Abstract
The Hsp90 chaperone cycle involves sequential assembly of different Hsp90-containing multiprotein complexes, the accessory proteins (“cochaperones”) that are associated with these complexes being exchanged as the cycle proceeds from its early to its late stages. To gain insight as to whether the 2-hybrid system could be used to probe the interactions of this Hsp90 system, yeast transformants were constructed that express the Gal4p deoxyribonucleic acid–binding domain (BD) fused to the 2 Hsp90 isoforms and the various Hsp90 system cochaperones of yeast. These “bait” fusions were then introduced by mating into other transformants expressing nearly all the 6000 proteins of yeast expressed as fusions to the Gal4p activation domain (AD). High throughput 2-hybrid screening revealed the ability of Hsp90 and Hsp90 system cochaperones to engage in stable interactions in vivo, both with each other and with the various other proteins of the yeast proteome. Consistent with the transience of most chaperone associations, interactions to Hsp90 itself were invariably weak and generally influenced by stress. Mutations within a Hsp90-BD bait fusion and an AD-Cdc37 “prey” fusion were used to provide in vivo confirmation of the in vitro data that shows that Cdc37p is interacting with the “relaxed” conformation of Hsp90 and also to provide indications that Cdc37p needs to be phosphorylated at its N-terminus for any appreciable interaction with Hsp90. A number of potentially novel cochaperone interactions were also identified, providing a framework for these to be analyzed further using other techniques.
INTRODUCTION
The Hsp90 molecular chaperone catalyses the final activation step of several of the key signaling and regulatory proteins of eukaryotic cells (targets hereafter designated as Hsp90 “clients”). Rather than acting at an early stage of protein folding, Hsp90 binds to these client proteins when they are already substantially folded. Then, often in response to an appropriate activatory signal, it facilitates the structural changes whereby they attain their full activity. This Hsp90-dependent activation process involves a number of cochaperones, proteins which associate with the Hsp90-based complexes at different stages of the chaperone cycle (reviewed in Csermely et al 1998; Pearl and Prodromou 2000; Young et al 2001; Picard 2002; Pratt and Toft 2003). For example, during the activation of certain steroid hormone receptors (SHRs) the cochaperone p60-Hop is present in the early-stage SHR complexes, before the attainment of the steroid-activatable state, whereas p23 and immunophilins such as Cyp40 are associated with the more mature Hsp90-SHR complexes where the receptor is capable of undergoing activation through the binding of the steroid (Pratt and Toft 2003). In some instances, cochaperone associations appear to be specific for a particular class of client. Thus, Hsp90 complexes with protein kinases often contain p50-Cdc37p, whereas those with SHRs generally do not (Stepanova et al 1996; Pratt and Toft 2003).
This study investigated the potential uses of the yeast 2-hybrid system for probing the interactions of Hsp90, the Hsp90 system cochaperones, and the proteome of yeast. This is a system that detects in vivo protein-protein association through this, leading to the reconstitution of transcription factor activity. The deoxyribonucleic acid (DNA)–binding domain (BD) and the activation domain (AD) of this transcription factor are expressed on different fusion proteins (“bait” and “prey” fusions, respectively), an active transcription factor being reconstituted only when the BD- and AD-bearing fusions associate noncovalently in the yeast nucleus. Yeast expressing fusions of the Gal4p BD to either Hsp90 or the individual cochaperones of yeast was mated to an array of other yeast cells expressing at least 80–90% of the proteins of yeast as fusions to the Gal4p AD (Uetz et al 2000). The diploids (now expressing both BD and AD fusions) were then transferred to selective plates, the colonies positive for activation of the reporter HIS3 gene being identified through their growth at defined positions on the arrays. The system was also optimized for the analysis of more transient protein-protein interactions by automated measurement of expression of an interaction-responsive LacZ gene in the tester yeast. It is shown that the latter system can allow in vivo monitoring of stress effects on an interaction and also the effects of mutations within the interacting fusions. These latter methods may be of more general use in the proteomics field.
This is the first study to apply genome-wide 2-hybrid analysis to the Hsp90 chaperone system. It describes the use of the Fields array of AD-yeast protein fusions (Uetz et al 2000), the potential uses and drawbacks of 2-hybrid analysis of this chaperone cycle, and also some potential ways of circumventing these problems by the introduction of defined mutations into the interacting fusions.
MATERIALS AND METHODS
Bait constructs comprising fusions of the Gal4p BD to Hsp90 and cochaperones
These fusion genes were generated by homologous recombination within yeast, essentially as described (Uetz et al 2000; Millson et al 2003). The open reading frame (ORF) of interest was initially amplified by 2 sequential polymerase chain reaction (PCR) amplifications, with the first PCR using primers that possess 3′ sequence homologies to the ORF of interest but 5′ homologies to plasmid pBDC and the second PCR a universal primer pair (see Millson et al 2003). Strain PJ69-4α (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ; James et al 1996) was then transformed with the product of this second PCR and either Nco1- or NruI-digested pBDC, thereby generating through homologous recombination within the yeast N-terminal and C-terminal fusions, respectively, of the Gal4p BD to the ORF of interest. Transformants were initially selected by plating on dropout medium (Adams et al 1997) lacking tryptophan. They were then checked for the presence of the correct fusion gene by colony PCR (Ling et al 1995) using the primers used for the second round of the above PCR amplification. They were also checked for expression of this gene by Western blotting using anti-Gal4p BD antiserum (Clontech).
Cells expressing a Hsp82-BD fusion with a point mutation in the Hsp82-coding region were constructed by slight modification of the procedure used to make the native, wild-type Hsp82-BD fusion (Millson et al 2003). Instead of using the wild-type HSP82 gene as the template in the initial PCR, well-characterized mutant hsp82 alleles from an earlier study were used (Prodromou et al 2000). Presence of the correct hsp82-BD fusion gene was checked by PCR reamplification from the PJ694α transformant, as above, followed by DNA sequencing.
Two-hybrid screening of protein interactions
Baits in PJ69-4α were checked for self-activation by plating onto minus tryptophan and histidine dropout medium (Adams et al 1997) containing increasing concentrations (0 to 8 mM) of 3-amino-1,2,4-triazole (3-AT). Baits that resulted in growth at 30°C in the absence of histidine but the presence of 2 mM or higher 3-AT were deemed self-activating and therefore not used to screen the array of AD-ORF fusions. The remaining baits in PJ69-4α were mated by pinning to the 16-plate, 384-well format array of yeast AD-protein fusions carried in the strain of opposite mating type (PJ69-4a [James et al 1996]). This array, described in an earlier report (Uetz et al 2000), was routinely maintained at 4°C except that once a month it was replica plated onto dropout agar medium lacking leucine and then incubated for 2 days at 30°C. All replications and inoculations were carried out using the 384-pin replicator of a Biomek® 2000 Laboratory Automation Workstation (Beckman), with movements programmed using the BioWorks™ Version software (Beckman).
After mating, the diploids were selected by plating on dropout medium lacking leucine and tryptophan. Screening for protein-protein interactions was by the pinning of these diploids onto dropout medium lacking leucine, tryptophan, and histidine, supplemented with increasing concentrations (0–4 mM) of 3-AT. Growth on the latter plates was scored after 10 days at 30°C. As with earlier screens of this array of yeast AD-protein fusions (Uetz et al 2000), 2 matings of each bait to the fusion array followed by selection for HIS3 activation were conducted, with the “double hits” identified in both these automated screens being counted as the positive interacting partners.
LacZ expression measurements
For measurement of the β-galactosidase activity resulting from expression of the interaction-responsive, GAL7 promoter-regulated LacZ gene of PJ69-4, the exponential phase cultures of diploid strains (grown at 25°C on dropout medium minus tryptophan and leucine) were initially aliquoted (8 × 100 μL) into the wells of replicate 96-well microtitre plates, using the 8-channel pipettor tool of Biomek. For stress experiments, 1 plate was maintained for 1 hour at 25°C, whereas the other was incubated for 1 hour at 39°C (heat shock) or 42°C (severe heat shock). For osmostress experiments, cells were diluted into sorbitol-containing medium (to a final concentration of 1.2 M) immediately before being subjected to the same regimen. The plates were centrifuged 1000 × g 5 minutes, the pellets resuspended in 100 μL Z-buffer (100 mM phosphate, pH 7.0, 10 mM MgSO4, 5 mM KCl, 50 mM β-mercaptoethanol), and the cells freeze-thawed twice. O-nitrophenyl-β-d-galactopyranoside (50 μL) (4 mg/mL in Z-buffer) was then added to each well, and the plates were incubated for 1 hour at 25°C. Fifty microliter 1 M sodium carbonate was then added and the cells were then collected by centrifugation for measurements of LacZ activity, as previously described (Harris et al 2001). Both optical density (600 nm) and total LacZ expression were then measured using a microtitre plate reader (Multiscan Ascent, ThermoLabsystems). The β-galactosidase data shown are the mean and standard deviations of each set of 8 individual assays. It is expressed relative to that of control diploid PJ69-4 cells containing both the empty pBDC (Millson et al 2003) with no gene insert and the plasmid for expression of the AD-fusion of interest (because the low basal expression of the GAL7 promoter-regulated LacZ gene in this system is generally attributable to the latter AD-protein fusion (unpublished data)).
RESULTS
Assessing the extent to which an Hsc82-BD fusion is functional as a chaperone
Hsp90 of Saccharomyces cerevisiae is encoded by 2 paralogous genes, HSC82 and HSP82, either of which can confer the essential Hsp90 function (Borkovich et al 1989). The encoded Hsp90 isoforms, denoted Hsc82 and Hsp82, share no less than 97% sequence identity. We have previously shown that the C-terminal fusion of the Gal4p BD to Hsc82 (Hsc82-BD) is functional as an Hsp90 in yeast, whereas the corresponding N-terminal fusion (BD-Hsc82) is not, by determining whether these BD fusions can confer the essential Hsp90 function in a hsc82Δ hsp82Δ double mutant strain (Millson et al 2003). Therefore, this study used 2-hybrid baits that comprise Hsc82 and Hsp82 with C-terminal BD extensions (Hsc82-BD and Hsp82-BD, respectively) on the premise that a bait that is substantially functional as a chaperone would be more likely to display authentic interactions than one that is nonfunctional.
Further studies revealed that the hsc82Δ hsp82Δ double mutant cells expressing the Hsc82-BD 2-hybrid bait as their sole form of Hsp90 (Millson et al 2003) were growth arrested when exposed to the mating pheromone α-factor. They are therefore not defective in this Hsp90-dependent response (Louvion et al 1998). Also, when rendered histidine prototrophs through the introduction of a HIS3 vector, these cells were capable of efficient growth at 30°C in the presence of 30 mM 3-AT (showing that they are not defective in the Hsp90-dependent activation of the Gcn2 kinase [Donze and Picard 1999]). These hsc82Δ hsp82Δ double mutant cells expressing the Hsc82-BD fusion as their sole form of Hsp90 are, though, greatly enlarged during 30°C growth (Fig 1). They are also slightly caffeine and temperature sensitive (not growing above 35°C; not shown). These latter defects are all corrected with an osmotic stabilization of the medium or with higher levels of expression of the Hsc82-BD fusion (yeast needs higher levels of Hsp90 to grow at high temperatures; Borkovich et al 1989). Swollen cell appearance, temperature sensitivity, and caffeine sensitivity are all characteristic phenotypes of mutants defective in cell wall maintenance (Gustin et al 1998; Martin et al 2000). Our preliminary growth tests indicate therefore that the addition of the Gal4p BD to the C-terminus of Hsc82 does not abrogate several functions, including the essential functions, of this chaperone in yeast. However, in those cells that express this fusion as their sole form of Hsp90, it appears to cause a partial defect in the maintenance of cell integrity (Fig 1).
Fig 1.
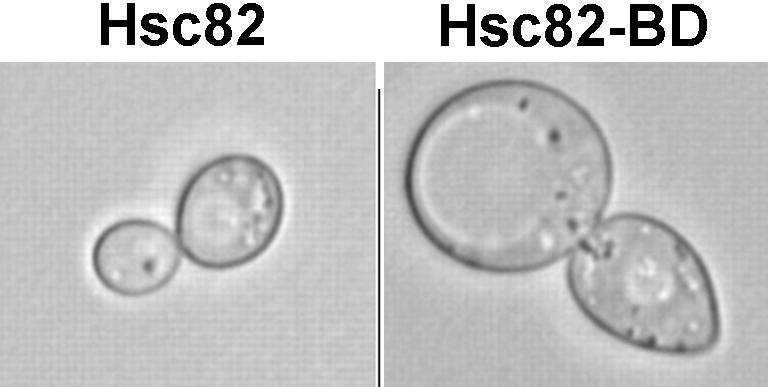
Phase-contrast images (100× magnification) of representative cells of the hsc82Δ hsp82Δ double mutant (Millson et al 2003) expressing either the wild-type Hsc82 (left) or the Hsc82-BD 2-hybrid bait fusion (right) as the sole form of Hsp90
Screening the Hsc82-BD and Hsp82-BD 2-hybrid bait fusions against the array of yeast AD-protein fusions
Robotic procedures were used to introduce the Hsc82-BD and Hsp82-BD 2-hybrid bait fusions, expressed in PJ694α, into the array of AD-yeast protein fusions expressed in the strain of opposite mating type, PJ694a (see Methods). This AD-fusion expression array consists of 6000 yeast transformants on 16 plates in 384-colony format (Uetz et al 2000). After mating, the cells were pinned onto the medium without tryptophan or leucine, so as to select the diploids expressing both Hsc82-BD (or Hsp82-BD) and an AD-yeast protein fusion. Diploid colonies were next transferred to the medium without tryptophan, leucine, or histidine but containing 1, 2, or 3 mM 3-AT. Then, after 10 days at 30°C, colonies scoring positive for activation of the GAL1 promoter-regulated HIS3 gene were identified from their positions on the arrays.
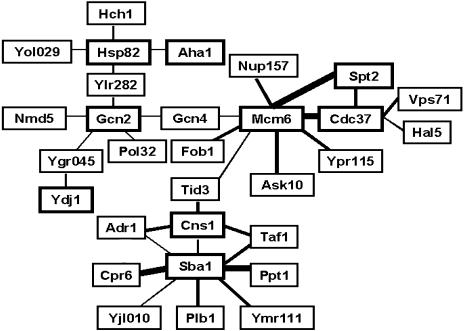
Discounting the known false positives in the array, no strong interactors to these Hsc82-BD and Hsp82-BD bait fusions were identified (interactions leading to growth in excess of 1 mM 3-AT). The strongest reproducible interactions (though permitting growth to only 0.5–1 mM 3-AT) were to known Hsp90 system cochaperones (Hch1p and Aha1p [Panaretou et al 2002], Cdc37p) and also to the products of 2 ORFs of unknown function, YLR282c and YOL029c (Fig 2; Table 1).
Fig 2.
Summary diagram of the Hsp90 (Hsp82) and Hsp90 system cochaperone interactions detected on the basis of HIS3 activation (growth during 10 days in the absence of histidine and the presence of 3-amino-1,2,4-triazole [3-AT]). Baits used (BD fusions) are boxed with a thick line, and their putative interactors (AD-protein fusions) are boxed in thinner lines. Only the interactions detected in 2 separate screens of the array are shown, the thickness of the line denoting the apparent strength of interaction (thin, growth to 1 mM 3-AT; medium, growth to 2 mM 3-AT; thick, growth at 3 mM or higher 3-AT)
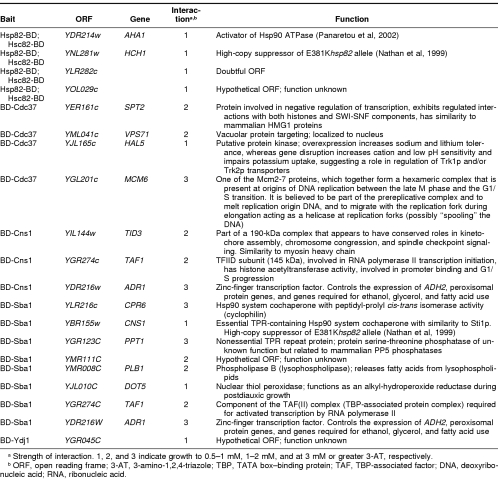
Table 1.
Summary of Hsp90 and Hsp90 cochaperone interactions detected on the basis of HIS3 activation
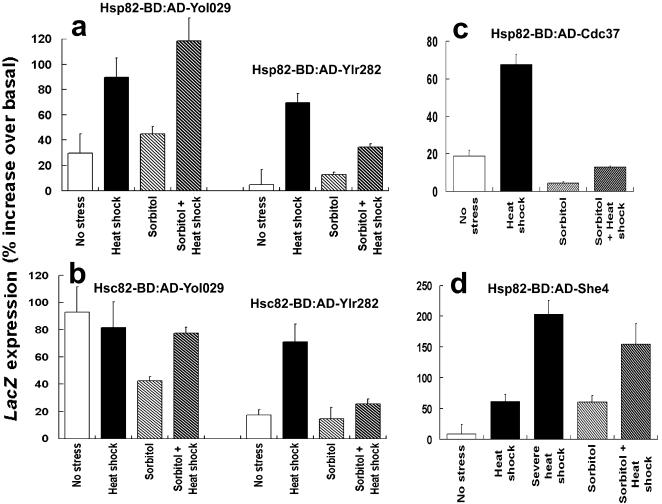
HIS3 activation, measured as growth during 10 days in the absence of histidine and presence of 3-AT, probably requires a relatively long-lived association of both bait and prey fusions (see Discussion). These results are therefore consistent with a general transience of the interactions involving the Hsp90 chaperone. The interaction-responsive, GAL7 promoter-regulated LacZ gene of the tester yeast strain, PJ69-4, is probably much more suitable for monitoring such transient (or stress-induced) interactions. Therefore, we automated the expression measurements of this LacZ reporter in 96- and 384-format arrayed yeast cultures (see Methods). This way we could conduct several replicate measurements for each relatively weak interaction and obtain quantitative LacZ data on how these interactions might be affected by stress. Figure 3 shows the use of this approach to monitor the effects of both heat and osmotic stress on the few, relatively weak Hsp82-BD and Hsc82-BD interactions detected in the above screens of the AD-protein fusion array. An interaction of these 2 baits to She4p (the yeast equivalent of the [Hsp90 interacting] UNC45 myosin-specific chaperone of metazoans [Barral et al 2002]) also was monitored. In yeast, Hsp90 interaction with this myosin-specific chaperone may be indirect (“bridged” through another protein) because She4p of fungi lacks the Hsp90-interacting tetratricopeptide repeat (TPR) domain of metazoan UNC45 (Hutagalung et al 2002).
Fig 3.
Stress effects on interactions involving Hsp90 bait fusions. (a) Interactions of Hsp82 with Yol029p and Ylr282p. (b) Interactions of Hsc82 with Yol029p and Ylr282p. (c) Interaction of Hsp82 with Cdc37p. (d) Interaction of Hsp82 with She4p. Cells were either unstressed or stressed for 1 hour under the conditions detailed in Methods. Because the low basal expression of the GAL7 promoter-regulated LacZ gene in this system is generally due to the AD-protein fusion, LacZ expression values are in all cases expressed relative to the control PJ69-4 cells containing the plasmid for expression of this AD-fusion and empty pBDC. Control experiments showed that the interaction signal for the Hsp82-BD or Hsc82-BD “bait” fusion plus empty AD vector was unaffected by all these stress treatments (mean and standard deviation in 4 separate experiments being 1.8 ± 0.3, 2.1 ± 0.3, 2.1 ± 0.5, and 1.5 ± 0.4 units of β-galactosidase per 1 × 106 cells for the unstressed, heat shocked, sorbitol-treated, and heat shocked plus sorbitol-treated samples, respectively)
As shown in Figure 3, heat shock generally leads to a stronger interaction signal of Hsp82-BD and Hsc82-BD with AD-Ylr282p, AD-Yol029p, AD-Cdc37p, and AD-She4p (an effect especially apparent with the AD-Cdc37p and AD-She4p prey fusions). With AD-Yol029p, only the interaction of with Hsp82-BD was strengthened by heat shock, not the interaction with Hsc82-BD (Fig 3a,b), even though these latter 2 bait fusions are expressing to similar levels in this yeast (Millson et al 2003). Because the growth defect of hsc82Δ hsp82Δ double mutant cells express the Hsc82-BD bait fusion as their sole Hsp90 can be largely corrected by osmotic stabilization of the medium (see above), and we also investigated how osmotic stress would affect these interactions. In the presence of 1.2 M sorbitol, the heat-induced increase of the interaction signal of Hsp82-BD with AD-She4p was still apparent (Fig 3d), but the heat-induced increase in the Hsp82-BD– AD-Cdc37p signal was largely abolished (Fig 3c).
Interaction of mutant Hsp82 forms and individual domains of Hsp82 with Cdc37p
We were intrigued as to whether such automated measurement of the interaction-responsive LacZ expression in the tester yeast strain PJ69-4 could reveal the conformation of Hsp90 participating in an interaction. Can the technique provide in vivo evidence for models of the chaperone cycle built on in vitro studies? We focused on the Hsp82-BD–AD-Cdc37p interaction because the direct binding of Hsp90 to Cdc37p is already well established (Abbas-Terki et al 2001; Siligardi et al 2002; Roe et al 2004) and the AD-Cdc37p prey fusion is largely functional in yeast (cells expressing this fusion as their sole Cdc37p being slightly temperature sensitive but otherwise normal in growth; S. Millson, unpublished data).
A pronounced conformation change is induced by adenosine triphosphate (ATP) binding to the N-terminal domains of the 2 Hsp90 molecules in the Hsp90 dimer. This appears to be driven by the self-association of a hydrophobic surface, exposed when a “lid” segment closes over the bound ATP (Prodromou et al 2000). The progression of Hsp90 from its “relaxed” conformation to this “tight” complex where the N-domains are associated is an integral part of the chaperone cycle and essential for client protein activation. In vitro studies indicate that Cdc37p, an adaptor protein that facilitates the interaction of protein kinase clients with the Hsp90 machine (Stepanova et al 1996; Lee et al 2002), only binds to the relaxed, non–ATP-bound conformation of Hsp90 (Siligardi et al 2002; Roe et al 2004).
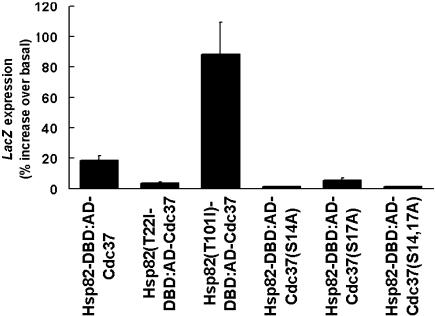
This ATP-induced switching of Hsp90 from its relaxed to its tight conformation is facilitated by the T22I mutation in yeast Hsp82 but hindered sterically by other mutations such as T101I (Prodromou et al 2000). We introduced these 2 mutations into the Hsp82-BD bait fusion (see Methods). We then measured their effects on the Hsp82-BD–AD-Cdc37p interaction in the 2-hybrid system. The T101I mutation substantially reinforced, whereas the T22I mutation greatly weakened, Hsp82-BD–AD-Cdc37p interaction (Fig 4). This result can be expected if Cdc37p binds to Hsp90 only when the latter is in its relaxed conformation. The data, therefore, provides in vivo corroboration that Cdc37p binds to the relaxed, non–ATP-bound state of Hsp90, a conclusion that was previously based only on the data of in vitro studies (Siligardi et al 2002; Roe et al 2004).
Fig 4.
The effects of mutations in the Hsp82-BD “bait” or AD-Cdc37p “prey” fusions on the strength of Hsp82-BD–AD-Cdc37p interaction, measured at 22°C (relative to LacZ expression values of the control PJ69-4 cells containing empty pBDC and the plasmid for expression of the corresponding AD-Cdc37p fusion)
Hsp90 binds a central region of Cdc37p, but it is the N-terminus of Cdc37p that is most conserved in sequence and the sole region necessary for the essential function of Cdc37p in yeast (see Lee et al 2002; MacLean and Picard, 2003; Roe et al 2004). It is thought that this N-terminal region interacts with protein kinases so as to facilitate their interaction with Hsp90 (Stepanova et al 1996). Conservative mutation of 2 serines within this conserved N-terminus (S14A, S17A) severely impairs Cdc37p function (Bandhakavi et al 2003). In parallel with other evidence, this indicates that the efficient operation of Cdc37p requires the phosphorylation of these serines in vivo by protein kinase CKII (Bandhakavi et al 2003). We tested how the same conservative mutations would affect the Hsp82-BD interaction with the AD-Cdc37p prey fusion. The S14A, S17A mutations, introduced either singly or in combination into the Cdc37p-coding part of the AD-Cdc37p fusion, substantially abolished any 2-hybrid interaction with Hsp82-BD (Fig 4). This indicates that Cdc37p, a cochaperone that actively participates in regulation of the Hsp90 chaperone cycle (Siligardi et al 2002), only binds to Hsp90 in vivo when these residues are capable of phosphorylation. This phosphorylation may be needed for the loading of protein kinase clients onto Cdc37p, before the association with Hsp90 in its non– ATP-bound relaxed state (Siligardi et al 2002; Bandhakavi et al 2003; Roe et al 2004).
Genome-wide screens for the interactions of different Hsp90 system cochaperone baits
Several cochaperone proteins assist the activation of client proteins by Hsp90. In many cases these may increase overall efficiency of Hsp90 function, in part, by stabilizing the discrete multiprotein complexes that are intermediates of the Hsp90 chaperone cycle. Cochaperones also alter several of the properties of Hsp90, including its ATPase activity, its conformation, and its interaction with substrates (Prodromou et al 1999; Young and Hartl, 2000; Barral et al 2002; Davies et al 2002; Panaretou et al 2002; Pratt and Toft 2003).
At least 9 cochaperones have now been firmly linked to Hsp90 function in yeast (Sti1p-p60-Hop, Cdc37p-p50, Cns1p, Sba1p(p23), Cpr6p, Cpr7p, Sse1p, Hch1p, and Aha1p (Chang et al 1997; Dolinski et al 1997; Liu et al 1999; Panaretou et al 2002; Siligardi et al 2002; Tesic et al 2003). Sti1p, Cpr6-7p, Cdc37p, and Sba1p correspond to the Hop, cyclophilin-40, p50Cdc37, and p23 of higher organisms. A few of these cochaperones are essential (eg, Cdc37p and Cns1p), though the loss of some (eg, the major Cpr6p cyclophilin) does not appear to be associated with any obvious phenotype. Losses of other cochaperones exert intermediate effects (eg, losses of the Cpr7p cyclophilin, Sti1p, or Sba1p(p23) result in temperature-sensitive growth) (Duina et al 1996; Chang et al 1997; Bohen, 1998; Dolinski et al 1998; Marsh et al 1998; Tesic et al 2003). Such mild phenotypic effects are often enhanced with multiple cochaperone defects (eg, combinatorial losses of both Sti1p and Sba1p, or both Hch1p and Aha1p, generate stronger temperature-sensitive phenotypes than are seen with the corresponding single gene deletes [Fang et al 1998; Panaretou et al 2002]).
Strain PJ469α was engineered to express N-terminal fusions of the Gal4p BD to the above cochaperones. These fusions did not self-activate except in the case of Cpr6p and Cpr7p, where it was necessary to construct the corresponding C-terminal BD fusions (Cpr6-BD; Cpr7-BD). Cells of strain PJ694α expressing these BD fusions were then mated to the array of AD-yeast protein fusions expressed in the strain of opposite mating type (PJ469a) (Uetz et al 2000). The diploids, now containing both BD- and AD-fusions, were next selected on medium lacking tryptophan and leucine. They were then transferred to medium without tryptophan, leucine, or histidine but containing 1, 2, or 3 mM 3-AT. After 10 days at 30°C, the colonies positive for activation of the reporter HIS3 gene were identified from their positions on the arrays. Duplicate matings of each cochaperone bait to the AD-yeast protein fusion array and then selection for HIS3 activation were performed, with the positive interacting partners being the double hits identified in both the identical screens.
Figure 2 summarizes the relatively small number of interactions detected for these cochaperone bait fusions on the basis of such HIS3 activation, together with their relative strengths. Table 1 in turn lists the interactors, their annotated functions in the Saccharomyces Genome Database (SGD) (www.yeastgenome.org), and the apparent strength of each interaction. It should be noted that, for several cochaperone baits, no interactors in the array were detected on the basis of HIS3 activation (BD-Sti1p, Sti1p-BD, Cpr6-BD, Cpr7-BD, BD-Sse1p, BD-Aha1, and BD-Hch1p) (no diploids growing in the presence of 1 mM or more 3-AT). With respect to the positive interaction data (Fig 2), Cdc37p forms a distinct “outlier” in the interaction diagram relative to all the other cochaperones. This BD-Cdc37p bait strongly selected a replication origin complex component, Mcm6p, and a transcriptional regulator, Spt2p. Therefore, we prepared BD-Mcm6p and BD-Spt2p baits as well as a BD-Gcn2p bait (the Gcn2p eIF2α kinase being a known Hsp90 client protein in yeast [Donze and Picard 1999]), all of which were also screened against the AD-fusion array (Fig 2). BD-Mcm6p strongly selected AD-Cdc37p, AD-Spt2p, the ribonucleic acid (RNA) polymerase II holoenzyme component, AD-Ask10p, and the nuclear pore complex component, AD-Nup157. In contrast, BD-Gcn2p only showed relatively weak interactions, although one of these was to AD-Gcn4p (Fig 2).
An appreciable fraction of these interactions of cochaperone bait fusions represent interactions between 2 Hsp90 system cochaperones. Thus, p23 (the BD-Sba1p bait) displayed strong interactions with no less than 2 TPR-domain proteins (AD-Cpr6p, AD-Ppt1p) and weak interaction with yet another (AD-Cns1p). The BD-Sba1p and BD-Cns1p baits in turn both selected 2 important components of the yeast transcriptional machinery, AD-Adr1p and AD-Taf1p. BD-Cns1p also showed a moderate strength interaction with AD-Tid3p (Ndc80p), a protein that interacts with Nuf2p within a 190-kDa complex important in kinetochore assembly and spindle checkpoint signaling (McCleland et al 2003).
DISCUSSION
This study set out to investigate the potential uses and drawbacks of the 2-hybrid approach when analyzing the interactions of the Hsp90 chaperone system. We commenced by monitoring HIS3 activation, a system that detects relatively stable interactions. Just a few, relatively weak Hsp90 interactions (interactions of the Hsc82-BD and Hsp82-BD baits) with the yeast proteome were detected by this approach (Fig 2; Table 1). Three of the latter interactions were with cochaperones already known either to bind Hsp90 directly (Aha1p [Panaretou et al 2002], Cdc37p [Abbas-Terki et al 2001; Siligardi et al 2002; Roe et al 2004]) or influence Hsp90 function genetically (Hch1p [Nathan et al 1999]). This is consistent with these being bona fide interactions of Hsp90. The remaining 2 Hsp82-Hsc82 interactions were with the products of ORFs of unknown function (YLR282c and YOL029c; Figs 2 and 3). Of these, the Hsp82-BD–AD-Ylr282c interaction may not be of functional relevance because while this work was in progress, YLR282c was reannotated as a “questionable” ORF in SGD.
Screening for HIS3 activation also uncovered a few candidate associations of Hsp90 system cochaperones with the yeast proteome (Fig 2; Table 1). An appreciable proportion of these are with other Hsp90 system cochaperones, again suggesting that some may be bona fide and paving the way for further experimentation, by other approaches for analyzing protein interactions, to determine if they represent cochaperone complexes of functional significance. It should be noted though that several of the well-established interactions of Hsp90 and its cochaperones (eg, interactions of Sti1p, a known interactor with Hsp90 [Chang et al 1997; Prodromou et al 1999]) were not detected by this 2-hybrid screening approach. It is not possible to attribute any significance to these negative findings of an in vivo screening approach because there are so many reasons as to why false negatives should arise (eg, the fusion protein is either misfolded, unstable, toxic, inactive, mislocalized, or lacking essential posttranslational modifications).
In considering those potential interactions of Hsp90 system cochaperones that were uncovered (Fig 2; Table 1), it is important to remember that these may in many cases not be direct but instead bridged through other proteins. Thus, the data are indicative of stable in vivo BD-Sba1p–AD-Cpr6p; BD-Sba1p–AD-Ppt1p and BD-Sba1p– AD-Cns1p associations. Many of these associations were, though, not apparent when the AD and BD of the respective fusions were transposed (eg, the Cpr6-BD and BD-Cns1p baits did not select AD-Sba1p). The BD-Sba1p and BD-Cns1p baits both selected components of the transcriptional machinery (interacting with AD-Adr1p and AD-Taf1p). Adr1p, one of the major transcription factors of yeast, is an important controller of several of the genes expressed in the absence of glucose repression (notably genes for alcohol dehydrogenase 2, peroxisomal proteins, as well as genes required for ethanol, glycerol, and fatty acid use [Denis and Audino 1991]). Taf1p is the 120-kDa component of TFIID, a complex that contains the TATA box–binding protein (TBP) and 12 TBP-associated factors (TAFs). As part of the general transcription machinery for RNA polymerase II, Taf1p binds to TBP directly (Takahata et al 2003) so as to generate a TFIID component that also possesses histone acetyltransferase and protein kinase activities. As part of the transcriptional machinery for ribosomal DNA, Taf1p also binds the activator UBF (Lin et al 2002). The Adr1p and Taf1p interactions in Fig 2 are therefore indicative of an involvement of Hsp90 system cochaperones in transcriptional control and chromatin remodeling. BD-Cns1p selected, in addition to Adr1p and Taf1p, Tid3p. The latter is part of a 190-kDa complex that, with Nuf2p, associates with the outer kinetochore from prometaphase through the anaphase stages of the cell cycle. This complex appears to have conserved roles in kinetochore assembly, chromosome congression, and spindle checkpoint signaling. In its absence, spindle checkpoint signaling is defective (McCleland et al 2003). Hsp90 is known to be involved in kinetochore-microtubule attachment and maintenance of the spindle assembly checkpoint, with Cdc37p-Hsp90 being required in the functioning of the aurora kinase regulators of centrosome function, bipolar spindle assembly, and chromosome segregation (Lange et al 2002).
Many of the protein associations of a chaperone cycle are transient and therefore not ideally suited for analysis by a system designed to detect stable protein-protein interaction. Interactions of Hsp90 client proteins probably mostly fall into this category. Nevertheless, this study shows that it is possible to adapt the 2-hybrid system to monitor the effects of stress on interaction in vivo (Fig 3) and also to obtain in vivo evidence for whether a mutation in a chaperone component might be acting to slow or accelerate a particular stage of a chaperone cycle. Such mutations might help to “freeze” or “unfreeze” particular intermediate complexes, thereby either stabilizing or destabilizing transient protein associations. We show that a mutation in Hsp82 that disfavors the ATP-induced conformational switch in the Hsp90 chaperone cycle (T101I) substantially reinforces Hsp82-BD–AD-Cdc37p interaction, whereas another mutation favoring this conformational switch (T22I) has the opposite effect (Fig 4). This effectively provides in vivo evidence that Cdc37p is interacting with the relaxed, non–ATP-bound conformation of Hsp90, thus confirming the conclusions emanating from recent in vitro studies (Siligardi et al 2002) and a crystal structure (Roe et al 2004). There may be other instances where the 2-hybrid system can provide useful evidence of the chaperone conformation or cochaperone phosphorylation state (Fig 4) engaging in an interaction in vivo. We are currently investigating whether the D79N and E33A mutations in the Hsp82-BD bait (mutations that substantially abolish either ATP binding to Hsp90 or the ATPase of Hsp90, respectively) affect interactions of this fusion with the yeast proteome in the 2-hybrid system.
Acknowledgments
We are indebted to Drs. C. Glover, G. Cagney, T. Hazbun, and S. Fields for their kind assistance in providing materials for this project and also to S. Fields for hosting the visit of S.M. to his laboratory. This work was supported by BBSRC Grant 31/C13023.
REFERENCES
- Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001.0270-7306(2001)021<7569:HIWHCI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A, Gottschling DE, Kaiser CA, and Stearns T 1997 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bandhakavi S, McCann RO, Hanna DE, Glover CV. A positive feedback loop between protein kinase CKII and Cdc37 promotes the activity of multiple protein kinases. J Biol Chem. 2003;278:2829–2836. doi: 10.1074/jbc.M206662200.0021-9258(2003)278<2829:APFLBP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–771. doi: 10.1126/science.1066648.0193-4511(2002)295<0669:ROTMAP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bohen SP. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330.0270-7306(1998)018<3330:GABAOP>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919.0270-7306(1989)009<3919:HIAEPT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Nathan DF, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318.0270-7306(1997)017<0318:IVAOTH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:1–39. doi: 10.1016/s0163-7258(98)00013-8.0163-7258(1998)079<0001:TKMCFS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Davies TH, Ning YM, Sanchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200.0021-9258(2002)277<4597:ANFSIA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Denis CL, Audino DC. The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet. 1991;229(3):395–399. doi: 10.1007/BF00267461.0026-8925(1991)229<0395:TCSASP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093.0027-8424(1997)094<13093:ACAFBP>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski KJ, Cardenas ME, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18:7344–7352. doi: 10.1128/mcb.18.12.7344.0270-7306(1998)018<7344:CEAESH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O, Picard D. Hsp90 binds and regulates Gcn2, the ligand-inducible kinase of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 1999;19:8422–8432. doi: 10.1128/mcb.19.12.8422.0270-7306(1999)019<8422:HBARGT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA, Chang HC, Marsh JA, Lindquist S, Gaber RF. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713.0193-4511(1996)274<1713:ACFIHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727.0270-7306(1998)018<3727:SEAYHC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998.1092-2172(1998)062<1264:MKPITY>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N, MacLean M, Hatzianthis K, Panaretou B, Piper PW. Increasing the stress resistance of Saccharomyces cerevisiae, by the overactivation of the heat shock response that results from Hsp90 defects, does not extend replicative life span but can be associated with a slower chronological ageing of nondividing cells. Mol Gen Genomics. 2001;265:258–263. doi: 10.1007/s004380000409.1617-4615(2001)265<0258:ITSROS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107.0021-9533(2002)115<3983:TUFOMC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425.0016-6731(1996)144<1425:GLAAHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Rebollo E, Herold A, Gonzalez C. Cdc37 is essential for chromosome segregation and cytokinesis in higher eukaryotes. EMBO J. 2002;21:5364–5374. doi: 10.1093/emboj/cdf531.0261-4189(2002)021<5364:CIEFCS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Rao J, Fliss A, Yang E, Garrett S, Caplan AJ. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J Cell Biol. 2002;159:1051–1059. doi: 10.1083/jcb.200210121.0021-9525(2002)159<1051:TCPKDI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Tuan J, Scalia P, Bui T, Comai L. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr Biol. 2002;12:2142–2146. doi: 10.1016/s0960-9822(02)01389-1.0960-9822(2002)012<2142:TCCRFT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ling M, Merante F, Robinson BH. A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 1995;23:4924–4925. doi: 10.1093/nar/23.23.4924.0305-1048(1995)023<4924:ARARDP>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654.0021-9258(1999)274<26654:TYHFMS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071.1059-1524(1998)009<3071:HIRFPS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean M, Picard D. Cdc37 goes beyond Hsp90 and kinases. Cell Stress Chaperones. 2003;8:114–119. doi: 10.1379/1466-1268(2003)008<0114:cgbhak>2.0.co;2.1466-1268(2003)008<0114:CGBHAK>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Kalton HM, Gaber RF. Cns1 is an essential protein associated with the hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Delta cells. Mol Cell Biol. 1998;18:7353–7359. doi: 10.1128/mcb.18.12.7353.0270-7306(1998)018<7353:CIAEPA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511.0021-9258(2000)275<1511:RMFMOS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903.0890-9369(2003)017<0101:THCNCI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson SM, Truman A, Piper PW. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. BioTechniques. 2003;35:60–64. doi: 10.2144/03351bm06.0736-6205(2003)035<0060:VFNOCP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. Identification of SSF1, CNS1, and HCH1 as multicopy suppressors of a Saccharomyces cerevisiae Hsp90 loss-of-function mutation. Proc Natl Acad Sci U S A. 1999;94:1409–1414. doi: 10.1073/pnas.96.4.1409.0027-8424(1999)094<1409:IOSCAH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, and Meyer P. et al. 2002 Activation of the ATPase activity of Hsp90 by AHA1 and other co-chaperones. Mol Cell. 10:1307–1318. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0.0959-440X(2000)010<0046:SAIVFO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491.1420-682X(2002)059<1640:HPACFF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, and Chohan S. et al. 2000 The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19:4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, and O'Brien R. et al. 1999 Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe SM, Ali MM, Meyer P, Vaughan CK, Panaretou B, Piper PW, Prodromou C, Pearl LH. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4.0092-8674(2004)116<0087:TMOHRB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, Piper PW, Pearl LH, Prodromou C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem. 2002;26:20151–20159. doi: 10.1074/jbc.M201287200.0021-9258(2002)026<20151:ROHAAB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491.0890-9369(1996)010<1491:MPIAPK>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Takahata S, Ryu H, Ohtsuki K, Kasahara K, Kawaichi M, Kokubo T. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J Biol Chem. 2003;278:45888–45902. doi: 10.1074/jbc.M306886200.0021-9258(2003)278<45888:IOANTE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tesic M, Marsh JA, Cullinan SB, Gaber RF. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J Biol Chem. 2003;278(35):32692–32701. doi: 10.1074/jbc.M304315200.0021-9258(2003)278<32692:FIBHAT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Uetz P, Cagney G, Lockshon D, Qureshi-Emili A, Conover D, Johnston M, Fields S. A protein array for genomewide screens of protein-protein interactions. Nature. 2000;403:623–627. doi: 10.1038/35001009.0028-0836(2000)403<0623:APAFGS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Young JC, Hartl FU. Polypeptide release by hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930.0261-4189(2000)019<5930:PRBHIA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–274. doi: 10.1083/jcb.200104079.0021-9525(2001)154<0267:HASBEP>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]