Abstract
In this study, a novel two-stage nanoparticle delivery platform was developed based on the dual functionalization of a liposome with moieties that have fundamentally different strengths of adhesion and binding kinetics. The essential concept of this system is that the nanoparticles are designed to loosely bind to the carrier cell until they come into contact with the target cell, to which they bind with greater strength. This allows the nanoparticle to be transferred from one cell to another, circulating for longer periods of time in the blood and delivering the therapeutic agent to the target circulating tumor cell. Liposomes were prepared using the lipid cake and extrusion technique, then functionalized with E-selectin (ES), anti-cell surface vimentin antibody fragments, and TRAIL via click chemistry. The binding of dual affinity (DA) liposomes was confirmed with the neutrophil-like cell line PLB985, the colorectal cancer cell line HCT116, and healthy granulocytes isolated from peripheral whole blood under physiologically relevant fluid shear stress (FSS) in a cone-and-plate viscometer. Transfer of the DA liposomes from PLB985 to HCT116 cells under FSS was greater compared to all of the control liposome formulations. Additionally, DA liposomes demonstrated enhanced apoptotic effects on HCT116 cells in whole blood under FSS, surpassing the efficacy of the ES/TRAIL liposomes previously developed by the King Lab.
Introduction
Delivering anticancer agents into the circulation is difficult as they are easily and quickly eliminated by the kidneys and can have toxic effects on other healthy cells in the circulation. The use of targeted nanoparticles as carriers for anticancer agents is a highly popular method as it enables the delivery of more concentrated doses of therapeutics to the affected area.1 This approach also helps minimize the cytotoxic effects that chemotherapies can have on healthy cells by reducing the amount of drug that reaches nonspecific targets.2 For example, the first instance of an FDA-approved nanomedicine is Doxil, the chemotherapeutic agent doxorubicin packaged in a liposome.3−5 Although such approaches have been widely pursued, nanoparticle systems still face limitations, including the brevity of circulation time and the specificity of targeting. Once nanoparticles enter the bloodstream, the body has several mechanisms to clear them, such as renal and hepatic clearance, as well as natural extravasation to other tissues.6−8 Furthermore, as nanoparticles traverse through the circulation, they encounter platelets, plasma proteins, coagulation factors, and blood cells, which may lead to immune responses and particle degradation. These challenges are magnified when the target cell is freely floating in the bloodstream, as is the case for circulating tumor cells (CTCs) in cancer.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is produced by natural killer (NK) cells and selectively kills cancer cells by binding to death receptors 4 and 5, thereby inducing apoptosis.9 A study published in 2013 by the King Lab demonstrated that fluid shear stress (FSS) sensitizes cancer cells to TRAIL-mediated apoptosis.10 Over the past 10 years, the King Lab has developed variations of a nanoparticle system aimed at inducing apoptosis in CTCs and cancer cells in the lymphatic and circulatory systems, leveraging this finding. Liposomes were first functionalized with TRAIL and E-selectin (ES), which enabled their tethering to healthy leukocytes in the circulation.11,12 Leukocytes naturally express E-selectin ligands (ESL) on their cell membrane, allowing them to adhere to the ES-rich endothelial tissue of the circulatory system during the inflammatory cascade, facilitating extravasation and trafficking to other parts of the body.13,14 This strategy extended the circulation time in the blood and resulted in high levels of colorectal cancer (CRC) cell apoptosis under physiological flow conditions. In vivo experiments using these liposomes were also successful in treating CRC. This nanoparticle design was also demonstrated to be effective against prostate and breast cancer.15−17
Two subsequent variations of the original liposomes were then successfully developed. The first variant replaced ES with an anti-NK cell antibody, aiming to target cancer cells flowing through the lymphatic system.18,19 The concept remained similar: the liposomes would passively target the cancer cells by attaching them to the surface of an NK cell. This liposome design effectively induced apoptosis in cancer cells and exhibited increased retention in the lymphatic system. The second variation focused back on the circulatory system again, but instead of leukocytes, the liposomes were designed to attach to healthy platelets.20,21 This was achieved by substituting ES with vWF-A1. This design demonstrated success in inducing apoptosis in CRC and breast cancer cells under physiological flow conditions.
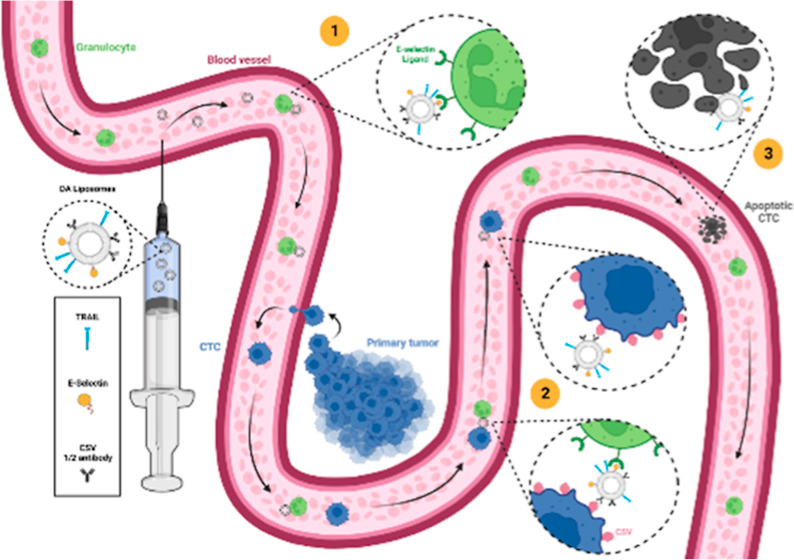
In this study, we developed a two-stage nanoparticle delivery platform relying on the dual functionalization of a liposome with moieties that exhibit fundamentally different strengths of adhesion and binding dissociation kinetics, allowing them to bind to a carrier cell (leukocyte) and then to the target cell (CTC) (Figure 1). As a proof of concept, cell surface vimentin (CSV) was chosen as the target protein due to the presence of vimentin on the cell surface of cancer cells along the EMT/MET axis, making it an attractive target for a nanoparticle-based drug delivery model for highly metastatic CTCs.22 It is selectively expressed on their surface and not on the surface of healthy cells.22,23 Additionally, it is an attractive target as it is only involved in endocytosis pathways in viral infections, making it a well-suited target for cell surface drug delivery (such as TRAIL therapies).24 The dual affinity (DA) liposomes are functionalized with TRAIL, ES, and anti-CSV half antibodies on their surface.
Figure 1.
Schematic detailing the two-step mechanism of the DA nanoparticle system. Step 1 shows the loose binding of the liposomes to healthy leukocytes (carrier cells) circulating in the bloodstream. Step 2 is the transfer of the nanoparticles from the carrier cell to the target cell (CTC). Step 3 is the delivery of the therapeutic agent to the target cell, in this case, resulting in cancer cell apoptosis. Created with http://BioRender.com.
The first stage of the delivery mechanism depends on the ES to ESL catch-slip bond, which is known to be weak and reversible.25 This bond rapidly forms within 0.5–16 s and ruptures under an applied force of about 30 pN.13,26,27 By incorporating ES molecules on the liposome, we take advantage of a healthy and abundant ligand present on the cell surface of leukocytes, employing them as our carrier cells in the bloodstream. Once the liposomes have entered the circulatory system, they tether themselves to healthy leukocytes through the ES/ESL bond. In theory, this allows the nanoparticles to be dynamically transported and protected by the carrier cell until they reach their target. The second stage of the system occurs when the leukocyte-tethered liposomes come into contact with the CTC; they detach from the leukocyte surface and bind to vimentin present on the CTC surface. Antigen-to-antibody bonds are considered strong and effectively irreversible in this setting, requiring over 1000–2000 pN of force to rupture.28,29 Once the liposome transfers from the surface of the leukocyte to the cancer cell surface, the therapeutic agent that it transports can take effect on the intended target. In this manner, CTCs can be actively targeted and eliminated while they circulate in the bloodstream, preventing the formation of metastatic lesions and disease progression.
Materials and Methods
Liposome Synthesis and Functionalization
Multilamellar liposomes were prepared via the thin lipid film method by mixing together l-α-lysophosphatidylcholine (Egg PC, 840051C-200 mg, Avanti Polar Lipids), egg sphingomyelin (Egg SM, 860061C-200 mg, Avanti Polar Lipids), ovine wool cholesterol (700000P-100 mg, Avanti Polar Lipids), 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] nickel salt (18:1 DGS-NTA(Ni), 860061C-200 mg, Avanti Polar Lipids), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide (polyethylene glycol)-2000] (DSPE-PEG(2000) Maleimide, 880126C-25 mg, Avanti Polar Lipids), and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindo-carbocyanine Perchlorate (DiI, D282, Thermo Fisher). The molar ratios were 47.5:28:20:2:2:0.5% for Egg PC/Egg SM/Cholesterol/DGS-NTA(Ni)/DSPE-PEG(2000) Maleimide/DiI.11,19 The lipid contents of the liposomes can be found in Table 1.
Table 1. Lipid Content of Fluorescent Liposomes.
| EggPC | EggSM | cholesterol | DGS-NTA(Ni) | DSPE PEG(2000) maleimide | DiD | |
|---|---|---|---|---|---|---|
| molar ratios (%) | 47.5 | 28 | 20 | 2 | 2 | 0.5 |
| volumes (μL) | 146 | 78.8 | 30.9 | 8.5 | 58.8 | 4.7 |
The solution was dried under vacuum overnight, and then the lipid cake was rehydrated in 1 mL of HBSS with Ca2+ and Mg2+ (14025–092, Gibco). Resulting multilamellar liposomes were extruded through membranes with pore sizes first of 400 nm (Nucleopore Track-Etch Membrane 0.4 μm, 800282, Whatman), then 200 nm (Nucleopore Track-Etch Membrane 0.2 μm, 10417004, Whatman), and lastly 100 nm to obtain unilamellar liposomes with a diameter of 100 nm (Nucleopore Track-Etch Membrane 0.1 μm, 800309, Whatman). Filter supports (610014-1EA, Avanti Polar Lipids) were used in each extrusion step to prevent membrane tearing.
To understand the binding of the DA liposome system to different cell types, four different species of functionalized liposomes were prepared: bare, anti-CSV half antibody, ES, and anti-CSV half antibody + ES (DA). Half antibodies were produced by incubating the unconjugated monoclonal human anti-cell-surface-vimentin antibody (H00007431-M08, Abnova) with the reducing agent 2-MEA (153770050, Acros Organics).30 A 2 M 2-MEA stock solution was prepared in a reaction buffer containing 150 mM potassium acetate (AM9610, Invitrogen), 10 mM EDTA (E57020–500.0, Research Products International), and 100 mM NaCl (S23020–1000.0, Research Products International). Antibodies were incubated at a volume ratio of 2:8 (mAb to 2-MEA stock solution) for 2 h at 37 °C. Desalting columns (Zeba spin desalting column 7k MWCO 0.5 mL, 89882, Thermo Fisher Scientific) were used to remove excess 2-MEA in the solution. Liposomes were incubated overnight at 4 °C on a rotator to achieve a final concentration of 6 half antibodies and 2 recombinant human his-tagged ES (724-ES-100, R&D Systems) on the surface, recognizing that the starting amount of lipids was 10 μmol and that 100 nm liposome are made up of roughly 80,050 lipid molecules.31 The number of proteins per liposomes was estimated via stoichiometry and is theoretical. Liposomes (in 10 μL aliquots) were flash frozen in liquid nitrogen for 15 s for long-term storage with 10 μmol of d-(+)-trehalose dihydrate (T9531–5G, Sigma-Aldrich) per μmol of lipids.32
To study the therapeutic efficacy of our delivery system, liposomes with TRAIL only, TRAIL + anti-CSV half antibody (t-CSV), TRAIL + ES (t-ES), and TRAIL + anti-CSV half antibody + ES (t-DA) were prepared. Lipid composition was altered to remove the fluorescent lipid dye DiI to allow for cell death assays using Annexin V and propidium iodide. The molar ratios were 48:28:20:2:2% for Egg PC: Egg SM: Cholesterol: DGS-NTA(Ni): DSPE-PEG(2000) Maleimide. Liposome synthesis and functionalization were carried out following the steps above. His-tag TRAIL was incubated with the anti-CSV half antibodies and ES to achieve approximately 20 TRAIL molecules (BML-SE721–0100, Enzo Life Sciences) on the surface of each liposome.
Liposome Characterization
Functionalization was confirmed by dynamic light scattering measurements of the liposome diameter on a Malvern Panalytical Advanced Series Ultra Zetasizer. Liposomal stability under FSS was also evaluated. Cone-and-plate viscometers (Brookfield) were equipped with a CP-41 spindle and blocked with 5% BSA (A1470–100G, Sigma-Aldrich) in HBSS with Ca2+ and Mg2+. 40 μL of liposomes was sheared at 188 s–1 for 2.5 h at RT to simulate the same shearing time as cells later in the study. The liposomes and a BSA-only control were measured after FSS exposure via dynamic light scattering, as described above.
Cell Culture and Reagents
The neutrophil-like cell line PLB985 (ACC-139, DSMZ) and the CRC cell line HCT116 (CCL-247, ATCC) were utilized throughout this study. PLB985 cells were cultured in RPMI medium with l-glutamine (RPMI) supplemented with 10% (v/v) HI FBS (16140-071, Gibco) and 1% penicillin–streptomycin solution (15140-122, Gibco). HCT116 cells were cultured in McCoy’s medium supplemented with 10% (v/v) FBS and 1% penicillin–streptomycin solution. Humidified culture conditions were maintained at 37 °C and 5% CO2 levels. PLB985 cells were washed and resuspended in HBSS with Ca2+ and Mg2+ at a rate of 106 cells per 1 mL. HCT116 cells were detached using trypsin (25200-056, Gibco), then washed and resuspended in HBSS with Ca2+ and Mg2+ at 106 cells per 1 mL.
Uniform FSS Experiments with a Single Cell Type
Cone-and-plate viscometers (Brookfield) equipped with a CP-40 spindle were used to recreate the FSS that immune cells experience in the circulation.33,34 Before shearing, the cone and plate of the viscometers were blocked with 5% BSA (A1470–100G, Sigma-Aldrich) in HBSS with Ca2+ and Mg2+. Then, 500,000 PLB985 cells were resuspended in 0.5 mL of HBSS with Ca2+ and Mg2+ and placed in the viscometers with 10 μL of liposomes. Uniform FSS was applied to the sample at 188 s–1 for 30 min at RT. This is equivalent to 5 dyn/cm2 of FSS. Static controls were performed by placing 0.5 mL of the cell solution with the liposomes into centrifuge tubes and placing them on a rocker for 30 min to ensure that the samples were properly mixed throughout the experiment. This is equivalent to less than 0.05 dyn/cm2 of FSS. The samples were washed twice in HBSS with Ca2+ and Mg2+ at 300g for 5 min and then fixed in 4% PFA (50-980-495, Fisher Scientific) for 15 min. These steps were repeated using the HCT116 cells to test for liposome binding to cancer cells.
Protocols using human subjects were approved by the Institutional Review Board of Vanderbilt University. After informed consent was obtained, peripheral blood from healthy volunteers was collected in Vacutainer tubes containing sodium citrate buffer (363083, BD). Tubes were stored at 4 °C on a rotator for a maximum of 24 h until used. To assess liposome binding to healthy leukocytes, 10 μL of liposomes were added to 2 mL of healthy whole blood and sheared in a Brookfield viscometer using a CP-41 spindle at 188 s–1 for 30 min at RT. Static controls were performed by placing 0.5 mL of healthy whole blood with the liposomes into centrifuge tubes and placing them on a rocker for 30 min. Blood separation was performed via gradient centrifugation. Two mL of one-step polymorphs (AN221725, Accurate Chemical) was warmed to 37 °C in a water bath, and then 2 mL of blood with liposomes was carefully layered on top. This was centrifuged for 47 min at 500g with acceleration = 4 and deceleration = 2. The buffy coat was carefully removed, fixed in 4% PFA for 15 min, and washed twice by centrifuging at 300g for 5 min.
Nanoparticle Transfer Experiments
First, the transfer efficacy between the PLB985 cells and the HCT116 cells was assessed using a cone-and-plate viscometer equipped with a CP-40 spindle. Before shearing, the cone and plate of the viscometers were blocked with 5% BSA in HBSS with Ca2+ and Mg2+. Then, 5 × 105 PLB985 cells were resuspended in 0.5 mL of HBSS with Ca2+ and Mg2+ and placed in the viscometers with 10 μL of liposomes. Uniform FSS was applied to the sample at 188 s–1 for 30 min at RT. Samples were washed to remove any unbound liposomes in the suspension via centrifugation at 300g for 5 min. Viscometers were also rinsed with HBSS, Ca2+, and Mg2+. The PLB985 cells decorated with liposomes were resuspended in 0.5 mL of HBSS with Ca2+ and Mg2+ and spiked with 5 × 105 HCT116 cells. The sample was then sheared for an additional 2 h at 188 s–1. Lastly, the samples were washed twice in HBSS with Ca2+ and Mg2+ at 300g for 5 min and then fixed in 4% PFA for 15 min at RT.
Cell Viability Assays in Whole Blood
Cone-and-plate viscometers (Brookfield) equipped with a CP-41 spindle were used to recreate the FSS that immune cells experience in the circulation.33,34 Before shearing, the cone and plate of the viscometers were blocked with 5% BSA (A1470–100G, Sigma-Aldrich) in HBSS with Ca2+ and Mg2+. 2 mL of peripheral whole blood from healthy volunteers was carefully pipetted into the cup of the viscometer, and 20 μL of each of the liposome suspensions was added. Soluble his-tagged TRAIL (BML-SE721–0100, Enzo Life Sciences) was added to reach a final concentration of 590 ng/mL, which is equivalent to the amount of TRAIL on the surface of 20 μL of the liposomes, and was added to one viscometer as a control for non-liposomal TRAIL-induced apoptosis. Another viscometer was left with no treatment as a control. Uniform FSS was applied to the sample at 188 s–1 for 30 min at RT.
All groups were spiked with 2 × 106 HCT116 cells in HBSS with Ca2+ and Mg2+ and sheared for 2 h. Leukocyte separation from whole blood was performed via gradient centrifugation. 2 mL of 1-step polymorphs (AN221725, Accurate Chemical) was warmed to 37 °C in a water bath, and then 2 mL of blood with liposomes was carefully layered on top. This was centrifuged for 47 min at 500g with acceleration = 4 and deceleration = 2. The buffy coat was carefully removed, washed once in HBSS with Ca2+ and Mg2+ and centrifuged at 300g for 5 min. The cell pellet was resuspended in 2 mL of complete McCoy’s media and incubated for 24 h in a 6-well plate (229106, CELLTREAT) in the cell culture conditions described above. Cell death was then quantified via flow cytometry using an annexin V and propidium iodide assay.
Flow Cytometry
To test for the presence of CSV in all of the cells used throughout the study, 5 × 105 PLB985 and HCT116 cells were fixed in 4% PFA for 15 min and then blocked in 5% BSA for 30 min. Cells were stained with either a monoclonal human anti-CSV antibody tagged with FITC (H00007431-MF08, Abnova) or a mouse IgG2b kappa isotype antibody tagged with FITC (400310, BioLegend) at a 3:100 ratio in 5% BSA for 1 h. Samples were run through a Guava EasyCyte 12HT flow cytometer by using the Green-B laser. Flow cytometry data were analyzed on FlowJo and normalized to the isotype control.
Binding of liposomes to PLB985 cells, HCT116 cells, and healthy leukocytes was confirmed by running the samples through the flow cytometer using the Red-B laser. Granulocyte and monocyte + lymphocyte populations were gated using the forward scatter and side scatter plot.35 In the transfer experiments from the PLB985 to the HCT116 cells, the PLB985 cells were stained with DAPI at a 1:1000 ratio in HBSS with Ca2+ and Mg2+ and then washed twice by centrifugation at 300g for 5 min before placing the samples in the flow cytometer. Samples for the transfer experiments were run through the flow cytometer using the Red-B laser to identify liposome-coated cancer cells and the Blue-V laser to gate out the PLB985 cells (DAPI).
To quantify cell death in the HCT116 cells after treatment in whole blood, an annexin V and propidium iodide assay was performed via flow cytometry. After the 24 h incubation, cells were lifted by trypsinization (keeping the supernatant for analysis), washed once in HBSS with Ca2+ and Mg2+, and centrifuged at 300g for 5 min. The cancer cell population was enriched by magnetic column separation. Cell pellets were resuspended in 80 μL of MACS buffer (130-091-376, Miltenyi Biotec), and 20 μL of anti-CD45 magnetic beads (130-045-801, Miltenyi Biotec) were added. These were incubated at 4 °C for 15 min, then passed through a magnetic column (130042201, Miltenyi Biotec) to negatively select for the HCT116 cells. Cells were centrifuged at 300g for 5 min and resuspended in 5% BSA for a 30 min blocking step. Cells were then stained with an anti-CD45 antibody preconjugated with the fluorophore eFluor450 (48-0459-42, Themo Scientific) at 1:100 in 5% BSA for 30 min. Additionally, cells were stained with annexin V (556419, BD) at 2:100 and propidium iodide (556463, BD) at 3:100 for 15 min. Samples were run through the flow cytometer using the Red-B laser to identify propidium iodide, the Green-B laser to identify annexin V, and the Blue-V laser to gate out the remaining immune cell population (CD45+ cells). No-stain and single-stain controls were used for gating and compensation.
Confocal Microscopy
For microscopy, PLB985 and HCT116 cells were fixed in 4% (v/v) PFA (50-980-495, Fisher Scientific) in HBSS with Ca2+ and Mg2+ for 15 min after FSS exposure. Healthy leukocytes were fixed in the same way after the buffy coat was extracted following gradient centrifugation. Then, cells were incubated in 2:1000 DAPI (D9542-5MG, Sigma-Aldrich) for 30 min at RT in the blocking serum (1 mL). Washing steps were performed between each step using HBSS with Ca2+ and Mg2+. The stained cells were spun onto glass slides using a CytoSpin3 (Shandon), and a drop of antifade mounting media (H-1000, Vectrashield) and coverslips were added. The slides were imaged on an LSM 900 Zeiss confocal microscope equipped with a 10× objective.
Statistical Analysis
Where appropriate, unpaired, nonparametric t tests, ordinary one-way ANOVAs with Tukey’s multiple comparisons test, and two-way ANOVAs with Šídák’s multiple comparisons test were used. Significance was shown as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. A sample size of n = 3 was used. All values are presented as mean ± SD. All statistical analysis and plot preparation were performed on GraphPad Prism.
Results
Successful Liposome Preparation via the Extrusion Synthesis Method and Click Chemistry Functionalization
Liposomes were prepared via the lipid cake and extrusion technique following the steps outlined in Figure 2A. Functionalization was achieved through click chemistry conjugation. His-tagged ES and his-tagged TRAIL readily bind to NTA(Ni) groups, while the free sulfur groups on cleaved half antibodies easily bind to maleimide. DA liposomes were obtained by incubating with both the his-tagged ES and the anti-CSV half antibodies. Three control liposome formulations were synthesized: bare liposomes with no surface groups, liposomes with only the CSV half antibodies (CSV liposomes), and liposomes with only ES on the surface (ES liposomes). Liposomes functionalized with only ES served as a direct comparison to the previously published liposomes developed by the King Lab. Liposomes functionalized with only CSV antibody fragments served as a single targeting control for direct binding to CTCs.
Figure 2.
Liposomal nanoparticle synthesis, functionalization, and characterization. (A) Diagram depicting the steps to synthesize liposomes and the functionalization click chemistry. (B) Measurements of liposomes without conjugated TRAIL. (C) Measurements of liposomes with conjugated TRAIL.
To assess the therapeutic efficacy of the liposomal system, these four liposome formulations were synthesized, and his-tagged TRAIL was added to their surface. The functionalization of the liposomes was confirmed by measuring the liposome size using dynamic light scattering. Bare liposomes exhibited a mean dynamic radius of 133.2 nm (Figure 2B). The addition of the half antibodies and the ES added 3 and 14 nm to the dynamic diameter, respectively. When both targeting moieties were present on the liposome surface, the diameter was 144.3 nm. Furthermore, the addition of TRAIL increased the dynamic diameter of all the liposome groups by an average of 10 nm (Figure 2C). Liposomes were also confirmed to be stable under FSS conditions. Dynamic light scattering data show the presence of peaks at the 100 nm mark before and after FSS exposure (Supplemental Figure 1). The peak at 7 nm following FSS was confirmed to be BSA.36
Testing CSV Expression of the Cell Lines Used
To evaluate the transfer efficacy of the DA nanoparticles from healthy leukocytes to cancer cells in the circulation, the neutrophil-like cell line PLB985 and the CRC cell line HCT116 were utilized. The expression of CSV on the PLB985 cell surface was investigated to confirm its suitability for serving as a model of the carrier cell, where attachment occurs through ES binding alone. These cells did not significantly express CSV, validating that they would be an appropriate cell line model for the transfer experiment (Figure 3A). In contrast, CSV expression on the HCT116 cells was confirmed to establish them as suitable target cells in our experiments (Figure 3B). Notably, a 67% increase of MFI compared to the isotype antibody control indicated significant CSV expression.
Figure 3.
Quantification of cell–surface vimentin on cell lines. (A) CSV quantification on the carrier cells (PLB985). (B) CSV quantification on the target cells (HCT116). Significance was determined by unpaired t tests. Graphs in this figure show mean ± SD for an n = 3, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Confirmation of Liposome Binding to Different Cell Types
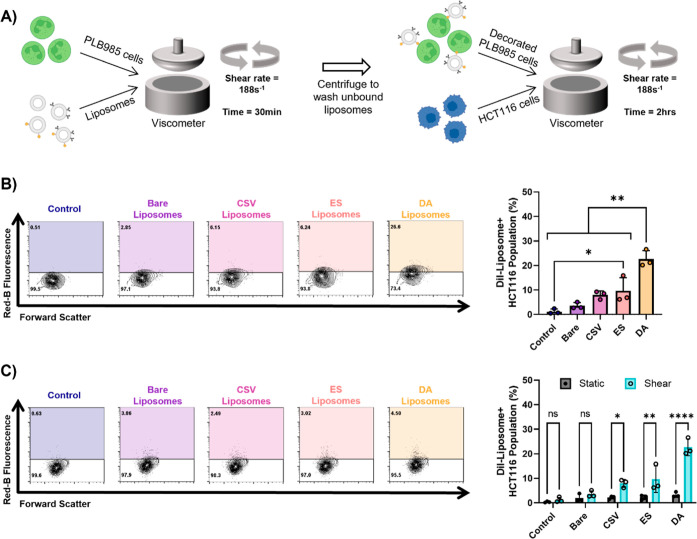
The binding capacity of the liposomes to the neutrophil model cell line PLB985 was tested under physiologically relevant FSS conditions by incubating them with nanoparticles in a cone-and-plate viscometer for 30 min. The percentage of PLB985 cells decorated with liposomes was quantified by using flow cytometry. Under FSS conditions, 36% of the cells in both the ES liposome group and the DA liposome group were decorated (Figure 4A). This represented a 6-fold increase compared with the bare liposome group.
Figure 4.
DA liposomes bind to PLB985 cells. (A) Flow cytometry plots and quantification of liposome binding to PLB985 cells under physiological FSS (statistical analysis: ordinary one-way ANOVA with Tukey’s multiple comparisons test). (B) Flow cytometry plots and quantification of liposome binding to PLB985 cells in static conditions compared to FSS results (statistical analysis: two-way ANOVA with Šídák’s multiple comparisons test). (C) Confocal microscopy images of PLB985 cells with bound liposomes showing DAPI in blue and the liposomes in red (scale bar = 20 μm). Graphs in this figure show mean ± SD for n = 3, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
No significant levels of binding were observed for the bare liposomes and the CSV liposomes compared to the control group, where no liposomes were present in the sample. When the experiment was repeated under static conditions, the percentage of cells decorated with liposomes decreased significantly by 15 fold and 8 fold in the ES and DA groups, respectively (Figure 4B). No significant changes in binding were observed between each of the liposome groups under static conditions (Supplementary Figure 2A). PLB985 cells decorated with liposomes under FSS were observed by using confocal microscopy (Figure 4C). In the micrographs, liposomes can be observed for the ES and DA conditions.
To confirm the attachment of DA liposomes to cancer cells, HCT116 cells were exposed to physiologically relevant FSS together with liposomes. The cells were sheared in a cone-and-plate viscometer with each of the liposome types for 30 min at a wall shear rate of 5 dyn/cm2. Flow cytometry analysis revealed an 8-fold increase in the percentage of HCT116 cells decorated with the CSV, ES, and DA liposomes compared to the bare liposome group (Figure 5A). There was no significant change in the level of binding between the bare liposome group and the control group without liposomes. When the liposomes were incubated with the HCT116 cells under static conditions, there was a significant decrease in binding to the CSV, ES, and DA liposomes (Figure 5B). No significant changes in binding were observed between each of the liposome groups under static conditions (Supplementary Figure 2B). The HCT116 cells decorated with liposomes under FSS were visualized by using confocal microscopy (Figure 5C). The signal in the red channel (DiI + liposomes) was particularly prominent in the cells treated with the CSV, ES, and DA liposomes. The liposomes were found to be localized to the outside of the cells.
Figure 5.
DA liposomes bind to HCT116 cells. (A) Flow cytometry plots and quantification of liposome binding to HCT116 cells under physiological FSS (statistical analysis: ordinary one-way ANOVA with Tukey’s multiple comparisons test). (B) Flow cytometry plots and quantification of liposome binding to HCT116 cells under static conditions compared to the FSS results (statistical analysis: two-way ANOVA with Šídák’s multiple comparisons test). (C) Confocal microscopy images of HCT116 cells with bound liposomes showing DAPI in blue and the liposomes in red (scale bar = 20 μm). Graphs in this figure show mean ± SD for n = 3 replicates, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Liposome Transfer Occurs under Physiological FSS
Liposome transfer between the neutrophil-like cell line PLB985 and the HCT116 CRC cells was investigated. First, the liposomes were incubated with the PLB985 cells for 30 min under the FSS conditions (Figure 6A). The cell suspension was washed to remove any unbound liposomes in the buffer and then spiked with the HCT116 cells. Once the cancer cells were added, samples were sheared in the viscometers for 2 additional hr. The DA liposomes showed significantly higher transfer efficacy compared with all of the other liposome formulations (Figure 6B). The percentage of HCT116 cells decorated with DA liposomes was 23%, representing a 3-fold increase compared to the percentage of decorated HCT116 cells incubated with the ES liposomes. Interestingly, ES liposomes did transfer under shear conditions but had a low efficacy in doing so, only transferring to 10% of the HCT116 cell population. As expected, when the transfer efficacy was tested under static conditions, there was a significant decrease in the transfer of the CSV, ES, and DA liposomes (Figure 6C). Under static conditions, there was a marginal but statistically significant increase in the transfer efficacy of the DA liposomes; however, the average percentage of the HCT116 cells that the DA liposomes transferred onto was only 3.3% (Supplemental Figure 2C).
Figure 6.
DA liposomes transfer from PLB985 cells to HCT116 cells under physiological FSS. (A) Overview of the experimental design. (B) Flow cytometry plots and quantification of liposome binding to HCT116 cells under physiological FSS (statistical analysis: ordinary one-way ANOVA with Tukey’s multiple comparisons test). (C) Flow cytometry plots and quantification of liposome binding to HCT116 cells in static conditions compared to the FSS results (statistical analysis: two-way ANOVA with Šídák’s multiple comparisons test). Graphs in this figure show mean ± SD for n = 3, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
DA Liposomes Bind to Primary Granulocytes
The binding ability of the nanoparticles to healthy human leukocytes was examined by using a cone-and-plate viscometer to model the FSS in the circulation. In this case, whole blood was used instead of the PLB985 cells in buffer. First, peripheral blood was collected from healthy volunteers after informed consent. Then, 2 mL samples of blood were sheared for 30 min at 188 s–1 with each of the liposome formulations. A control group without liposomes was included. Afterwards, the leukocytes were isolated from whole blood by gradient centrifugation and analyzed using flow cytometry.
Consistent with the results from the PLB985 experiments, the ES and DA liposomes exhibited the highest binding affinity with the healthy granulocytes (Figure 7A). These liposomes decorated 48% and 40% of the healthy granulocyte population, respectively. There was no significant difference in binding between these two formulations. In contrast, the CSV and bare liposomes showed significantly lower binding to the healthy granulocytes. Under shear stress, no significant binding was observed to occur with the healthy monocytes and lymphocytes compared to the liposome-free control group (Supplemental Figure 3A).
Figure 7.
DA liposomes bind to healthy primary human granulocytes. (A) Flow cytometry plots and quantification of liposome binding to healthy granulocytes under physiological FSS (statistical analysis: ordinary one-way ANOVA with Tukey’s multiple comparisons test). (B) Flow cytometry plots and quantification of liposome binding to primary granulocytes under static conditions compared to the FSS results (statistical analysis: two-way ANOVA with Šídák’s multiple comparisons test). (C) Confocal microscopy images of healthy granulocytes with bound liposomes showing DAPI in blue and the liposomes in red (scale bar = 20 μm). Graphs in this figure show mean ± SD for n = 3, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
When the experiment was repeated under static conditions, no significant binding was observed for the healthy granulocytes or for the monocyte and lymphocyte populations (Supplemental Figure 2D&E). Comparatively, the binding to healthy granulocytes decreased significantly under static conditions compared to the shear condition (Figure 7B). No changes in binding affinity were observed between the static and shear conditions in the monocyte and lymphocyte populations (Supplemental Figure 3B). The healthy blood cells decorated with liposomes under FSS were observed by using confocal microscopy (Figure 7C). The signal in the red channel (DiI + liposomes) was especially visible in the cells treated with the ES, and DA liposomes, localized on the cell surface.
DA Liposomes Conjugated with TRAIL Induce Apoptosis in Cancer Cells under FSS Conditions
The experimental set-up for testing the capacity of the DA liposomes to induce apoptosis in HCT116 cells in whole blood under FSS conditions can be seen in Figure 8A. First, whole blood was sheared in the viscometers with each of the liposome groups with TRAIL (TRAIL-only, CSV-TRAIL, ES-TRAIL, and DA-TRAIL). Additionally, a control sample was prepared with no liposomes, and a sixth group was prepared with soluble his-tagged TRAIL (unbound). The amount of soluble TRAIL added was equivalent to the amount of liposomal TRAIL used in each treatment in terms of nanograms per milliliter of blood. Next, all the samples were spiked with HCT116 cells and sheared for an additional 2 h. Afterward, the buffy coat was separated by gradient centrifugation, and the cells were plated for 24 h before performing an annexin V and propidium iodide assay. In this way, the change in cell viability after exposure to the liposomes under FSS was quantified, and the primary route for cell death was identified.
Figure 8.
Quantification of cancer cell death in whole blood after liposome treatments under FSS. (A) Overview of experimental design. (B) Flow cytometry plots and quantification of HCT116 cell viability (left), apoptosis (middle), and necrosis (right). Statistical analysis: ordinary one-way ANOVA. Graphs in this figure show mean ± SD for n = 3 replicates, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
In general, the DA liposome groups caused significant cell death compared to all of the other treatments tested. After treatment with the DA liposomes, there was a 51% decrease in viable HCT116 cells compared to the no liposome control (Figure 8B). In comparison, the ES liposome group decreased only 23% in viability compared to the group with no treatment. A significant decrease of 28% on average in cell viability was observed between the ES and DA liposome groups as well. Interestingly, the CSV and DA groups were not significantly different from each other. Lastly, the majority of cell death was seen to occur via apoptosis, while the percentage of necrotic cells in all groups was negligible and did not significantly change over the course of this experiment. The HCT116 cells in the control group (where no liposomes were added) maintained a cell viability of 80% throughout the experiment.
Discussion
In this study, we successfully designed and validated a nanoparticle system capable of transferring itself between a carrier cell and a target cell under physiologically relevant FSS conditions, as found in the circulation (Figure 1). Liposomal nanoparticles were decorated with ES and half of an antibody directed against CSV. ES was chosen because it is expressed in healthy adult granulocytes, and the ES-ESL bond is easily ruptured, providing an ideal tethering mechanism for the carrier cells. An anti-CSV antibody was utilized to target and irreversibly adhere to the target cell, as these bonds require very high forces to rupture. For the surface functionalization chemistry of the liposomes, reactions that could be performed via click chemistry were chosen. ES with a polyhistidine tag was attached to the liposome surface by incorporating a lipid with NTA(Ni) into the formulation (Figure 2). This same approach was used to conjugate the his-tagged TRAIL. Furthermore, a lipid with a maleimide group was utilized to synthesize the liposomes, enabling easy binding to the free sulfur group of a cleaved antibody.37 Changes in the liposome diameter indicated the successful conjugation of the surface moieties. Additionally, the specificity of ES and DA liposome binding to the PLB985 cells and healthy granulocytes compared to the naked liposomes confirms successful ES protein conjugation onto the liposome surface. This is also observed in the HCT116 cells for the anti-CSV and DA liposomes compared to the naked liposomes, confirming successful anti-CSV antibody fragment functionalization of the liposomes.
To validate that an anti-CSV antibody would be an appropriate targeting moiety for the CRC cell line HCT116, these cells were examined via flow cytometry (Figure 3). The binding capacity of the DA liposomes to the PLB985 cells under FSS conditions was confirmed through flow cytometry and confocal microscopy (Figure 4). Additionally, there was no significant difference in binding between the ES liposomes and the DA liposomes, indicating that the addition of the half antibodies did not have a negative impact on the nanoparticle binding affinity. No significant binding of the naked liposomes to any cell type occurred compared to the liposome-free controls, showing that positive liposome signals were due to specific recognition and not membrane fusion during collision events between the cells and liposomes. Under static conditions, no liposomes adhered to the PLB985 cells, highlighting the importance of FSS on ES-ESL 2 (Supplemental Figure 2).25,26 These results were also observed in a healthy granulocyte population (Figure 7). No binding was seen in the healthy monocyte and leukocyte populations (Supplemental Figure 3). Significant binding of the DA liposomes to the surface of the HCT116 cancer cells was observed after the FSS exposure (Figure 5). We confirmed that there was no significant difference in the binding of DA liposomes to the surface of the HCT116 cells compared to the CSV liposomes. Again, this shows that the addition of different targeting moieties did not affect the binding capacity of the DA liposomes. Interestingly, the ES liposomes exhibited the same degree of binding to the HCT116 cells as the CSV liposomes. It is worth noting that healthy leukocytes are not the only cells expressing Sialyl LewisX ligands. Many cancers also express ESLs, which can play a role in rolling/adhesion to facilitate CTC extravasation, migration, increased shear resistance, and enhanced evasion of immune cells in the circulation.38−43
To investigate the transfer efficiency of the DA liposomes, experiments were conducted using the neutrophil-like cell line PLB-985. This is a cell line that was derived from HL-60 and serves as a model for human neutrophils, which are the most prevalent leukocyte in the peripheral blood, and the immune cells with the highest levels of ESL expression.44−49 Since white blood cell counts can vary among healthy donors, utilizing a cell line provides a clearer picture of the binding affinity and transfer efficacy of the different liposome groups, separate from those other factors. Furthermore, for the transfer efficiency experiments, it aided in the interpretation of the removal of unbound liposomes from the cell suspension before introducing the cancer cells into the sample. Leukocyte isolation by gradient centrifugation is a time-consuming process that separates leukocytes from other components of whole blood (Figure 6). For the transfer experiment, having a constant binding environment before and after the incorporation of cancer cells was helpful for understanding how the collisions under FSS could facilitate nanoparticle binding and transfer between cell types. For the transfer experiment, a concentration of 5 × 105 PLB985 cells/mL was utilized, as this represents the average density of polymorphonuclear cells/mL that are found in healthy human blood.50 Liposome transfer was highly significant in the DA liposome group compared with all of the other liposome formulations and the liposome-free control (Figure 6). Minimal transfer was observed in the ES liposome group, but it was significantly lower than in the DA liposome group. This indicates that the presence of the CSV half antibodies is necessary to achieve the transfer of the liposomal nanoparticles from the carrier cell, to which they are loosely bound, to the target cells under physiologically relevant shear conditions.
Cell viability after FSS exposure using an annexin V and propidium iodide assay was tested with TRAIL added to each of the liposome formulations (Figure 8). Cell viability was significantly decreased in the DA liposome group compared with the other liposome formulations and the soluble TRAIL control. This demonstrates the effectiveness of nanoparticle transfer in facilitating the induction of cell apoptosis in the target cell. Cell viability experiments were only performed in whole blood and not with the PLB985 cell line, as these are cancer cells that overexpress DR4 and DR5, making them susceptible to TRAIL-mediated apoptosis.51,52 Additionally, the current model of FSS is limited as a cone-and-plate viscometer applies a constant shear stress to the sample, whereas FSS in the circulation varies from 1 dyn/cm2 of laminar flow to thousands of dyn/cm2 of turbulent FSS in the heart.33 In future studies, it would be interesting to test the liposome treatment in a mouse model of metastasis while focusing on circulating tumor cell killing using cell lines with differing levels of CSV expression.11 This could be done by intravenously injecting the liposome solution, and after 30 min, the HCT116 cells could be injected intravenously. After 2 h, blood could be removed via a cardiac puncture, and cell viability could be investigated following the same steps.
This nanoparticle system provides a platform for an easily customizable targeted delivery mechanism for CTCs. Aside from CSV, many other markers for CTCs have been explored in the literature for targeted delivery of therapeutics in circulation and could be substituted or added as synergistic targets on the CTC surface. These include, but are not limited to, CD44, CD133, EpCAM, and PSMA.37,53−55 The carrier cell could also be switched. In 2020, the King Lab validated a liposomal delivery system using platelets as a carrier cell for TRAIL to CTCs by functionalizing the liposome surface with the von Willebrand Factor A1 domain (vWF-A1).21 This is a plasma protein which binds to platelet glycoproteins during inflammatory processes in the bloodstream.56 Interactions between von Willebrand factor proteins and platelet glycoproteins follow the same dynamics as catch-slip bonds, in a similar manner to the ES-to-ESL bond.57 Additionally, the force required to rupture these bonds is low, ranging from 50 to 110 pN, which is an order of magnitude lower than the force required to rupture an antigen-to-antibody bond.58 Antiphagocytosis signals could also be added to the liposome surface if desired.
Our liposomes were designed to carry the cancer apoptosis signal TRAIL, which selectively induces death in cancer cells when it binds to the cell surface death receptors 4 and 5.12,59,60 While TRAIL is a promising therapeutic option, cancer cells with a more mesenchymal phenotype have increased resistance to TRAIL-induced apoptosis.61,62 CSV-positive CTCs have a more mesenchymal phenotype, which likely makes them more resistant to TRAIL, yet they are the most important population to target with this liposomal system. In future studies, it is important to explore the codelivery of a sensitizing agent of TRAIL encapsulated within the DA liposomes to enhance the killing of mesenchymal CTCs in the circulation. One option to consider is Yoda1, the agonist of the calcium channel Piezo1, which sensitizes cancer cells to TRAIL-mediated apoptosis.60 Additionally, other drugs can be delivered or codelivered in this liposomal system that have been validated by the King Lab, such as aspirin, piperlongumine, curcumin, and low-dose chemotherapies.63−69
Very few comparable technologies beyond those developed by the King Lab currently exist in the literature. One method investigated the transfer of nanoparticles between macrophages and cancer cells in the primary tumor.70 In that study, mesoporous silica nanoparticles were loaded with doxorubicin and incubated with macrophages. When these cells came into contact with cancer cells, the nanoparticles were transferred to neighboring cells via microtubules, resulting in an effective but low-throughput approach. Another study explored the efficacy of T cells in carrying nanoparticles (similar to the ES liposomes) to the tumor site.71 This method effectively reduced the solid tumor burden but lacked an active targeting component. Erythrocytes have also been explored as an option for tethering nanoparticles to prevent metastasis.72,73 However, these approaches lacked an active targeting moiety for the nanoparticles, and additionally, red blood cells are not as robust as other immune cells as they are prone to lysis.
The goal of this study was to have the liposome loosely bind to carrier cells under FSS conditions, and when the decorated cell collided with a target cell, the liposome would transfer from the carrier cell to the target cell. In this manner, nanoparticles would avoid clearance via the renal filtration system, and a higher therapeutic dose of the drug could be delivered via active targeting moieties. This system was successful in binding to neutrophils and the HCT116 CRC cell line and resulted in transfer from the first to the second under FSS. Additionally, when TRAIL was attached to the DA liposome surface, significant cell apoptosis was induced in the HCT116 cells compared with all other treatment groups. By achieving this transfer technology, we provide a nanoparticle design that can both actively target the CTC and increase circulation time by tethering to healthy leukocytes.
Acknowledgments
Liposome characterization was conducted in the Analytical Lab at the Vanderbilt Institute of Nanoscale Science and Engineering. We are grateful for support from Dr. Joshua Greenlee, Dr. Jacob Hope, and Dr. Dmitry Koktysh. This work was funded by the National Institute of Health National Cancer Institute grant number CA203991.
Data Availability Statement
All data in this study will be shared upon reasonable request by the lead contact.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c05605.
The authors declare no competing financial interest.
Supplementary Material
References
- Gu F. X.; Karnik R.; Wang A. Z.; Alexis F.; Levy-Nissenbaum E.; Hong S.; Langer R. S.; Farokhzad O. C. Targeted Nanoparticles for Cancer Therapy. Nano Today 2007, 2 (3), 14–21. 10.1016/S1748-0132(07)70083-X. [DOI] [Google Scholar]
- Yao Y.; Zhou Y.; Liu L.; Xu Y.; Chen Q.; Wang Y.; Wu S.; Deng Y.; Zhang J.; Shao A. Nanoparticle-Based Drug Delivery in Cancer Therapy and Its Role in Overcoming Drug Resistance. Front. Mol. Biosci. 2020, 7, 193. 10.3389/fmolb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz Y. (C.). Doxil-The First FDA-Approved Nano-Drug: Lessons Learned. J. Controlled Release 2012, 160 (2), 117–134. 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Gabizon A.; Shmeeda H.; Barenholz Y. Pharmacokinetics of Pegylated Liposomal Doxorubicin. Clin. Pharmacokinet. 2003, 42 (5), 419–436. 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- Safra T.; Muggia F.; Jeffers S.; Tsao-Wei D. D.; Groshen S.; Lyass O.; Henderson R.; Berry G.; Gabizon A. Pegylated Liposomal Doxorubicin (Doxil): Reduced Clinical Cardiotoxicity in Patients Reaching or Exceeding Cumulative Doses of 500 Mg/M2. Ann. Oncol. 2000, 11 (8), 1029–1034. 10.1023/A:1008365716693. [DOI] [PubMed] [Google Scholar]
- Longmire M.; Choyke P. L.; Kobayashi H. Clearance Properties of Nano-Sized Particles and Molecules as Imaging Agents: Considerations and Caveats. Nanomedicine 2008, 3 (5), 703–717. 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen A. B.; Guan C. M.; Ferrer J. R.; Barnaby S. N.; Merkel T. J.; Mirkin C. A. Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev. 2015, 115, 10530–10574. 10.1021/acs.chemrev.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon W.; Zhang Y.-N.; Ouyang B.; Kingston B. R.; Wu J. L. Y.; Wilhelm S.; Chan W. C. W. Elimination Pathways of Nanoparticles. ACS Nano 2019, 13 (5), 5785–5798. 10.1021/acsnano.9b01383. [DOI] [PubMed] [Google Scholar]
- von Karstedt S.; Montinaro A.; Walczak H. Exploring the TRAILs Less Travelled: TRAIL in Cancer Biology and Therapy. Nat. Rev. Cancer 2017, 17 (6), 352–366. 10.1038/nrc.2017.28. [DOI] [PubMed] [Google Scholar]
- Mitchell M. J.; King M. R. Fluid Shear Stress Sensitizes Cancer Cells to Receptor-Mediated Apoptosis via Trimeric Death Receptors. New J. Phys. 2013, 15 (1), 015008. 10.1088/1367-2630/15/1/015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J.; Wayne E.; Rana K.; Schaffer C. B.; King M. R. TRAIL-Coated Leukocytes That Kill Cancer Cells in the Circulation. Proc. Natl. Acad. Sci. U.S.A. 2014, 111 (3), 930–935. 10.1073/pnas.1316312111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee J. D.; Lopez-Cavestany M.; Ortiz-Otero N.; Liu K.; Subramanian T.; Cagir B.; King M. R. Oxaliplatin Resistance in Colorectal Cancer Enhances TRAIL Sensitivity via Death Receptor 4 Upregulation and Lipid Raft Localization. eLife 2021, 10, e67750 10.7554/eLife.67750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M.; Zhao H.; Huang K.-S.; Zhu C. Kinetic Measurements of Cell Surface E-Selectin/Carbohydrate Ligand Interactions. Ann. Biomed. Eng. 2001, 29 (11), 935–946. 10.1114/1.1415529. [DOI] [PubMed] [Google Scholar]
- Wild M. K.; Huang M.-C.; Schulze-Horsel U.; van der Merwe P. A.; Vestweber D. Affinity, Kinetics, and Thermodynamics of E-Selectin Binding to E-Selectin Ligand-1. J. Biol. Chem. 2001, 276 (34), 31602–31612. 10.1074/jbc.M104844200. [DOI] [PubMed] [Google Scholar]
- Ortiz-Otero N.; Marshall J. R.; Glenn A.; Matloubieh J.; Joseph J.; Sahasrabudhe D. M.; Messing E. M.; King M. R. TRAIL-Coated Leukocytes to Kill Circulating Tumor Cells in the Flowing Blood from Prostate Cancer Patients. BMC Cancer 2021, 21 (1), 898. 10.1186/s12885-021-08589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyotsana N.; Zhang Z.; Himmel L. E.; Yu F.; King M. R. Minimal Dosing of Leukocyte Targeting TRAIL Decreases Triple-Negative Breast Cancer Metastasis Following Tumor Resection. Sci. Adv. 2019, 5 (7), eaaw4197 10.1126/sciadv.aaw4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne E. C.; Chandrasekaran S.; Mitchell M. J.; Chan M. F.; Lee R. E.; Schaffer C. B.; King M. R. TRAIL-Coated Leukocytes That Prevent the Bloodborne Metastasis of Prostate Cancer. J. Controlled Release 2016, 223, 215–223. 10.1016/j.jconrel.2015.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J.; King M. R. Unnatural Killer Cells to Prevent Bloodborne Metastasis: Inspiration from Biology and Engineering. Expet Rev. Anticancer Ther. 2014, 14 (6), 641–644. 10.1586/14737140.2014.916619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S.; Chan M. F.; Li J.; King M. R. Super Natural Killer Cells That Target Metastases in the Tumor Draining Lymph Nodes. Biomaterials 2016, 77, 66–76. 10.1016/j.biomaterials.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Otero N.; Mohamed Z.; King M. R. Platelet-Based Drug Delivery for Cancer Applications. Adv. Exp. Med. Biol. 2018, 1092, 235–251. 10.1007/978-3-319-95294-9_12. [DOI] [PubMed] [Google Scholar]
- Ortiz-Otero N.; Marshall J. R.; Lash B. W.; King M. R. Platelet Mediated TRAIL Delivery for Efficiently Targeting Circulating Tumor Cells. Nanoscale Adv. 2020, 2 (9), 3942–3953. 10.1039/D0NA00271B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A.; Batth I.; Brownlee Z.; Mitra A.; Zhou S.; Noh H.; Rojas C. R.; Li H.; Meng Q. H.; Li S. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 2017, 8 (30), 49329–49337. 10.18632/oncotarget.17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A.; Li S. Vimentin in Cancer and Its Potential as a Molecular Target for Cancer Therapy. Cell. Mol. Life Sci. 2011, 68 (18), 3033–3046. 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surolia R.; Antony V. B. Pathophysiological Role of Vimentin Intermediate Filaments in Lung Diseases. Front. Cell Dev. Biol. 2022, 10, 872759. 10.3389/fcell.2022.872759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms G.; Dasanna A. K.; Schwarz U. S.; Lanzer M. Modeling Cytoadhesion of Plasmodium Falciparum-Infected Erythrocytes and Leukocytes-Common Principles and Distinctive Features. FEBS Lett. 2016, 590 (13), 1955–1971. 10.1002/1873-3468.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook J. H.; Guilford W. H. The Effects of Load on E-Selectin Bond Rupture and Bond Formation. Cell. Mol. Bioeng. 2010, 3 (2), 128–138. 10.1007/s12195-010-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau A. D.; Cao T. M.; Takitani T.; King M. R. Comparison of Human and Mouse E-Selectin Binding to Sialyl-Lewisx. BMC Struct. Biol. 2016, 16 (1), 10. 10.1186/s12900-016-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S.; Chen X.; Davies J.; Davies M. C.; Dawkes A. C.; Edwards J. C.; Roberts C. J.; Sefton J.; Tendler S. J. B.; Williams P. M. Detection of Antigen-Antibody Binding Events with the Atomic Force Microscope. Biochemistry 1997, 36 (24), 7457–7463. 10.1021/bi962531z. [DOI] [PubMed] [Google Scholar]
- Dammer U.; Hegner M.; Anselmetti D.; Wagner P.; Dreier M.; Huber W.; Güntherodt H. Specific Antigen/Antibody Interactions Measured by Force Microscopy. Biophys. J. 1996, 70 (5), 2437–2441. 10.1016/S0006-3495(96)79814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake S.; Yamada Y.; Ishikawa E.; Masseyeff R. Conjugation of Glucose Oxidase from Aspergillus Niger and Rabbit Antibodies Using N-Hydroxysuccinimide Ester of N-(4-Carboxycyclohexylmethyl)-Maleimide. Eur. J. Biochem. 1979, 101 (2), 395–399. 10.1111/j.1432-1033.1979.tb19731.x. [DOI] [PubMed] [Google Scholar]
- Dennison S. M.; Stewart S. M.; Stempel K. C.; Liao H.-X.; Haynes B. F.; Alam S. M. Stable Docking of Neutralizing Human Immunodeficiency Virus Type 1 Gp41 Membrane-Proximal External Region Monoclonal Antibodies 2F5 and 4E10 Is Dependent on the Membrane Immersion Depth of Their Epitope Regions. J. Virol. 2009, 83 (19), 10211–10223. 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N. K.; Roy I. Effect of Trehalose on Protein Structure. Protein Sci. 2009, 18 (1), 24–36. 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee J. D.; Liu K.; Lopez-Cavestany M.; King M. R. Piezo1Mechano-Activation Is Augmented by Resveratrol and Differs between Colorectal Cancer Cells of Primary and Metastatic Origin. Molecules 2022, 27 (17), 5430. 10.3390/molecules27175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J. M.; Bersi M. R.; Dombroski J. A.; Clinch A. B.; Pereles R. S.; Merryman W. D.; King M. R. Circulating Prostate Cancer Cells Have Differential Resistance to Fluid Shear Stress-Induced Cell Death. J. Cell Sci. 2021, 134 (4), 251470. 10.1242/jcs.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M.; Drummond M.; Doig A.. Practical Flow Cytometry in Haematology Diagnosis; John Wiley & Sons, Incorporated: Hoboken, United, Kingdom, 2013. [Google Scholar]
- Li Y.; Yang G.; Mei Z. Spectroscopic and Dynamic Light Scattering Studies of the Interaction between Pterodontic Acid and Bovine Serum Albumin. Acta Pharm. Sin. B 2012, 2 (1), 53–59. 10.1016/j.apsb.2011.12.001. [DOI] [Google Scholar]
- Lourenço B. N.; Pereira R. F.; Barrias C. C.; Fischbach C.; Oliveira C.; Granja P. L. Engineering Modular Half-Antibody Conjugated Nanoparticles for Targeting CD44v6-Expressing Cancer Cells. Nanomaterials 2021, 11 (2), 295. 10.3390/nano11020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin-Karim S.; King M. R.; Messing E. M.; Lee Y.-F. E-Selectin Ligand-1 Controls Circulating Prostate Cancer Cell Rolling/Adhesion and Metastasis. Oncotarget 2014, 5 (23), 12097–12110. 10.18632/oncotarget.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-A.; Blache C. A.; Bajana S.; Hasan N.; Kamal M.; Morita Y.; Gupta V.; Tsolmon B.; Suh K. S.; Gorenstein D. G.; Razaq W.; Rui H.; Tanaka T. The Effect of Soluble E-Selectin on Tumor Progression and Metastasis. BMC Cancer 2016, 16, 331. 10.1186/s12885-016-2366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsig L. Selectins in Cancer Immunity. Glycobiology 2018, 28 (9), 648–655. 10.1093/glycob/cwx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strell C.; Entschladen F. Extravasation of Leukocytes in Comparison to Tumor Cells. Cell Commun. Signal. 2008, 6, 10. 10.1186/1478-811X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirure V. S.; Reynolds N. M.; Burdick M. M. Mac-2 Binding Protein Is a Novel E-Selectin Ligand Expressed by Breast Cancer Cells. PLoS One 2012, 7 (9), e44529 10.1371/journal.pone.0044529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N.; d’Água B. B.; Ridley A. J. Crossing the Endothelial Barrier during Metastasis. Nat. Rev. Cancer 2013, 13 (12), 858–870. 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- Munro J. M.; Lo S. K.; Corless C.; Robertson M. J.; Lee N. C.; Barnhill R. L.; Weinberg D. S.; Bevilacqua M. P. Expression of Sialyl-Lewis X, an E-Selectin Ligand, in Inflammation, Immune Processes, and Lymphoid Tissues. Am. J. Pathol. 1992, 141 (6), 1397–1408. [PMC free article] [PubMed] [Google Scholar]
- Hauert A. B.; Martinelli S.; Marone C.; Niggli V. Differentiated HL-60 Cells Are a Valid Model System for the Analysis of Human Neutrophil Migration and Chemotaxis. Int. J. Biochem. Cell Biol. 2002, 34 (7), 838–854. 10.1016/S1357-2725(02)00010-9. [DOI] [PubMed] [Google Scholar]
- Millius A.; Weiner O. D. Manipulation of Neutrophil-Like HL-60 Cells for the Study of Directed Cell Migration. Methods Mol. Biol. 2010, 591, 147–158. 10.1007/978-1-60761-404-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Fang Y.; Ding X.; Wu J. Force-Dependent Bond Dissociation Govern Rolling of HL-60 Cells through E-Selectin. Exp. Cell Res. 2012, 318 (14), 1649–1658. 10.1016/j.yexcr.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Stroud M. R.; Handa K.; Salyan M. E. K.; Ito K.; Levery S. B.; Hakomori S.; Reinhold B. B.; Reinhold V. N. Monosialogangliosides of Human Myelogenous Leukemia HL60 Cells and Normal Human Leukocytes. 2. Characterization of E-Selectin Binding Fractions, and Structural Requirements for Physiological Binding to E-Selectin. Biochemistry 1996, 35 (3), 770–778. 10.1021/bi952461g. [DOI] [PubMed] [Google Scholar]
- Huang J.; Chen J.; Chesla S. E.; Yago T.; Mehta P.; McEver R. P.; Zhu C.; Long M. Quantifying the Effects of Molecular Orientation and Length on Two-Dimensional Receptor-Ligand Binding Kinetics*. J. Biol. Chem. 2004, 279 (43), 44915–44923. 10.1074/jbc.M407039200. [DOI] [PubMed] [Google Scholar]
- Kuhns D. B.; Priel D. A. L.; Chu J.; Zarember K. A. Isolation and Functional Analysis of Human Neutrophils. Curr. Protoc. Im. 2015, 111, 7.23.1–7.23.16. 10.1002/0471142735.im0723s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Rossin A.; Clifford J. L.; Gronemeyer H. Co-Resistance to Retinoic Acid and TRAIL by Insertion Mutagenesis into RAM. Oncogene 2006, 25 (26), 3735–3744. 10.1038/sj.onc.1209410. [DOI] [PubMed] [Google Scholar]
- Altucci L.; Rossin A.; Hirsch O.; Nebbioso A.; Vitoux D.; Wilhelm E.; Guidez F.; De Simone M.; Schiavone E. M.; Grimwade D.; Zelent A.; de Thé H.; Gronemeyer H. Rexinoid-Triggered Differentiation and Tumor-Selective Apoptosis of Acute Myeloid Leukemia by Protein Kinase A-Mediated Desubordination of Retinoid X Receptor. Cancer Res. 2005, 65 (19), 8754–8765. 10.1158/0008-5472.CAN-04-3569. [DOI] [PubMed] [Google Scholar]
- Faltas B. Cornering Metastases: Therapeutic Targeting of Circulating Tumor Cells and Stem Cells. Front. Oncol. 2012, 2, 68. 10.3389/fonc.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. R.; King M. R. Therapeutic Targeting of Circulating Tumor Cells: An Important Problem That Deserves Careful Study. Cell. Mol. Bioeng. 2015, 8 (4), 527–529. 10.1007/s12195-015-0418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E.; Cao T.; Nagrath S.; King M. R. Circulating Tumor Cells: Diagnostic and Therapeutic Applications. Annu. Rev. Biomed. Eng. 2018, 20 (1), 329–352. 10.1146/annurev-bioeng-062117-120947. [DOI] [PubMed] [Google Scholar]
- Denorme F.; Vanhoorelbeke K.; De Meyer S. F. Von Willebrand Factor and Platelet Glycoprotein Ib: A Thromboinflammatory Axis in Stroke. Front. Immunol. 2019, 10, 2884. 10.3389/fimmu.2019.02884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T.; Lou J.; Wu T.; Yang J.; Miner J. J.; Coburn L.; López J. A.; Cruz M. A.; Dong J.-F.; McIntire L. V.; McEver R. P.; Zhu C. Platelet glycoprotein Ibα forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J. Clin. Invest. 2008, 118 (9), 3195–3207. 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J.; Ling Y.; Westfield L. A.; Sadler J. E.; Shao J.-Y. Unfolding the A2 Domain of Von Willebrand Factor with the Optical Trap. Biophys. J. 2010, 98 (8), 1685–1693. 10.1016/j.bpj.2009.12.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke J.; von Karstedt S.; Zinngrebe J.; Walczak H. Getting TRAIL Back on Track for Cancer Therapy. Cell Death Differ. 2014, 21 (9), 1350–1364. 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J. M.; Lopez-Cavestany M.; Wang W.; Reinhart-King C. A.; King M. R. Activation of Piezo1 Sensitizes Cells to TRAIL-Mediated Apoptosis through Mitochondrial Outer Membrane Permeability. Cell Death Dis. 2019, 10 (11), 837. 10.1038/s41419-019-2063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyre L.; Meyer M.; Hofman P.; Roux J. TRAIL Receptor-Induced Features of Epithelial-to-Mesenchymal Transition Increase Tumour Phenotypic Heterogeneity: Potential Cell Survival Mechanisms. Br. J. Cancer 2021, 124 (1), 91–101. 10.1038/s41416-020-01177-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey D. J.; Choi W.; Marquis L.; Martin F.; Williams M. B.; Shah J.; Svatek R.; Das A.; Adam L.; Kamat A.; Siefker-Radtke A.; Dinney C. Role of Epithelial-to-Mesenchymal Transition (EMT) in Drug Sensitivity and Metastasis in Bladder Cancer. Cancer Metastasis Rev. 2009, 28 (3–4), 335–344. 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Patel S. B.; King M. R. Micelle-in-Liposomes for Sustained Delivery of Anticancer Agents That Promote Potent TRAIL-Induced Cancer Cell Apoptosis. Molecules 2020, 26 (1), 157. 10.3390/molecules26010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey C. C.; Li J.; Roy S.; Wu Q.; King M. R. Two-Stage Nanoparticle Delivery of Piperlongumine and Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand (TRAIL) Anti-Cancer Therapy. Technology 2016, 04 (01), 60–69. 10.1142/S2339547816500011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Sharkey C. C.; King M. R. Piperlongumine and Immune Cytokine TRAIL Synergize to Promote Tumor Death. Sci. Rep. 2015, 5, 9987. 10.1038/srep09987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardana K. W.; Jyotsana N.; Zhang Z.; King M. R.. Loading of Piperlongumine to Liposomes after Complexation with SS- Cyclodextrin and Its Effect on Viability of Colon and Prostate Cancer Cells. 2018, bioRxiv:314161. [Google Scholar]
- Li J.; Sharkey C. C.; Huang D.; King M. R. Nanobiotechnology for the Therapeutic Targeting of Cancer Cells in Blood. Cell. Mol. Bioeng. 2015, 8 (1), 137–150. 10.1007/s12195-015-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana K.; Reinhart-King C. A.; King M. R. Inducing Apoptosis in Rolling Cancer Cells: A Combined Therapy with Aspirin and Immobilized TRAIL and E-Selectin. Mol. Pharm. 2012, 9 (8), 2219–2227. 10.1021/mp300073j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J.; Chen C. S.; Ponmudi V.; Hughes A. D.; King M. R. E-Selectin Liposomal and Nanotube-Targeted Delivery of Doxorubicin to Circulating Tumor Cells. J. Controlled Release 2012, 160 (3), 609–617. 10.1016/j.jconrel.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S.; Noureddine A.; Guo J.; Keth J.; Paffett M. L.; Brinker C. J.; Serda R. E. Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells. Cancers 2020, 12 (10), 2892. 10.3390/cancers12102892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; Abraham W. D.; Zheng Y.; Bustamante López S. C.; Luo S. S.; Irvine D. J. Active Targeting of Chemotherapy to Disseminated Tumors Using Nanoparticle-Carrying T Cells. Sci. Transl. Med. 2015, 7 (291), 291ra94. 10.1126/scitranslmed.aaa5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Ukidve A.; Gao Y.; Kim J.; Mitragotri S. Erythrocyte Leveraged Chemotherapy (ELeCt): Nanoparticle Assembly on Erythrocyte Surface to Combat Lung Metastasis. Sci. Adv. 2019, 5 (11), eaax9250 10.1126/sciadv.aax9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alapan Y.; Yasa O.; Schauer O.; Giltinan J.; Tabak A. F.; Sourjik V.; Sitti M. Soft Erythrocyte-Based Bacterial Microswimmers for Cargo Delivery. Sci. Robot. 2018, 3 (17), eaar4423 10.1126/scirobotics.aar4423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study will be shared upon reasonable request by the lead contact.