Abstract
Background
Helicobacter pylori (H. pylori) infection is a major risk factor for gastric diseases, including gastritis and gastric cancer. Heat shock protein 60 (HSP60) is a chaperone protein involved in various cellular processes and has been implicated in the immune response to bacterial infections. Extracellular vesicles (EVs) containing various protein components play important roles in cell communication. In the present study, a systematic proteomic analysis of EVs obtained from H. pylori infected cells was performed and the EV-derived HSP60 function was studied.
Methods
EVs were evaluated by nanoparticle tracking analysis, transmission electron microscopy and western blotting. The recognized protein components were quantified by label-free proteomics and subjected to bioinformatics assays. The expression of HSP60 in EVs, host cells and gastric cancers infected by H. pylori was determined by western blotting and immunohistochemical, respectively. In addition, the apoptotic regulation mechanisms of HSP60 in H. pylori infection were analyzed by western blotting and flow cytometry.
Results
A total of 120 important differential proteins were identified in the EVs from H. pylori-infected cells and subjected to Gene Ontology analysis. Among them, CD63, HSP-70 and TSG101 were verified via western blotting. Moreover, HSP60 expression was significantly increased in the EVs from H. pylori-infected GES-1 cells. H. pylori infection promoted an abnormal increase in HSP60 expression in GES-1 cells, AGS cells, gastric mucosa and gastric cancer. In addition, knockdown of HSP60 suppressed the apoptosis of infected cells and the expression of Bcl2, and promoted the upregulation of Bax.
Conclusion
This study provides a comprehensive proteomic profile of EVs from H. pylori-infected cells, shedding light on the potential role of HSP60 in H. pylori infection. The findings underscore the significance of EV-derived HSP60 in the pathophysiology of H. pylori-associated diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-023-03131-1.
Keywords: H. pylori, Extracellular vesicle, Proteomics, HSP60
Background
Helicobacter pylori (H. pylori), a main Gram-negative bacterium, asymptomatically colonizes more than 50% of the global population and is associated with numerous gastrointestinal diseases, such as gastritis, gastric ulcer and gastric cancer (GC) [1]. The identification of the involvement of H. pylori infection causes gastrointestinal diseases was a landmark [2, 3]. H. pylori, as an inducer of GC, is considered a group 1 carcinogen by the World Health Organization [4]. Although 10% of peptic ulcer disease is associated with H. pylori infection, 1–3% of those infected patients will develop to GC, and survival is < 5 years [5]. Another 0.1% of patients will suffer mucosa-related lymphoid tissue lymphoma [6]. The fact that H. pylori infection causes gastritis and GC has been recognized worldwide, but the specific molecular mechanism remains unclear.
Extracellular vesicles (EVs) are small, membrane-bound vesicles that are released by all cells in the body [7–9]. EVs can contain a variety of biomolecules, including proteins, nucleic acids, and lipids [11–13]. EVs play a role in cell communication and can also be used by pathogens to deliver virulence factors to host cells [10]. Studies have shown that EVs from H. pylori-infected gastric epithelial cells can contain a variety of virulence factors, including the cytotoxin-associated gene A (CagA) protein [14]. CagA is a major virulence factor of H. pylori and is thought to play a role in the development of gastric cancer [15]. EVs from H. pylori-infected can also interact with host cells and modulate their immune response. For example, H. pylori EVs can induce the production of inflammatory cytokines and suppress the activity of T cells [16, 17]. This can lead to the development of chronic inflammation in the stomach, which is a risk factor for gastric cancer [16, 17]. In addition to their role in the pathogenesis of H. pylori infection, EVs from H. pylori-infected cells may also be used as diagnostic and therapeutic targets. For example, H. pylori EVs can be detected in the blood of patients with H. pylori infection, which could be used to develop a non-invasive diagnostic test for H. pylori infection [16]. Overall, research on the relationship between EVs from H. pylori-infected cells and gastric diseases is still in its early stages, and in particular, there is a lack of systematic studies on the profiling of these EVs and the function of EV components.
Heat shock protein 60 (HSP60) is a highly conserved protein that plays a vital role in protein folding and assembly. It is also involved in other cellular processes, such as stress response, immunity, and apoptosis [18]. There is growing evidence that human HSP60 plays a role in the pathogenesis of a variety of diseases, including cancer, autoimmune diseases, and infectious diseases [19–21]. HSP60 has been shown to promote tumor growth and progression, suppress the anti-tumor immune response, and promote the development of gastric cancer stem cells [22]. However, the specific mechanism underlying these activities of HSP60 remains to be further explored.
Proteomics offers complementary information to genomics and transcriptomics, and is essential for the understanding of complex biochemical processes at the molecular level [23]. Mass spectrometry (MS) has emerged as the most important and popular tool to identify, characterize and quantify proteins and their post-translational modifications (PTMs) with high throughput and on a large scale [24]. The present study screened by proteomics and bioinformatics analysis certain differentially expressed proteins in H. pylori-infected gastric epithelial cells. These candidate differential proteins in EVs were further confirmed. Furthermore, the expression of heat shock protein (HSP) 60 in clinical samples was evaluated, and its functions in the H. pylori-infection process were investigated.
Methods
Ethics statement
Gastric mucosa samples of 30 normal individual and 30 patients with H. pylori infection were obtained from the Kunshan First People's Hospital from February to December 2021. The samples were then clipped by endoscopy and verified by urease test. All samples were kept at − 80 °C for further experiments. GC tissue microarray (66 paracancerous and 68 complete GC tissues, including 26 H. pylori-infected GC tissues; cat. no. HStmA180su30-T-113 RG-K-16-C13) was purchased from Shanghai Xinchao Biotechnology Co., Ltd.
Ethics approval was obtained from the institutional Ethics Committee of Kunshan first people's Hospital (Suzhou, China, approval no. 2021-06-014-K01). The experiments were conducted as per the Declaration of Helsinki, and all participants offered written informed consent.
Cell culture and H. pylori infection
GES-1 and AGS cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). GES-1 and AGS cells were cultivated in culture dishes with DMEM supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37 °C with 5% CO2. Both GES-1 and AGS cells were used in the current study for H. pylori infection and EV production.
H. pylori (NCTC 11637 strain) was donated by Marshall International Digestive Disease Diagnosis and Treatment Center of Shanghai Oriental Hospital. H. pylori was cultured on Columbia blood agar plates and incubate in a microaerophilic chamber at 37 °C for 3–4 days. Infection was conducted at a multiplicity of cells: H. pylori = 1:50 for 24 h, and cleaned with PBS three times.
Isolation of EVs
Cell culture supernatants from either uninfected or H. pylori-infected GES-1 cells for 48 h were collected. EVs from culture supernatants were centrifuged at 300×g for 10 min, 3000×g for 15 min, and 10,000×g for 60 min to remove cells, cell debris, bacteria and microvesicles respectively. Then pellets therefrom were resuspended with 200 μl of 9% sucrose containing protease inhibitors and centrifuged at 100,000g for 2 h. Each step was followed by two washes with PBS at 4 °C. EVs were resuspended with 50 μl of 9% sucrose containing protease inhibitors and stored at − 80 °C before further experiments.
Transmission electron microscopy (TEM)
An EVs diluted solution (10 μl) was put onto the copper wire at 25 °C for 5 min, and washed three times with ultrapure water in triple. After air drying, 10 μl of a phosphotungstic acid solution was added to the copper mesh coloring for 2 min for coloring, and then air dried at ambient temperature. The morphology of the EVs was observed under a Hitachi H-600 transmission electron microscope (Hitachi, Ltd., Tokyo, Japan).
Nanoparticle size analysis
EVs were diluted to 106 EVs/ml with 100 μl PBS. Upon homogenization, EVs (10 μl) were diluted to 1 ml with ultrapure water, and placed into a NanoSight NS300 nanoparticle size analyzer for measurement of particle size distribution and concentration following standard procedures at 4 °C. The laser beam passed through EVs outside the sample room; achieved particle visualization through a microscope equipped with a camera; captured the Brownian motion of EVs; and used software to calculate the concentration and hydrodynamic diameter according to their motion. The diameter and concentration of the EVs were obtained through NanoSight Nanoparticle tracking analysis (NTA) 3.2 software analysis.
Protein isolation and digestion
EVs samples were isolated with an SDT buffer (4% SDS, 100 mM Tris–HCl and 1 mM DTT, pH 7.6). Protein levels were detected with a BCA assay kit (Bio-Rad Laboratories, Inc.). Next, the EV proteins were digested with trypsin as per the filter-aided sample preparation (Matthias Mann). The obtained peptides were desalted by using Empore SPE Cartridges C18 [standard density, bed internal diameter (I.D.), 7 mm; volume, 3 ml; Sigma-Aldrich; Merck KGaA], vacuum-centrifuged and reconstructed in 40 µl 0.1% (v/v) formic acid. Proteins (200 μg) were placed into 30 μl another SDT buffer (4% SDS, 100 mM DTT and 150 mM Tris–HCl pH 8.0). The detergent, DTT and other low-molecular-weight chemicals were ultrafiltered numerous times with a UA buffer (8 M urea and 150 mM Tris–HCl, pH 8.0) (Microcon; 10 kDa). Next, the reduced cysteine residues were blocked with 100 μl iodoacetamide (100 mM in UA buffer) and incubated for 30 min in the dark. The filters were washed first with 100 μl UA buffer three times, and then with 100 μl 25 mM NH4HCO3 twice. Finally, the protein suspensions were digested with 4 μg trypsin (Promega Corporation) in 40 μl NH4HCO3 buffer at 37 °C overnight, and the final peptides were collected as a filtrate.
The ultraviolet density at 280 nm of the peptides was detected with an extinction coefficient of 1.1 of a 0.1% (g/l) solution, which was calculated based on the frequency of tryptophan and tyrosine in vertebrate proteins.
Liquid chromatography with tandem mass spectrometry (LC–MS/MS)
LC–MS/MS was conducted on a Q-Exactive MS meter with EASY nLC for 120 min (Thermo Fisher Scientific, Inc.). The peptides were placed into a PepMap100 reverse trap column (Acclaim, 100 μm × 2 cm, nanoViper C18; Thermo Fisher Scientific, Inc.) connected to a C18 reversed column (Easy, 10-cm long, 75 μm I.D., 3 μm resin) in buffer A (0.1% formic acid), and separated with a linear gradient of buffer B (84% acetonitrile, 0.1% formic acid) at a rate of 300 nl/min controlled on IntelliFlow. MS data in the positive ion mode were acquired using a data-based top10 method to dynamically select the richest precursor ions from 300 to 1800 m/z for hierarchical cellular decomposition (HCD) decomposition. The largest injection time, gain auto-control target and dynamic exclusion duration were 10 ms, 3e6 and 40.0 s, respectively. Survey scanning resolution, HCD resolution and isolation width were 70,000 at m/z 200, 17,500 at m/z 200, and 2 m/z, respectively. The normalized collision energy was 30 eV. The under fill ratio, which was the lowest percentage of the target value to be met at the longest fill time, was 0.1%. The peptide recognition mode was adopted. The raw MS data were combined and searched on MaxQuant 1.5.3.17 for identification and quantification.
Bioinformatic analysis
HCD was conducted with Cluster 3.0 (http://bonsai.hgc.jp/mdehoon/software/cluster/software.htm). Tree diagrams, heat maps and volcano plot were adopted. The subcellular positioning of EV proteins was predicted with a Cell Ontology system (http://cello.life.nctu.edu.tw/) [25]. With the KEGG database (http://geneontology.org/) [26], the EV differential proteins were blasted to identify their KEGG orthology, and then mapped to the path in KEGG path.
Total quantitative protein was used as the background dataset in the enrichment analysis based on Fisher’s exact test. P < 0.05 was considered to indicate a statistically significant difference. Protein–protein interactions (PPIs) were represented to express the molecular mechanisms and signaling pathways of cell processing. The PPIs of EV proteins were explored with Search Tool for the Retrieval of Interacting Genes/Proteins (http://string.embl.de/) [27], and then was visualized with Cytoscape (https://cytoscape.org/) [28].
Cell transfection
HSP60 small interfering RNA (siRNA) and scrambled siRNA (50 nM; GenePharma Co., Ltd.) were transfected into GES-1 cells cultured in 6-well plates at 75% confluence using Lipofectamine 3000® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocols at 37 °C for 24 h. The HSP60 siRNA sense sequence was 5′-GACGAUGCCAUGCUCUUAAT-3′, and the antisense sequence was 5′-UUAAGAGCAUGGCAUCGUCTT-3′, while the scrambled siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′.
Apoptosis assay
Four treated cells groups (untreated GES-1 cell control, scrambled siRNA, siRNA and siRNA + Hp) were collected and washed twice with PBS. Cells were re-suspended in 100 μl binding buffer, and then incubated for 15 min with Annexin-V and PI (each 5 μl) (BD Biosciences Company, USA) in the dark at 37 °C. Subsequently, cells in 500 μl binding buffer were detected on a FACS Canto II flow cytometer (BD Biosciences Company). The results of apoptosis were analyzed by FlowJo 7.6 software (Tree Star, Inc., Ashland, OR, USA) and GraphpadPrism5 software (GraphPad Software; Dotmatics).
Western blotting
Total EV or cellular proteins were separated and lysed using a RIPA peptide lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Proteins (30 μg) were loaded onto 10% SDS-PAGE gels, electrophoresed, and transferred to PVDF membranes (MilliporeSigma, Burlington, MA, USA). Next, the membranes were incubated with primary antibodies against CD63 (1:10,000), HSP70 (1:10,000), ERK1/2 (1:1400), HSP60 (1:10,000), Bcl2 (1:6000), Bax (1:8000), GAPDH (1:200,000), and TSG101 (1:20,000) (all from ProteinTech Group Inc., Chicago, USA) at 4 °C overnight. Next, the membranes were washed three times with TBS-Tween 20 under shaking, incubated with a secondary antibody solution (mouse anti-rabbit IgG-HRP at 1:6,000 dilution) at room temperature under mild shaking for 1.5 h, and re-washed with TBS-T under shaking three times. Protein bands were photographed and examined with a chemiluminescent substrate system (Thermo Fisher Scientific, Inc.). The intensity of the western blot was determined with ImageJ software and analyzed by GraphPad Prism 5 software (GraphPad Software; Dotmatics).
Multicolor immunohistochemistry (IHC)
For multicolor staining, GC tissue arrays with 66 paracancerous and 68 complete GC tissues, including 26 H. pylori-infected GC tissues (Shanghai Xinchao Biological Technology Co., Ltd.) were processed for IHC according to the instructions of the manufacturer of the Opal 2-Color Anti-Rabbit Automation IHC Kit (cat. no. NEL830001KT; Akoya Biosciences). Slides were incubated at 4 °C overnight with primary anti-HSP60 (Signaling Technology, Inc.) antibody. Next, the array was incubated with the secondary antibody solution for 50 min at room temperature under light agitation, followed by three washed with PBS, and imaged using a digital microscope camera (Nikon Corporation).
Statistical analysis
Data are expressed as the mean ± standard error. Statistical analysis was performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). Groups were compared via one-way ANOVA or paired Student's t test. P < 0.05 was considered to indicate a statistically significant difference.
Results
Biological features of EVs from H. pylori-infected cells
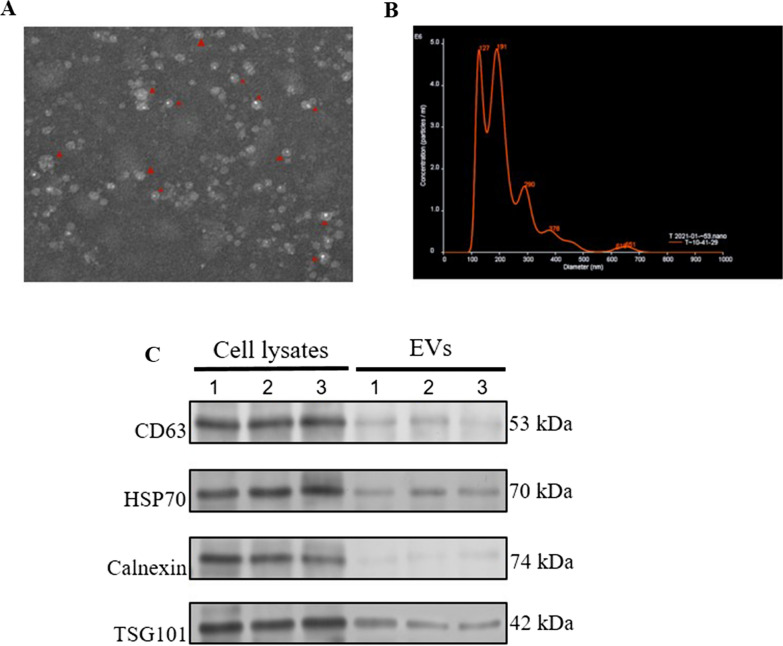
EVs were first purified from H. pylori infected or uninfected GES-1 cells, and then the morphology, size and purity of EVs were determined by transmission election microscopy, nanoparticle tracking analysis and western blot, respectively. As shown in Fig. 1A, EVs were in the typical cup-like vesicles under TEM. Nanoparticle tracking analysis (NTA) showed that the size of the majority of particles was between 30 and 150 nm, and the concentration of particles reached to 5.63 × 109 particles/ml (Fig. 1B and Additional file 1: Fig. S1A-B). In addition, the purity of the EVs were assessed by measuring the presence of 3 positive markers (CD63, HSP70 and TSG101) and 1 negative marker (Calnexin). Our data in Fig. 1F showed that purified EVs had high levels of all 3 positive markers but very low level of the negative marker Calnexin, indicating that the purified EVs were in good purity. Taken together, our data here indicates that purified EVs from H. pylori infected or uninfected cells had expected morphology, size, and purity.
Fig. 1.
The biological properties of EVs purified from H. pylori infected GES-1 cells. A The morphological character of EVs under the transmission electron microscopy (magnification of × 200). B The size distribution of EVs were determined with NTA. C The purity of EVs was determined by western blotting
Proteomics analysis of EVs
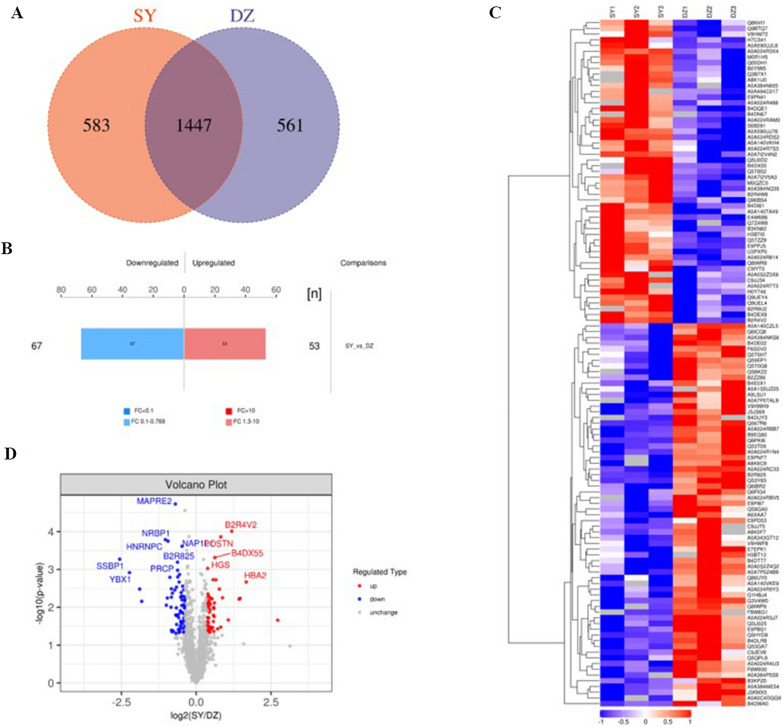
To evaluate the EV protein profiles of H. pylori-infected GES-1 cells, total EV proteins were quantified via label-free quantitative proteomics analysis. The results of MS/MS analysis revealed that there were 1447 common EV proteins in the two groups investigated (SY: H. pylori-infected GES-1 cells vs. DZ: H. pylori-uninfected GES-1 cells), as indicated in Venn diagram show in Fig. 1A. Bioinformatics analysis of the proteomics data indicated that there were 120 EV differential proteins including 53 up-regulated proteins and 67 down-regulated proteins were cluster analysis with 1.5 fold change (Fig. 2B). The heat map and volcano plot results represented the expression levels of 120 EV differential proteins sufficiently (Fig. 2C, D), with red indicating high expression and blue indicating low expression. The 13 upregulated and 14 downregulated proteins of interest in are summarized in Table 1.
Fig. 2.
EV differential proteins were analyzed by bioinformatics. A The venn diagram illustrated 1447 overlapping proteins in GES-1 infected with and without H. pylori (SY vs. DZ). B 53 up-regulated and 67 down-regulated EV proteins in two groups were indicated by the histogram of quantitative difference. C The heat map displayed the expression intensity of EV differential proteins between two groups (SY vs. DZ). D The volcano diagram represented 53 up-regulated and 67 down-regulated EV proteins in two groups and some EV proteins were marked with the unique and stable entry identifier of protein. Note DZ: Hp uninfected group, SY: Hp infected group. Red: high expression, blue: low expression
Table 1.
List of 27 EV differential proteins with significant interesting from H. pylori infected GES-1 cells
| Number | Protein | Protein name | Gene | Proteins | Peptides | Unique peptides | Coverage | Mol/weight | Expression pattern |
|---|---|---|---|---|---|---|---|---|---|
| 1 | E9PPJ5 | Midkine (Fragment) | MDK | 2 | 5 | 5 | 39.7 | 14.374 | Up |
| 2 | A0A0S2Z3X8 | Rab GDP dissociation inhibitor (Fragment) | GDI1 | 6 | 13 | 7 | 43 | 50.582 | Up |
| 3 | Q5TZZ9 | Annexin | ANXA1 | 29 | 21 | 21 | 64.2 | 38.714 | Up |
| 4 | A0A590UJ76 | Deleted in malignant brain tumors 1 protein | DMBT1 | 5 | 3 | 3 | 6 | 260.63 | Up |
| 5 | Q9UEY4 | Anion exchange protein | SLC4A2 | 16 | 16 | 16 | 23.5 | 135.58 | Up |
| 6 | A0A024R3X4 | 60 kDa chaperonin (Fragment) | HSPD1 | 24 | 19 | 19 | 46.8 | 61.054 | Up |
| 7 | E9PN41 | Tetraspanin-4 (Fragment) | TSPAN4 | 7 | 4 | 4 | 47.7 | 15.195 | Up |
| 8 | B4DN67 | Kin of IRRE-like protein 1 | KIRREL1 | 3 | 7 | 7 | 16.3 | 72.721 | Up |
| 9 | A0A024R7S3 | Clathrin light chain | CLTB | 3 | 6 | 6 | 26.1 | 23.181 | Up |
| 10 | H3BTI0 | Cysteine-rich secretory protein LCCL domain-containing 2 | CRISPLD2 | 4 | 7 | 7 | 21.6 | 55.79 | Up |
| 11 | Q6NVI1 | MARCKS protein (Fragment) | MARCKS | 2 | 8 | 8 | 83.4 | 14.859 | Up |
| 12 | B4DEX8 | S-adenosylmethionine synthase | MAT2A | 5 | 9 | 7 | 37.6 | 39.71 | Up |
| 13 | A0A494C017 | Myeloid-associated differentiation marker (Fragment) | MYADM | 7 | 1 | 1 | 18.9 | 13.153 | Up |
| 14 | F6SDV2 | Tubulointerstitial nephritis antigen-like | TINAGL1 | 1 | 11 | 3 | 56.4 | 40.867 | Down |
| 15 | Q86UY0 | TXNDC5 protein | TXNDC5 | 4 | 8 | 8 | 33.1 | 40.369 | Down |
| 16 | Q6FIG4 | RAB1B protein | RAB1B | 2 | 8 | 0 | 52.7 | 22.198 | Down |
| 17 | E9PNF7 | Lysosomal Pro-X carboxypeptidase | PRCP | 11 | 5 | 5 | 20.8 | 49.902 | Down |
| 18 | A0A024RC33 | Microtubule-associated protein RP/EB family member 2 | MAPRE2 | 3 | 9 | 2 | 36.7 | 37.031 | Down |
| 19 | Q1HBJ4 | Mitogen-activated protein kinase | MAPK1 | 9 | 8 | 8 | 27.2 | 41.389 | Down |
| 20 | A0A1S5UZ25 | Mediator of ErbB2-driven cell motility 1 | Memo1 | 2 | 5 | 1 | 29.2 | 30.051 | Down |
| 21 | Q53Y83 | Alpha-galactosidase | GLA | 12 | 8 | 8 | 24.9 | 48.766 | Down |
| 22 | A8K0F7 | Vitamin-K-epoxide reductase (warfarin-sensitive) | 2 | 2 | 2 | 11.9 | 19.775 | Down | |
| 23 | F8W6G1 | Nuclear receptor-binding protein | NRBP1 | 3 | 5 | 5 | 13.8 | 60.829 | Down |
| 24 | Q6PKI6 | YBX1 protein (Fragment) | YBX1 | 6 | 11 | 3 | 62 | 29.374 | Down |
| 25 | F6SDV2 | Tubulointerstitial nephritis antigen-like | TINAGL1 | 1 | 11 | 3 | 56.4 | 40.867 | Down |
| 26 | Q86UY0 | TXNDC5 protein | TXNDC5 | 4 | 8 | 8 | 33.1 | 40.369 | Down |
| 27 | Q6FIG4 | RAB1B protein | RAB1B | 2 | 8 | 0 | 52.7 | 22.198 | Down |
Bioinformatics analyses of EVs derived from H. pylori-infected cells
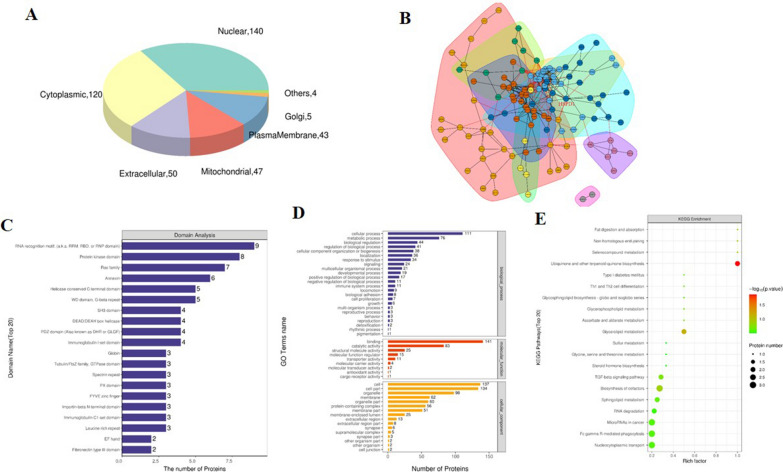
Bioinformatics analysis of protein function showed that subcellular positioning of EV differential proteins was enriched in the nuclear (140), cytoplasmic (120), extracellular (50), mitochondrial (47) and plasma membrane (43) and other (9) (Fig. 3A). The interaction relationship of EV differential proteins was represented in a network diagram (Fig. 3B), and proteins with the same or similar functions are divided into different clusters with different colors. The interaction network of EV differential proteins showed a complex interaction between EV differential proteins, with the nodes of the HSP60 protein in the network are marked and the proteins interacting with HSP60 is clearly displayed in the diagram.
Fig. 3.
Bioinformatics analysis of EV differential proteins. A Subcellular localization pie chart of EV differential proteins in two groups (SY vs. DZ). B The interaction network of EV differential proteins. C Domain analysis revealed that EV differential proteins involved in many intracellular domains. D GO enrichment map including BP, MF and CC showed that EV differential proteins involved in various biological functions, molecular functions and cell structures. E Bubble diagram of KEGG pathway enrichment indicated EV differential proteins participated in various intracellular metabolic responses. Note DZ: Hp uninfected group, SY: Hp infected group
Domain analysis suggested that EV differential proteins were associated with RNA binding protein domain, protein kinase domain, Ras family and annexin etc. (Fig. 3C). The Gene Ontology (GO) analysis divided the results into biological processes (BP), cellular components (CC) and molecular functions (MF) (Fig. 3D). The results of GO analysis showed that EV differential protein participate in cellular process, metabolic process and biological regulation etc. (BP); binding, catalytic activity and structural molecular activity (MF); and cell, cell part and organelle etc. (CC). The KEGG pathway analysis demonstrated the EV proteins were enriched in ubiquinone and the terpenoid-quinone biosynthesis, glycerolipid metabolism, biosynthesis of cofactors and miRNAs in cancer (Fig. 3E). Bioinformatics analysis revealed that these identified EV proteins were associated with the physiology and pathophysiology of H. pylori-infected cells.
Confirmation of HSP60 expression in H. pylori-infected cells and gastric mucosa
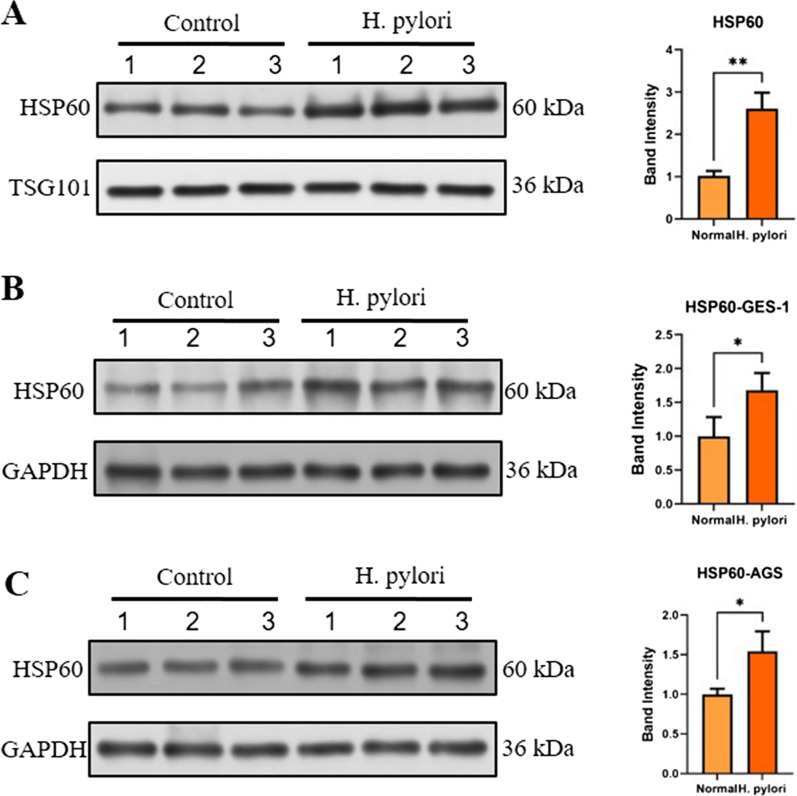
The upregulation of HSP60 in EVs from H. pylori-infected cells was further confirmed by western blot. In consistent with the proteomics data, HSP60 expression was significantly higher in EVs from H. pylori-infected cells compared to that from control cells (Fig. 4A, B). HSP60 is active in protecting cell physiology under the stimulation of fever, inflammation and other conditions. We next investigated whether the expression of HSP60 was also increased in H. pylori-infected cells. GES-1 and AGS cells infected or noninfected by H. pylori was tested for HSP60 expression by western blot, and results showed that HSP60 increased significantly in H. pylori infected cells relative to non-infected cells (Fig. 4C–F).
Fig. 4.
EV differential proteins HSP60 were analyzed in H. pylori infected cells and GC tissues. A, B EV-derived HSP60 in H. pylori uninfected group and H. pylori infected group (Hp vs control) were detected by western blotting. B The relative expression of HSP60 was semiquantified. C–F The expression levels of HSP60 in H. pylori infected (C, D) GES-1 and (E, F) AGS cells were determined via western blotting. All western blotting experiments were repeated three times (three panels 1, 2, 3 presents repeated three times). *p < 0.05; **p < 0.01
HSP60 expression in H. pylori-infected GC tissues
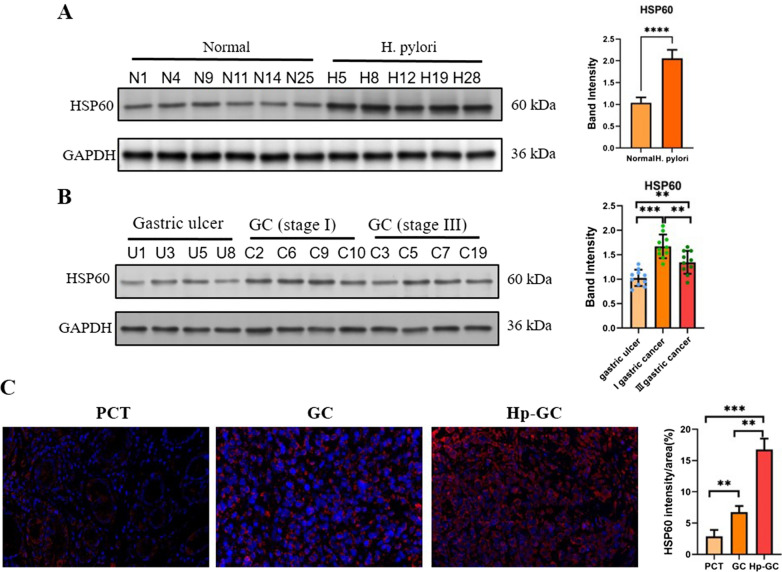
To further investigate the association between HSP60 and H. pylori infection in vivo, we next tested the expression of H. pylori in various gastric tissues. First, the HSP60 level between normal gastric mucosal tissue and H. pylori-infected tissue was compared. As shown in Fig. 5A, HSP60 level was around 2-folds higher in H. pylori-infected gastric mucosal tissue than in normal tissue. Next, we also checked whether the level HSP60 expression was gastric cancer (GC) stage related. GC stage I and III samples together with gastric ulcer samples as control were collected and the expression of HSP60 was determined by western blot. Our data showed that HSP60 expression was elevated in GC compared to gastric ulcer, but stage III GC did no show further increase in HSP60 expression than stage I GC (Fig. 5B and Additional file 1: Fig. S2). Third, HSP60 expression in paracancerous tissues (PCT), H. pylori-negative GC and H. pylori-positive GC was also compared. As shown in Fig. 5C, the HSP60 level was significantly upregulated in GC compared to PCT samples, while H. pylori-positive GC showed a further drastic increase comparing to H. pylori-negative GC.
Fig. 5.
The infection of H. pylori regulates the expression of HSP60 in GC. A The expression of HSP60 in normal and H. pylori-infected mucosal tissues was determined by western blot. The relative expression of HSP60 was semiquantified. B The expression of HSP60 in gastric ulcer, and stage I and III GC mucosal tissues was determined by western blot. The relative expression of HSP60 was semiquantified. C The expression of HSP60 in PCT, GC and Hp-GC was determined by IHC, and the MFI of HSP60 was also determined. Magnification: 40×. ns, not statistically significant; *p < 0.05; **p < 0.01; ***p < 0.001
HSP60 regulates cell apoptosis
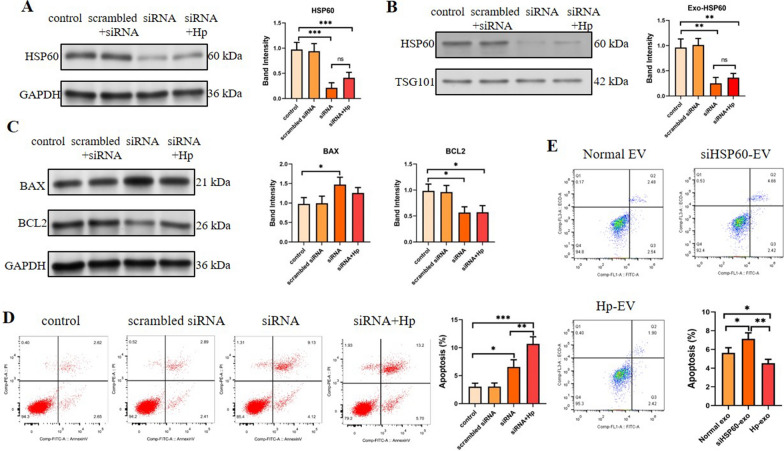
To explore the function of HSP60 protein in H. pylori infection, HSP60 in GES-1 cells was knocked down with siRNA, and cell apoptosis and apoptosis-related proteins BCL1 and Bax in HSP60 knockdown cells were analyzed by flow cytometry and western blotting. The results indicated that the expression of HSP60 was knocked down by siRNA successfully, which resulted in EVs with reduced HSP60 loading (Fig. 6A, B). The knockdown of HSP60 inhibited the expression of the anti-apoptotic protein Bcl2 while enhanced the expression of the apoptotic protein Bax (Fig. 6C). In consistent, apoptosis analysis showed that HSP60 knockdown increased the apoptosis of GES-1 cells (Fig. 6D).
Fig. 6.
The apoptosis of GES-1 cells regulated by HSP60. A–C GES-1 cells were first treated with or without HSP60-siRNA, and then either mock-infected or infected with H. pylori. Following infection, cells were harvested and the expression of A HSP60 in cells, B HSP60 and TSG101 in EVs, C BAX and BCL2 and D apoptosis were measured by western blot and flow cytometry, respectively. E GES-1 cells were either untreated, or treated with HSP60-siRNA or infected with H. pylori, and then EVs were isolated and their impact on the cell viability of GES-1 cells were determined by flow cytometry. *p < 0.05; **p < 0.01; ***p < 0.001
To further explore the function of EV-derived HSP60, EVs from normal cells, HSP60-knocked-down cell and H. pylori-infected cells were isolated and their impact on cell viability was assessed. As shown in Fig. 6E, compared with the normal EV group, EVs from HSP60 knocked-down cell demonstrated induced more cell apoptosis, while EVs from H. pylori-infected cells showed reduced cell apoptosis. This data indicate that EV-derived HSP60 from H. pylori-infected cells could reduce cell apoptosis.
Discussion
Our bioinformatics analysis identified 120 differentially expressed proteins between the EVs isolated from H. pylori-infected and uninfected cells. These proteins are involved in a variety of cellular processes, including metabolism, cell signaling, and immune response. Some of the most notable differentially expressed proteins include peptidylprolyl isomerase, annexin and chaperonin, which have all been shown to be overexpressed in a variety of cancers, including gastric cancer [29–32]. The overexpression of these proteins in the H. pylori-infected EV samples suggests that they may play a role in the cancer development and progression. Further research is needed to understand the role of these proteins in gastric cancer and to develop new diagnostic and therapeutic strategies targeting these proteins.
The domain analysis in our current study revealed that the majority of differentially regulated proteins in EVs isolated from H. pylori-infected cells were involved in RNA recognition. RNA recognition is the process by which proteins bind to RNA molecules. This process is essential for many cellular functions, including gene expression, RNA processing, and protein translation [33]. A number of RNA recognition proteins have been implicated in the development and progression of gastric cancer [34]. Although beyond the scope of our current study, further research to explore the effects of RNA recognition proteins in EVs from H. pylori-infected cells would be warranted. In addition, we observed that HSP60 expression was enhanced in GC tissues compared to gastric ulcer tissues. However, stage III GC did not show higher expression of HSP60 than stage I GC. Because we could not obtain GC samples of intestinal and diffuse types, the expression of HSP60 was not determined in these samples. It would be valuable for future studies to check if there is difference of HSP60 expression in these GC types.
Heat shock proteins (HSPs) participate in protein homeostasis under stresses and heat shock amid normal physiology [35, 36]. HSP60, a nuclear-encoded mitochondrial protein associated with co-chaperonin HSP10, facilitates the folding of proteins to promote the degradation of misfolded or denatured proteins, and participates in immune system activation. It also plays chronic inflammatory and pro-survival or pro-apoptotic roles, and enhances cancer metastasis [37–39]. Chmiela et al. [40] found that HSPB of H. pylori and human HSP60 share homology and have immunogenicity, which may cause a cross reaction and further damage to the gastric mucosa. EV differential protein interaction network revealed that HSP60 protein was a key node in the PPI network, and participated in the energy metabolism of mitochondria, the regulation of membrane function and purine metabolism. Furthermore, HSP60 protein interacted with other proteins in the interaction network to regulate cell energy metabolism and maintain cell homeostasis.
According to the results of HSP60 protein in EVs identified by MS, HSP60 proteins were confirmed in H. pylori-infected gastric epithelial cells, GC cells and gastric mucosa tissues. Chen et al. [41] reported that HSP60 was an advanced biomarker for digestive system cancer, and its abnormal expression may have implications for early diagnosis in the screening of GC. Notably, HSP60 protein expression increased considerably in GES-1 and AGS cells, and in H. pylori-infected gastric mucosa tissues. Additionally, H. pylori infection could promote the abnormal increase of HSP60 in GC tissues relative to that in H. pylori non-infected GC tissues. HSP60 protein was significantly upregulated in EVs, GES-1 and AGS cells, and GC tissues under H. pylori infection. Inactivation of HSP60 affected the key functions of mitochondria, and reduced the respiratory capacity and ATP level of cells [42]. In addition, the release of HSP60 may activate the innate immune system, promoting a proinflammatory state, including an increase in TNF-α [43]. According to the literature, and GO and KEGG analyses, it was concluded that H. pylori infection may prevent bacterial infection by upregulating the expression of HSP60 to induce the inflammatory response of cells, and regulate the energy metabolism of cells to maintain normal cell homeostasis.
HSP60 in mitochondria can act on apoptosis-related factors and inhibit the activation of mitochondrial apoptosis pathway, and can reduce the production of oxygen free radicals in mitochondria. A low level of HSP60 in the cytoplasm can also inhibit cell apoptosis through interaction with apoptosis-related factors [44, 45]. Sandeep et al. [40, 46] found that knocking down HSP60 decreased IL-8 expression and its release in prostate cancer cell xenograft tumors in SCID mice, indicating that the HSP60 and IL-8 axis promoted apoptosis resistance in cancer. In the present study, after HSP60 was knocked down in GES-1 cells, the anti-apoptotic protein Bcl2 decreased and the apoptotic protein Bax increased significantly in GES-1 cells. Simultaneously, knockdown of HSP60 promoted cell apoptosis indicating that HSP60 could regulate apoptosis during H. pylori infection. Previous studies have indicated that HSP60 may regulate cell apoptosis through BCL2 and BAX [47, 48]. However, the detailed mechanism underlying HSP60-EVs on cell apoptosis remains to be further investigated.
The profiles of the EV proteins in H. pylori-infected host cells were further investigated, and the roles of significant differential proteins were explored. The differential proteins associated with H. pylori infection, such as HSP60, may be involved in infection, inflammatory responses and cancer induced by H. pylori. However, the underlying functions and mechanisms of HSP60 protein, which is highly expressed in EVs and cells under H. pylori infection, require further studies.
Supplementary Information
Additional file 1: Fig. S1. NTA analysis of purified exosomes from H. pylori infected GES-1 cells. (A) The particle diameter and (B) concentration of exosomes were determined with NTA. Fig. S2. The infection of H. pylori regulates the expression of HSP60 in GC. The expression of HSP60 in gastric ulcer, and stage I and III GC mucosal tissues was determined by western blot.
Acknowledgements
Not applicable.
Author contributions
JJW and HZ conceived, designed, discussed the work, and revised the manuscript. YJL, HC and, DWQ did the experiments and wrote the manuscript. NW and TTW edited the manuscript. All authors read and approved the final version.
Funding
This work was supported by grants from the Suzhou Key Discipline-Experimental Diagnosis (No. SZXK202124), the National Natural Science Foundation of China (No. 82002111), the Science and Technology Development Plan Project of Suzhou (No. SKY2022078), Young Scientists Foundation of Changzhou No.2 People's Hospital (NO.2022K006).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study involving human participants was reviewed and approved by the Ethics Committee of Affiliated Kunshan Hospital of Jiangsu University. The patients/participants provided their written informed consent to participate in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yujie Li, Hui Cao and Qiude Wen contributed equally to this work.
Contributor Information
Jianjun Wang, Email: wangjianjun0520@163.com.
Hong Zhu, Email: emeraldzh@sina.com.
References
- 1.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Pereira-Marques J, Ferreira RM, Pinto-Ribeiro I, Figueiredo C. Helicobacter pylori infection, the gastric microbiome and gastric cancer. Adv Exp Med Biol. 2019;1149:195–210. doi: 10.1007/5584_2019_366. [DOI] [PubMed] [Google Scholar]
- 3.Johnson KS, Ottemann KM. Colonization, localization, and inflammation: the roles of H. pylori chemotaxis in vivo. Curr Opin Microbiol. 2018;41:51–57. doi: 10.1016/j.mib.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alipour M. Molecular mechanism of Helicobacter pylori-induced gastric cancer. J Gastrointest Cancer. 2021;52:23–30. doi: 10.1007/s12029-020-00518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 6.Peek RM, Jr, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208(2):33–48. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Xu C, Gong R, Hu T, Zhang X, Xie X, et al. EV CagA from Helicobacter pylori aggravates intestinal epithelium barrier dysfunction in chronic colitis by facilitating Claudin-2 expression. Gut Pathog. 2022;14:13. doi: 10.1186/s13099-022-00486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Yao Y, Chen X, Wu J, Gu T, Tang X. Host derived EVs-pathogens interactions: potential functions of EVs in pathogen infection. Biomed Pharmacother. 2018;108:1451–1459. doi: 10.1016/j.biopha.2018.09.174. [DOI] [PubMed] [Google Scholar]
- 9.Marote A, Teixeira FG, Mendes-Pinheiro B, Salgado AJ. MSCs-derived EVs: cell-secreted nanovesicles with regenerative potential. Front Pharmacol. 2016;7:231. doi: 10.3389/fphar.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L. EVs in pathogen infections: a bridge to deliver molecules and link functions. Front Immunol. 2018;9:90. doi: 10.3389/fimmu.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded EVs from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meckes DG, Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes EVs for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimoda A, Ueda K, Nishiumi S, Murata-Kamiya N, Mukai SA, Sawada S, et al. EVs as nanocarriers for systemic delivery of the Helicobacter pylori virulence factor CagA. Sci Rep. 2016;6:18346. doi: 10.1038/srep18346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia X, Zhang L, Chi J, Li H, Liu X, Hu T, et al. Helicobacter pylori infection impairs endothelial function through an EV-mediated mechanism. J Am Heart Assoc. 2020;9:e014120. doi: 10.1161/JAHA.119.014120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González MF, Díaz P, Sandoval-Bórquez A, Herrera D, Quest AFG. Helicobacter pylori outer membrane vesicles and extracellular vesicles from Helicobacter pylori-infected cells in gastric disease development. Int J Mol Sci. 2021;22:4823. doi: 10.3390/ijms22094823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HI, Choi JP, Seo J, Kim BJ, Rho M, Han JK, et al. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp Mol Med. 2017;49:e330. doi: 10.1038/emm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bethke K, Staib F, Distler M, Schmitt U, Jonuleit H, Enk AH, et al. Different efficiency of heat shock proteins (HSP) to activate human monocytes and dendritic cells: superiority of HSP60. J Immunol. 2002;169:6141–6148. doi: 10.4049/jimmunol.169.11.6141. [DOI] [PubMed] [Google Scholar]
- 19.Merendino AM, Bucchieri F, Campanella C, Marcianò V, Ribbene A, David S, et al. Hsp60 is actively secreted by human tumor cells. PLoS ONE. 2010;5:e9247. doi: 10.1371/journal.pone.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N, Li N, Wen S, Li B, Zhang Y, Liu Q, et al. HSP60 regulates lipid metabolism in human ovarian cancer. Oxid Med Cell Longev. 2021;2021:6610529. doi: 10.1155/2021/6610529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappello F, de Macario EC, Zummo G, Macario AJ. Immunohistochemistry of human Hsp60 in health and disease: from autoimmunity to cancer. Methods Mol Biol. 2011;787:245–254. doi: 10.1007/978-1-61779-295-3_18. [DOI] [PubMed] [Google Scholar]
- 22.Tong WW, Tong GH, Kong H, Liu Y. The tumor promoting roles of HSP60 and HIF2α in gastric cancer cells. Tumour Biol. 2016;37:9849–9854. doi: 10.1007/s13277-015-4783-2. [DOI] [PubMed] [Google Scholar]
- 23.Smith LM, Kelleher NL, Consortium for Top Down Proteomics Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Wu S, Stenoien DL, Paša-Tolić L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif). 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 25.Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, et al. CELLO2GO: a web server for protein subcelllular localization prediction with functional gene ontology annotation. PLoS ONE. 2014;9:e99368. doi: 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43D:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Shen L, Li Y, Shen L, Li N. Identification of pyroptosis-related gene prognostic signature in head and neck squamous cell carcinoma. Cancer Med. 2022;11:5129–5144. doi: 10.1002/cam4.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, He Y, Lu LL, Zhou ZY, Wan NB, Li GP, et al. miRNA-192-5p impacts the sensitivity of breast cancer cells to doxorubicin via targeting peptidylprolyl isomerase A. Kaohsiung J Med Sci. 2019;35:17–23. doi: 10.1002/kjm2.12004. [DOI] [PubMed] [Google Scholar]
- 31.Xu XH, Pan W, Kang LH, Feng H, Song YQ. Association of annexin A2 with cancer development (Review) Oncol Rep. 2015;33:2121–2128. doi: 10.3892/or.2015.3837. [DOI] [PubMed] [Google Scholar]
- 32.Carr AC, Khaled AS, Bassiouni R, Flores O, Nierenberg D, Bhatti H, et al. Targeting chaperonin containing TCP1 (CCT) as a molecular therapeutic for small cell lung cancer. Oncotarget. 2017;8:110273–110288. doi: 10.18632/oncotarget.22681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 34.Masuda K, Kuwano Y. Diverse roles of RNA-binding proteins in cancer traits and their implications in gastrointestinal cancers. Wiley Interdiscip Rev RNA. 2019;10:e1520. doi: 10.1002/wrna.1520. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Syldath U, Bellmann K, Burkart V, Kolb H. Human 60-kDa heat-shock protein: a danger signal to the innate immune system. J Immunol. 1999;162:3212–3219. doi: 10.4049/jimmunol.162.6.3212. [DOI] [PubMed] [Google Scholar]
- 37.Ren D, Fan M, Sun C, Zhou C, Li Y. Capillary electrophoresis with laser induced fluorescence detection for study of the association of HSP60 gene polymorphism with gouty arthritis. J AOAC Int. 2019;102:810–814. doi: 10.5740/jaoacint.18-0242. [DOI] [PubMed] [Google Scholar]
- 38.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gröbe A, Blessmann M, Hanken H, Friedrich RE, Schön G, Wikner J, et al. Prognostic relevance of circulating tumor cells in blood and disseminated tumor cells in bone marrow of patients with squamous cell carcinoma of the oral cavity. Clin Cancer Res. 2014;20:425–433. doi: 10.1158/1078-0432.CCR-13-1101. [DOI] [PubMed] [Google Scholar]
- 40.Chmiela M, Kupcinskas J. Review: pathogenesis of Helicobacter pylori infection. Helicobacter. 2019;24:e12638. doi: 10.1111/hel.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Li X, Shao S. The clinical value of HSP60 in digestive system cancers: a systematic review and meta-analysis. Clin Lab. 2019; 65. [DOI] [PubMed]
- 42.Fan F, Duan Y, Yang F, Trexler C, Wang H, Huang L, et al. Deletion of heat shock protein 60 in adult mouse cardiomyocytes perturbs mitochondrial protein homeostasis and causes heart failure. Cell Death Differ. 2020;27:587–600. doi: 10.1038/s41418-019-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L, Kim SC, Wang Y, Gupta S, Davis B, Simon SI, et al. HSP60 in heart failure: abnormal distribution and role in cardiac myocyte apoptosis. Am J Physiol Heart Circ Physiol. 2007;293:H2238–2247. doi: 10.1152/ajpheart.00740.2007. [DOI] [PubMed] [Google Scholar]
- 44.Huang YH, Yeh CT. Functional compartmentalization of HSP60-survivin interaction between mitochondria and cytosol in cancer cells. Cells. 2019;9:23. doi: 10.3390/cells9010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caruso Bavisotto C, Alberti G, Vitale AM, Paladino L, Campanella C, Rappa F, et al. Hsp60 post-translational modifications: functional and pathological consequences. Front Mol Biosci. 2020;7:95. doi: 10.3389/fmolb.2020.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, O'Malley J, Chaudhary AK, Inigo JR, Yadav N, Kumar R, et al. Hsp60 and IL-8 axis promotes apoptosis resistance in cancer. Br J Cancer. 2019;121:934–943. doi: 10.1038/s41416-019-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song E, Tang S, Xu J, Yin B, Bao E, Hartung J. Lenti-siRNA Hsp60 promote bax in mitochondria and induces apoptosis during heat stress. Biochem Biophys Res Commun. 2016;481:125–131. doi: 10.1016/j.bbrc.2016.10.153. [DOI] [PubMed] [Google Scholar]
- 48.Shan YX, Liu TJ, Su HF, Samsamshariat A, Mestril R, Wang PH. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J Mol Cell Cardiol. 2003;35:1135–1143. doi: 10.1016/S0022-2828(03)00229-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. NTA analysis of purified exosomes from H. pylori infected GES-1 cells. (A) The particle diameter and (B) concentration of exosomes were determined with NTA. Fig. S2. The infection of H. pylori regulates the expression of HSP60 in GC. The expression of HSP60 in gastric ulcer, and stage I and III GC mucosal tissues was determined by western blot.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.