Abstract
The genes encoding the aflatoxin biosynthetic pathway enzymes have been localized as a cluster to a 75-kb DNA fragment. The enzymatic functions of the products of most of the genes in the cluster are known, but there are a few genes that have not yet been characterized. We report here the characterization of one of these genes, a gene designated aflJ. This gene resides in the cluster adjacent to the pathway regulatory gene, aflR, and the two genes are divergently transcribed. Disruption of aflJ in Aspergillus flavus results in a failure to produce aflatoxins and a failure to convert exogenously added pathway intermediates norsolorinic acid, sterigmatocystin, and O-methylsterigmatocystin to aflatoxin. The disrupted strain does, however, accumulate pksA, nor-1, ver-1, and omtA transcripts under conditions conducive to aflatoxin biosynthesis. Therefore, disruption of aflJ does not affect transcription of these genes, and aflJ does not appear to have a regulatory function similar to that of aflR. Sequence analysis of aflJ and its putative peptide, AflJ, did not reveal any enzymatic domains or significant similarities to proteins of known function. The putative peptide does contain three regions predicted to be membrane-spanning domains and a microbodies C-terminal targeting signal.
Aflatoxins are toxic polyketide secondary metabolites produced by Aspergillus flavus and Aspergillus parasiticus. Both A. flavus and A. parasiticus infect corn, peanut, cottonseed, and tree nuts (21). In efforts to control preharvest aflatoxin contamination of commodities, research has focused on understanding the biosynthesis of aflatoxin. Although aflatoxin has no known role in the ecology of the fungus, the pathway is induced and appears to be under tight regulatory control. Thus, understanding the ecology of the fungus and the genetic cues that stimulate aflatoxin production may be the key to elimination of aflatoxin in the field.
The biosynthetic pathway for aflatoxin has been studied for a number of years, and a biosynthetic scheme that is accepted by most researchers has been proposed (2, 9, 28). Recent research has focused on the molecular genetics of aflatoxin formation and on the identification of biosynthetic genes. Several genes whose functions are known or implied have been characterized. These genes include nor-1 (4), norA (3), ver-1 (26), omtA (33), vbs (25), avnA (35), and avf-1 (23). Recently, three genes which are involved in the early steps of the pathway, pksA, fas1A, and fas2A, have been characterized (6, 16, 28). In addition to these biosynthetic genes, a pathway-specific regulatory gene, aflR, has been identified in A. flavus and A. parasiticus and characterized (5, 22, 31). This gene is required for transcription of all of the known pathway genes. It is now clear that most, if not all, of the pathway genes and the pathway-specific regulatory gene are clustered in 75 kb of DNA (25, 27, 32, 34).
In addition to the genes in the cluster whose functions have been characterized, there are additional genes of unknown function whose transcription coincides with aflatoxin biosynthesis. One such gene, aflJ, is adjacent to the pathway regulatory gene, aflR. These two genes are transcribed in opposite directions and share a 737-bp intergenic region from their translational start sites. Because of the profile of aflJ transcription and the location of aflJ adjacent to aflR in the gene cluster (22), we postulated that aflJ may be involved in aflatoxin biosynthesis, as well as in aflatoxin biosynthetic pathway regulation. The objectives of this study were to characterize aflJ and to determine its role in aflatoxin biosynthesis.
MATERIALS AND METHODS
Fungal strains and media.
The aflatoxin-producing strain A. flavus 86 (w arg7) = (ATCC 60041) (20) was provided by S. V. Peterson, National Center for Agricultural Utilization Research, Peoria, Ill. Strain 86-10 (w arg pyrG) was obtained by UV mutagenesis of A. flavus 86 by using methods previously described by Woloshuk et al. (29). Colonies mutated at the pyrG locus were directly selected by plating conidia onto YUG medium (0.5% yeast extract, 2.0% glucose, 10 mM uridine, 2% agar) containing 1 mg of fluoroorotic acid per ml (29). Colonies resistant to 5-fluoroorotic acid were subsequently characterized to confirm their uracil auxotrophy. One strain, designated 86-10, was selected for this study. All fungal strains were stored as lyophilized cultures. Fungal strains were cultured on potato dextrose agar (Difco Laboratories, Detroit, Mich.) and Czapek solution agar for production of conidia. The media were supplemented as needed with 10 mM arginine and 10 mM uracil.
Aflatoxin analysis.
Coconut agar was used to screen for presumptive aflatoxin production (7). The presence of aflatoxin production on this medium was determined by the bright blue fluorescence of aflatoxin when it was exposed to UV light (29). Aflatoxin concentrations were determined by growing the fungus in liquid culture by using sucrose low-salts (SLS) medium (22) or potato dextrose broth (PDB) and assaying filtrates by an enzyme-linked immunosorbent assay (ELISA). Aflatoxin B1 monoclonal antibodies and aflatoxin B1-horseradish peroxidase conjugates were purchased from Sigma Chemical Co. (St. Louis, Mo.). Peptone mineral salts (PMS) medium was used as a nonconducive medium for aflatoxin production (30). The abilities of strains 86-10 and 86D to produce aflatoxin were compared by growing them on PDB at 28°C. Medium and tissue were harvested after 5 and 11 days. The culture methods used for 86D transcript analysis were the culture resuspension methods described by Flaherty et al. (10). Briefly, all cultures were grown on PMS medium for 3 days and resuspended in either SLS medium to stimulate aflatoxin production or PMS medium, which served as a negative control. Tissue and media were collected at 6-h intervals for a 24-h period after resuspension.
Isolation and analysis of DNA and RNA.
Total genomic DNA was isolated from fungal tissue as previously described (29). All plasmid constructs were purified by using spin columns obtained from 5 Prime 3 Prime Inc. (Boulder, Colo.). Zeta Probe membrane filters obtained from Bio-Rad (Richmond, Calif.) were used for Southern blot, RNA slot blot, and Northern hybridization analyses. Probes for aflJ, omtA, pksA, and nor-1 were made by using an oligolabeling kit obtained from Pharmacia LKB Biotechnology (Piscataway, N.J.). RNAs were isolated from lyophilized mycelia of 86-10, an aflatoxin-producing strain, and 86D, a disrupted strain, by using Genosys RNA isolator (Genosys Biotechnology’s Inc). For RNA slot blot hybridization analysis, 20 μg of RNA was loaded directly onto a Zeta Probe membrane filter and hybridized with 32P-labeled DNA probes. A PCR in which genomic DNA was used as the template was performed to determine the presence of constructs in 86C, a complemented strain. Strains 86-10 and 86D served as controls.
Cloning and sequencing of aflJ.
Both strands of a 1.7-kb (1,719-bp) region of GAP 20 containing aflJ (22), three partial cDNA clones, and a full-length cDNA clone were sequenced by using a Circumvent Thermal Cycle Dideoxy DNA sequencing kit (NE Biolabs) with primers that spanned the length of the sequence. A DNA analysis was performed by using the MacDNAsis Pro 3.5 software. Additional database searches were performed by using programs available on the Expasy Molecular Biology page (expasy.hcuge.ch/www/tools.html) on the worldwide web.
Plasmid constructs and fungal transformation.
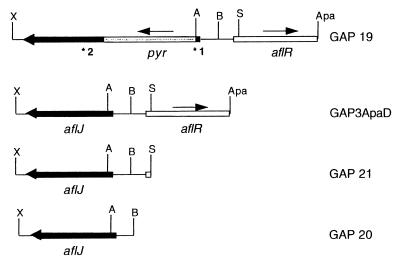
Transformations were carried out by previously described methods (29). The plasmid constructs used are shown in Fig. 1. A 4.6-kb XbaI-ApaI fragment from cosmid B9 (22), containing both the aflJ gene and the aflR gene, was subcloned into Bluescript SK to create plasmid GAP3ApaD. GAP3ApaD was used to make all subsequent constructs. An aflJ disruption vector (GAP 19) was made by inserting a PvuI-SmaI fragment containing the pyr4 gene of Neurospora crassa (from plasmid PRG1 29) into a single AspI site found in the open reading frame of aflJ. The direction of the pyr4 gene was determined by restriction mapping. An additional vector (GAP 21) was used to complement the aflJ disrupted strain. This vector was created by deleting the SmaI-ApaI fragment containing the open reading frame of aflR from GAP3ApaD, which left the coding region of aflJ and the entire intergenic region between aflJ and aflR intact. Vector GAP 20 containing the coding region of aflJ and 324 bp of the putative promoter region was used to sequence aflJ. GAP 20 was created by deleting a 2,162-bp BamHI-ApaI fragment containing aflR from GAP3ApaD. The DNA primers used in this study are shown in Fig. 1. The sequences of primers *1 and *2 were 5′ AGTCAAAGGTTGAATACC 3′ and 5′ GCTCAGCCATGACCTTGACTG 3′, respectively. Taq DNA polymerase was purchased from Boehringer Mannheim (Indianapolis, Ind.).
FIG. 1.
Plasmid constructs and primers. GAP3ApaD is an XbaI-ApaI fragment of cosmid B9 (22) cloned into Bluescript SK+ and contains the open reading frames for aflJ and aflR. GAP 19, the construct used for gene disruption, contains the pyr4 gene of N. crassa inserted into a single AspI site in the open reading frame of aflJ. GAP 21 contains aflJ and the entire intergenic region between aflJ and aflR and was derived from GAP3ApaD by deletion of the SmaI-ApaI fragment containing aflR. GAP 20 contains aflJ and 324 bp of the intergenic region between aflR and aflJ and was created by deleting a BamHI-ApaI fragment from GAP3ApaD. The sites of the two primers used for PCR analysis are indicated by *1 and *2. A, ApaI; B, BamHI; S, SmaI; X, XbaI.
Metabolite conversion studies.
Aflatoxin pathway intermediates were converted by whole fungal cells by using previously described methods (22). Cultures of wild-type strain 86-10 and the aflJ disrupted strain, 86-D, were amended with the pathway intermediates norsolorinic acid, sterigmatocystin, and O-methylsterigmatocystin, and then an assay for aflatoxin accumulation and the production of pathway intermediates in which thin-layer chromatography was used was performed.
Nucleotide sequence accession number.
The nucleotide sequence of A. flavus aflJ has been deposited in the GenBank database under accession no. AF077975.
RESULTS
Disruption of the aflJ locus.
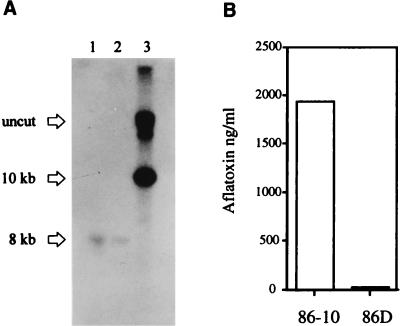
GAP 19, which contains a functional pyr4 gene inserted in the open reading frame of aflJ, was linearized with XbaI and transformed into strain 86-10. Uracil prototrophs were selected and screened for their ability to produce aflatoxin. Thirty non-aflatoxin-producing transformants were examined by Southern blot hybridization analysis for the presence of GAP 19. Figure 2A shows a DNA hybridization blot obtained with strains 86, 86-10, and 86-D, a representative GAP 19 transformant that did not produce aflatoxin. Hybridization of a labeled aflJ probe to BamHI-digested genomic DNA (BamHI cuts the disruption vector once but does not cut within aflJ) revealed different hybridization patterns. Hybridization of the aflJ probe to DNA from strains 86 and 86-10 revealed a single hybridizing fragment at 8 kb (Fig. 2A, lanes 1 and 2). In contrast, hybridization of DNA from 86D revealed no 8-kb fragment; instead, there was a 10-kb fragment not present in strain 86 or 86-10 (Fig. 2A, lane 3). This 10-kb fragment was the predicted size of the disruption construct GAP 19 if it was successfully inserted at the native site in the fungal chromosome by double crossover replacement.
FIG. 2.
(A) Southern analysis of strains 86 (lane 1), 86-10 (lane 2), and 86D (lane 3). (B) Aflatoxin concentrations in strains 86-10 and 86D. Cultures were grown for 5 days in PDB at 28°C. Genomic DNA was extracted and digested with BamHI and probed by using an aflJ radioactive probe. The aflatoxin concentrations in culture media were determined by ELISA.
Analysis of aflJ mutant. Aflatoxin production and transcript accumulation in the mutant strain (86-10) followed the typical profile observed in wild-type strains (11). Aflatoxin appeared in the cultures after 6 h, and the aflatoxin concentration peaked at 12 and 18 h and then declined. No aflatoxin was detected in cultures grown on the nonconducive medium, PMS medium. Transcripts of several aflatoxin genes, including aflR, omtA, and fas-1, appeared after 12, 18, and 24 h under aflatoxin-inducing conditions in SLS medium, as expected (11). ELISA analysis of filtrates from strains grown on medium conducive for aflatoxin formation showed that the disruptant strain produced only 20 ng of aflatoxin per ml, whereas strain 86-10 produced 2,000 ng of aflatoxin per ml (Fig. 2B). There were no obvious morphological differences between 86-10 and 86D, except that 86D produced a large number of sclerotia in culture.
Complementation of the aflJ locus in the knockout strain.
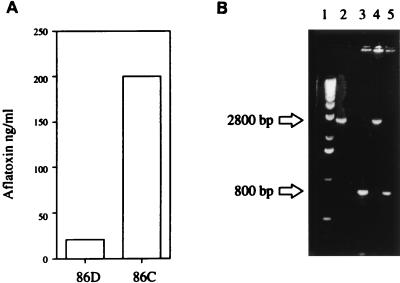
To confirm that the lack of aflatoxin production by 86D was due to disruption of aflJ, 86D was cotransformed with a functional copy of aflJ (GAP 21) and a 3.4-kb DNA fragment containing the arg7 gene of A. flavus (12). Thirty-seven transformants were screened for arginine prototrophy and aflatoxin production. Nine transformants that were highly fluorescent on coconut agar were grown in PDB for 7 days, and the aflatoxin concentrations were determined by ELISA. Transformant 86C and strain 86D were grown in PDB for 5 days and then assayed for aflatoxin production (Fig. 3A). Strain 86D produced only 20 ng of aflatoxin per ml, but the complemented strain, 86C, produced 200 ng/ml.
FIG. 3.
(A) Aflatoxin concentrations in strains 86D and 86C. (B) PCR analysis of strains 86-10 (lane 3), 86D (lane 4), and 86C (lane 5). Lane 1 contained a molecular weight marker, and lane 2 contained control plasmid GAP 19. Cultures were grown in PDB at 28°C for 5 days, and aflatoxin concentrations were determined by ELISA. Genomic DNA was extracted and used as a template in PCR performed with probes *1 and *2 (Fig. 1).
PCR analysis was used to confirm the presence of GAP 21 in strain 86C. Primers were designed to flank the AspI site (the insertion site for pyr4 in the disruption construct) in aflJ (Fig. 1). In the native gene, the primer sites are 800 bp apart, but in the disruption construct the primer sites are 2,800 bp apart. Figure 3B shows the fragment sizes of the PCR products obtained from DNA preparations of 86-10, 86D, and 86C. Strain 86-10 (lane 3) produced a predicted single fragment at 800 bp that was indicative of a wild-type copy of aflJ. Strain 86D (lane 4) produced a 2,800-bp fragment, the predicted size of the disruption construct, and did not produce the 800-bp fragment. The complemented disrupted strain, 86C (lane 5), produced the 800-bp fragment but not the 2,800-bp fragment. Apparently, GAP 21 replaced the disrupted copy of aflJ in strain 86C. Nutritional analysis confirmed that 86C is a transformant of 86D because it is a uracil and arginine prototroph. Thus, restoration of aflatoxin production was associated with a wild-type copy of aflJ.
Intermediate feeding studies.
Cultures of strains 86-10 and 86D were fed the early pathway intermediate norsolorinic acid and the late precursors sterigmatocystin and O-methylsterigmatocystin. Strain 86-10 produced aflatoxin when it was fed any of the intermediates. In contrast, strain 86D was unable to convert any of the exogenously added intermediates to aflatoxin (data not shown). Furthermore, no colored or fluorescent pathway intermediates accumulated during thin-layer chromatography of extracts of 86D in the feeding studies. We also observed that strain 86D grown on coconut agar or in liquid media without added precursors does not accumulate any colored compounds that are conducive to aflatoxin biosynthesis. Thus, it appears that the enzymatic activities necessary to convert pathway intermediates to aflatoxin are not active in a strain with a disrupted copy of aflJ.
Transcript analysis of aflJ knockout strain 86D.
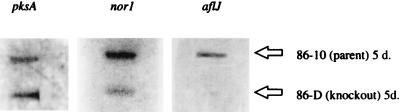
Because disruption of aflJ appeared to affect several enzymatic functions involved in aflatoxin biosynthesis, we suspected that aflJ may be involved in transcriptional control of the biosynthetic pathway. To determine the effect of aflJ disruption on transcription of the pathway genes, strains 86D and 86-10 were grown in continuous PDB cultures at 28°C, and RNA isolated from the cultures were assayed for transcripts of the early pathway genes pksA and nor-1. A slot blot hybridization analysis (Fig. 4) showed that pksA and nor-1 transcripts were present in 5-day-old cultures of 86-10 and 86D. As expected, no aflJ transcript was found in 86D. The aflatoxin concentrations in these cultures were determined; 86-10 produced 3,191 ng of alfatoxin per ml, and 86D produced 22 ng of aflatoxin per ml. These aflatoxin levels are similar to results shown in Fig. 2B.
FIG. 4.
RNA slot blot analysis of nor-1, pksA, and aflJ. Strains 86-10 and 86D were grown in PDB at 28°C for 5 days. Total RNA was extracted and probed for nor-1, pksA, and aflJ. d, days.
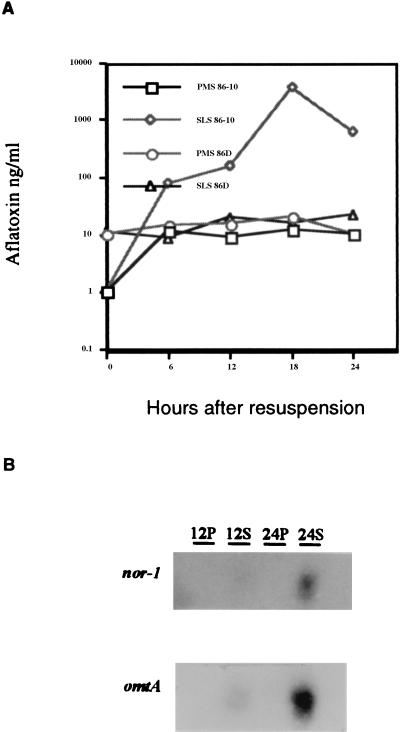
To confirm these results, aflatoxin time course and Northern analyses were performed. Strain 86D was grown under conducive and nonconducive conditions for 24 h. To ensure that conditions were conducive for aflatoxin production, 86-10 was cultured under the same conditions. Media collected at 6-h intervals were analyzed for aflatoxin production (Fig. 5A). As expected, strain 86D produced only low levels of aflatoxin. The concentrations of aflatoxin in the media did not reach levels greater than 25 ng/ml. Strain 86-10 produced high concentrations of aflatoxin under inducing conditions, as expected (Fig. 5A). Northern analysis of RNA extracted from strain 86D at 12 and 24 h revealed the presence of an early pathway gene, nor-1, and a late pathway gene, omtA (Fig. 5B).
FIG. 5.
(A) Aflatoxin B1 concentrations in A. flavus 86-10 and 86D. (B) Northern blot analysis of nor-1 and omtA in A. flavus 86D. Cultures were grown for 3 days in PMS (P) medium and resuspended in PMS medium or SLS (S) medium. Media were collected when the cultures were resuspended and every 6 h for 24 h. Media were assayed to determine aflatoxin concentrations by ELISA. Total RNA was extracted from 12- and 24-h culture samples and probed for nor-1 and omtA.
Characterization of the aflJ gene.
To determine if aflJ had similarities to previously described genes, genomic and cDNA clones were sequenced, and the DNA and putative protein sequences were compared with sequences in the database. Three partial cDNA clones, a full-length cDNA clone, and a 1.8-kb genomic fragment from GAP 20 containing aflJ were sequenced. Alignment of cDNA sequence with the genomic sequence of aflJ revealed two introns (76 and 60 bp). Both the sizes and the consensus sites of the introns are consistent with data for other known fungal introns. The introns are smaller than 100 bp, and the splice sites follow the gt...ag rule (14, 17, 24). The predicted start codon was chosen based on the long open reading frame that followed the start codon. A computer-generated protein based on this start codon was predicted to be 438 amino acids long.
Two cDNA clones that span the length of the region coding for the gene were identified. Each of these clones had a different polyadenylation cleavage site; one of these sites was at position 1832, and the other was at position 2000, suggesting that there was differential polyadenylation in this gene. Only one possible polyadenylation signal was found. The sequence ATTAAA at position 1769 exhibited homology with a human laminin A noncanonical polyadenylation signal; the position of this signal 60 bp upstream of the poly(A) tail is also consistent with the information obtained for the laminin A gene (13).
BLAST analysis of the DNA and protein sequences revealed no significant similarities to previously described GenBank entries. Several other analysis programs, such as Sbase, FastA, Blitz, and Propsearch, were also used. None of these programs identified motifs or domains that indicated that aflJ has an enzymatic function. A Prosite scan of the amino acid sequence did reveal a microbodies C-terminal targeting signal (CMTS) (1). The amino acid sequence for the CMTS is NRY, which corresponds to the consensus pattern [STAGCN]-[RKH]-(LIVMAFY] (8). An analysis of the putative peptide by TMpred revealed three regions that scored high as possible membrane-spanning regions; these regions were at amino acids 139 to 164, 232 to 252, and 306 to 326 (15). These three regions and the CMTS are the only landmarks that we could identify within the protein. A final analysis of AflJ with the program Psort revealed that AflJ may be associated with the mitochondrial outer membrane or peroxisomes (microbodies) (19), further validating the CMTS sequence. We compared aflJ in all three possible reading frames with the genes that encode several other proteins that do not exhibit high levels of sequence homology as a group but share functions, such as hydrophobins and peroxisome transporter genes. We did not find any significant relationship between these genes and aflJ.
DISCUSSION
The aflatoxin biosynthetic pathway is a well-characterized pathway of secondary metabolism. The basic biosynthetic scheme is known, and several genes involved in the biosynthetic steps have been cloned and characterized. The functions of the known genes have been determined by complementation of characterized mutants blocked in the pathway or by sequence homology with genes whose functions are known. Because the biosynthetic scheme is known, the functions of many genes in the pathway can be predicted. We were surprised to find no homology between the putative peptide of aflJ and the peptides encoded by known genes with enzymatic domains.
It is clear from the metabolite feeding studies that disruption of aflJ results in a block very early in the pathway or a block that affects many steps in the pathway. Disrupted strain 86D does not accumulate any pathway intermediates and does not convert the three known intermediates, norsolorinic acid, sterigmatocystin, and O-methylsterigmatocystin, to aflatoxin. This phenotype is very similar to the phenotype of strains with mutations at the aflR locus. A functional aflR locus is required for transcriptional activation of all of the known aflatoxin biosynthetic genes. Thus, our initial hypothesis was that aflJ interacts with aflR to transcriptionally regulate aflatoxin biosynthesis. The results of a transcript analysis of 86D, a disrupted strain, indicate that this hypothesis is not valid. Under conditions conducive for aflatoxin biosynthesis, 86D accumulates transcripts of the pathway genes pksA, nor-1, and omtA, even though no transcript of aflJ is present and no aflatoxin accumulates. Thus, aflJ does not appear to be required for the transcription of these pathway genes. If aflJ is involved in the regulation of aflatoxin biosynthesis, it does not appear to be at the level of transcription.
Our data indicate that the involvement of aflJ in aflatoxin biosynthesis is more complex. At this time we cannot assign a definitive role to AflJ in the aflatoxin biosynthetic pathway based on sequence homologies, but the predicted peptide has several interesting features. First, there are three possible membrane-spanning regions and the CMTS sequence. Second, analysis of the peptide showed that there is a moderate probability that the peptide is associated with peroxisomes or the mitochondrial outer membrane. Although the data are by no means conclusive, they suggest that aflJ may be localized to a cellular organelle.
We realize that it may be premature to speculate on the function of AflJ based on the information available from its sequence. One hypothesis is that AflJ is involved either in transmembrane transport of intermediates through intercellular compartments or in the localization of pathway enzymes to an organelle. The localization of aflatoxin biosynthesis is not known, but some enzymatic reactions during penicillin biosynthesis in Aspergillus nidulans occur in microbodies. Thus, it is easy to envision the need for a gene in the pathway which codes for the formation of the microbodies associated with this function or targets the enzymes to these organelles (18). Such gene functions could be determined by localizing AflJ in the cell and localizing pathway enzymes in strains with and without a functional copy of aflJ.
ACKNOWLEDGMENTS
A full-length cDNA for aflJ was provided by C. Brown-Jenco.
Support for this research was provided by USDA/NRI grant 9601295 and USDA-SCA grant 58-6435-075.
REFERENCES
- 1.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatnagar D, Ehrlich K C, Cleveland T E. Oxidation-reduction reactions in biosynthesis of secondary metabolites. In: Bhatnagar D, Lillehoj E B, Arora D K, editors. Handbook of applied mycology. 5. Mycotoxins in ecological systems. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 255–286. [Google Scholar]
- 3.Cary J W, Wright M, Bhatnagar D, Lee R, Chu F S. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl Environ Microbiol. 1996;62:360–366. doi: 10.1128/aem.62.2.360-366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang P K, Skory C D, Linz J E. Cloning of a gene associated with aflatoxin B1 biosynthesis in Aspergillus parasiticus. Curr Genet. 1992;21:231–233. doi: 10.1007/BF00336846. [DOI] [PubMed] [Google Scholar]
- 5.Chang P-K, Cary J W, Bhatnagar D, Cleveland T E, Bennett J W, Linz J E, Woloshuk C P, Payne G A. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang P K, Cary J W, Yu J, Bhatnagar D, Cleveland T E. The Aspergillus parasiticus polyketide synthase gene, pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol Gen Genet. 1995;248:270–277. doi: 10.1007/BF02191593. [DOI] [PubMed] [Google Scholar]
- 7.Davis N D, Iyer S K, Diener U L. Improved method of screening for aflatoxin with coconut agar medium. Appl Environ Microbiol. 1987;53:1593–1595. doi: 10.1128/aem.53.7.1593-1595.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Hoop M J, AB G. Import of proteins into peroxisomes and other microbodies. Biochem J. 1992;286:657–669. doi: 10.1042/bj2860657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutton M F. Enzymes and aflatoxin biosynthesis. Microbiol Rev. 1988;52:274–295. doi: 10.1128/mr.52.2.274-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty J E, Weaver M A, Payne G A, Woloshuk C P. A β-glucuronidase reporter gene construct for monitoring aflatoxin biosynthesis in Aspergillus flavus. Appl Environ Microbiol. 1995;61:2482–2486. doi: 10.1128/aem.61.7.2482-2486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flaherty J E, Payne G A. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microbiol. 1997;63:3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foutz K R, Woloshuk C P, Payne G A. Cloning and assignment of linkage group loci to a karyotypic map of the filamentous fungus Aspergillus flavus. Mycologia. 1995;87:787–794. [Google Scholar]
- 13.Gottschling C, Huber J, Oberbaumer I. Expression of the laminin A chain is down regulated by a non-canonical polyadenylation signal. Eur J Biochem. 1993;216:293–299. doi: 10.1111/j.1432-1033.1993.tb18144.x. [DOI] [PubMed] [Google Scholar]
- 14.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: I. R. L. Press; 1987. pp. 93–139. [Google Scholar]
- 15.Hoffman K, Stoffel W. TMBASE—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 16.Mahanti N, Bhatnagar D, Cary J W, Joubran J, Linz J E. Structure and function of fas-1A, a gene encoding a putative fatty acid synthase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1996;62:191–195. doi: 10.1128/aem.62.1.191-195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller W H, van der Krift T P, Krouwer A J J, Wosten H A B, van der Voort L H M, Smaal E B, Verkleij A J. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO. 1991;10:489–495. doi: 10.1002/j.1460-2075.1991.tb07971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakai K. Predicting various targeting signals in amino acid sequences. Bull Inst Chem Res Kyoto Univ. 1991;69:691–702. [Google Scholar]
- 20.Papa K E. Genetics of Aspergillus flavus: linkage of aflatoxin. Can J Microbiol. 1984;30:68–73. doi: 10.1139/m84-012. [DOI] [PubMed] [Google Scholar]
- 21.Payne G A. Aflatoxin in maize. Crit Rev Plant Sci. 1992;10:423–440. [Google Scholar]
- 22.Payne G A, Nystrom G J, Bhatnagar D, Cleveland T E, Woloshuk C P. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto R, Yousibova G L, Woloshuk C P. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl Environ Microbiol. 1996;62:3567–3571. doi: 10.1128/aem.62.10.3567-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seip E R, Woloshuk C P, Payne G A, Curtis S E. Isolation and sequence analysis of a β-tubulin gene from Aspergillus flavus and its use as a selectable marker. Appl Environ Microbiol. 1990;56:3686–3692. doi: 10.1128/aem.56.12.3686-3692.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva J C, Minto R E, Barry E E, Holland K A, Townsend C A. Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. J Biol Chem. 1996;271:13600–13608. doi: 10.1074/jbc.271.23.13600. [DOI] [PubMed] [Google Scholar]
- 26.Skory C D, Chang P K, Cary J, Linz J E. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1992;58:3527–3537. doi: 10.1128/aem.58.11.3527-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trail F, Mahanti N, Linz J E. Molecular biology of aflatoxin biosynthesis. Microbiology. 1995;141:755–765. doi: 10.1099/13500872-141-4-755. [DOI] [PubMed] [Google Scholar]
- 28.Trail F, Mahanti N, Rarick M, Mehigh R, Liang S H, Zhou R, Linz J E. Physical and transcriptional map of an aflatoxin gene cluster in Aspergilus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl Environ Microbiol. 1995;62:191–195. doi: 10.1128/aem.61.7.2665-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woloshuk C P, Seip E R, Payne G A, Adkins C R. Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl Environ Microbiol. 1989;55:86–90. doi: 10.1128/aem.55.1.86-90.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woloshuk C P, Payne G A. The alcohol dehydrogenase gene adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:670–676. doi: 10.1128/aem.60.2.670-676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woloshuk C P, Foutz K R, Brewer J F, Bhatnagar D, Cleveland T E, Payne G A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woloshuk C P, Prieto R. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol Lett. 1998;160:169–176. doi: 10.1111/j.1574-6968.1998.tb12907.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, Cary J W, Bhatnagar D, Cleveland T E, Keller N P, Chu F S. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3564–3571. doi: 10.1128/aem.59.11.3564-3571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Chang P-K, Wright M, Bhatnagar D, Cleveland T E, Payne G A, Linz J E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol. 1995;61:2365–2371. doi: 10.1128/aem.61.6.2365-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Chang P K, Cary J W, Bhatnagar D, Cleveland T E. avnA, a gene encoding a cytochrome P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1997;63:1349–1356. doi: 10.1128/aem.63.4.1349-1356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]