ABSTRACT
Paenibacillus sp. strain RC67 was isolated from the Harvard Forest long-term soil warming experiment. The assembled genome is a single contig with 7,963,753 bp and 99.4% completion. Genome annotation suggests that the isolate is of a novel bacterial species.
KEYWORDS: soil microbiology, microbial ecology, bioinformatics
ANNOUNCEMENT
Soil microbes mediate nutrient cycling, but it remains elusive how climate warming impacts microorganisms and their metabolism. The ongoing Harvard Forest soil warming experiment investigates the influence of warming temperature on soils (1). Paenibacillus sp. strain RC67 was isolated from the Harvard Forest in Petersham, Massachusetts and sequenced to understand the impact of warmer climate on soil bacterial genomes. The genome sequence indicates that this isolate is a novel bacterial species belonging to the Paenibacillus genus.
RC67 was isolated from 1 g of mineral soil collected 10 cm below surface at an elevation of 355 m with a steel corer in 2022 from a heated plot (43°N, 72.18°W) using ISP2 (2) medium in aerobic conditions. For gDNA extraction, RC67 was grown on 10% tryptic soy broth (TSB) at 30°C with shaking at 150 rpm until an OD of 0.5 was reached. Cells were pelleted using centrifugation at 4,000 rpm for 15 minutes, and genomic DNA was extracted using CTAB method (3). The library was prepared using Ligation Sequencing Kit SQK-LSK-109 from Oxford Nanopore Technologies (4). The DNA was not sheared or size selected. The genome was sequenced using Oxford Nanopore sequencing technology at SeqCenter (Pittsburgh, PA). R9.4.1 flowcells were run on GridION platform, and Guppy v4.5.5 was used for high-accuracy basecalling to archive Q20 performance and 288,137,203 bp.
The genome was assembled, annotated, and analyzed as part of the Bioinformatics Lab (MICROBIO 590B) course at the University of Massachusetts Amherst (5). Default parameters were used for all software unless otherwise specified. To estimate the genome size, the 16S rRNA gene was sequenced (3), and BLAST (6) determined that the closest related organism with an available genome is Paenibacillus rigui (accession number: NR_116517 [97.03% similarity]) with a 7.173-Mb genome size. Filtlong v0.2.1 (7) filtered 85% of the highest quality reads with minimum length of 1,000 bp to target 40× coverage, which yielded 577,588,430 bp. De novo assembly was performed using Flye v2.8.1 (8). A consensus assembly was generated using Minimap2 v2.17 (9) and Racon v1.4.3 (10), followed by a final polishing using Medaka v1.5.0 (11). The genome was not trimmed, rotated, or circularized.
The RC67-assembled genome was uploaded to KBase (12) for annotation, and quality was assessed using QUAST v4.4 (13). The RC67 genome was annotated using Prokka v1.14.5 (14). The genome assembled into a single contig with an N50 of 7,963,753 bp. CheckM v1.018 (15) indicated a completion of 99.4% and a contamination of 2.07%. Prokka annotation indicated the presence of 23S, 16S, and 5S rRNA genes with 99 tRNA genes for 38 tRNAs, a high-quality assembly (16). Classify microbes with GTDB-Tk v1.7.0 (17) matched RC67 to the Bacteria domain, Bacillus phylum, Bacilli class, Bacillales order, Paenibacillaceae family, and Paenibacillus genus. The closest sequenced genome from the RefSeq database to RC67 was an unclassified Paenibacillus sp. UNC451MF (Fig. 1). FastANI v.0.1.3 (18, 19) determined the ANI of RC67 to Paenibacillus sp. UNC451MF to be 85.91%. The novel isolate may provide a further understanding on the impact of warming climate on Paenibacillus.
Fig 1.
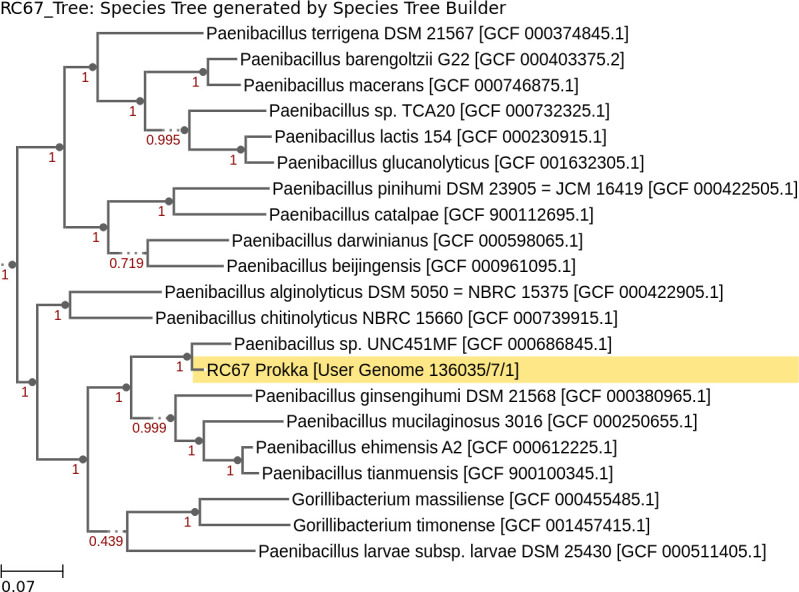
Phylogenetic tree constructed through estimating the approximate maximum likelihood of phylogeny from the concatenated multiple sequence alignments (MSAs). The phylogenetic tree was generated based on default parameters on the Insert Genome Into Species Tree v2.2.0 (20), which uses MSAs for each 49 core universal genes defined by Clusters of Orthologous Groups, and relatedness is determined by alignment similarity.
ACKNOWLEDGMENTS
We are grateful to Drs. Serita Frey and Mel Knorr for maintaining this infrastructure for our community. We thank Ashley Eng for her support and expertise in assisting with the genome assembly.
This project was conducted with support by a grant from the National Science Foundation (No. DEB-1749206). The soil warming experiments at Harvard Forest are maintained with support from the National Science Foundation Long-Term Ecological Research Program (DEB-1832110) and a Long-Term Research in Environmental Biology grant (DEB-1456610).
Contributor Information
Kristen M. DeAngelis, Email: deangelis@microbio.umass.edu.
Irene L. G. Newton, Indiana University, Bloomington, Indiana
DATA AVAILABILITY
The 16S rRNA gene sequence accession number is OQ581834. The raw whole-genome sequence reads are available in GenBank under the BioProject accession number PRJNA949990. The Sequence Read Archive (SRA) accession number is SRR24091469 with a BioSample accession number of SAMN33969015. Public KBase narrative of Paenibacillus sp. strain RC67 genome annotation is available on Kbase under the title “Draft Genome Sequence of Paenibacillus sp. strain iRC67, an Isolate from a Long-Term Forest Soil Warming Experiment in Petersham, Massachusetts.”
REFERENCES
- 1. Melillo JM, Frey SD, DeAngelis KM, Werner WJ, Bernard MJ, Bowles FP, Pold G, Knorr MA, Grandy AS. 2017. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science 358:101–105. doi: 10.1126/science.aan2874 [DOI] [PubMed] [Google Scholar]
- 2. Shirling EB, Gottlieb D. 1966. Methods for characterization of streptomyces species1. Int J Syst Evol Microbiol 16:313–340. doi: 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- 3. DeAngelis KM, Pold G, Topçuoğlu BD, van Diepen LTA, Varney RM, Blanchard JL, Melillo J, Frey SD. 2015. Long-term forest soil warming alters microbial communities in temperate forest soils. Front Microbiol 6:104. doi: 10.3389/fmicb.2015.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eng AY, Narayanan A, Alster CJ, DeAngelis KM. 2023. Thermal adaptation of soil microbial growth traits in response to chronic warming. bioRxiv. doi: 10.1101/2023.05.19.541531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeAngelis K, Choudoir M, Eng A, Morrow M. 2023. Bioinformatics lab: a course-based undergraduate research experience curriculum. Microbiol Educ Mater. 10.7275/g0fg-5939. [DOI] [Google Scholar]
- 6. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 7. Foster-Nyarko E, Cottingham H, Wick RR, Judd LM, Lam MMC, Wyres KL, Stanton TD, Tsang KK, David S, Aanensen DM, Brisse S, Holt KE. 2023. “Corrigendum: 'nanopore-only assemblies for genomic surveillance of the global priority drug-resistant pathogen, Klebsiella pneumoniae'” Microb Genom 9:mgen001084. doi: 10.1099/mgen.0.001084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. 5. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 9. Li H, Birol I. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. 2023. Medaka. Python. Oxford Nanopore Technologies. [Google Scholar]
- 12. Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, Sneddon MW, Henderson ML, Riehl WJ, Murphy-Olson D, Chan SY, Kamimura RT, Kumari S, Drake MM, Brettin TS, Glass EM, Chivian D, Gunter D, Weston DJ, Allen BH, Baumohl J, Best AA, Bowen B, Brenner SE, Bun CC, Chandonia J-M, Chia J-M, Colasanti R, Conrad N, Davis JJ, Davison BH, DeJongh M, Devoid S, Dietrich E, Dubchak I, Edirisinghe JN, Fang G, Faria JP, Frybarger PM, Gerlach W, Gerstein M, Greiner A, Gurtowski J, Haun HL, He F, Jain R, Joachimiak MP, Keegan KP, Kondo S, Kumar V, Land ML, Meyer F, Mills M, Novichkov PS, Oh T, Olsen GJ, Olson R, Parrello B, Pasternak S, Pearson E, Poon SS, Price GA, Ramakrishnan S, Ranjan P, Ronald PC, Schatz MC, Seaver SMD, Shukla M, Sutormin RA, Syed MH, Thomason J, Tintle NL, Wang D, Xia F, Yoo H, Yoo S, Yu D. 2018. Kbase: the United States department of energy systems biology knowledgebase. 7. Nat Biotechnol 36:566–569. doi: 10.1038/nbt.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mikheenko A, Valin G, Prjibelski A, Saveliev V, Gurevich A. 2016. Icarus: visualizer for de novo assembly evaluation. Bioinformatics 32:3321–3323. doi: 10.1093/bioinformatics/btw379 [DOI] [PubMed] [Google Scholar]
- 14. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 15. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Schriml L, Banfield JF, Hugenholtz P, Woyke T. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. 2019. GTDB-TK: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- 20. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16S rRNA gene sequence accession number is OQ581834. The raw whole-genome sequence reads are available in GenBank under the BioProject accession number PRJNA949990. The Sequence Read Archive (SRA) accession number is SRR24091469 with a BioSample accession number of SAMN33969015. Public KBase narrative of Paenibacillus sp. strain RC67 genome annotation is available on Kbase under the title “Draft Genome Sequence of Paenibacillus sp. strain iRC67, an Isolate from a Long-Term Forest Soil Warming Experiment in Petersham, Massachusetts.”


