Abstract
Homologous recombination (HR) repairs DNA double-strand breaks and maintains genome stability. HR between linked, direct repeats can occur by gene conversion without an associated crossover that maintains the gross repeat structure. Alternatively, direct repeat HR can occur by gene conversion with a crossover, or by single-strand annealing (SSA), both of which delete one repeat and the sequences between the repeats. Prior studies of different repeat structures in yeast and mammalian cells revealed disparate conversion:deletion ratios. Here, we show that a key factor controlling this ratio is the distance between the repeats, with conversion frequency increasing linearly with the distances from 850 to 3800 bp. Deletions are thought to arise primarily by SSA, which involves extensive end-processing to reveal complementary single-strands in each repeat. The results can be explained by a model in which strand-invasion leading to gene conversion competes more effectively with SSA as more extensive end-processing is required for SSA. We hypothesized that a transcription unit between repeats would inhibit end-processing and SSA, thereby increasing the fraction of conversions. However, conversion frequencies were identical for direct repeats separated by 3800 bp of transcriptionally silent or active DNA, indicating that end-processing and SSA are not affected by transcription.
INTRODUCTION
DNA double-strand breaks (DSBs) are potentially lethal events that can be repaired by homologous or non-homologous repair pathways. If left unrepaired, DSBs can lead to chromosome loss or cell death. DSBs are induced by ionizing radiation, X-rays, free radicals, chemicals, nucleases, and they also arise at stalled replication forks (1). DSB repair can occur by non-homologous end-joining (NHEJ) or homologous recombination (HR). Although DSB repair by NHEJ or HR can be accurate, misrepair can have serious genetic consequences. Genomic rearrangements associated with the misrepair of DSBs may lead to carcinogenesis through the activation of proto-oncogenes or inactivation of tumor suppressor genes (1,2). The critical role for HR is underscored by the marked genome instability observed in cells with defects in HR proteins, including BRCA1, BRCA2 and the RAD51 paralogs XRCC2, XRCC3, RAD51B, RAD51C and RAD51D (3–8).
Unlike single-strand breaks and other single-strand damage for which a repair template is readily available, the repair of DSBs by HR requires a search for a homologous template. In genomes with large quantities of repeated sequences, there may be many possible homologous templates. Potential interaction partners include homologous chromosomes, sister chromatids and ectopic sequences linked to the damaged locus or at unlinked sites on homologous or heterologous chromosomes. HR can result in significant genomic changes, including localized or large-scale loss of heterozygosity (LOH), gene deletion and duplication, inversions and translocations. The particular outcome depends on the type of HR event and the arrangement of the interacting regions (2).
HR can occur by conservative and non-conservative mechanisms. Gene conversion is conservative, involving non-reciprocal transfer between donor and recipient loci; for DSB-induced events, the broken locus is almost always the recipient. Gene conversions without crossovers preserve the gross structure of the genome, leading only to localized LOH. However, conversions with associated crossovers in homologous chromosomes result in LOH of all genes from the point of the crossover to the telomere in 50% of subsequent mitotic divisions. Crossovers between linked direct repeats result in deletion of one repeat and sequences between repeats as a circular molecule that is usually mitotically unstable. Crossovers between sister chromatids (unequal sister chromatid exchange) yield the same deletion in one daughter cell and a triple-repeat structure in the other daughter cell. Single-strand annealing (SSA) in direct repeats is a non-conservative HR mechanism that also deletes one repeat and sequences between repeats, but in this case the deleted DNA is degraded. SSA between unlinked loci can result in translocations, but this is thought to require DSBs at both loci, as for NHEJ-mediated translocations (9). Because crossovers are suppressed in mammalian cells (9–11), most direct repeat deletions are thought to result from SSA. For simplicity we describe gene conversions without associated crossovers as ‘conversions’ and deletions by any mechanism as ‘deletions.’
Several factors may influence the direct repeat conversion:deletion ratio. For example, in yeast, conversions accounted for 20–50% of DSB-induced HR between 1.2 kb repeats (12–14), but 94% with 6.5 kb repeats (15). Similarly in mammalian cells, conversions accounted for 17–60% of events with two different repeats ∼0.7 kb in length (16–18), but 97% with 1.4 kb repeats (19). These results suggest that conversion is favored with longer repeats. However, the yeast repeat systems also differed in that the longer repeats flanked an autonomously replicating sequence (15). To date, there have been no systematic studies of the effects of the composition of sequences between repeats on the conversion:deletion ratio. It should also be noted that increasing repeat size effectively increases the distance between homologous sequences within repeats. In a yeast plasmid-based system it was shown that the conversions increase with the increasing distance between repeats (20). However, plasmid-based systems may not mimic chromosomal systems, particularly when HR occurs immediately after plasmids are introduced into cells. For example, essentially all DSB-induced HR between extrachromosomal direct repeats in mammalian cells results in deletions (21,22), but chromosomal repeats yield conversions as a common or predominant outcome (16–19).
In the present study we tested the effect of varying the distance between otherwise identical 1.4 kb neo direct repeats in human cells. All HR substrates were targeted to a specific locus to eliminate potential position effects. We show that the conversion:deletion ratio increases with increasing distance between repeats. However, even when repeats were separated by 3.8 kb, the conversion frequency was only ∼40%, whereas a related HR substrate in CHO cells with repeats separated by the same distance gave 97% conversions (19). One difference between the low and high conversion substrates was the presence of an active transcription unit between repeats in the latter substrate. We hypothesized that an active transcription unit between repeats inhibits end-processing and SSA, and thereby increases conversions. However, adding a transcription unit did not affect the conversion frequency. These results indicate that the relative frequencies of direct repeat conversion and deletion in human cells are influenced by the distance between repeats, and by factors other than the transcriptional status of the intervening sequences, such as chromosomal position or cell type.
MATERIALS AND METHODS
Plasmid DNAs
Plasmids were manipulated and prepared as described previously (23). Plasmids pcDNA5/FRT and pOG44 were purchased from Invitrogen (Carlsbad, CA). pCDNA5/FRT was modified such that the Flp-recombinase target site (FRT) and hygromycin-resistance gene (hyg) were flanked by MfeI sites, creating plasmid p2XMfeI. Plasmid pOG44 carries the Flp-recombinase gene regulated by the CMV promoter. Plasmids pDR1/FRT, pDR2/FRT and pDR3/FRT each contain 1.4 kb neo direct repeats. The distance between neo repeats differs in these plasmids; in pDR1/FRT and pDR2/FRT, the DNA between repeats is from pSV2neo, and in pDR3/FRT there is an additional 2.2 kb fragment carrying the yeast LEU2 gene. Plasmid pDR4/FRT is identical to pDR3/FRT except that LEU2 was replaced by an active transcription unit comprising an SV40 promoter driving the Escherichia coli gpt gene (SVgpt). Plasmid pDP/FRT is identical to pDR1/FRT except it carries a third copy of neo. The pDR and pDP plasmids all have the FRT site and hyg from p2XMfeI. One copy of neo (recipient) is regulated by the SV40 promoter but is inactivated by a 29 bp insertion containing the I-SceI cleavage site within the natural BanII site (19). The other copies of neo (donors) have wild-type coding capacity but are inactive because they lack promoters. Donor neos have either a wild-type BanII site or a silent mutation that converts the BanII site to a BsaI site; the silence of the BsaI mutation was confirmed in direct transfection experiments and in HR experiments in which both the donors were present (E. Schildkraut and J. A. Nickoloff, unpublished data). Because the wild-type BanII site and silent BsaI mutation occur opposite of the I-SceI insertion, these donors present identical homologous regions and are interchangeable. The I-SceI expression vector pCMV(3xNLS)I-SceI, and negative control vector pCMV(I-SceI−) with the I-SceI nuclease in reverse orientation, were described previously (24,25).
Targeting HR substrates to human 293 cells
Flp-In 293 (human embryonic kidney) cells (Invitrogen) were cultured in DMEM with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 6% CO2 at 37°C. Plasmids with neo direct repeats and hyg were targeted to Flp-In 293 cells by co-transfection with pOG44. Correct targeting activates the hyg gene and interrupts an endogenous zeocin-resistance gene (zeo). Stable transfectants were selected in medium with 100 μg/ml hygromycin and then tested for sensitivity to zeocin (Invitrogen). Correct targeting was confirmed by PCR using primers P1 (5′-GGCCTCTGAGCTATTCCAGA) and P2 (5′-GACCAATGCGGAGCATATAC) (Figure 1). Southern hybridization was used to ensure that HR substrates were intact and also to further verify targeting. Genomic DNA was digested with EcoRI, separated on 0.8% agarose gels, transferred to nylon membranes and hybridized with a 32P-labeled neo fragment (Figure 2).
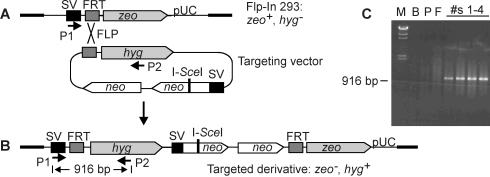
Figure 1.
Targeting strategy. (A) The parent Flp-In 293 cells have a single integrated copy of a vector with an FRT target site between an SV40 promoter and zeo. Each targeting vector has an FRT site upstream of a promoterless hyg gene and neo repeats (not to scale). (B) Flp-mediated targeting links the SV40 promoter to hyg, creating hyg+, zeo− derivatives. (C) Confirmation of targeting by PCR. Primers P1 and P2 amplify a 916 bp fragment in correctly targeted substrates. Lane M = λ/HindIII size marker; lane B = blank (no template) control; lane P = parent human 293 DNA; lane F = Flp-In 293 DNA; and #s1–4 = four targeted derivatives.
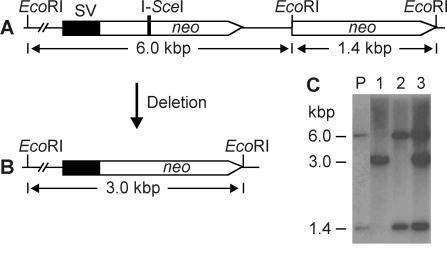
Figure 2.
Southern blot strategy. (A) Map of neo direct repeats. In this example, neo genes are separated by 1.6 kb. Parent and gene conversion structures both yield 1.4 and 6.0 kb EcoRI fragments when probed with neo. (B) Deletion produces a single 3.0 kb EcoRI fragment. (C) Representative Southern blot patterns with the 1.6 kb spacer. Lane P = parental DNA. G418r HR products are shown in lane 1 (deletion), lane 2 (gene conversion) and lane 3 (mixed deletion + gene conversion). Analogous patterns were obtained with 0.85 and 3.8 kb spacers (data not shown).
Recombination assays
DSB-induced HR was assayed as follows. For each experiment, 5 × 105 cells were seeded into 3.5 cm (diameter) wells, incubated for 24 h and lipofected with 2 μg of either pCMV(3xNLS)I-SceI to induce DSBs or the negative control vector pCMV(I-SceI−). Twenty-four hours post-transfection, 105 cells were seeded into each of the four 10 cm dishes. After an additional 24 h, G418 was added to a final concentration of 750 μg/ml and the cells were incubated for 14 days. Cell viability was measured by seeding appropriate numbers of transfected cells into non-selective medium and scoring colony formation after 12–14 days. HR frequencies were calculated as the ratio of the total number of G418-resistant (G418r) colonies to the number of viable cells plated in the selective medium. G418r colonies were expanded in 10 cm dishes and genomic DNA was prepared using DNeasy tissue kits (Qiagen Inc., Valencia, CA). The recipient allele was amplified by PCR using primers specific for the SV40 promoter (5′-GCCCAGTTCCGCCCATTCTC) and the 3′ end of neo (5′-CGAAATCTCGTGATGGCAGG), and HR was confirmed for each G418r product by showing that the resulting PCR products were cleaved by BanII or BsaI, and resistant to cleavage by I-SceI. Southern hybridization of EcoRI digested genomic DNA, as described above, distinguishes gene conversions from deletions (Figure 2). For each cell line, G418r products were isolated from 1 to 3 independent experiments. In cases where products were isolated from more than one experiment, similar product spectra were observed and all data for a single cell line were pooled.
RESULTS
Experimental design
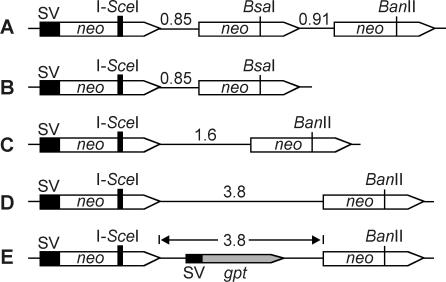
To investigate factors that regulate conversion:deletion ratios for DSB-induced HR events in human cells, we constructed cell lines with one of the five HR substrates shown in Figure 3. Each substrate had 1.4 kb neo repeats. One neo was regulated by the SV40 promoter and inactivated by insertion of an I-SceI recognition site; the other copies had wild-type coding potential but were inactive because they lacked promoters. In four of the HR substrates the sequences between the neo repeats included vector DNA or vector DNA plus a 2.2 kb fragment carrying the yeast LEU2 gene; hence these intervening sequences were not transcriptionally active in human cells. In the fifth substrate, the intervening sequences included an active transcription unit (SVgpt). All HR substrates were targeted to an FRT site in human 293 kidney cells; targeting eliminates variation in HR outcome caused by chromosome position effects and allows direct comparisons among the HR substrates. HR was stimulated by transient expression of I-SceI nuclease and G418r HR products were selected, and HR frequencies and outcomes were determined as described in Materials and Methods. HR frequencies with the five HR substrates are shown in Table 1. Variations in spontaneous and DSB-induced HR frequencies among HR substrates probably reflect differences in the number of pre-existing recombinants in each population, and day-to-day variation in transfection efficiency. Nonetheless, HR was increased by ∼10-fold or more upon I-SceI expression with each HR substrate, thus ensuring that the resulting HR product spectra reflect primarily DSB-induced events.
Figure 3.
Structures of HR substrates. Each HR substrate contains a recipient neo regulated by the SV40 promoter and inactivated by an I-SceI site insertion. Donors lack a promoter and contain either the wild-type BanII site or a silent BsaI mutation in place of BanII. Distances (in kb) between neo repeats are indicated. Each neo repeat is 1.4 kb. (A) Triple repeat substrate. (B–D) Direct repeat substrates with increasing amounts of non-transcribed DNA between neo genes. (E) Direct repeat substrate with a transcriptionally active SVgpt gene between neo repeats.
Table 1.
HR Frequencies in neo direct repeat strains
| HR substratea | HR frequency (×103)b | |
|---|---|---|
| Spontaneous | DSB-induced | |
| A | 0.38 | 11.2 |
| B | 0.31 | 3.2 |
| C | 1.4 | 14.7 |
| D | 2.4 | 18.9 |
| E | 0.45 | 12.3 |
aShown in Figure 1.
bHR frequencies for cells transfected with control vector (spontaneous) or I-SceI expression vector (DSB-induced).
Conversion:deletion ratios with double and triple repeat HR substrates
We previously reported that DSB-induced HR between 1.4 kb neo direct repeats in CHO cells resulted in 97% conversions (19). In that study, the repeats were separated by 3.8 kb. In a separate study of donor preference using HR substrates with three 1.4 kb neo repeats Flp-targeted to human 293 cells, including the pDP/FRT substrate with one recipient and two donors (Figure 3A), we found that only 2 of the 14 products (14%) arose by conversion. These markedly different conversion frequencies could be due to several factors including differences in the number of repeats, the distance or composition of the DNA between repeats, chromosomal position and cell type. To test if the third neo enhances deletions, we examined HR products from a related HR substrate with two neo genes targeted to the same locus in 293 cells (Figure 3B). Of 38 HR products from this strain, only 1 arose by conversion (2.6%). Thus, the low conversion frequency with the triple repeat HR substrate was not due to the third copy of neo.
The conversion:deletion ratio is proportional to the distance between direct repeats
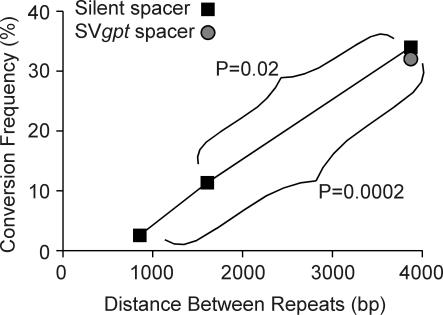
The low frequency of conversion when neo repeats were separated by 855 bp (Figure 4) is consistent with a general trend in the literature indicating that conversion rates are proportional to distances between repeats in mammalian cells (Table 2). However, the systems used in these studies differ in several other respects, such as repeat size, chromosomal position and cell type. We therefore tested whether the conversion frequency would increase with increasing distance between repeats by targeting to human 293 cells two HR substrates with neo repeats separated by 1.6 kb (Figure 3C) and 3.8 kb (Figure 3D). We analyzed 37 G418r products from the 1.6 kb strain and found two arose by complex rearrangement, and four of the remaining 35 (11%) arose by conversion. We then analyzed 53 HR products from the 3.8 kb strain and found 18 (34%) arose by conversion. Thus, conversion increases as a linear function of distance between direct repeats (Figure 4).
Figure 4.
Conversion frequency increases with the increase in distance between the repeats. Significant differences are indicated (P values calculated with Fisher's exact tests).
Table 2.
Reported DSB-induced HR outcomes in mammalian chromosomal direct repeats
The conversion:deletion ratio is not affected by a transcription unit between direct repeats
Although increasing the distance between repeats increased conversion rates, dissimilar conversion rates were obtained with 3.8 kb separating 1.4 kb neo repeats in CHO cells (97%) (19) and human 293 cells (34%), indicating that conversion frequency is influenced by additional factors. In the CHO HR substrate, the neo genes were separated by an active transcription unit (SVgpt) whereas in the substrate used here the neo genes were separated by SV40 DNA and yeast LEU2, which are not transcriptionally active in human cells. Because most or all deletions arise by SSA, and SSA requires extensive strand resection to expose complementary single strands in the neo repeats, we reasoned that a transcription unit between repeats might slow resection, for example, if the transcription machinery interfered with the enzyme(s) responsible for end-resection. Reduced end-resection would inhibit SSA and increase the frequency of gene conversion. To test this concept, we replaced the 2.2 kb LEU2 fragment with a 2.2 kb SVgpt fragment (Figure 3E) and targeted the resulting HR substrate to human 293 cells. We inserted the SVgpt fragment so that gpt was transcribed in the same direction as neo to allow comparisons with our prior substrates and because most published systems use analogous configurations. Among 25 HR events from the SVgpt strain, 8 (32%) arose by gene conversion (Figure 4), 17 arose by deletion and no complex events were detected. Thus, the addition of a transcription unit between repeats had no effect on the conversion frequency, suggesting that the transcription machinery does not affect end-resection. Other factors that may control the relative frequencies of conversions and deletions are discussed below.
DISCUSSION
HR between direct repeats has been examined with a variety of substrates in plasmid or chromosomal contexts in a variety of cell types. Each system gives a characteristic conversion:deletion ratio, but because systems differ in more than one parameter, it has been difficult to pinpoint factors that control this ratio. Early studies of transfected plasmids with DSBs within or outside direct repeats revealed that essentially all HR in these substrates resulted in deletions, and that HR efficiency depended on the position of the DSB relative to the repeats. Thus, HR is very efficient when the DSB is equidistant from the two repeats, and it decreases as the DSB is moved to more asymmetric positions (21,22). This observation led to the model that these HR events arise by SSA which requires end processing to expose single-stranded regions in both repeats to allow complementary strands to anneal. In this model, dependence of SSA efficiency on DSB symmetry reflects the requirement for simultaneous exposure of complementary single strands in the two repeats. However, results with transfected plasmids cannot be extrapolated to chromosomal repeats because plasmid-borne repeats are linked by two segments, 5′→3′ single strand resection eventually degrades both strands, and DNA ends in transfected (naked) plasmid DNA and chromatin are likely to be processed at different rates and/or to different extents. The first analysis of DSB-induced HR products in mammalian chromosomal direct repeats underscored these differences, with 97% of HR occurring by gene conversion (19).
Subsequent studies suggested that conversion frequencies are proportional to the distance between repeats and/or the length of the repeats themselves (Table 2). In yeast chromosomal direct repeats, the fraction of conversions increases with increasing distance between repeats (26). The present study is the first systematic examination of the effect of repeat separation on the conversion:deletion ratio in a mammalian chromosomal context. The decrease in deletions with longer distances between repeats (Figure 4) is consistent with the view that SSA requires simultaneous exposure of complementary single strands in the two repeats, as proposed for plasmid-borne repeats (21,22). Note, however, that even with widely spaced repeats or highly asymmetric DSBs, transfected plasmid substrates yield essentially 100% deletions (albeit at low frequency). Thus, the chromosomal context appears to enhance conversion; this enhances genome stability, although the effect is minimal when repeats are separated by <1000 bp (Figure 4). The position of the DSB within a repeat may also control the conversion frequency as this too would influence the timing of complementary single-strand exposure.
In yeast, gene conversion requires Rad51, Rad52 and Rad54, and is facilitated by the Rad51 paralogs Rad55 and Rad57. In contrast, SSA is independent of all of these proteins except Rad52 (27,28), presumably because of Rad52's strand annealing activity (29). The role of mammalian RAD52 has been questioned because mouse rad52 knockouts show minimal sensitivity to DNA-damaging agents, minimal HR defects, and do not show the serious cell growth defect characteristic of mouse rad51 knockouts (30,31). It is presently unclear whether RAD52 is required for SSA in mammalian cells, but this can be addressed using the HR substrates described here coupled with RNAi knockdown of endogenous RAD52. In any case, the observed linear relationship between conversion rates and the distance separating repeats (Figure 4) suggests a dynamic competition between RAD51-dependent strand invasion leading to gene conversion and SSA. Both mechanisms depend on strand resection from broken ends. As the distance between repeats increases, the extent of strand resection required for SSA increases, and this in turn increases the likelihood that processed ends will invade a donor template and complete repair by gene conversion before complementary single-strands are exposed and annealed.
Although our data show that conversion increases with distance between repeats, it is clear that additional factors control the conversion:deletion ratio. An HR substrate with 1.4 kb neo repeats separated by 3.8 kb in CHO cells gave 97% conversions (19), but the frequency in a closely related substrate in human cells was only 34%. We ruled out the possibility that a transcription unit between repeats inhibited deletions. Although we did not test whether the orientation of a central transcription unit influences the conversion frequency, this cannot account for the distinct results in our CHO and human cell experiments as SVgpt was in the same orientation in these substrates. There are several other factors that may influence the conversion:deletion ratio, including cell species, cell type and structural/environmental differences. It is difficult to account for the distinct conversion rates on the basis of species differences because the HR substrate with a high conversion frequency in CHO cells had similarly high frequency when randomly integrated into human HT1080 cells (98%) (32). This does not rule out potential effects from other cell type differences, such as p53 status. p53 is known to suppress HR (33–38), and p53 is active in HT1080 cells (39,40) but inactive in human 293 cells (41). However, although a direct analysis of p53 effects on the conversion:deletion ratio has not yet been reported, it is unlikely that the low conversion frequency we observed in human 293 cells is due to the p53 defect, because wild-type p53 strongly suppresses conservative HR (conversion) (38). Thus, conversion would be predicted to be relatively efficient in p53 defective cells, yet the opposite was observed.
There are several structural differences between the HR substrates with high and low conversion rates that might account for their distinct behavior. The high conversion HR substrate has 12 single-base heterologies in the donor neo but these are absent in the low conversion substrate. It is not clear how heterologies would preferentially inhibit SSA and/or promote conversion. In yeast, a single heterology affects spontaneous HR frequencies (42), but the efficiency of DSB-induced gene conversion is not affected by similar heterology densities (∼1 single-base change per 100 bp) (43). Another difference is that the transcriptionally active neo genes in the high and low conversion substrates were regulated by mouse mammary tumor virus and SV40 promoters, respectively, but again, it is not clear how promoters might influence conversion frequencies. In fact, it is best to consider the promoter differences as part of a larger set of differences owing to the distinct chromosomal environments of these HR substrates. It is possible that differences in chromatin structure at different loci influence the rate of strand excision, which in turn would influence the frequencies of conversion and SSA. Differences in chromatin appear to influence spontaneous gene-conversion rates (44). Nonetheless, at any particular locus, conversion frequencies are likely to increase with distance between repeats, as observed in the present study.
Although the additional factors that regulate conversion rates in direct repeats remain obscure, it is clear that the conversion:deletion ratio is under genetic control. Thus, BRCA2 mutant cells, which display severe genome instability (5,45) and deficient HR-mediated DSB repair (5,46), also display a marked shift in direct repeat HR from conversion toward deletions (17). These results suggest the possibility that other HR proteins regulate conservative and non-conservative HR mechanisms, and underscore the importance of such control in tumor suppression.
Acknowledgments
We thank Kimberly Paffett for assistance with plasmid construction and Alan Waldman for helpful comments on the manuscript. This research was supported by grant CA77693 to J.A.N. from the National Cancer Institute of the NIH. Funding to pay the Open Access publication charges for this article was provided by NCI grant CA77693.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kanaar R., Hoeijmakers J.H.J., van Gent D.C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 2.Nickoloff J.A. Recombination: Mechanisms and roles in tumorigenesis. In: Bertino J.R., editor. Encyclopedia of Cancer, Second Edition. Vol. 4. San Diego: Elsevier Science (USA); 2002. pp. 49–59. [Google Scholar]
- 3.Brenneman M.A., Wagener B.M., Miller C.A., Allen C., Nickoloff J.A. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 4.Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynahan M.E., Pierce A.J., Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 6.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R.D., Liu N., Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 8.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 9.Richardson C., Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R.D., Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickoloff J.A., Chen E.Y.C., Heffron F. A 24-base-pair sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl Acad. Sci. USA. 1986;83:7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickoloff J.A., Singer J.D., Hoekstra M.F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J. Mol. Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 14.Cho J.W., Khalsa G.J., Nickoloff J.A. Gene conversion tract directionality is influenced by the chromosome environment. Curr. Genet. 1998;34:269–279. doi: 10.1007/s002940050396. [DOI] [PubMed] [Google Scholar]
- 15.Ray A., Siddiqi I., Kolodkin A.L., Stahl F.W. Intrachromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J. Mol. Biol. 1988;201:247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- 16.Dronkert M.L.G., Beverloo H.B., Johnson R.D., Hoeijmakers J.H.J., Jasin M., Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tutt A., Bertwistle D., Valentine J., Gabriel A., Swift S., Ross G., Griffin C., Thacker J., Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang F., Han M.G., Romanienko P.J., Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghian D.G., Nickoloff J.A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fishman-Lobell J., Rudin N., Haber J.E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin F.-L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin F.-L., Sperle K., Sternberg N. Extrachromosomal recombination in mammalian cells as studied with single- and double-stranded DNA substrates. Mol. Cell. Biol. 1987;7:129–140. doi: 10.1128/mcb.7.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taghian D.G., Nickoloff J.A. Subcloning strategies and protocols. In: Harwood A., editor. Basic DNA and RNA Protocols. Vol. 58. Totowa, NJ: Humana Press; 1996. pp. 221–235. [DOI] [PubMed] [Google Scholar]
- 24.Choulika A., Perrin A., Dujon B., Nicolas J.-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim P.M., Allen C., Wagener B.M., Shen Z., Nickoloff J.A. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugawara N., Ira G., Haber J.E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartsch S., Kang L.E., Symington L.S. Rad51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov E.L., Sugawara N., Fishman-Lobell J., Haber J.E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortensen U.H., Bendixen C., Sunjevaric I., Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rijkers T., Vandenouweland J., Morolli B., Rolink A.G., Baarends W.M., Vansloun P.P.H., Lohman P.H.M., Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim D.-S., Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H., Guo X., Meng X., Liu J., Wray J., Allen C., Nickoloff J.A., Shen Z. The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol. Cell. Biol. 2005;25:1949–1957. doi: 10.1128/MCB.25.5.1949-1957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mekeel K.L., Tang W., Kachnic L.A., Luo C.M., DeFrank J.S., Powell S.N. Inactivation of p53 results in high-rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 34.Linke S.P., Sengupta S., Khabie N., Jeffries B.A., Buchhop S., Miska S., Henning W., Pedeux R., Wang X.W., Hofseth L.J., et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- 35.Romanova L.Y., Willers H., Blagosklonny M.V., Powell S.N. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23:9025–9033. doi: 10.1038/sj.onc.1207982. [DOI] [PubMed] [Google Scholar]
- 36.Willers H., McCarthy E.E., Hubbe P., Dahm-Daphi J., Powell S.N. Homologous recombination in extrachromosomal plasmid substrates is not suppressed by p53. Carcinogenesis. 2001;22:1757–1763. doi: 10.1093/carcin/22.11.1757. [DOI] [PubMed] [Google Scholar]
- 37.Willers H., McCarthy E.E., Wu B., Wunsch H., Tang W., Taghian D.G., Xia F., Powell S.N. Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene. 2000;19:632–639. doi: 10.1038/sj.onc.1203142. [DOI] [PubMed] [Google Scholar]
- 38.Akyuz N., Boehden G.S., Susse S., Rimek A., Preuss U., Scheidtmann K.-H., Wiesmuller L. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol. Cell. Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labrecque S., Matlashewski G.J. Viability of wild-type p53-containing and p53-deficient tumor cells following anticancer treatment: the use of human papillomavirus E6 to target p53. Oncogene. 1995;11:387–392. [PubMed] [Google Scholar]
- 40.Slebos R.J.C., Taylor J.A. A novel host cell reactivation assay to assess homologous recombination capacity in human cancer cell lines. Biochem. Biophys. Res. Commun. 2001;281:212–219. doi: 10.1006/bbrc.2001.4335. [DOI] [PubMed] [Google Scholar]
- 41.Yu Q., Rose J.H.L., Zhang H., Pommier Y. Antisense inhibition of Chk2/hCds1 expression attenuates DNA damage-induced S and G2 checkpoints and enhances apoptotic activity in HEK-293 cells. FEBS Lett. 2001;505:7–12. doi: 10.1016/s0014-5793(01)02756-9. [DOI] [PubMed] [Google Scholar]
- 42.Datta A., Hendrix M., Lipsitch M., Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl Acad. Sci. USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickoloff J.A., Sweetser D.B., Clikeman J.A., Khalsa G.J., Wheeler S.L. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics. 1999;153:665–679. doi: 10.1093/genetics/153.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raynard S.J., Baker M.D. Cis-acting regulatory sequences promote high-frequency gene conversion between repeated sequences in mammalian cells. Nucleic Acids Res. 2004;32:5916–5927. doi: 10.1093/nar/gkh926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donoho G., Brenneman M.A., Cui T.X., Donoviel D., Vogel H., Goodwin E.H., Chen D.J., Hasty P. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36:317–331. doi: 10.1002/gcc.10148. [DOI] [PubMed] [Google Scholar]
- 46.Xia F., Taghian D.G., DeFrank J.S., Zeng Z.C., Willers H., Iliakis G., Powell S.N. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl Acad. Sci. USA. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sargent R.G., Brenneman M.A., Wilson J.H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]