Abstract
Post-transcriptional gene silencing (PTGS) involving small interfering RNA (siRNA)-directed degradation of RNA transcripts and transcriptional silencing via DNA methylation have each been proposed as mechanisms of genome defence against invading nucleic acids, such as transposons and viruses. Furthermore, recent data from plants indicates that many transposons are silenced via a combination of the two mechanisms, and siRNAs can direct methylation of transposon sequences. We investigated the contribution of DNA methylation and the PTGS pathway to transposon control in the filamentous fungus Neurospora crassa. We found that repression of the LINE1-like transposon, Tad, requires the Argonaute protein QDE2 and Dicer, each of which are required for transgene-induced PTGS (quelling) in N.crassa. Interestingly, unlike quelling, the RNA-dependent RNA polymerase QDE1 and the RecQ DNA helicase QDE3 were not required for Tad control, suggesting the existence of specialized silencing pathways for diverse kinds of repetitive elements. In contrast, Tad elements were not significantly methylated and the DIM2 DNA methyltransferase, responsible for all known DNA methylation in Neurospora, had no effect on Tad control. Thus, an RNAi-related transposon silencing mechanism operates during the vegetative phase of N.crassa that is independent of DNA methylation, highlighting a major difference between this organism and other methylation-proficient species.
INTRODUCTION
In many organisms, transposon-related sequences make up a large portion of the genome. There is evidence that some of these sequences have evolved to play important roles in heterochromatin formation, centromere function and gene regulation (1–3). On the other hand, the cell has developed mechanisms to limit the expansion of these elements in order to prevent excessive genome instability. The relatively recent discovery and subsequent dissection of post-transcriptional gene silencing mechanisms (RNAi, cosuppression, quelling) that can act to silence repeated sequences has led to a model in which a double-stranded RNA intermediate, homologous to the targeted gene is processed by the RNAseIII molecule Dicer into siRNAs of 21–25 nt in length (4). siRNAs are subsequently used as guides by the RNA-induced silencing complex (RISC) to degrade homologous transcripts (5). This general model is conserved in a wide range of organisms, thus one proposed explanation for the retention of post-transcriptional gene silencing (PTGS) has centred on a role in limiting the expansion of naturally occurring repeated sequences, such as transposable elements (6). A number of observations support this hypothesis: in Caenorhabditis elegans and Chlamydomonas, several genes are essential to both RNAi- and transposon control-pathways (7–9); siRNAs against transposon sequence have been cloned in both Arabidopsis and Drosophila (10,11). These findings all suggest that there is an ongoing control of transposon proliferation in the host.
In addition to PTGS, transcriptional gene silencing (TGS) mechanisms such as DNA and histone methylation have also been implicated in transposon control. Large swathes of the human and Arabidopsis genomes are methylated, and these methylated areas are rich in transposon-related sequence (12–14). Moreover, mutations which block DNA methylation lead to increased transcript levels and increased transposition rate of both DNA transposons and retrotransposons (12,15–18). Remarkably, in fission yeast and plants, it has been demonstrated that heterochromatin formation is directed by siRNAs in an Argonaute complex with similarities to the RISC involved in RNAi, suggesting that the processes of PTGS and TGS are intertwined (2,16,19–21).
The phenomenon of ‘quelling’ in Neurospora displays most of the characteristic features of PTGS observed in other organisms (22). In addition to quelling however, Neurospora possesses additional homology-dependent gene silencing mechanisms such as repeat-induced point mutation (RIP) and meiotic silencing by unpaired DNA (MSUD) (23,24). In particular, RIP is able to inactivate duplicated DNA sequences by causing multiple C:G to T:A mutations and cytosine methylation at the pre-meiotic, dikaryotic sexual phase of the life cycle. That the Neurospora genome is littered with RIP-mutated relics of a wide range of transposon families is testament to the efficiency of RIP. Indeed, the Neurospora genome does not reveal the presence of a single active transposon (25).
Despite this, transposition of the LINE1-like, non-LTR retrotransposon Tad has been detected in an African strain, Adiopodoumé, which contains up to 40 copies of this element (26,27). Subsequent analysis showed that this strain was RIP-proficient and that the Tad elements in this strain were fully susceptible to RIP (28,29). To search for an explanation as to why Tad remained active in the RIP-proficient Adiopodoumé strain, we considered the possibility that additional silencing mechanisms, such as quelling, might also be employed in transposon control in Neurospora. To explore this hypothesis, we examined whether Adiopodoumé was quelling-proficient and also monitored the rate of Tad transposition in diverse quelling-defective mutants. A role for DNA methylation in limiting the expansion of artificially introduced Tad elements in Neurospora has also been documented (30). Therefore, in light of the above observations (in Schizosaccharomyces pombe and Arabidopsis) of a link between PTGS and TGS mechanisms, we investigated the relative effects of quelling and DNA methylation on transposon control in Neurospora, and whether these two mechanisms are connected in this species.
We found that the expansion of an experimentally introduced Tad element was particularly pronounced in a qde-2 null background, lacking an Argonaute protein thought to constitute part of the RISC in Neurospora (31). This was evidenced both in terms of increased transcript levels and increased copy number of the Tad elements. Importantly, a similar effect was confirmed when we introduced a qde-2− allele into progeny of a strain harbouring a natural invasion of TAD. Additionally, siRNAs against Tad sequence were found, suggesting that a quelling-like mechanism is responsible for the taming of transposable elements in N.crassa. In support of this, Tad transcripts accumulated heavily in strains lacking both of the Neurospora DICER-LIKE proteins. However, in contrast to quelling, QDE1 (an RdRP) and QDE3 (a DNA helicase) were not required suggesting that, although similar, the quelling- and transposon control-pathways do not entirely overlap (32,33). Notably, we found that the major bulk of introduced Tad elements were not significantly methylated, and furthermore the absence of the DNA methylation machinery had no obvious effect on Tad expression, indicating that in Neurospora, unlike several other organisms, there is no RNAi-directed DNA methylation mechanism for transposon control and instead suggesting that the major determinant is post-transcriptional.
MATERIALS AND METHODS
N.crassa strains: The wild-type (WT) strain 74-OR23A and Adiopodoumé were obtained from the Fungal Genetics Stock Center, University of Kansas, Kansas City (strains FGSC 987 and FGSC 430, respectively). The dim-2− strain (34) was a gift from Eric Selker, University of Oregon. The qde and dcl mutant strains used in this study were obtained by insertional disruption by plasmid insertion, have been previously described in detail elsewhere, and were as follows: 627 (qde-3−), 820 (qde-2−), 107 (qde-1−) DCL1ko (dcl-1−), DCL2ko (dcl-2−) (31–33,35). The double Dicer mutant strain was obtained by the crossing of dcl-1− and dcl-2− strains.
Transformation and growth conditions
Strains were grown in Vogel's minimal medium for Neurospora (NMM), as described elsewhere (36). Ascospores from crosses were heat-activated to induce germination at 60°C for 30 min, as previously described (37). For Tad transformation experiments, spheroplasts were prepared from the mutant and wild-type strains using the method of Orbach et al. (38). Each strain was co-transformed with 1 μg of the plasmid pCSN44.1, which contains the hph hygromycin-resistance gene as a selectable marker (39), and 5 μg of the plasmid pTad1-1, containing a full-length cloned Tad element (40). Transformations were plated on NMM containing 0.2 mg/ml hygromycin and at least 10 individual colonies from each transformed strain were then purified by three serial platings to purify for homokaryons. Purified colonies were then transferred to solid slant NMM media and arbitrarily referred to as the first asexual ‘generation’. Subsequent generations were propagated by serial transfer of conidia after three days of vegetative growth on slants. Each generation was then grown in liquid NMM for three days, with shaking, at 28°C to produce sufficient mycelia for nucleic acid extraction. Differences in the Tad load between the transformed mutant strains were compared using the Student's t-test (two-tailed).
Northern and small RNA analysis
Small RNA purification was performed as described previously (41). A riboprobe was transcribed using T3 RNA polymerase (Promega) on a Tad PCR template that contained a T3 promoter site at the 5′ end. To produce a template for ORF1, the primers used were ORF1aT3 (5′-CGCGAATTAACCCTCACTAAAGGGAGAAACTGCTTT-3′) and ORF1b (5′-CCAAGGCAGCAACAGTAC-3′), while for ORF2 the primers used were ORF2aT3 (5′-CGCGAATTAACCCTCACTAAAGGGATACTGTATTGGAACGTG-3′) and ORF2B (5′-GGTCTCGTCTGCGAAGCCG-3′), where T3 promoter sites (including adaptor region) are underlined. Prior to hybridization, labelled transcripts were hydrolysed to an average size of 50 nt, 15 volumes of 80 mM sodium bicarbonate and 120 mM sodium carbonate were added to the transcriptional reaction and incubated at 60°C for 3 h. To stop the hydrolysis, 20 μl of 3 M sodium acetate (pH 5.0) was added. Pre-hybridization and hybridization were at 35°C in 50% deionized formamide, 7% SDS, 250 mM NaCl, 125 mM sodium phosphate (pH 7.2), and sheared, denatured, salmon sperm DNA (100 mg/ml). After overnight hybridization, membranes were washed twice in 2× SSC and 0.2% SDS at 35°C for 30 min and once in 20 mM Tris–HCl (pH 7.5), 5 mM EDTA, 60 mM sodium chloride and 10 μg/ml RNase A at 37°C for 1 h to remove unspecific background. Northern analysis of Tad expression was performed using standard protocols (42). A PCR product comprising nucleotides 54–1569 of Tad used as a template to create a 32P-labelled DNA probe. In each blot, ∼5 μg of total RNA was loaded per lane. Equal loading was ensured by controlling ethidium bromide staining of the constitutively expressed 28S ribosomal RNA. In comparing transcript levels between different Tad containing samples, densitometric analysis of the hybridization signal (minus control background) corresponding to the full-length 7 kb Tad transcript was performed using the Instant Imager Analysis software programme (Canberra Packard). The values obtained from this analysis were then expressed as a function of Tad copy number to compensate for unequal segregation of Tad elements during the crossing procedure and values were compared between strains using Student's t-test (two-tailed).
Southern blot analysis
Genomic DNA was isolated from frozen mycelia and ∼4 μg were digested and run according to standard protocols. For methylation analysis, the isoschizomer pair of Dpn11 (methylation-insensitive) and Sau3A1 (methylation-sensitive) were used. To analyse copy number, we digested either with Nde1 and used an internal 602 bp Nde1 Tad fragment in order to produce a single hybridizing band whose intensity was proportional to the copy number of Tad (Figure 1), or we digested with EcoR1 and used a PCR probe comprising nucleotides 54–1569 of Tad that hybridizes to a band unique for each Tad insertion (not shown). An estimate of copy number was calculated by comparing the total signal in each lane [minus control (non-transformed) background] to that of Adiopodoumé DNA, which is reported to have 40 copies of the Tad element (26). As a loading control, all blots were stripped and re-hybridized with either a probe corresponding to a 1397 bp Sph1 fragment upstream of the single copy dcl-1 gene or a 1213 bp Nde1–BamH1 fragment of the single copy al-2 gene. The relative strength of the signal was used to adjust the estimated copy number in each strain.
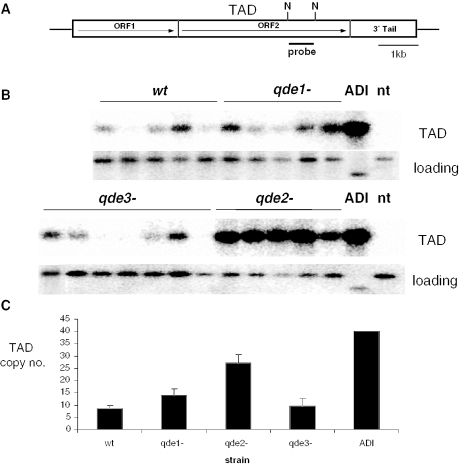
Figure 1.
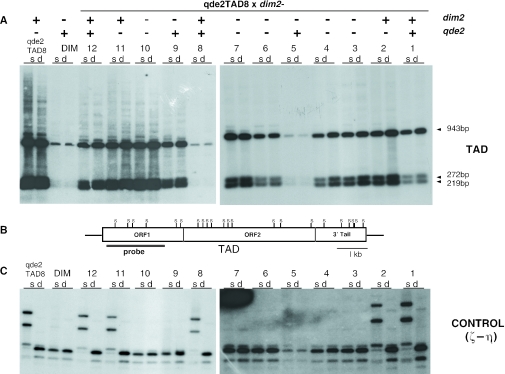
Tad transposition shows a marked increase specifically in a qde2 mutant background. (A) Schematic representation of the Tad element. Nde1 (N) sites and the probe used in the Southern-blot analysis in (B) are indicated. (B) Representative Southern blots in Tad-transformed wild-type (wt), qde-1−, qde-3− and qde-2− strains showing hybridization to the Tad probe or the single copy al-2 gene used as a loading control (this gene shows a restriction polymorphism in Adiopodoumé). DNA from a non-transformed wild-type strain (nt) and the Adiopodoumé strain (ADI) containing 40 Tad copies were included as controls. (C) The mean copy number of Tad in each strain after 15 asexual generations is shown. A minimum of 10 independent transformants were analysed for each strain. Error bars represent standard error of the mean.
Tad hybridizations were performed at 68°C in a solution containing 1 M NaCl, 1% SDS, 10% dextran sulphate, 100 μg/ml sheared salmon sperm DNA. After hybridization, two very stringent washes of 40 min at 68°C in 0.1× SSC, 1% SDS were included in order to minimize hybridization to RIPed Tad relics. In crosses of the mutant strains with either qde2TAD8 or Adiopodoumé, the mutant status of the daughter progeny was checked in each case using probes for qde-1, dcl-1, dcl-2, qde-2, as previously described (31,32,35). The probe used to hybridize the ζ–η region when controlling the dim-2 status was amplified from genomic DNA using the primers zeta-F (5′-CGATTAGCGAATCCTAAGTG-3′) and zeta-R (5′-TTTCTACCATCTATAGCCG-3′).
RESULTS
Adiopodoumé is quelling proficient
To explore the hypothesis that a deficiency in quelling might explain the apparent expansion of Tad in Adiopodoumé, we tested for quelling proficiency by using an assay that involves transformation with a segment of the al-1 gene involved in carotenoid biosynthesis. In quelled strains, silencing of the endogenous gene leads to an albino phenotype. The frequency of quelling in Adiopodoumé was comparable to a wild-type strain (Table 1), ruling out the possibility that Tad expansion in Adiopodoumé could be accounted for by a quelling deficiency.
Table 1.
Adiopodoumé is quelling proficient
| Wild-type | Adiopodoumé | |
|---|---|---|
| Orange colonies | 222 | 231 |
| White colonies | 99 | 85 |
| Quelling efficiency (%) | 31 | 28 |
Tad expansion in the qde mutants
To ascertain whether the quelling pathway contributed to control of Tad transposition, we investigated the expansion of a cloned Tad element in various qde mutants. Previous work in our lab had identified three genes essential for quelling: qde-1, encoding an RNA-dependent RNA polymerase (RdRP) thought to produce a dsRNA template for Dicer (32); qde-2, encoding a PAZ-Piwi domain protein constituting part of RISC (31); qde-3, encoding a recQ DNA helicase thought to be upstream of qde-1 (33). We used a cloned full-length version of the Tad element, Tad1-1 (40), to transform strains that contained insertional disruptions of each of the qde genes. As a control, the wild-type reference strain ORS74a was also transformed. We grew single transformants until conidiation, at which point they were germinated on fresh media and allowed to grow again until conidiation and the cycle was repeated. We referred to each of these vegetative cycles as an asexual ‘generation’. Tad-transformed strains were propagated in this way for up to 15 generations, and the copy number and expression levels of Tad were analysed at various timepoints. We performed Southern blots using a probe hybridizing to a single internal Nde1 fragment of the Tad element (Figure 1A), the intensity of which is proportional to number of Tad elements in the genome. A representative blot is shown in Figure 1B. We found that in the wt strain, the mean number of Tad copies increased over the course of 15 asexual generations to ∼10 copies per genome (Figure 1C). A similar situation was observed in the qde-1− and qde-3− mutants. However, in a qde-2− mutant background Tad showed a very significant increase in transposition rate (P < 0.01), approaching 30 copies per genome. These results show that while Tad is able to achieve a limited rate of transposition in a wt background, this transposition is normally restricted by qde-2. Similar results were confirmed when we repeated the experiment using independently created u.v mutants (data not shown). We further analysed the transformed strains by looking at levels of Tad transcripts in each. Again, in the wt background we found a basal level of Tad transcription that was similar in qde-1− and qde-3− backgrounds, while in the qde-2− mutants the level of transcription was markedly increased, as expected from the high copy number in this strain (not shown).
Specific role for qde-2 in transposon control in Neurospora
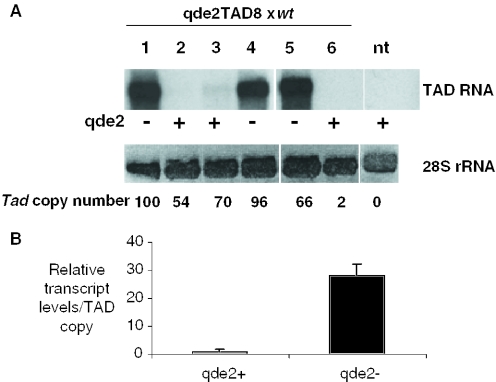
Although the above results suggested a role for qde-2 in the control of Tad, we could not rule out that differences in expression and transposition rate were accentuated by varying initial transformation efficiencies with the cloned Tad element between the different mutant strains, either in terms of number or arrangement of transgenes, or possible insertions in transposition ‘hotspots’. In order to definitively confirm a role for the qde-2 gene in Tad control, we took a Tad-transformed qde-2− strain and crossed it with a wt strain in order to re-introduce a qde-2+ allele. Since RIP inactivates ∼50% of unlinked repeated sequences during a cross, we chose a strain (qde2TAD8) that contained a high Tad load (>80 copies) in order to increase the likelihood that intact elements would survive into the daughter progeny.
We noticed an obvious disparity in the levels of Tad expression in the first ‘generation’ growth of daughter progeny that was dependent on the segregation of the qde-2 allele; in qde-2− progeny Tad expression was consistently high (Figure 2A, lanes 1, 4 and 5) whereas in qde-2+ progeny, the reverse was true (lanes 2, 3 and 6), confirming that qde-2 plays an important role in transposon taming in Neurospora. In this and subsequent crosses, we have plotted transcription as a function of Tad copy number in order to normalize for uneven segregation of elements during the cross (Figure 2B).
Figure 2.
Release of Tad repression segregates with the qde-2− allele. (A) A Tad-transformed qde2− strain (qde2TAD8) was crossed with a wild-type strain in order to re-introduce a qde-2+ allele into a Tad background. Expression of the full-length TAD transcript was monitored by northern blot in cultures deriving from six different ascospores (1–6). RNA form a non-transformed qde-2− strain (nt) was used as a negative control. The qde-2 status of each ascospore is indicated as ‘+’ (wild-type allele) or ‘−’ (mutant allele). Ethidium bromide staining of the ribosomal 28S RNA was used as a loading control. The estimated copy number of Tad in each ascospore is indicated under each lane. (B) Levels of Tad transcripts were expressed as a function of copy number in order to normalize for differences due to uneven segregation of elements during the cross and the mean calculated for each type of allele. Error bars display standard error of the mean.
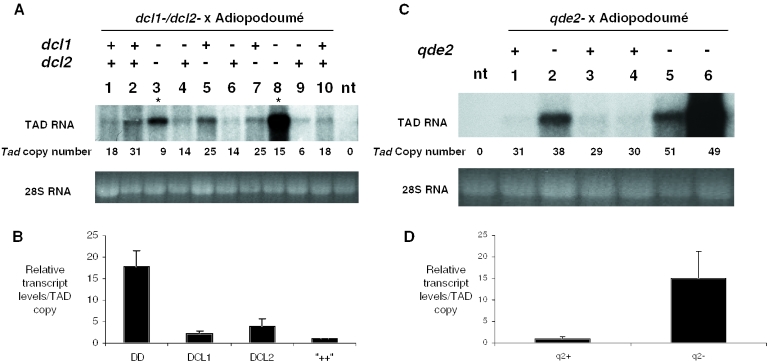
Notwithstanding the role demonstrated above for qde-2 in controlling Tad elements introduced initially by transformation, there remained the possibility that this effect might not be representative of a natural invasion of Tad. To answer this point, we crossed a qde-2− strain with a naturally infected strain (Adiopodoumé) in order to segregate wt and mutant alleles of qde-2 into the progeny. Again, we observed a significant de-repression of Tad only in qde-2− progeny (P < 0.05) (Figure 3C and D). This demonstrates that our observations on Tad control are representative of a natural scenario of Tad invasion. Furthermore, introducing a mutant qde-1− allele into an Adiopodoumé-derived background mirrored the situation observed with introduced cloned Tad elements, in that no derepression was observed in the absence of qde-1+ (data not shown). This finding is consistent with our previous failure to detect a significant expansion of a cloned TAD element in the qde-1− strain (Figure 1B) indicating that the RdRP required for transgene silencing has no obvious role in transposon control in Neurospora.
Figure 3.
The two Neurospora dcl genes are mutually redundant in Tad control. Expression of the full-length Tad transcript was monitored by northern blot in cultures grown from individual ascospores deriving from the cross of (A) a double dcl mutant or (C) a qde-2− strain with Adiopodoumé. Double dcl− progeny are indicated with asterisks. Mutant strains not tran sformed with Tad (nt) were included as negative controls. (B) The mean relative TAD transcript levels in each of the segregating genotypes from (C) the double dcl− cross or (D) the qde-2− cross. Representative blots are shown, a total of 15 ascospores were analysed in the double dcl− cross, and 16 ascospores in the qde-2− cross. Error bars display standard error of the mean. DD, double dcl−.
The two Neurospora Dicer genes (dcl1 and dcl2) are mutually redundant in Tad control
The Dicer enzymes DICER-LIKE1 and DICER-LIKE2 are mutually redundant in the quelling pathway during the silencing of transgenes (35). However, recent reports have demonstrated that in organisms with multiple Dicers, different enzymes are each responsible for processing of different substrates such as microRNA precursors, hairpin dsRNA structures and viral RNA templates (43,44). To explore further similarities between quelling and the Tad control pathway, and also to investigate the possibility of a transposon-specific role for either of the Dicers in Neurospora, we performed a cross between a double-dcl mutant and Adiopodoumé, and analysed Tad expression in the progeny that contained either wild-type, single- or double-dcl mutant genotypes (Figure 3A and B). As a control comparison, we also crossed a qde-2− strain with Adiopodoumé (Figure 3C). Similar to the situation in quelling of transgenes (35), we only noticed an obvious increase in Tad expression in the double-dcl mutant (Figure 3A, lanes 3 and 8; Figure 3B), whereas no effect was observed in either of the single mutants. This elevation was similar in magnitude (∼15-fold) to that observed in qde-2− progeny (cf. Figure 3B and D) from a similar cross, in agreement with the role of DICER-LIKE and QDE2 in the same pathway that controls Tad.
siRNAs against Tad suggest a quelling-related silencing mechanism
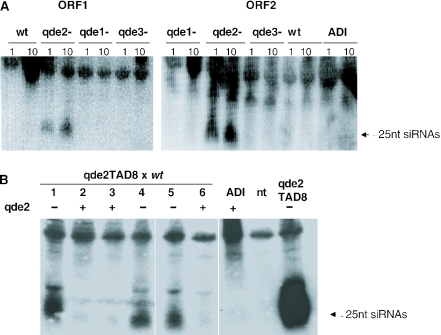
The involvement of qde-2 in reducing Tad expression suggested similarities with a quelling-like mechanism. Another feature of quelling is the production of siRNAs of 21–25 nt that correspond to the targeted sequence (41). We isolated total RNA and enriched for the small RNA fraction in order to look for Tad siRNAs in our strains. We found that siRNAs accumulated in both the qde-2− strains transformed with Tad (Figure 4A), and all of the Tad qde2− progeny resulting from the cross of qde2TAD8 with wt (Figure 4B, lanes 1, 4 and 5). siRNAs were detected using either probes homologous to the ORF1 (proximal end) or ORF2 (central region) of Tad, suggesting that the siRNAs were distributed along most of the element (Figure 4A and data not shown). This observation is consistent with the fact that QDE2 is downstream of the production of siRNAs in the quelling pathway (41). However, our inability to detect siRNAs in either the Tad-transformed wt strain, the Adiopodoumé isolate or the qde-2+ progeny from the wt cross (Figure 4A and B, compare lanes 2, 3 and 6 with 1, 4 and 5) are not consistent with the usual features of quelling, in which siRNAs accumulate in a quelling-proficient background.
Figure 4.
siRNAs against Tad accumulate only in a qde2 mutant background. (A) In the Tad-transformed wild-type (wt), qde-1−, qde-3− and qde-2− strains, Tad siRNAs could only be detected in the qde-2− strain. siRNAs were detected in both the first (1) and tenth (10) asexual generation after transformation with the cloned Tad element. No siRNAs were detected in the Adiopodoumé strain (ADI). Probes recognizing sequence from both ORF1 of Tad (left panel) and ORF2 of Tad (right panel). (B) In a cross of qde2TAD8 with a wild-type strain, the presence of Tad siRNAs segregates strictly with the absence of QDE2. The cultures used in this analysis derive from the same ascospores analysed in Figure 2A. A non-transformed qde-2− strain (nt) was used as a negative control.
To explain the apparent lack of siRNAs in the presence of QDE2, we considered the idea that QDE2 might function in a complex with siRNAs to direct a form of TGS, in a manner similar to S.pombe, where a negative feedback loop exists in the production of siRNAs (2,20,21).
A QDE2/Dicer-dependent silencing pathway, rather than DNA methylation, is the major contributor to Tad control
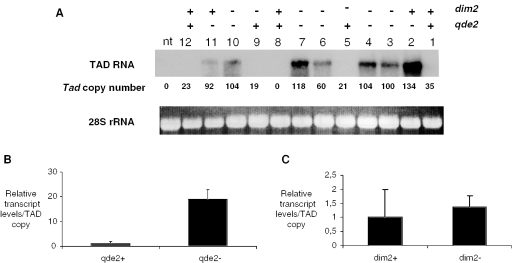
To investigate the possibility of a form of TGS that might be directed by a QDE2/siRNA complex, we considered DNA methylation; evidence from Arabidopsis has shown that DNA methylation can be targeted to transposon loci via an siRNA/Argonaute complex. Moreover, a role for DNA methylation in the control of Tad elements has previously been demonstrated; an artificially introduced Tad element became de novo methylated and showed an increased transposition rate in the absence of methylation (30). Therefore, both to evaluate the relative contributions of qde-2 and methylation to transposon taming in Neurospora, and to see if indeed the two might be linked via a TGS mechanism, we introduced qde-2− and dim-2− mutations into similar Tad backgrounds by crossing qde2TAD8 with a dim-2− strain; the dim-2 gene is responsible for all known methylation in Neurospora (45). To confirm the dim-2 status of the segregants, we performed Southern blots digested with either the methylation-sensitive enzyme Sau3A1, or its methylation-insensitive isoschizomer DpnII and probed for the control ζ–η region, a RIP'd locus that is constitutively methylated in a dim-2+ background (Figure 5B) (23). In the progeny of the cross, all possible combinations of the qde2 and dim2 alleles segregated. Interestingly, when we used a probe covering the first 1.5 kb of Tad (Figure 5B) to check the methylation status of the elements in the progeny, we noticed that there was very little, if any, methylation, judging by the near identity of Sau3A1 and DpnII digests in all of the dim-2+ segregants, regardless of qde2 status (Figure 5A, lanes 1, 2, 8, 11 and 12). The absence of methylation was similarly observed when we controlled a 0.6 kb region comprising the proximal portion of ORF2 of Tad (not shown). Furthermore, DIM2 had no obvious effect on the expression levels of Tad. In contrast, Tad expression correlated only with the qde-2− allele, similar to the situation in the qde2TAD8 × wt cross (Figure 6A, lanes 2–4, 6, 7, 10 and 11), indicating that it is the QDE2-based pathway, rather than methylation, that plays the major role in the control of this transposon. We similarly failed to detect methylation of Tad elements that were introduced into a wild-type background, ruling out the possibility that the qde-2 gene might be required only in the initiation of Tad methylation at the time of transformation (data not shown).
Figure 5.
The major bulk of Tad elements are not significantly methylated. (A) The methylation status of Tad elements in the progeny of a qde2TAD8 × dim-2− cross was determined by digesting genomic DNA with either the methylation-insensitive enzyme DpnII (d) or its methylation-sensitive isoschizomer Sau3AI (s). The digested DNA was hybridized with the probe shown in (B), covering the first four Sau3AI (s) sites of the Tad element. The sizes of fully digested hybridizing bands are indicated at the right of (A). (C) The blot shown in (A) was stripped and re-probed for the constitutively methylated control region ζ–η to check the dim-2 status of the progeny. The dim-2 and qde-2 status of each ascospore is indicated by a ‘+’ (wild-type allele) or ‘−’ (mutant allele) above each lane (DIM, non-transformed dim-2− parent strain).
Figure 6.
QDE2, not DNA methylation, is the major determinant in limiting Tad expression. (A) Expression of the full-length Tad transcript was monitored by northern blot in cultures grown from individual ascospores deriving from the cross of qde2TAD8 × dim-2−. The cultures used in this analysis derive from the same ascospores analysed in Figure 5A. Mean relative transcript levels were plotted based on segregation of either (B) the qde-2− allele or (C) the dim-2− allele.
Our failure to detect significant Tad methylation also rules out the possibility of any QDE2-directed TGS mechanism functioning via the DIM2 methyltransferase.
DISCUSSION
Transposition can represent a threat to genome stability by causing gene disruptions, illegitimate recombination and unregulated transcription. Indeed, in most species the major bulk of transposons are transcriptionally silent due to either DNA methylation and/or histone modifications (13,15,17,19). In addition to these TGS mechanisms, PTGS pathways similar to RNAi are also involved in the silencing of transposons. Moreover, it has been shown recently that DNA methylation of repeated sequences can be directed by the PTGS machinery, providing a direct link between the two mechanisms (16,19,46).
In our analysis, we evaluated the contribution of both DNA methylation and components of the PTGS pathway in the control of Tad expression. The principle determinant of Tad expression and expansion in our strains was qde-2, and the presence of Tad siRNAs hints at a quelling-related mechanism. In mutants lacking QDE2, an introduced Tad element showed a rapid expansion over the course of 15 asexual generations and, in outcrosses, the mutant allele segregated strictly with a release of Tad repression. Moreover, this dependence on the qde-2 gene was also observed in a naturally infected, Adiopodoumé-derived strain. On the contrary, we did not observe a significant level of methylation of the Tad element in our strains. Strengthening this finding, Tad expression levels were unaffected by the absence of DIM2, the DNA methyltransferase responsible for all known methylation in Neurospora. This highlights a major difference between Neurospora and other DNA methylation-proficient species in which blocking methylation leads to a large increase in transcription of diverse transposon families (15–17). Our data suggest that in the vegetative phase of Neurospora, a QDE2/siRNA-based mechanism, rather than DNA methylation, is mainly responsible for Tad control. The fact that this QDE2-based mechanism is able to limit Tad expression by itself, and without recourse to methylation, is again in contrast to other methyltransferase-containing species where siRNAs and methylation of several classes of transposon DNA are inexorably linked. Our data are in agreement with the recent observation that heterochromatin formation can occur in Neurospora in the absence of the RNAi machinery, suggesting that in this species the two silencing pathways are self-contained mechanisms (47,48).
Previous reports have shown that a Tad element at the am locus could be methylated and, in the absence of methylation, there is an increased mobilization of the element (30). However, methylation was rare and sporadic and indeed the major bulk of Tad elements are similarly not methylated in Adiopodoumé (28), suggesting that any role for methylation should be minor in comparison to the qde-2-silencing pathway.
In addition to QDE2, the requirement for DICER-LIKE activity in the repression of Tad highlights further similarities to quelling. On the other hand, the superfluity of the RdRP QDE1 and the RecQ DNA helicase QDE3 in this repression, and the accumulation of siRNAs only in the absence of the QDE2 protein are uncharacteristic of quelling. These features could point to a transposon-specific control pathway in addition to quelling. Recent evidence from Drosophila and Arabidopsis has demonstrated that different Dicer homologues can provide distinct substrate specificity (43,44). However, this does not seem to be the case in Neurospora as we have demonstrated that each of the two DICER-LIKE enzymes are able to repress Tad, and this mutual redundancy is also seen in the silencing of transgenes during quelling (35). It is possible that other RdRP homologues of the qde-1 gene, such as sad-1 (involved in MSUD) and RdRP-3 (function unknown), neither of which is necessary for quelling, might be involved in a transposon-specific pathway (24). Similarly, the qde-3 homologue, recQ-2, might have a role in transposon silencing (49).
An explanation for our failure to detect siRNAs in a qde-2+ background might rely on the degradation by siRNA-loaded RISC of Tad precursors upstream of the formation of dsRNA transcripts, thereby creating a negative feedback loop in which the siRNA pool could be reduced to levels below our detection limits. However, this does not appear to be the case in quelling of transgenes where siRNAs accumulate in both the presence and absence of QDE2, perhaps suggesting either greater abundance of the precursors upstream of dsRNA or lower susceptibility to degradation by RISC.
Again, a transcriptional silencing mechanism, other than DNA methylation, guided by an siRNA/QDE2 complex similar to the RNA-induced initiation of transcriptional silencing complex (RITS) in yeast or AGO4-containing complexes in plants would have a similar effect in terms of siRNA prevalence. While our data do not exclude this possibility, recent evidence indicates that both maintenance and establishment of heterochromatin (both histone methylation and DNA methylation) at silenced transgenes and endogenous sequences can occur independently of qde-2/RNAi in Neurospora, meaning that, as yet, there is no precedent for siRNA-based transcriptional silencing in this species (16,19,20,48).
Models of transposon silencing need to provide a mechanism by which these elements are recognized. In the control of certain transposable elements for example, one such mechanism might rely on the presence of inverted repeat (IR) sequences contained at their termini. Sijen and Plasterk found that in C.elegans read-through transcription of these IRs is able to trigger silencing of the Tc1 family of transposons by forming a hairpin dsRNA structure that can enter the RNAi pathway (50). However, Tad and many other transposons, such as non-LTR retrotransposons that include the human and mouse LINE elements, do not contain IR sequences. Thus, an alternative explanation is needed in these cases. One possible reason why Tad triggers a QDE2-dependent silencing pathway may rely on the very complex transcriptional profile of the element that, in addition to a full-length transcript includes at least two shorter anti-sense transcripts that originate from its 3′ end (51). Inter-molecular base pairing of sense and anti-sense transcripts would lead to a dsRNA intermediate, providing a template for Dicer cleavage and subsequent RISC-mediated degradation of Tad templates. In support of this, the direct production of dsRNA has been shown to bypass the requirement of both QDE1 and QDE3, but is upstream of QDE2 in the silencing of transgenes, a situation similar to that which we observed (52). However, an explanation based on intermolecular pairing of sense transcripts and 3′-originating anti-sense transcripts needs to explain the presence of siRNAs from ORF1, into which the main anti-sense transcripts do not extend (51). An auxiliary role for one of the three Neurospora RdRP genes in extending a paired dsRNA molecule in a 3′ direction could speculatively explain this observation. Alternatively, other uncharacterized more extensive anti-sense transcripts might be responsible.
The fact that transposon control via QDE2 is not linked to DNA methylation would appear to set apart Neurospora from other methylation-proficient species. In addition to the QDE2-based mechanism that we have identified for the silencing of Tad, Neurospora also possesses RIP that mutates repeated sequences during the pre-meiotic sexual phase of the life cycle. Strikingly, the methylated component of the genome largely represents RIP-inactivated sequences (53). Indeed, almost any RIP-inactivated sequence is able to function as a de novo methylation signal when re-transformed into Neurospora (54). It is therefore possible that, in Neurospora, methylation of repeated sequences is largely signalled by previous RIP inactivation, obviating the requirement for an RNAi-directed methylation mechanism in this species.
Since Adiopodoumé appears to be proficient for both mechanisms, we prefer the previously proposed hypothesis that the presence of Tad in Adiopodoumé represents a recent invasion (29) that should eventually be cleared by a combination of the two silencing pathways. It would appear likely that Neurospora, during the process of a Tad invasion, uses a qde-2/quelling-related mechanism to limit the expansion of Tad during vegetative growth whilst the resulting restricted set of Tad elements is gradually eliminated by RIP in successive cycles. It seems reasonable to expect that many of the transposon relics that litter the Neurospora genome will have at one time been subject to a similar concert of silencing mechanisms, possibly also involving MSUD, that function to ensure their deactivation and maintain genome integrity.
Acknowledgments
We are very grateful to Eric Selker for the dim-2− strain and to John Kinsey for the pTad1-1 plasmid. This work was supported by grants from the European Union Research Training Network (HPRN-CT-2002–00257), the Instituto Pasteur Fondazione Cenci Bolognetti and FIRB-MIUR 2001 (RBNEO15MPB_001/RBNE01KXC9_006). Funding to pay the open access publication charges for this article was provided by the European Union Research Training Network (grant no. HPRN-CT-2002-00257).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hall I.M., Noma K., Grewal S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 3.Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 6.Waterhouse P.M., Wang M.B., Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 7.Wu-Scharf D., Jeong B., Zhang C., Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- 8.Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 9.Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 10.Llave C., Kasschau K.D., Rector M.A., Carrington J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 12.Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Carrington J.C., Doerge R.W., Colot V., Martienssen R. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 13.Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 14.Martienssen R.A., Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 15.Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 16.Lippman Z., May B., Yordan C., Singer T., Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 18.Hirochika H., Okamoto H., Kakutani T. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 20.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schramke V., Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 22.Pickford A.S., Cogoni C. RNA-mediated gene silencing. Cell Mol. Life Sci. 2003;60:871–882. doi: 10.1007/s00018-003-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selker E.U., Jensen B.C., Richardson G.A. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science. 1987;238:48–53. doi: 10.1126/science.2958937. [DOI] [PubMed] [Google Scholar]
- 24.Shiu P.K., Raju N.B., Zickler D., Metzenberg R.L. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 25.Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 26.Kinsey J.A. Restricted distribution of the Tad transposon in strains of Neurospora. Curr. Genet. 1989;15:271–275. doi: 10.1007/BF00447042. [DOI] [PubMed] [Google Scholar]
- 27.Kinsey J.A. Tad, a LINE-like transposable element of Neurospora, can transpose between nuclei in heterokaryons. Genetics. 1990;126:317–323. doi: 10.1093/genetics/126.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinsey J.A., Garrett-Engele P.W., Cambareri E.B., Selker E.U. The Neurospora transposon Tad is sensitive to repeat-induced point mutation (RIP) Genetics. 1994;138:657–664. doi: 10.1093/genetics/138.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson C., Tang Q., Kinsey J.A. Elimination of active tad elements during the sexual phase of the Neurospora crassa life cycle. Fungal Genet. Biol. 2001;33:49–57. doi: 10.1006/fgbi.2001.1267. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y., Cambareri E.B., Kinsey J.A. DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Genet. Genomics. 2001;265:748–754. doi: 10.1007/s004380100472. [DOI] [PubMed] [Google Scholar]
- 31.Catalanotto C., Azzalin G., Macino G., Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 32.Cogoni C., Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 33.Cogoni C., Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 34.Foss H.M., Roberts C.J., Claeys K.M., Selker E.U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993;262:1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- 35.Catalanotto C., Pallotta M., ReFalo P., Sachs M.S., Vayssie L., Macino G., Cogoni C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell Biol. 2004;24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis R.H., De Serres F.J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- 37.Westergaard M.M., Mitchell H.K. ‘Neurospora V: a synthetic medium favoring sexual reproduction’. Am. J. Bot. 1947:573–577. [Google Scholar]
- 38.Orbach M.J., Porro E.B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol. Cell. Biol. 1986;6:2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staben C., Jensen B., Singer M., Pollock J., Schechtman M., Kinsey J., Selker E. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newslett. 1989;36:79. [Google Scholar]
- 40.Cambareri E.B., Helber J., Kinsey J.A. Tad1-1, an active LINE-like element of Neurospora crassa. Mol. Gen. Genet. 1994;242:658–665. doi: 10.1007/BF00283420. [DOI] [PubMed] [Google Scholar]
- 41.Catalanotto C., Azzalin G., Macino G., Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor; 1989. [Google Scholar]
- 43.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 45.Kouzminova E., Selker E.U. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 47.Chicas A., Cogoni C., Macino G. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 2004;32:4237–4243. doi: 10.1093/nar/gkh764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freitag M., Lee D.W., Kothe G.O., Pratt R.J., Aramayo R., Selker E.U. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- 49.Pickford A., Braccini L., Macino G., Cogoni C. The QDE-3 homologue RecQ-2 co-operates with QDE-3 in DNA repair in Neurospora crassa. Curr. Genet. 2003;42:220–227. doi: 10.1007/s00294-002-0351-6. [DOI] [PubMed] [Google Scholar]
- 50.Sijen T., Plasterk R.H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 51.Sewell E., Kinsey J.A. Tad, a Neurospora LINE-like retrotransposon exhibits a complex pattern of transcription. Mol. Gen. Genet. 1996;252:137–145. [PubMed] [Google Scholar]
- 52.Goldoni M., Azzalin G., Macino G., Cogoni C. Efficient silencing by expressing double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 2004;41:1016–1024. doi: 10.1016/j.fgb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Selker E.U., Tountas N.A., Cross S.H., Margolin B.S., Murphy J.G., Bird A.P., Freitag M. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- 54.Singer M.J., Marcotte B.A., Selker E.U. DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol. Cell. Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]