2 H). 31P NMR (121 MHz, D2O/10 mM EDTA, pH = 8): δ = −21.69 (t, Pβ), −10.97 (d, Pα), −6.86 (d, Pγ). MS (ESI): 516.9 [M-H]−. UV [380 μM in H2O]: λmax = 370 nm (1842).
Polymerization assays with KF exo−
General preparation of primer/templates duplexes
5′-32P-labeled primer were annealed to template oligonucleotides (2 equivalents) in 50 mM Tris (pH 7.5), 10 mM MgCl2, 5 mM 2-mercaptoethanol by heating at 70°C for 5 min then cooling to room temperature >2 h.
Primer extension by KF exo− opposite d(7-Ni) or d(5-Ni) present in template
Reactions were conducted on modified (X) [X = d(7-Ni), sequence 1; X = d(5-Ni), sequence 2] or unmodified (X = T, sequence 4) template (100 nM) hybridized to 5′ end-labeled primer (sequence 3, 50 nM). Primer extension reaction catalyzed by KF exo− (0.01 U/μl) were carried out in buffer hybridization in the presence of single dNTP (20 μM) or all four dNTPs (20 μM each) for 20 min at 30°C in a final volume of 10 μl. Reactions were quenched in formamide loading buffer (30 μl) (95% formamide, 20 mM EDTA pH 8, 0.1% bromophenol blue, 0.1% xylene cyanol) heating at 70°C for 5 min. Aliquots (2 μl) were loaded on a 20% denaturing PAGE.
Insertion kinetics by Klenow exo− opposite d(7-Ni) or natural base
The 5′ end-labeled 18mer (sequence 12, 50 nM) was annealed to modified template containing either d(7-Ni) or d(5-Ni) (sequences 10 or 11, 0.1 μM) or unmodified template (sequences 13–16, 0.1 μM). Duplexes were incubated with various amounts of dNTP (10–1000 μM) and KF exo− (0.005–1 U per sample depending on the template and dNTP) in a final volume of 10 μl for 2 min at 30°C in order to obtain <20% elongation of primer. The reactions were quenched by adding 20 μl formamide dye mixture and heating at 70°C for 5 min. The reaction mixtures were resolved by 20% denaturing PAGE and the radioactivity was quantified by means of an Imager (Typhoon 9400) and the ImageQuant program. A plot of the initial velocity versus [dNTP] was fit to a Michaelis–Menten equation using the program Kaleida-graph (Synergy Software). The data presented are averages of three independent determinations.
d(7-Ni) triphosphate incorporation opposite d(7-Ni) or normal base in template followed by full-length primer extension
Reactions were conducted on d(7-Ni) modified template (sequences 1 or 11–13, 100 nM) or unmodified template (sequences 4–10, 100 nM) annealed to 5′ end-labeled primer (sequence 3, 50 nM). The first step relating to d(7-Ni)MP addition to the 3′ primer terminus was carried out using KF exo− (0.0625 U/μl) in presence of d(7-Ni)TP (1 mM) yielding a total reaction volume of 10 μl, for 5 min at 30°C. Reaction was stopped by heating at 70°C for 5 min. The second step of reaction consisting in adding one more monophosphate complementary to the base in 5′ position of the d(7-Ni) in the template or in full-length primer extension was catalyzed by KF exo− (0.0125 U/μl) in presence of either one dNTP (500 μM) or the four dNTPs (500 μM each) in a final volume of 20 μl. Reactions were incubated at 30°C for 20 min, then quenched with formamide loading buffer (30 μl) and analyzed by 20% denaturing PAGE.
RESULTS
Preparation of the 7-nitroindole nucleoside triphosphate 1
The 7-nitroindole nucleoside triphosphate 1 was prepared using the Ludwig–Eckstein method (29), which allows one-pot triphosphate formation starting from 3′-protected nucleosides (see Figure 4). Thus the 5′-trityl nucleoside 2 was first converted to 3′-protected derivative 3, which was then treated successively (i) with 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one (4), (ii) with tributylammonium pyrophosphate, (iii) with iodine–aqueous pyridine and (iv) with aqueous ammoniac solution. Purification of crude mixture was carried out by reverse-phase chromatography. The triphosphate 1 was obtained with 20% overall yield and characterized by 1H and 31P NMR and by ES-MS spectrometry.
Hybridization properties
UV thermal melting studies were performed in order to assess the effects of d(7-Ni) on duplex stability relative to the natural base pairs. Studies were performed on a series of 11mer duplexes containing the modified d(7-Ni) nucleoside in the middle of the sequence in one strand (sequence 1) with the complementary strand containing successively the four natural bases facing the 7-Ni base (sequences 3–6). For comparison, we also determined the behavior of duplexes containing the d(5-Ni) nucleoside analog inserted in the same sequence context (sequence 2). The unnatural self-pair 7-Ni:7-Ni containing duplex was also examined (sequence 1:sequence 9). The sigmoidal curves α = f(T) obtained from the standard UV melting curves for the different duplexes exhibited melting cooperativity and were analyzed to derive the Tm values and the Van't Hoff transition enthalpy ΔH, entropy ΔS and free energy ΔG°. The thermodynamics of melting were determined with the base pairs as indicated in the boxed region (Figure 3). The resulting data are listed in Table 2.
Table 2.
Melting temperatures and thermodynamic parameters of 11mer duplexes containing natural or unnatural base pairs (X:Y) in the middle of the sequence
| Duplex | X | Y | Tm (°C) | ΔS° (cal/mol/K) | ΔH° (kcal/mol) | ΔG° (kcal/mol) |
|---|---|---|---|---|---|---|
| 3′-CGCACXCACGC-5′ | Unnatural base pair | |||||
| 5′-GCGTGYGTGCG-3′ | ||||||
| 7-Ni | A | 49 | 91 | 37 | 10 | |
| 7-Ni | T | 46 | 170 | 62 | 11 | |
| 7-Ni | G | 51 | 194 | 71 | 13 | |
| 7-Ni | C | 46 | 127 | 48 | 11 | |
| A | 7-Ni | 48 | 170 | 62 | 11 | |
| G | 7-Ni | 45 | 159 | 58 | 11 | |
| 7-Ni | 7-Ni | 51 | 157 | 59 | 12 | |
| 5-Ni | A | 50 | 178 | 66 | 13 | |
| 5-Ni | T | 49 | 211 | 76 | 13 | |
| 5-Ni | G | 52 | 227 | 82 | 14 | |
| 5-Ni | C | 50 | 182 | 67 | 13 | |
| Natural base pair | ||||||
| A | T | 59 | 231 | 85 | 16 | |
| G | C | 61 | 240 | 89 | 17 | |
| Mismatch | ||||||
| A | C | 46 | 185 | 67 | 12 | |
Experiments were run in triplicate. See Materials and Methods for details.
A first observation is that the two modified nucleosides induce strong destabilization of the duplexes as compared to duplexes containing the natural bases, the d(7-Ni) nucleoside being slightly more destabilizing than the d(5-Ni) analog. Decrease of the melting temperatures ΔTm = 13 and 15°C were measured when natural base pairs A:T (sequence 7:sequence 6) and G:C (sequence 8:sequence 4) were replaced by the unnatural pairs 7-Ni:T (sequence 1:sequence 6) and 7-Ni:C (sequence 1:sequence 4), corresponding to loss in free energy of 5 and 6 kcal/mol, respectively. By comparison, 5-Ni present in the same sequence context led to Tm decrease of 10 and 11°C corresponding to loss of free energy of 3 and 4 kcal/mol, respectively. The destabilization caused by the unnatural bases is of the same order of magnitude as that caused by a mismatch [see for example, the 46°C value determined for the duplex containing one A:C mismatch in the same sequence context (sequence 7:sequence 4)]. However, when present in each of the complementary strands in opposite positions, the Tm value for the duplex containing the 7-Ni:7-Ni pair (sequence 1:sequence 9) exhibits Tm value = 51°C, i.e. in the same range as measured for the natural duplex.
Second observation relates to discrimination of 7-Ni and 5-Ni for the different natural bases. The two bases have comparable behavior although 7-Ni shows more discrimination than 5-Ni. Tm values are in the range 46–51°C for duplexes containing 7-Ni and all four natural bases in opposite position in complementary strand. Values vary in the range 49–52°C for the analogous duplexes containing 5-Ni.
Circular dichroism (CD) spectra were also measured (data in Supplementary Material). The CD-spectra obtained for the various unnatural base pair containing duplexes showed similarities to that of the natural parent duplex, indicating a right-handed. Thus, the unnatural base pair does not appear to induce major structural distortions of the B-form duplex.
Replication—enzymatic recognition of the unnatural bases 5-Ni and 7-Ni by KF exo−
In general, enzyme efficiency is defined by the ability to base-pair synthesis (e.g. incorporation of a triphosphate opposite a natural or unnatural base in the template), and second, by the aptitude to extend a natural or unnatural base pair (synthesis of the next base pair). The efficiency and the selectivity of unnatural DNA synthesis involving the d(7-Ni) nucleoside by KF exo− were examined both in the context of base-pair synthesis and base-pair extension.
Insertion of dNMP opposite d(7-Ni) and d(5-Ni)
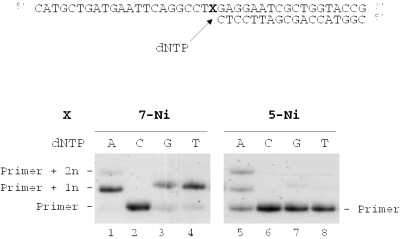
Duplexes containing the 7-Ni nucleobase and for comparison 5-Ni were evaluated as substrates for the exonuclease-deficient Klenow fragment of E.coli DNA polymerase I (KF exo−). Studies were investigated using templates containing in the middle of the sequence one of the unnatural bases 7-Ni (sequence 10) or 5-Ni (sequence 11) hybridized to the 18mer primer (sequence 12). For comparison, a template containing the natural base T (sequence 13) hybridized to the same 18mer primer was also examined. The resulting duplexes were subjected to primer extension in the presence of one single dNTP (see Figure 5). DNA synthesis on template containing the natural base (sequence 13) led to the expected incorporation of dAMP opposite T (data not shown). When using the 7-Ni containing strand as template (sequence 10) dAMP, dGMP and dTMP were incorporated opposite the unnatural base, however, with quite different efficiencies (Figure 5, lanes 1, 3 and 4), while insertion of dCMP could not be detected (Figure 5, lane 2). The same experiment conducted with the template containing 5-Ni indicated that dAMP was incorporated opposite the unnatural base (Figure 5, lane 5).
Figure 5.
Nucleotide incorporation opposite 7-Ni or 5-Ni in the presence of one single dNTP. Reactions were investigated on 40mer templates containing the unnatural base X (X = 7-Ni), sequence 10 (lanes 1–4) or 5-Ni, sequence 11 (lanes 5–8); 100 nM primed with 32P-labeled 18mer (sequence 12, 50 nM) in the presence of one single dNTP (20 μM) and KF exo− (0.1 U). Reactions were incubated for 20 min at 30°C before processing as described in Materials and Methods.
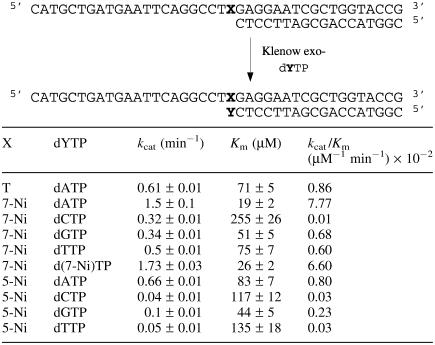
Kinetic parameters for insertion of dNMP opposite the modified sites were determined. The initial velocities were measured and plotted versus [dNTP] and fitted to the Michaelis–Menten equation to determine the Km and kcat constants. To allow comparison of the kinetic parameters for all substrates, the specificity constants kcat/Km were calculated. The steady-state kinetic constants for single nucleotide incorporation are reported in Table 3.
Table 3.
Efficiency of KF exo− incorporation of the triphosphates opposite natural or unnatural bases (X) in the template
With template containing d(7-Ni) (sequence 10), dAMP was preferentially inserted opposite the modified site. The kcat/Km value for dAMP incorporation was respectively 11 times, 13 times and 777 times higher than measured for dGMP, dTMP and dCMP incorporation. In addition, the d(7-Ni) nucleoside was incorporated with almost the same frequency as dAMP. Thus, the efficiency for nucleotide insertion opposite d(7-Ni) follows the order dAMP ∼ d(7-Ni)MP >> dGMP ∼ dTMP > dCMP.
The same experiments performed with the template containing d(5-Ni) (sequence 11) also showed a preference for dAMP incorporation opposite the modified base, and the incorporation frequency opposite d(5-Ni) followed the same order as determined for d(7-Ni). However, kcat/Km for dAMP incorporation was only three times higher than for dGMP and 27 times higher than for dTMP and dCMP.
Effect of the downstream template base on dNMP incorporation opposite d(7-Ni)
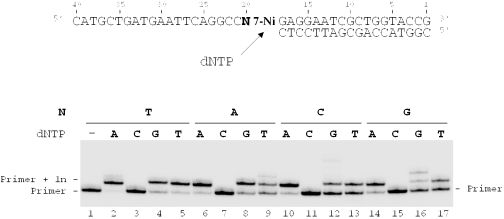
The primer-templates used in these experiments were designed in order to examine the effect of changing the template base at the 5′ side of d(7-Ni) on the initiation of the extension of the primer. The 18mer primer (sequence 12) was hybridized successively to 40mer templates differing by the nature of the base N = T, A, C, G at the 5′ side of d(7-Ni) (sequences 10, 17, 18 and 19, respectively) (Figure 6). Extension of the primer in the four duplexes was conducted in the presence successively of each of the natural dNTPs. For each duplex, dAMP (Figure 6, lanes 2, 6, 10 and 14), dGMP (Figure 6, lanes 4, 8, 12 and 16) and dTMP (Figure 6, lanes 5, 9, 13 and 17) were incorporated opposite d(7Ni). Only dCMP was not inserted by the enzyme (lanes 3, 7, 11 and 15) even when guanosine was present at the 5′ side of d(7-Ni) (Figure 6, lane 15). It thus appears that changing a single downstream template base does not modify nucleotide incorporation opposite the unnatural 7-Ni base.
Figure 6.
Influence of the nature of the downstream template base N20 on nucleotide incorporation opposite 7-Ni. Reactions were performed on modified templates containing 7-Ni, but varying by the nature of the natural base N20 in 5′ side of 7-Ni, (N20 = T, sequence 10; N20 = A, sequence 17; N20 = C, sequence 18 and N20 = G, sequence 19G; 100 nM) hybridized to 18mer primer (sequence 12, 50 nM). Polymerization reactions were conducted in the presence of one single dNTP (20 μM) and KF exo− (0.1 U) for 20 min at 30°C.
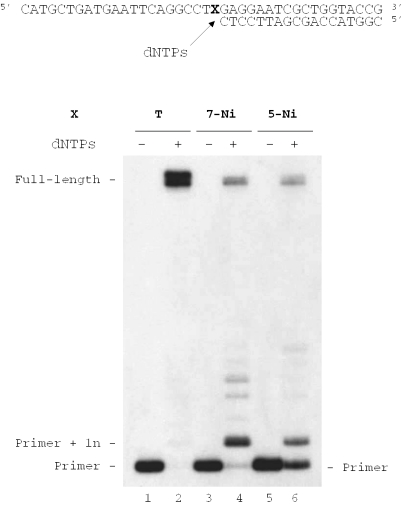
Full-length primer extension on DNA templates containing d(7-Ni) or d(5-Ni)
Modified templates containing one unnatural base 7-Ni (sequence 10) or 5-Ni (sequence 11) were annealed to the 18mer primer (sequence 12) and subjected to extension in the presence of the four dNTPs. For comparison, measurements were also made by using an otherwise identical template that contained dT (sequence 13) instead of the modified nucleoside (Figure 7). As expected, KF exo− led to fully extended 40mer products when experiments were conducted on the natural template (template: sequence 13, primer: sequence 12) (Figure 7, lane 2). Employing identical enzyme concentrations and dNTP quantities as used for the unmodified duplex, KF exo− synthesized full-length DNA starting from both modified duplexes containing either d(7-Ni) or d(5-Ni) in the template (Figure 7, lanes 4 and 6). However, extension was more efficient with template containing d(7-Ni). The additional major band seen in lanes 4 and 6 results from single nucleotide extension of the primer through incorporation of a native triphosphate opposite the modified base. The accumulation of this intermediate implies that the continued synthesis after 1 nt incorporation is slow.
Figure 7.
Full-length extension of primer hybridized to modified templates. Reactions were carried out at 30°C for 20 min using a natural template (X = T) (lanes 1 and 2) or modified templates containing one unnatural base X (X = 7-Ni), sequence 10 [lanes 3 and 4) or 5-Ni, sequence 11 (lanes 5 and 6); 100 nM] primed with 32P-labeled 18mer (sequence 12, 50 nM) in the presence of the four dNTPs (at a final concentration of 20 μM each). Reactions were catalyzed by KF exo− (0.1 U).
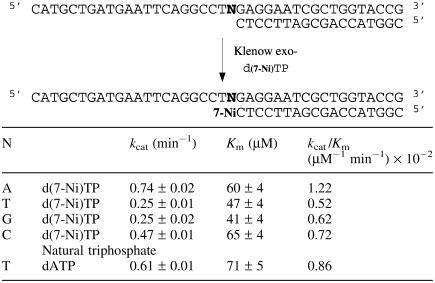
Incorporation of d(7-Ni) triphosphate
Efficiency of insertion of the triphosphate of d(7-Ni) against each of the natural bases in the template has been characterized. Experiments were run using the 18mer primer (sequence 12) hybridized successively to the four templates (sequences 13–16) varying by the nature of the base N at the initiation site of the polymerization. The kinetic parameters for insertion of the unnatural triphosphate opposite each natural base are reported in Table 4. Incorporation appeared to be more efficient opposite adenine (kcat/Km = 1.22 × 10−2 min−1 mM−1). However, the kcat/Km value was only two times higher than observed for incorporation opposite the three other bases that in addition showed quite remarkably comparable values. Furthermore, efficiency of incorporation opposite each natural base was of the same order of magnitude as incorporation of natural dAMP opposite thymine.
Table 4.
Efficiency of KF exo− incorporation of unnatural triphospate [d(7-Ni)TP] opposite natural bases in the template
Extension of the primer terminated by d(7-Ni)
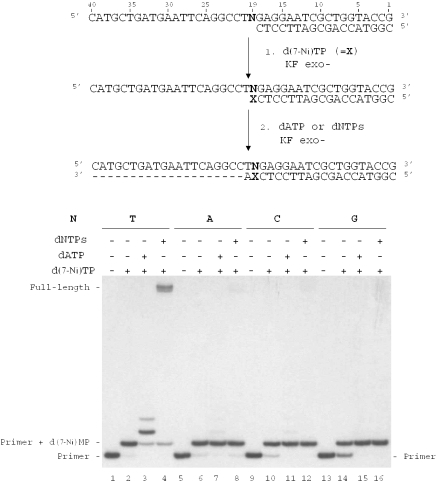
The ability of KF exo− to extend a primer terminated by the unnatural 7-Ni base opposite the different natural bases N in the template was studied (Figure 8). Reactions were conducted on the 18mer primer (sequence 12) annealed to the 40mer templates in which base N19 was successively a thymine (sequence 13), an adenine (sequence 16), a cytosine (sequence 14) or a guanine (sequence 15). In a first step, primer extension was run in the presence of the triphosphate d(7-Ni)TP (Figure 8, lanes 2, 6, 10 and 14). Incorporation of one d(7-Ni) nucleoside was observed for all duplexes, permitting to obtain all possible unnatural base pairs 7-Ni:N. In a second step, polymerization was continued on these extended primers in the presence either of dATP (the next complementary base in the template) (Figure 8, lanes 3, 7, 11 and 15) or in the presence of the four dNTPs (Figure 8, lanes 4, 8, 12 and 16). No extension could be observed for the duplexes in which d(7-Ni) was facing adenine, cytosine or guanine, neither in the presence of dATP (Figure 8, lanes 7, 11 and 15) nor in the presence of the mixed dNTPs (Figure 8, lanes 8, 12 and 16). Only the 7-Ni:T base pair was substrate for extension of the modified primer. Moreover, in this case addition of two adenines was detected in the presence of dATP (Figure 8, lane 3) and formation of a full-length product was observed in the presence of the four natural triphosphates (Figure 8, lane 4).
Figure 8.
Chain extension reaction catalyzed by KF exo− in the presence of the modified triphosphate d(7-Ni)TP. Experiments were investigated on different natural templates (100 nM) varying by the nature of the base N19 (N = T, sequence 13; N = C, sequence 14; N = G, sequence 15; N = A, sequence 16) hybridized to 18mer primer (sequence 12, 50 nM). In a first step, chain extension reactions were conducted in the presence of d(7-Ni)TP (1 mM) and KF exo− (0.0625 U/μl) for 5 min at 30°C (lanes 2, 6, 10 and 14). In the second step, reactions were carried out using KF exo− (0.0125 U/μl) in the presence of dATP (lanes 3, 7, 11 and 15) or in the presence of a mixture of the four dNTPs (lanes 4, 8, 12 and 16) for 20 min at 30°C.
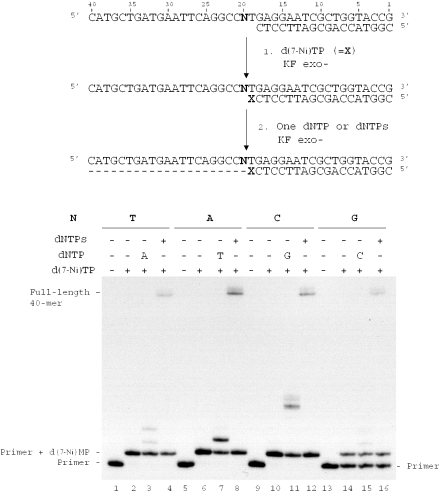
Influence of the downstream base on the extension of a primer terminated by the unnatural 7-Ni base
In order to interpret the previous results, we investigated the possible influence of the nature of the downstream template base (N20) on the extension of the primer terminated by the 7-Ni unnatural base. Experiments were conducted on the same 18mer primer (sequence 12) hybridized successively to 40mer templates (sequences 13, 20–22) varying only by the nature of base N20 (Figure 9). The primers elongated by one 7-Ni base were prepared as described in the previous experiment (Figure 9, lanes 2, 6, 10 and 14). It should be noted that the 18mer primer annealed to template in which N20 was a guanine was only partially converted in the conditions used. In a second step, elongation was continued either in the presence of the triphosphate complementary to base N20 located 5′ to thymine in the template (Figure 9, dATP: lane 3, dTTP: lane 7, dGTP: lane 11, dCTP: lane 12) or in the presence of the four dNTPs (Figure 9, lanes 4, 8, 12 and 16). For each duplex, primers terminated by 7-Ni were extended in the presence of a unique triphosphate. When dATP was added (duplex in which N20 is thymine) the 7-Ni primer was elongated by one or two units (Figure 9, lane 3). In the presence of dTTP or dCTP, the 7-Ni primer was extended by 1 nt (Figure 9, lanes 7 and 15). However, primer extension in the presence of dCTP only led to small amounts of reaction product. Incorporation of three guanines was observed for the duplex in which the template base N was a cytosine. Full-length extension (40mer) was observed in the presence of the mixture of the four dNTPs for all duplexes.
Figure 9.
Effect of the downstream template base N20 on chain extension reaction catalyzed by KF exo− starting from the unnatural base pair 7-Ni(in primer):T(in template). Experiments were investigated on different natural templates (100 nM) varying by the nature of the base N20 (N = T, sequence 13; N = A, sequence 20; N = C, sequence 21; N = G, sequence 22) hybridized to 18mer primer (sequence 12, 50 nM). Addition of one d(7-Ni) to the primer was carried out using KF exo− (0.0625 U/μl) in presence of d(7-Ni)TP (1 mM) for 5 min at 30°C (lanes 2, 6, 10 and 14). The second step of reaction was performed using KF exo− (0.0125 U/μl) in the presence of one single dNTP (lanes 3, 7, 11 and 15) or in the presence of the mixture of the four dNTPs (lanes 4, 8, 12 and 16) for 20 min at 30°C.
DISCUSSION
We have studied the impact on DNA duplex stability of the presence of the d(7-Ni) unnatural base opposite each native base. For comparison, we also measured the stability of duplexes containing the 5-Ni analog that is generally considered as a universal base. Thermal denaturation experiments showed that the two unnatural bases lead to strong decrease in duplex stability comparable to that caused by the presence of a mismatch. Destabilization is slightly more important for the duplex containing the 7-Ni base as compared to the 5-Ni isomer. Another criterion required for a universal base concerns absence of discrimination when the unnatural base in one strand faces any of the four natural bases in the complementary strand. The two nitroindole bases have comparable behavior, exhibiting slight discrimination, the 7-Ni base being slightly more discriminating. From a structural point of view, the CD spectra of the various duplexes containing the 7-Ni base showed the typical shape of B-DNA, indicating the absence of major distortions induced by the unnatural base. It can thus be concluded that as far as hybridization properties are concerned, the 7-Ni base exhibits behavior comparable to that of the 5-isomer although it cannot be considered as a true universal base according to this criterion.
Replication was next examined involving the unnatural nucleobase 7-Ni both in template DNA and as triphosphate. For comparison, recognition by KF exo− of d(5-Ni) present in the template was also studied. In the sequence context examined, we observed that dAMP was preferentially inserted opposite both 7-Ni and 5-Ni, with rates varying by a factor close to 10. The frequency for incorporation opposite both unnatural bases followed the order: dAMP ≫ dGMP = dTMP > dCMP. However, dAMP, dGMP and dTMP were more efficiently incorporated when facing d(7-Ni) than d(5-Ni). dCMP was almost not inserted by the enzyme opposite each of the unnatural bases. These results suggest that d(7-Ni) is recognized by KF exo− more efficiently than d(5-Ni). We also investigated the effect of the sequence on nucleoside incorporation opposite d(7-Ni). We observed that the nature of the downstream template base was without effect on the selectivity of the reaction: insertion of dAMP opposite the unnatural base was favored over dGMP, dTMP and dCMP. The effect of 7-Ni on KF exo− seems similar to that exhibited by a large spectrum of DNA lesions, such as the abasic site for which the ‘A-rule’ has been defined (30), or the 2′-deoxyribonolactone damage. Consequently, 7-Ni, as well as 5-Ni cannot be considered as a true universal base when present in the template, since the four natural triphosphates are not incorporated equally well in front of these unnatural bases. Primer extension experiments investigated in the presence of the four natural triphosphates showed that d(7-Ni) like d(5-Ni) when present in the template led to a decrease of the polymerase activity. Indeed, after addition of 1 nt, primer extension was very slow although full-length primers were obtained.
We also examined incorporation of the d(7-Ni) triphosphate opposite each natural base in the template. Quite remarkably, the kinetic parameters determined do not reveal significant differences for insertion in front of the four natural bases, behaving in this aspect as a universal nucleotide. The triphosphate was inserted opposite each natural base with at least the same efficiency as dAMP opposite thymine. In addition, the d(7-Ni) triphosphate was also inserted efficiently opposite itself in the template. Indeed, Romesberg reported that several hydrophobic aromatic nucleosides were preferentially incorporated opposite themselves, or similar structures rather than opposite natural nucleosides (26).
We also studied the extension of primers terminated by the unnatural base 7-Ni. We found that 7-Ni behaves as chain terminator when facing adenine, guanine or cytosine in the template. On the other hand, when facing thymine the 7-Ni primer was elongated by two adenines in the presence of dAMP and full-length elongation occurred in the presence of the four triphosphates. This elongation was not dependent on the nature of the base downstream to thymine in the template. The fact that extension of the primer terminated by 7-Ni is possible when opposed to thymine suggests recognition of 7-Ni by KF exo− as an adenine. It thus appears that the d(7-Ni) triphosphate behaves differently from the d(5-Ni) triphosphate. Indeed, Brown reported that once incorporated opposite natural bases, 5-nitroindole (like 3-nitropyrrole, also considered as universal base) caused chain termination (31).
All those hybridization and enzymatic data indicate that this newly introduced 7-nitroindole nucleoside can be considered as a photochemically cleavable nucleoside analog. It was first designed as a precursor to prepare DNA fragments containing the highly labile deoxyribonolactone lesion. Further work showed that it furnishes a highly efficient approach to generate DNA backbone cleavage at preselected position in the sequence. The present work shows that 7-Ni presents some of the biophysical and enzymatic recognition properties required for a universal base analog as it shares many common features with its 5-nitroindole isomer. It can thus be considered as a cleavable base analog, although not fulfilling strictly all requirements for a ‘universal’ base analog. Indeed as pointed out in a recent review (32), none of the analogs reported so far does fulfill all criteria and it may be that a single analog cannot.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Prof. Jean Lhomme for helpful discussion regarding these analyses and critical reading of the manuscript. The ‘Institut Universitaire de France’ is acknowledged for financial support. Funding to pay the Open Access publication charges for this article was provided by Université Joseph Fourier.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sigman D.S. Chemical nucleases. Biochemistry. 1990;29:9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- 2.Pitie M., Bernadou J., Meunier B. Oxidation at carbon-1′ of DNA deoxyriboses by the Mn-TMPyP/KHSO5 system results from a cytochrome P-450-type hydroxylation reaction. J. Am. Chem. Soc. 1995;117:2935–2936. [Google Scholar]
- 3.Pratviel G., Bernadou J., Meunier B. Carbon–hydrogen bonds of DNA sugar units as targets for chemical nucleases and drugs. Angew. Chem. Int. Ed. Engl. 1995;34:746–769. [Google Scholar]
- 4.Meijler M.M., Zelenko O., Sigman D.S. Chemical mechanism of DNA scission by (1,10-phenantroline) copper. Carbonyl oxygen of 5-methylenefuranone is derived from water. J. Am. Chem. Soc. 1997;117:1135–1136. [Google Scholar]
- 5.Pratviel G., Bernadou J., Meunier B. DNA and RNA cleavage by metal complexes. Adv. Inorg. Chem. 1998;45:251–312. [Google Scholar]
- 6.Armitage B. Photocleavage of nucleic acids. Chem. Rev. 1998;98:1171–1200. doi: 10.1021/cr960428+. [DOI] [PubMed] [Google Scholar]
- 7.Peoc‘h D., Meyer A., Imbach J.-L., Rayner B. Efficient chemical synthesis of oligodeoxynucleotides containing a true abasic site. Tetrahedron Lett. 1991;32:207–210. [Google Scholar]
- 8.Ordoukhanian P., Taylor J.-S. Design and synthesis of a versatile photocleavable DNA building block. Application to phototriggered hybridization. J. Am. Chem. Soc. 1995;117:9570–9571. [Google Scholar]
- 9.Zhang K., Taylor J.-S. A caged ligatable DNA strand break. J. Am. Chem. Soc. 1999;121:11579–11580. [Google Scholar]
- 10.Ordoukhanian P., Taylor J.-S. Caged single and double strand breaks. Bioconjug. Chem. 2000;11:94–103. doi: 10.1021/bc9900993. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K., Taylor J.-S. Phototriggered formation and repair of DNA containing a site specific single strand break of the type produced by ionizing radiation or AP-lyase activity. Biochemistry. 2001;40:153–159. doi: 10.1021/bi001781j. [DOI] [PubMed] [Google Scholar]
- 12.Lenox H.J., McCoy C.P., Sheppard T.L. Site-specific generation of deoxyribonolactone lesions in DNA oligonucleotides. Org. Lett. 2001;3:2415–2418. doi: 10.1021/ol016255e. [DOI] [PubMed] [Google Scholar]
- 13.Dussy A., Meyer C., Quennet E., Bickle T.A., Giese B. New light-sensitive nucleosides for caged DNA strand breaks. Chembiochem. 2002;3:54–60. doi: 10.1002/1439-7633(20020104)3:1<54::AID-CBIC54>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Kotera M., Bourdat A.-G., Defrancq E., Lhomme J. A highly efficient synthesis of oligodeoxyribonucleotides containing the 2′-deoxyribonolactone lesion. J. Am. Chem. Soc. 1998;120:11810–11811. [Google Scholar]
- 15.Averbuch-Pichot M.T., Bourdat A.G., Defrancq E., Durif A., Kotera M., Lhomme J. Crystal structure of 7-nitro-1(2′-deoxy-β-D-ribofuranosyl)-indole. Zeitschrift fur Kristallographie. 1998;213:181–182. [Google Scholar]
- 16.Kotera M., Roupioz Y., Defrancq E., Bourdat A.-G., Garcia J., Coulombeau C., Lhomme J. The 7-nitroindole nucleoside as a photochemical precursor of 2′-deoxyribonolactone: access to DNA fragments containing this oxydative abasic lesion. Chem. Eur. J. 2000;6:4163–4169. doi: 10.1002/1521-3765(20001117)6:22<4163::aid-chem4163>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 17.Loakes D., Brown D.M. 5-Nitroindole as an universal base analogue. Nucleic Acids Res. 1994;22:4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loakes D., Brown D.M., Linde S., Hill F. 3-Nitropyrrole and 5-nitrindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loakes D., Hill F., Linde S., Brown D.M. Nitroindoles as universal bases. Nucleosides Nucleotides. 1995;14:1001–1003. [Google Scholar]
- 20.Seela F., Debelak H. The N8-(2′-deoxyribofuranoside) of 8-aza-7-deazaadenine: a universal nucleoside forming specific hydrogen bonds with the four canonical DNA constituents. Nucleic Acids Res. 2000;28:3224–3232. doi: 10.1093/nar/28.17.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J., Seela F. Base pairing of 8-aza-7-deazapurine-2,6-diamine linked via the N(8)-position to the DNA backbone: universal base-pairing properties and formation of highly stable duplexes when alternating with dT. Helv. Chim. Acta. 2002;85:1340–1355. [Google Scholar]
- 22.McMinn D.L., Ogawa A.K., Wu Y., Liu J., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- 23.Berger M., Ogawa A.K., McMinn D.L., Schultz P.G., Romesberg F.E. Stable and selective hybridization of oligonucleotides with unnatural hydrophobic bases. Angew. Chem. Int. Ed. Engl. 2000;39:2940–2942. doi: 10.1002/1521-3773(20000818)39:16<2940::aid-anie2940>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Berger M., Wu Y., Ogawa A.K., McMinn D.L., Schultz P.G., Romesberg F.E. Universal bases for hybridization, replication and chain termination. Nucleic Acids Res. 2000;28:2911–2914. doi: 10.1093/nar/28.15.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Ogawa A.K., Berger M., McMinn D.L., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: optimization of interbase hydrophobic interactions. J. Am. Chem. Soc. 2000;122:7621–7632. [Google Scholar]
- 26.Ogawa A.K., Wu Y., McMinn D.L., Liu J., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 27.Henry A.A., Yu C., Romesberg F.E. Determinants of unnatural nucleobase stability and polymerase recognition. J. Am. Chem. Soc. 2003;125:9638–9646. doi: 10.1021/ja035398o. [DOI] [PubMed] [Google Scholar]
- 28.Pirrung M.C., Zhao X., Harris S.V. A universal, photocleavable DNA base: nitropiperonyl 2′-deoxyriboside. J. Org. Chem. 2001;66:2067–2071. doi: 10.1021/jo001594r. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig J., Eckstein F.J. Rapid and efficient synthesis of nucleoside 5′-0-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 30.Goodman M.F., Creighton S., Bloom L.B., Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 31.Smith C.L., Simmonds A.C., Felix I.R., Hamilton A.L., Kumar S., Nampalli S., Loakes D., Hill F., Brown D.M. DNA polymerase incorporation of universal base triphosphates. Nucleosides Nucleotides. 1998;17:541–554. [Google Scholar]
- 32.Loakes D. The applications of universal DNA base analogues. Nucleic Acids Res. 2001;29:2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.