Abstract
Heat shock protein 40 (Hsp40) family proteins are known to bind to Hsp70 through their J-domain and regulate the function of Hsp70 by stimulating its adenosine triphosphatase activity. In the endoplasmic reticulum (ER), there are 5 Hsp40 family proteins known so far, 3 of which were recently identified. In this report, one of the novel Hsp40 cochaperones, ERdj3, was characterized in terms of its subcellular localization, stress response, and stress tolerance of cells. By using ERdj3-specific polyclonal antibody, endogenous ERdj3 protein was shown to reside in the ER as gene transfer–mediated exogenous ERdj3. Analysis of the expression level of endogenous ERdj3 protein revealed its moderate induction in response to various ER stressors, indicating its possible action as a stress protein in the ER. Subsequently, we analyzed whether this molecule was involved in ER stress tolerance of cells, as was the case with the ER-resident Hsp70 family protein BiP. Although overexpression of ERdj3 by gene transfection could not strengthen ER stress tolerance of neuroblastoma cells, reduction of ERdj3 expression by small interfering ribonucleic acid decreased the tolerance of cells, indicating that ERdj3 might have just a marginal role in the ER stress resistance of neuroblastoma cells. In contrast, overexpression of ERdj3 notably suppressed vero toxin–induced cell death. These data suggest that ERdj3 might have diverse roles in the ER, including that of the molecular cochaperone of BiP and an as yet unknown protective action against vero toxin.
INTRODUCTION
Approximately one-third of all cellular proteins are transported into the lumen of the endoplasmic reticulum (ER), where posttranslational modification, folding, and oligomerization occur (Fink 1999; Kaufman 1999; Imaizumi et al 2001; Ma and Hendershot 2002). BiP (immunoglobulin heavy chain–binding protein) is a member of the 70-kDa heat shock protein 70 (Hsp70) family in the ER and plays an essential role in the folding and maturation of newly synthesized proteins in the secretory pathway (Bertolotti et al 2002). Molecular chaperone activity of various members of Hsp70 proteins is regulated by Hsp40 cochaperones, which contain DnaJ homology domain (J-domain) and stimulate adenosine triphosphatase (ATPase) activity of Hsp70 proteins because the intrinsic ATPase activity of Hsp70 is extremely weak (Cyr et al 1994; Cheetham and Caplan 1998; Greene et al 1998; Kelley 1998; Ohtsuka and Hata 2000; Abdul et al 2002). Of the mammalian Hsp40 family proteins, 5 molecules have been identified as possible ER-resident members (Brightman et al 1995; Skowronek et al 1999; Yu et al 2000; Shen et al 2002; Hosoda et al 2003; Cunnea et al 2003; Gu et al 2003). All of them are suggested to have importance in a wide variety of physiological and pathological functions in the ER.
ERdj3/HEDJ gene was initially isolated in the process of a phenotypic cloning approach to identify genes that confer resistance on Vero cells to vero toxin (Yu et al 2000). It was shown that HEDJ was capable of binding to BiP and stimulating its ATPase activity in vitro, suggesting its role as a cochaperone in the ER. Although exogenously expressed ERdj3 with fusion tag was shown in the ER, another research group suggested the possible subcellular localization to the nucleus and partly to the cytosol (Lau et al 2001). Because all the previous studies have been conducted to determine the localization of exogenously introduced ERdj3, it is still uncertain whether endogenous ERdj3 is an ER-resident protein. To address this issue, we developed a polyclonal anti-ERdj3 antibody, and analyzed it by confocal laser microscopy and a biochemical assay using the rat microsome.
Cellular responses to unfolded proteins in the ER have been termed ER stress. ER stress is induced by glucose starvation, disturbance of intracellular stores of calcium, inhibition of protein glycosylation, and disturbance of disulfide bond formation, leading to accumulation of incorrectly folded proteins in the ER lumen (Lee 2001). Recently, ERdj4, one of the ER-resident Hsp40 family proteins, which has a close structural similarity to ERdj3, has been reported to protect mammalian cells from ER stress–induced cytotoxicity (Kurisu et al 2002). This was the first report showing that this ER-resident Hsp40 family protein was involved in the ER stress tolerance in mammalian cells. Although ERdj3 was identified as a vero toxin–resistant gene, it remains ambiguous if the molecule is involved in the ER stress tolerance.
By using anti-ERdj3 antibody, this study demonstrates for the first time that endogenous ERdj3 is located in the ER. SH-SY5Y neuroblastoma cells and Vero cells, which have a low level of endogenous ERdj3 expression, were used to assess whether ERdj3 expression was altered by ER stress such as tunicamycin and thapsigargin treatment. ERdj3 protein levels were upregulated in response to ER stress treatment but not to vero toxin. Next, we examined if altered expression levels of ERdj3 might have any effect on ER stress tolerance. Gene transfer–mediated overexpression of ERdj3 had no effect on the ER stress tolerance of neuroblastoma cells. However, small interfering ribonucleic acid (siRNA)–mediated reduction of endogenous ERdj3 protein level led to decreased ER stress tolerance, indicating that ERdj3 might have a marginal effect on the ER stress tolerance. In contrast, overexpression of ERdj3 significantly enhanced the resistance to vero toxin cytotoxicity. This study revealed the distinct roles played by ER-resident Hsp40 family chaperone ERdj3 in ER stress tolerance and vero toxin resistance of mammalian cells.
MATERIALS AND METHODS
Cell culture, expression plasmids, and antibodies
Human breast cancer cell line, MCF7, human cervical cancer cell line, HeLa, human prostate cell line, LNCaP, human B cell lymphoma cell line, Raji, simian renal cell line, Vero, and mouse myeloma cell line, NS-1, were purchased from American Type Culture Collection (Manassas, VA, USA). Rat insulinoma cell line, RIN-m, was purchased from Dainippon Pharmaceutical Co (Osaka, Japan). Human neuroblastoma SH-SY5Y cells were kindly provided by Dr. Nakagawara (Chiba Cancer Center, Chiba, Japan) and mouse MIN-6 insulinoma cells were kindly provided by Dr. Miyazaki (Osaka University, Osaka, Japan).
MCF7, HeLa, and Vero cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS) (GIBCO Invitrogen Co, Grand Island, NY, USA). SH-SY5Y cells were cultured in minimal essential medium containing 10% FCS. LNCaP cells were cultured in Rosewell Park Memorial Institute (RPMI)-1640 medium containing 10% FCS, 0.1 mM minimum essential medium (MEM) nonessential amino acids solution (GIBCO-BRL, Gaithersburg, MD, USA), and 1 mM MEM sodium pyruvate (GIBCO-BRL). Raji and NS-1 cells were cultured in RPMI-1640 medium containing 10% FCS. MIN-6 and RIN-m cells were cultured in RPMI-1640 medium containing 10% FCS and 0.05 mM 2-mercaptoethanol (Wako, Tokyo, Japan).
To construct expression plasmids, total RNA from human peripheral blood cells was extracted, and complementary deoxyribonucleic acid (cDNA) mixture was synthesized from the RNA by reverse transcription. A full-length, C-terminal myc epitope–tagged ERdj3 (ERdj3-myc) was amplified by using the specific forward and reverse primers including BamHI and EcoRI restriction sites, respectively. The polymerase chain reaction (PCR) product was purified and cloned into pcDNA3 mammalian expression vector (Invitrogen, Carlsbad, CA, USA). Similarly, the PCR product of ERdj3 amplified by using the specific forward and reverse primers including XhoI and HindIII restriction sites, respectively, was cloned into pIRES2-EGFP vector (BD Clontech, Palo Alto, CA, USA). The nucleotide sequence of each insert was analyzed by an ABI analyzer PRISM 310 and an AmpliCycle sequencing kit (Perkin-Elmer, Norwalk, CT, USA).
Rabbit antisera were raised against a synthetic peptide (Sigma Genosis, Hokkaido, Japan) corresponding to residues 196–211 of human ERdj3 protein (Fig 1A) because the region was hydrophilic and conserved among humans, mice, and rats. Mouse monoclonal anti-KDEL and anti–β-actin antibodies were purchased from Stressgen (Victoria, BC, Canada) and Sigma (St Louis, MO, USA), respectively. Anti-BiP antibody was purchased from BD Biosciences (San Jose, CA, USA).
Fig 1.
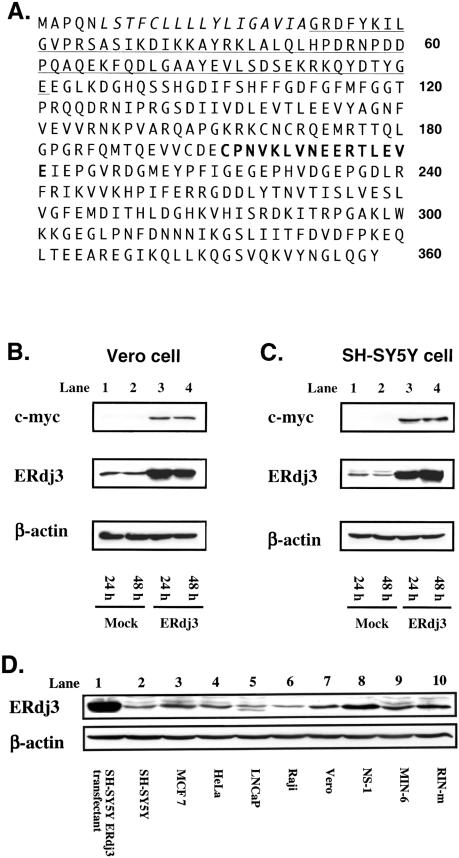
The structure and expression of ERdj3. (A) The deduced amino acid sequence of ERdj3. ERdj3 is composed of 358 amino acids. Italic letters indicate the predicted signal peptide. J-domain is underlined. The peptide sequence that the antibody was raised against is indicated in bold type. (B, C) Western blotting analysis of ERdj3 protein. ERdj3-myc expression vector or mock vector was transfected into Vero cells (B) and SH-SY5Y cells (C). Twenty-four or 48 hours after transfection, Western blotting was performed using the anti-myc, anti-ERdj3, or anti–β-actin antibody. β-Actin was detected as an internal control. (D) Western blotting analysis of total cellular protein isolated from various cell lines
Transfection
Expression vectors were transfected into cells by using Lipofectamin2000 reagent (Invitrogen) as recommended by the manufacturer's protocol. In brief, 5 μg of deoxyribonucleic acid and 15 μL of Lipofectamin2000 reagent were each diluted into 250 μL Opti-MEM (Invitrogen), mixed, and added to cells cultured in 6-well plates at 1 million cells/well. After 4 hours, the cells were washed and cultured in a fresh medium for 4 hours. Subsequently, the cells were plated onto 96-well flat-bottom plates for 3(4,5)-dimethylthiazol-2,5-diphenyl tetrazolium bromide (MTT) assay or onto 10-cm-diameter dishes for Western blotting.
Western blotting
Cultured cells were washed in ice-cold phosphate-buffered saline (PBS), lysed by incubation on ice in a lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, protease inhibitor cocktail [Complete, Roche Diagnostics Inc, Indianapolis, IN, USA]), and clarified by centrifugation at 15 000 rpm for 20 minutes at 4°C. The whole-cell lysates were boiled for 5 minutes in the presence of sodium dodecyl sulfate (SDS) sample buffer, resolved by 10% SDS–polyacrylamide gel electrophoresis (PAGE), and electrophoretically transferred to polyvinylidene fluoride membranes (Immobilon-P, Millipore, Bedford, MA). The membranes were then incubated with blocking buffer (5% nonfat dry milk, PBS) for 1 hour at room temperature and incubated for 40 minutes with rabbit anti-ERdj3 antibody, mouse anti–c-myc antibody (Kamiguchi et al 1999), mouse anti-BiP antibody, or mouse anti–β-actin antibody, followed by incubation with horseradish peroxidase–conjugated anti-rabbit immunoglobulin G (IgG) antibody or anti-mouse IgG antibody (KPL). Finally, the reaction was made visible with an enhanced chemiluminescence Western blotting kit (Amersham Pharmacia Biotech (Piscataway, NJ, USA) according to the manufacturer's protocol. Signal intensities were quantified using Image Reader LAS-1000 (Fuji Film, Tokyo, Japan). Relative expression levels of ERdj3 or BiP were calculated as: relative expression = (ERdj3 or BiP signal density/actin signal density).
Confocal microscopy
MCF7 cells or SH-SY5Y cells cultured on a glass coverslip (Fisher Scientific, Pittsburgh, PA, USA) were fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100. After blocking of cells with 10% goat serum for 1 hour at room temperature, cells were incubated with polyclonal anti-ERdj3 antibody and monoclonal anti-KDEL antibody to detect the subcellular localization of endogenous ERdj3 protein, whereas polyclonal anti–myc-tag antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and monoclonal anti-KDEL antibody were used to determine the localization of exogenously introduced myc-tagged ERdj3 protein. After washing with PBS, cells were immunostained with fluorescein isothiocyanate– conjugated anti-rabbit IgG and rhodamine-conjugated anti-mouse IgG antibodies (KPL), followed by visualization using confocal laser microscopy (LSM510, Carl Zeiss, Tokyo, Japan).
Proteinase K digestion of ERdj3 and BiP
A male rat liver microsome fraction was purchased from BD Biosciences. Protease treatment was performed by adding 10 μg/mL proteinase K to the microsome suspended in PBS with or without 1% Triton X-100. After incubating at 4°C for 30 minutes, the samples were mixed with SDS sample buffer, subjected to SDS-PAGE, and analyzed by Western blotting.
Treatment of vero toxin and induction of ER stress
Cells were incubated with the indicated concentrations of tunicamycin or thapsigargin (Wako) to induce ER stress or with recombinant vero toxin (VT1) for 18 hours and then analyzed for cell viability by MTT assay or by propidium iodide (PI) staining (Annexin-V-FLUOS Staining Kit; Roche, Mannheim, Germany). Tunicamycin inhibits N-linked glycosylation of nascent proteins, and thapsigargin is an ER calcium ATPase inhibitor depleting intraluminal calcium. Tunicamycin and thapsigargin were stored at −30°C as stock solutions of 10 mg/mL in 5% dimethyl sulfoxide and 1 mM in water, respectively.
MTT assay
The assay for viable cells using MTT (Sigma, St. Louis, MO, USA) was described previously (Asanuma et al 1999). In brief, Vero cells or SH-SY5Y cells were transiently transfected with ERdj3-myc expression vector in 6-well plates for 8 hours. Subsequently, 40 000 cells plated on 96-well flat-bottom plates were incubated at 37°C for 16 hours, followed by washing and culture in a medium containing vero toxin, tunicamycin, or thapsigargin. After incubation for 18 hours, MTT reagent was added into the culture medium. A purple formazan product was then formed by the action of mitochondrial enzymes in the living cells. This product was solubilized by the addition of acidic isopropanol. The absorbance of each well was quantified by optical density (OD) analyzed using a microplate reader (NovaPath, BioRad, Tokyo, Japan). The test wavelength was 570 nm, and the reference was 630 nm. Relative cell viability was calculated according to the formula: % cell viability = (OD of experimental sample with reagent/OD of control sample without reagent) × 100. All the MTT assays were performed in triplicate.
Small interfering RNA
Synthetic ready-to-use siRNA (21 nucleotides) complementary to a region of ERdj3 exon 6 and nonsilencing control siRNA targeting to green fluorescence protein (GFP) gene were custom synthesized by Qiagen (Tokyo, Japan).
SH-SY5Y cells were transfected with 8 μg of ERdj3-siRNA using the Lipofectamin2000 reagent. In brief, 8 μg of siRNA and 15 μL of Lipofectamin2000 reagent were diluted with Opti-MEM to a volume of 250 μL, mixed, and added to cells in 6-well plates that had been grown to 60% confluency. After 4 hours, the cells were washed and cultured in fresh media for 12 hours. Then, the cells were plated onto 10-cm-diameter dishes, cultured for a further 24 hours, and replated onto 96-well flat-bottom plates, followed by incubation at 37°C for 32 hours. Seventy-two hours after the transfection, cells were washed and cultured in media containing the indicated concentrations of thapsigargin or tunicamycin for 48 hours, followed by MTT assay.
RESULTS
Establishment of anti-ERdj3 antibody
To characterize the subcellular localization and function of endogenous ERdj3, we established rabbit antisera against ERdj3 by immunization of ERdj3 polypeptide (residues 196–211), as shown in Figure 1A. The specificity of the polyclonal antibody was examined by Western blotting. An immunoreactive band of approximately 40 kDa was detected in Vero cells transfected with ERdj3-myc expression vector (Fig 1B, lanes 3 and 4) but not in mock-transfected cells (Fig 1B, lanes 1 and 2) when stained with anti–c-myc antibody, indicating that ERdj3-myc protein was successfully expressed by the transfection. When stained with anti-ERdj3 antibody, a strong band with the same size was detected in the ERdj3-myc– transfected cells, whereas no band was detected in the same cells with preimmune serum (data not shown). Furthermore, a weak band with an almost identical size to the band in ERdj3-myc–transfected cells was also detected in the mock transfectant shown in Figure 1B (lanes 1 and 2), indicating that the bands were endogenous ERdj3 protein. A similar result was obtained using SH-SY5Y neuroblastoma cells as shown in Figure 1C. Staining with anti–β-actin antibody confirmed that the amount of proteins loaded in each lane was almost identical in Figure 1B,C. Thus, it was demonstrated that the polyclonal anti-ERdj3 antibody specifically reacted to both endogenously and exogenously expressed ERdj3 protein. To determine the species specificity of this antibody in mammals, we performed Western blotting using various cell lines. As shown in Figure 1D, ERdj3 band was detected in total cell lysates from human cells (lanes 2–6), monkey cells (lane 7), mouse cells (lanes 8 and 9), and rat cells (lane 10). These data indicate that this antibody can react with ERdj3 proteins derived from a variety of mammalian species.
Subcellular localization of ERdj3
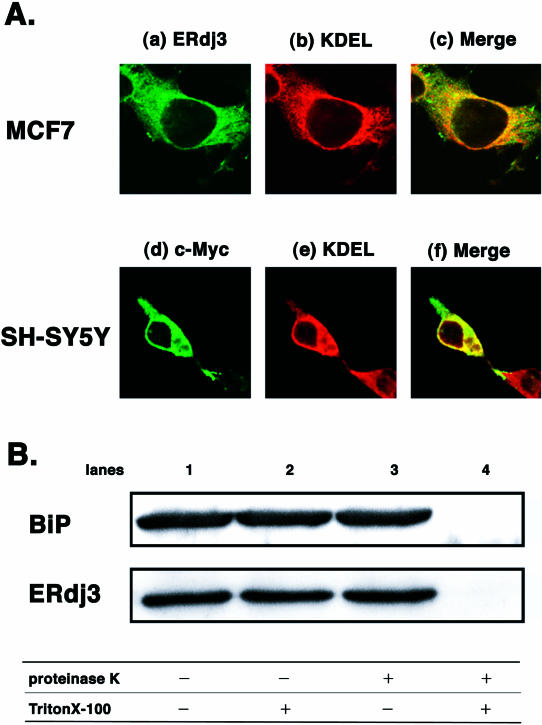
There have been discrepancies as to the subcellular localization of ERdj3 protein. One article reported that epitope-tagged exogenous ERdj3 was located in the ER (Yu et al 2000), but another suggested the possibility that exogenous ERdj3 might reside in the nucleus and cytosol (Lau et al 2001). Therefore, we examined the subcellular localization of endogenous ERdj3 in MCF7 cells, which expressed this protein at a considerable level (Fig 1D), by using anti-ERdj3 antibody and confocal laser microscopy. Immunostaining of MCF7 cells with polyclonal anti-ERdj3 antibody (Fig 2A, a) and monoclonal anti-KDEL antibody (Fig 2A, b), which detected ER-resident molecular chaperones such as BiP and GRP94, revealed that the perinuclear pattern of anti-ERdj3 antibody reactivity completely overlapped in the most part with that of anti-KDEL antibody (Fig 2A, c). Control experiments performed in the absence of the primary antibodies demonstrated only negligible background staining (data not shown). These data strongly suggested that ERdj3 protein was located in the ER but not in the nucleus. The data were further supported by an immunohistochemical study in which ERdj3 immunostaining showed a nonnuclear pattern in a wide variety of human tissues examined (data not shown).
Fig 2.
Subcellular localization of endogenous ERdj3. (A) MCF7 cells were immunostained with polyclonal anti-ERdj3 antibody followed by fluorescein isothiocyanate–conjugated anti-rabbit immunoglobulin G (IgG) antibody or monoclonal anti-KDEL antibody followed by rhodamine-conjugated anti-mouse IgG antibody. After mounting, cells were visualized by confocal laser microscopy. (a) Immunostaining of ERdj3, (b) immunostaining of KDEL, and (c) merge. SH-SY5Y cells were transfected with ERdj3-myc and immunostained with polyclonal anti-myc antibody followed by fluorescein isothiocyanate–conjugated anti-rabbit antibody or monoclonal anti-KDEL antibody followed by rhodamine-conjugated anti-mouse IgG antibody. After mounting, cells were visualized by confocal laser microscopy. (d) Immunostaining of myc tag, (e) immunostaining of KDEL, and (f) merge. (B) Proteinase K digestion of ERdj3 and BiP. A rat microsome was incubated in phosphate-buffered saline supplemented with (lanes 3 and 4) or without (lanes 1 and 2) 10 μg/mL proteinase K in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 1% Triton X-100 at 4°C for 30 minutes, followed by sodium dodecyl sulfate–polyacrylamide sulfate gel electrophoresis and Western blotting with anti-BiP or anti-ERdj3 antibody
Next, we examined the subcellular localization of exogenously introduced ERdj3, which was used in the following overexpression experiments. ERdj3-myc expression vector was transfected into SH-SY5Y cells, and transient expression of myc-tagged ERdj3 was detected by immunostaining with both polyclonal anti-myc antibody and monoclonal anti-KDEL antibody. The immunostaining with anti-myc antibody showed a perinuclear and reticular pattern (Fig 2A, d), and it was largely superimposable on that with anti-KDEL antibody (Fig 2A, e and f), suggesting that exogenously introduced ERdj3-myc was also localized in the ER.
To further examine the localization of ERdj3, we performed protease susceptibility assays (Yu et al 2000). Rat microsomes were exposed to proteinase K in the presence or absence of Triton X-100. Because proteinase K cannot cross the intact microsomal membrane, luminal proteins should not be digested by the protease. Addition of Triton X-100 disrupts the membrane, leading to access of the protease to intraluminal proteins. As shown in Figure 2B, ERdj3 was clearly detected by anti-ERdj3 antibody in a microsome fraction derived from the rat liver (lane 1). In the presence of proteinase K, ERdj3 protein as well as BiP protein in the microsome was completely digested when Triton X-100 disrupted the membrane (lane 4). In contrast, they were protected from the digestion in the absence of Triton X-100 (lane 3). These results further confirm that ERdj3 is localized within the lumen of the ER.
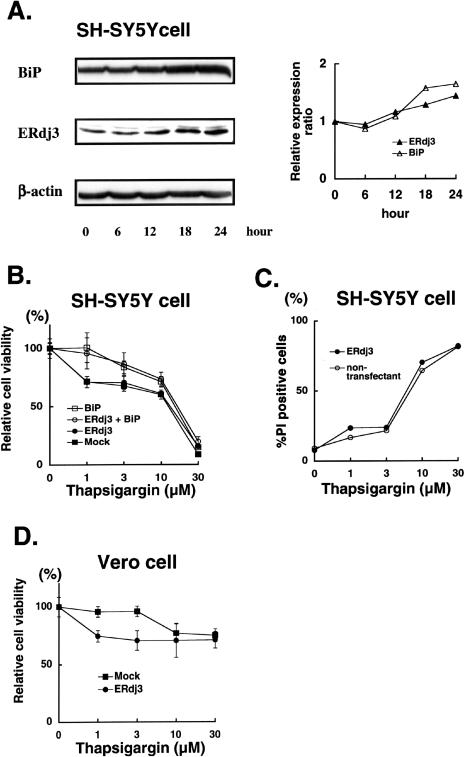
Overexpression of ERdj3 enhances resistance to vero toxin cytotoxicity in Vero cells
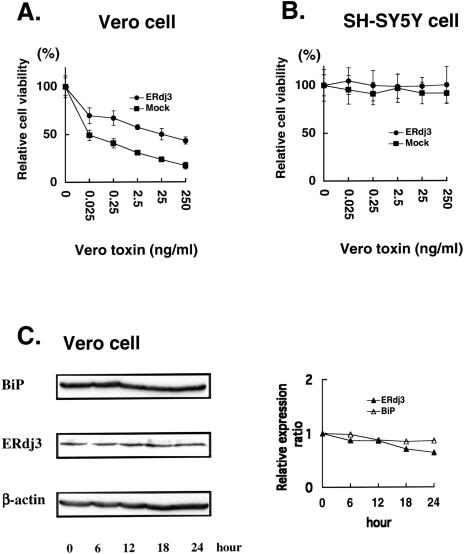
Yu et al (2000) originally isolated ERdj3/HEDJ gene by a functional cloning experiment in which genes conferring resistance to vero toxin cytotoxicity on Vero cells were screened. We examined whether overexpression of ERdj3 would actually increase resistance to VT1 in our cells. As shown in Figure 3A, relative cell viability was decreased in a dose-dependent manner in Vero cells after treatment with the indicated concentrations of vero toxin. Cell viability of ERdj3-transfected Vero cells was kept significantly higher than that of mock-transfected cells at all the concentrations of VT1. In SH-SY5Y neuroblastoma cells, however, cell viability was not changed after vero toxin treatment (Fig 3B), indicating that neuroblastoma cells were not sensitive to vero toxin. Next, we performed Western blotting to determine whether expression of endogenous ERdj3 could be upregulated by vero toxin in Vero cells. As shown in Figure 3C, ERdj3 protein levels were not changed after treatment with 0.25 ng/mL of vero toxin for the indicated time periods. Another ER chaperone, BiP, also showed no change in the protein level. Similar results were obtained when cells were treated with higher doses of vero toxin (data not shown), indicating that vero toxin could not induce the expression of ERdj3. These results suggested that ERdj3 was involved in the mechanism of vero toxin cytotoxicity as a cellular protective factor (Wesche et al 1999; Sandvig et al 2002; Sekino et al 2002), and the low constitutive level of ERdj3 expression and the lack of its induction in Vero cells might explain, at least in part, the high toxin sensitivity of this cell.
Fig 3.
Cellular response to vero toxin treatment. Cells were transiently transfected with ERdj3 expression vector or mock vector. Twenty-four hours after transfection, cells were treated with the indicated concentrations of vero toxin for 18 hours followed by 3(4,5)-dimethylthiazol-2,5-diphenyl tetrazolium bromide assay to assess cell viability. (A) Vero cells. (B) SH-SY5Y cells. Vero cells were treated with 0.25 ng/mL of vero toxin, and cell lysates were collected at the indicated time points after treatment. (C) The lysates were subjected to Western blotting using anti-ERdj3, anti-BiP, or anti–β-actin antibody. (D) Signal intensities were quantified, and relative ERdj3 or BiP expression level was calculated as: relative expression ratio = (sample ERdj3 or BiP signal intensity/sample actin signal intensity)
Effect of ERdj3 overexpression on ER stress tolerance
Given that ERdj3 could serve as a cochaperone of BiP in the ER, it may be possible that ERdj3 expression is induced by ER stress and its overexpression can enhance ER stress tolerance as reported in BiP-overexpressed cells (Katayama et al 1999). Therefore, we next examined changes in the ERdj3 expression after ER stress and the effect of its overexpression on ER stress tolerance of cells. In this study, tunicamycin and thapsigargin, which disturb N-linked glycosylation of ER proteins and intraluminal calcium homeostasis, respectively, were used as ER stressors.
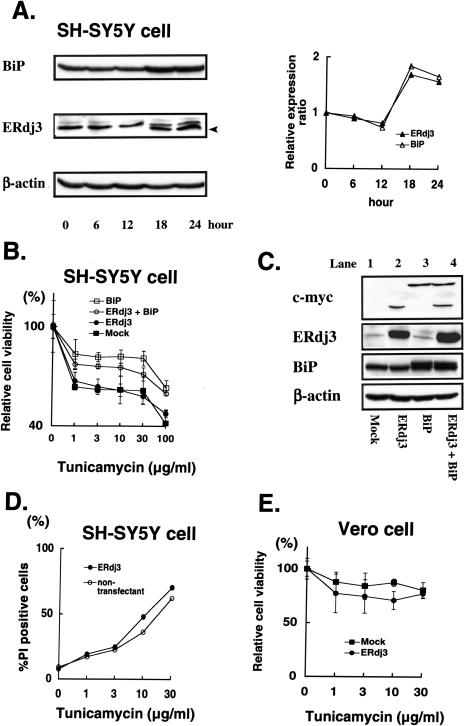
As shown in Figure 4A, treatment for 18 hours with 1 μg/mL tunicamycin could lead to mild upregulation of endogenous ERdj3 expression as well as BiP expression in SH-SY5Y cells, indicating the characteristic induction as observed in most stress protein families. Note that a band of the aglycosylated form of ERdj3 appeared after an 18-hour treatment of tunicamycin (Fig 4A, arrowhead). A similar induction was observed in Vero cells (data not shown). However, as shown in Figure 4B, relative cell viability of SH-SY5Y cells with constitutive ERdj3 overexpression (Fig 4C, lane 2) was almost identical to that of mock-transfected control cells with the marginal expression (Fig 4C, lane 1) after treatment with the indicated concentrations of tunicamycin. On the other hand, transient overexpression of BiP by the transfection of BiP cDNA (Fig 4C, lanes 3 and 4) conferred resistance to tunicamycin stress on either cells. To exclude the possibility that these results were due to the clonal variation of the ERdj3-transfected SH-SY5Y cells, we next performed transient transfection of pERdj3-IRES-EGFP vector that could express both ERdj3 and enhanced GFP (EGFP) in a single cell. Because EGFP-positive cells represent the ERdj3-overexpressing cells in this experiment system, we examined the rate of PI-positive cells in the total EGFP-positive cells (ERdj3-transfected cells) and that of PI-positive cells in the EGFP-negative cells (nontransfectant) by using fluorescent microscopy (Fig 4D). The rate of the PI-positive cells in the EGFP-positive cells was almost similar to that in the EGFP-negative cells after treatment with the indicated concentrations of tunicamycin. These results further confirmed that overexpression of ERdj3 could not confer resistance to the tunicamycin-induced ER stress on SH-SY5Y cells. In Vero cells, overexpression of ERdj3 rather decreased cellular viability as compared with mock-transfected cell (Fig 4E).
Fig 4.
Cellular response to tunicamycin treatment. (A) SH-SY5Y cells were treated with 1 μg/mL of tunicamycin, and cell lysates were collected at the indicated time points after the treatment. The lysates were subjected to Western blotting using anti-BiP, anti-ERdj3, or anti–β-actin antibody. An arrowhead indicates the aglycosylated form of ERdj3. (B) An ERdj3-transfected SH-SY5Y clone and a mock-transfected clone were transfected with BiP expression vector or a mock vector. Twenty-four hours after the transfection, cells were treated with the indicated concentrations of tunicamycin for 18 hours, followed by 3(4,5)-dimethylthiazol-2,5-diphenyl tetrazolium bromide (MTT) assay to assess cell viability. (C) ERdj3-transfected SH-SY5Y clone (lanes 2 and 4) or mock-transfected clone (lanes 1 and 3) was transiently transfected with BiP expression vector (lanes 3 and 4) or mock vector (lanes 1 and 2), followed by Western blotting with anti-myc, anti-ERdj3, anti-BiP, or anti–β-actin antibody. (D) SH-SY5Y cells were transfected with pERdj3-IRES-EGFP vector. The rate of propidium iodide–positive cells in the enhanced green fluorescence protein (EGFP)–expressing cells (closed circles) or that in the EGFP-negative cells (open circles) was assessed. (E) Vero cells were transfected with ERdj3 expression vector or mock vector. Twenty-four hours after transfection, cells were treated with the indicated concentrations of tunicamycin for 18 hours, followed by MTT assay to assess cell viability
Similar results were obtained when cells were treated with thapsigargin, another ER stressor. Levels of endogenous ERdj3 protein were slightly increased in response to 1 μM thapsigargin treatment of SH-SY5Y cells and Vero cells (Fig 5A, data not shown). However, gene transfer–mediated overexpression of ERdj3 could not change the tolerance against the thapsigargin-induced ER stress in SH-SY5Y cells as shown in Figure 5B,C. Unexpectedly, it was noted that cell viability was rather decreased in the ERdj3-overexpresssed Vero cells as compared with mock-transfected cells, indicating that forced expression of ERdj3 might decrease ER stress tolerance in the case of Vero cells (Fig 5D). Taken together, our results indicated that overexpression of ERdj3 could not enhance the resistance to the ER stress on either cell, whereas overexpression of BiP could increase it.
Fig 5.
Cellular response to thapsigargin treatment. (A) SH-SY5Y cells were treated with 1 μM of thapsigargin, and cell lysates were collected at the indicated time points after the treatment. The lysates were subjected to Western blotting using anti-BiP, anti-ERdj3, or anti–β-actin antibody. (B) An ERdj3-transfected SH-SY5Y clone and a mock-transfected clone were transfected with BiP expression vector or a mock vector, respectively. Twenty-four hours after the transfection, cells were treated with the indicated concentrations of thapsigargin for 18 hours, followed by 3(4,5)-dimethylthiazol-2,5-diphenyl tetrazolium bromide (MTT) assay to assess cell viability. (C) SH-SY5Y cells were transfected with pERdj3-IRES-EGFP vector. The rate of propidium iodide–positive cells in the enhanced green fluorescence protein (EGFP)–expressing cells (closed circles) or that in the EGFP-negative cells (open circles) was assessed. (D) Vero cells were transfected with ERdj3 expression vector or mock vector. Twenty-four hours after the transfection, cells were treated with the indicated concentrations of tunicamycin for 18 hours, followed by MTT assay to assess cell viability
Effect of endogenous ERdj3 knockdown on ER stress tolerance of cells
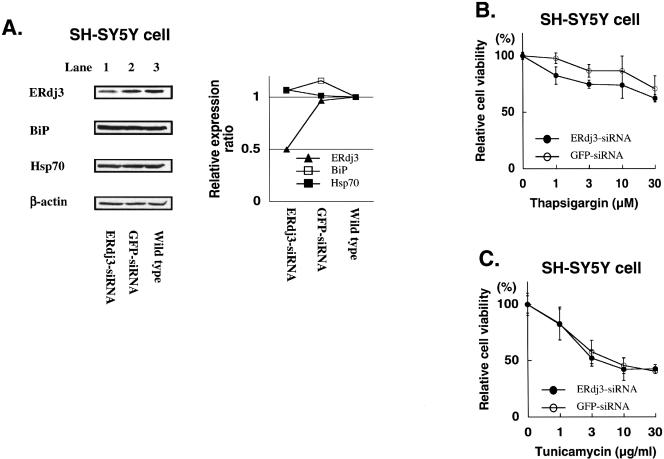
It was possible that endogenous ERdj3 expression level might have been high enough to ensure the ER stress tolerance, explaining that its overexpression in SH-SY5Y cells had no effect. Therefore, we next examined the effect of downregulation of endogenous ERdj3 protein level by siRNA. Transfection of ERdj3-siRNA into SH-SY5Y cells efficiently reduced endogenous ERdj3 protein level, in which approximately 50% of endogenous ERdj3 was suppressed (Fig 6A). ERdj3-specific reduction was confirmed because ERdj3-siRNA could not change the levels of BiP, Hsp70, or actin proteins. GFP-specific siRNA was used as a control siRNA, and it did not affect protein levels of ERdj3 or the other cellular proteins. As shown in Figure 6B, cell viability of SH-SY5Y cells treated with ERdj3-siRNA was reduced as compared with those treated with GFP-siRNA after incubation with thapsigargin.
Fig 6.
Reduction of ERdj3 level by small interfering ribonucleic acid (siRNA) and cellular response against ER stress treatment. (A) SH-SY5Y cells were transfected with either ERdj3-siRNA or green fluorescence protein (GFP)–siRNA. Seventy-two hours after the transfection, cell lysates were collected and subjected to Western blotting using anti-ERdj3, anti-BiP, anti-Hsp70, or anti–β-actin antibody. Signal intensities were quantified, and relative ERdj3, BiP, or Hsp70 expression level was calculated as: relative expression ratio = (sample ERdj3, BiP, or Hsp70 signal intensity/sample actin signal intensity). (B, C) SH-SY5Y cells were transfected with either ERdj3-siRNA or GFP-siRNA. Seventy-two hours after transfection, cells were cultured in media containing the indicated concentrations of thapsigargin (B) or tunicamycin (C) for 48 hours, followed by 3(4,5)-dimethylthiazol-2,5-diphenyl tetrazolium bromide assay to assess cell viability
However, no obvious change in the cell viability was observed after the tunicamycin treatment in SH-SY5Y cells with reduced ERdj3 protein by siRNA (Fig 6C). Taken together, these data indicate that ERdj3 might be involved in the tolerance against thapsigargin-induced ER stress but not against tunicamycin-induced ER stress in SH-SY5Y neuroblastoma cells.
DISCUSSION
We established polyclonal anti-ERdj3 antibody by immunization with the peptide fragment derived from ERdj3 protein. Specificity of the antibody was confirmed by Western blotting. The antibody reacted with exogenously overexpressed ERdj3 protein as well as endogenous ERdj3 proteins derived from a variety of species and effectively immunostained formalin-fixed, paraffin-embedded tissue specimens (data not shown). In this study, the ERdj3-specific antibody was used to probe endogenous ERdj3 proteins.
Because of discrepancy among reports (Yu et al 2000; Lau et al 2001), in this study we investigated whether endogenous ERdj3 is located in the ER or the nucleus. We demonstrated that both endogenous ERdj3 and exogenously expressed ERdj3 were located in the ER. This result was further confirmed by the proteinase K digestion assay using the microsome fraction (Fig 2B) and by an immunohistochemical study, which showed a nonnuclear pattern of ERdj3 staining in a wide variety of human tissues examined. Lau et al (2001) examined the subcellular localization by introducing an expression vector encoding N-terminally GFP-fused ERdj3 protein. Because there exists a signal sequence in the N-terminus of ERdj3 protein (Fig 1A, an italic portion), N-terminally fused GFP might mask the signal sequence and cause altered sorting and subcellular localization of the exogenous ERdj3 fusion protein.
Recently, increasing evidence has suggested important roles of molecular chaperones in the ER when cells are under a condition of ER stress. ERdj4, one of the mammalian Hsp40 family cochaperones in the ER and a structurally homologous molecule to ERdj3, has been reported to enhance ER stress tolerance of cells. In this study, however, overexpression of ERdj3 failed to confer resistance to standard ER stresses such as tunicamycin-induced stress and thapsigargin-induced stress on either Vero cells or SH-SY5Y cells. Besides the ER stressors used in this study, we induced ER stress by using the other stressors, including glucose starvation, 2-mercaptoethanol, lactacystin, heavy metal, or brefeldin A (data not shown). However, overexpression of ERdj3 failed to increase the cell viability after any of these stress treatments, although the levels of endogenous ERdj3 protein were mildly upregulated in SH-SY5Y cells on treatment. These results lead us to hypothesize that the endogenous expression level of ERdj3 in SH-SY5Y cells might be enough in concert with BiP to exert a full tolerance potential.
Therefore, we next tried to reduce the level of endogenous ERdj3 protein by using ERdj3 siRNA. Specific reduction of ERdj3 expression was observed by the siRNA. When the ERdj3 level was reduced in SH-SY5Y cells, cells became more sensitive to the thapsigarigin treatment but not to the tunicamycin treatment. The increased sensitivity of siRNA-treated cells to thapsigargin was not due to a nonspecific effect of the ERdj3 siRNA because tunicamycin treatment did not affect the cell viability. Sensitivity against calcium ionophore–induced stress was also increased in the ERdj3 siRNA–treated cells (data not shown). Thus, we speculated that ERdj3 might play a role in protecting cells from the disruption of calcium homeostasis but not from the inhibition of N-linked glycosylation in the ER. Considering that the Hsp40 family has a greatly divergent region outside the J-domain that may contribute to the varied actions of Hsp70 and BiP (Cheetham and Caplan 1998), it is likely that ERdj3 might play distinct roles in the cellular responses against different stresses.
Acknowledgments
We thank Dr Nakagawara (Chiba Cancer Institute) and Dr Miyazaki (Osaka University) for generously providing human SH-SY5Y neuroblastoma cells and MIN-6 cells, respectively, used in this study.
REFERENCES
- Abdul KM, Terada K, Gotoh T, Hafizur RM, Mori M. Characterization and functional analysis of a heart-enriched DnaJ/ Hsp40 homolog dj4/DjA4. Cell Stress Chaperones. 2002;7:156–166. doi: 10.1379/1466-1268(2002)007<0156:cafaoa>2.0.co;2.1466-1268(2002)007<0156:CAFAOA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma H, Takahashi S, Ishikawa M, Kamiguchi K, Sato N, Poppema S, Fujimoto JI, Kikuchi K. A monoclonal antibody, 3G12, reacts with a novel surface molecule, Hal-1, with high expression in CD30-positive anaplastic large cell lymphomas. Br J Haematol. 1999;106:55–63. doi: 10.1046/j.1365-2141.1999.01520.x.0007-1048(1999)106<0055:AMAGRW>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded protein response. Nat Cell Biol. 2002;2:326–332. doi: 10.1038/35014014.1465-7392(2002)002<0326:DIOBAE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Brightman SE, Blatch GL, Zetter BR. Isolation of a mouse cDNA encoding MTJ1, a new murine member of the DnaJ family of proteins. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a.0378-1119(1995)153<0249:IOAMCE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2.1466-1268(1998)003<0028:SFAEOD>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnea PM, Miranda-Vizuete A, and Bertoli G. et al. 2003 ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem. 278:1059–1066. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x.0376-5067(1994)019<0176:DPMCAS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425.0031-9333(1999)079<0425:CPF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108.0027-8424(1998)095<6108:ROTJIT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SH, Chen JZ, and Ying K. et al. 2003 Cloning and identification of a novel cDNA which encodes a putative protein with a DnaJ domain and a thioredoxin active motif, human macrothioredoxin. Biochem Genet. 41:245–253. [DOI] [PubMed] [Google Scholar]
- Hosoda A, Kimata Y, Tsuru A, Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J Biol Chem. 2003;278:2669–2676. doi: 10.1074/jbc.M208346200.0021-9258(2003)278<2669:JANERP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Imaizumi K, Miyoshi K, Katayama T, Yoneda T, Taniguchi M, Kudo T, Tohyama M. The unfolded protein response and Alzheimer's disease. Biochim Biophys Acta. 2001;1536:85–96. doi: 10.1016/s0925-4439(01)00049-7.0006-3002(2001)1536<0085:TUPRAA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kamiguchi K, Tachibana K, Iwata S, Ohashi Y, Morimoto C. Cas-L is required for beta 1 integrin-mediated costimulation in human T cells. J Immunol. 1999;163:563–568.0022-1767(1999)163<0563:CIRFBI>2.0.CO;2 [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, and Sato N. et al. 1999 Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1:479–485. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211.0890-9369(1999)013<1211:SSFTLO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8.0376-5067(1998)023<0222:TJFATR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kurisu J, Honma A, Miyajima H, Kondo S, Okumura M, Imaizumi K. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells. 2002;8:189–202. doi: 10.1046/j.1365-2443.2003.00625.x.1356-9597(2002)008<0189:EAEDFM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lau PP, Villanueva H, Kobayashi K, Nakamuta M, Chang BH, Chan L. A DnaJ protein, Apobec-1-binding protein-2, modulates apolipoprotein B mRNA editing. J Biol Chem. 2001;276:46445–46452. doi: 10.1074/jbc.M109215200.0021-9258(2001)276<46445:ADPAPM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9.0376-5067(2001)026<0504:TGPSIA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress Chaperones. 2002;7:222–229. doi: 10.1379/1466-1268(2002)007<0222:tmeraa>2.0.co;2.1466-1268(2002)007<0222:TMERAA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2.1466-1268(2000)005<0098:MDHCON>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Grimmer S, Lauvrak SU, Torgersen ML, Skretting G, van Deurs B, Iversen TG. Pathways followed by ricin and Shiga toxin into cells. Histochem Cell Biol. 2002;117:131–141. doi: 10.1007/s00418-001-0346-2.0948-6143(2002)117<0131:PFBRAS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sekino T, Kiyokawa N, and Taguchi T. et al. 2002 Inhibition of Shiga toxin cytotoxicity in human renal cortical epithelial cells by nitrobenzylthioinosine. J Infect Dis. 185:785–796. [DOI] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200.0021-9258(2002)277<15947:IACOAN>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Skowronek MH, Rotter M, Haas IG. Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol Chem. 1999;380:1133–1138. doi: 10.1515/BC.1999.142.1431-6730(1999)380<1133:MCOANM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wesche J, Rapak A, Olsnes S. Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J Biol Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443.0021-9258(1999)274<34443:DORTOT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200.0021-9258(2000)275<24984:HAHCLT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]