Abstract
We studied the role of the regulatory gene aflR and its product, AflR, in the biosynthesis of aflatoxin in Aspergillus. Western blot and enzyme-linked immunosorbent assay analyses revealed that aflatoxin B1 accumulation was directly related to AflR expression and was regulated by various environmental and nutritional conditions, including temperature, air supply, carbon source, nitrogen source, and zinc availability. Expression of an aflatoxin biosynthetic pathway structural gene, omtA, was regulated by the presence of AflR. Induction patterns for aflR mRNA and AflR were correlated with that for omtA mRNA in an aflatoxin-producing strain of Aspergillus parasiticus. Analysis of non-aflatoxin-producing strains of A. flavus, A. sojae, and A. oryzae grown in medium suitable for aflatoxin B1 production showed that both aflR mRNA and AflR production were present; however, omtA mRNA production was not detected in any of these examined strains. AflR in the A. oryzae strain was regulated by carbon source and temperature in a manner similar to that seen with A. parasiticus.
Aflatoxin B1 (AFB1) is one of the most potent naturally occurring carcinogens and mutagens and is produced primarily by Aspergillus flavus and A. parasiticus (26). Several genes and enzymes involved in aflatoxin biosynthesis have been identified, cloned, and purified (2); they include a regulatory gene locus, aflR, from A. flavus and A. parasiticus (4, 25). Metabolite feeding studies suggest that aflR regulates gene expression and the activity of other enzymes in the aflatoxin biosynthetic pathway (4, 25). Northern hybridization analysis with A. parasiticus has indicated that aflR mRNA is required for the transcriptional expression of several aflatoxin biosynthetic structural genes, including nor-1, ver-1, and omtA (6). The aflR gene encodes a 47-kDa protein, AflR, which has a putative DNA-binding domain of the GAL4-type binuclear zinc finger motif (20, 32). Southern analyses have shown that genomic DNA from species in Aspergillus section Flavi, e.g., A. flavus, A. parasiticus, A. oryzae, and A. sojae, hybridizes with genomic DNA probes derived from aflR and omtA (18, 32). Because A. oryzae and A. sojae are widely used in the food industry, it is important to know whether aflR and omtA can be expressed in the nonaflatoxigenic Aspergillus species under conditions that support aflatoxin production. Recently, Klich et al. (19) found that RNA from some A. sojae isolates hybridizes to aflR but not to omtA.
The production of aflatoxin by toxigenic aspergilli is regulated by environmental and nutritional factors, such as temperature, oxygen availability, carbon source, and nitrogen source (21). Our objective was to determine whether aflR expression is regulated by these factors. We used polyclonal antibodies specific for AflR protein (20) to examine the relationship between AflR level and aflatoxin formation. In addition, we also examined the relationship between aflR expression and the appearance of omtA, a gene that encodes the 42-kDa sterigmatocystin (ST) O-methyltransferase (O-MTase), an enzyme involved in the final stages of the aflatoxin biosynthetic pathway (17, 34).
MATERIALS AND METHODS
Materials.
A. parasiticus NRRL 2999 and NRRL 3386; A. flavus NRRL 3357, NRRL 5565, NRRL 6341, and NRRL 482; A. oryzae NRRL 451 and NRRL 1911; A. sojae NRRL 5596 and NRRL 6271; and A. nomius NRRL 3161 were obtained from the National Center for Agricultural Utilization Research, U.S. Department of Agriculture, Peoria, Ill. A. flavus SRRC 2111 and A. versicolor SRRC 111 were provided by the Southern Regional Research Center, U.S. Department of Agriculture, New Orleans, La. A. nidulans FGSC 26 and Cochliobolus sativus ATCC 24081 were provided by Nancy Keller, Texas A & M University, College Station. All strains were maintained on potato dextrose agar at 4°C. Tween 20, Ponceau S, 4-chloro-1-naphthol, proteinase K, and RNase A were obtained from Sigma, St. Louis, Mo. Goat anti-rabbit immunoglobulin G–horseradish peroxidase conjugate was obtained from Boehringer Mannheim Biochemicals, Indianapolis, Ind. Acrylamide, N,N′-methylene-bis-acrylamide, ammonium persulfate, and nitrocellulose membranes were from BioRad, Richmond, Calif. Prestained broad-range protein molecular weight markers were purchased from New England Biolabs, Beverly, Mass. Except as otherwise described, all restriction enzymes and plasmid DNAs were obtained from Promega, Madison, Wis. T7 RNA polymerase and pT7 18S RNA antisense control template were purchased from Ambion, Austin, Tex. All other chemicals and organic solvents used were of reagent grade or better.
Culture medium.
Peptone-mineral salts (PMS) medium served as the non-aflatoxin-supporting medium (1). Glucose-mineral salts (GMS) medium (peptone was replaced by glucose in PMS medium) and sucrose–low-salt (SLS) medium (12) were used as the aflatoxin-supporting media. Zinc-deficient medium (GMS−Zn) had the same composition as GMS medium except that the concentration of ZnSO4 · 7H2O was 0.25 mg liter of medium−1 (11). Experiments on the nitrogen source were conducted with GMS medium containing either 4 g of ammonium nitrate liter−1 (GMS+NH4NO3) or 4 g of sodium nitrate liter−1 (GMS+NaNO3) instead of 4 g of (NH4)2SO4. The nitrogen levels with GMS+NH4NO3, GMS+NaNO3, and (NH4)2SO4 were 1.4, 0.66, and 0.8 g liter−1, respectively.
Culture method.
To perform the nutritional and temperature shift assays (9), frozen spores (kept at −20°C in 15% glycerol for up to 1 month) were inoculated into PMS medium and grown at 29°C for 48 or 64 h on a rotatory shaker (150 rpm). The resulting mycelia were filtered through two layers of cheesecloth and washed with sterile 0.85% NaCl. The wet weight of the mycelia was determined after they were squeezed and blotted with paper towels. A portion of the mycelia (0.8 to 1 g [wet weight]) was transferred to a flask containing 100 ml of fresh PMS or GMS medium and cultured on a rotatory shaker (150 rpm) at 29 or 37°C. Duplicate flasks were removed at each harvest, and mycelia were collected by filtration through cheesecloth and then immediately frozen in liquid nitrogen. Culture filtrates were used in a direct competitive enzyme-linked immunosorbent assay for the determination of AFB1 concentrations (8). For the batch fermentation culture, fungal spores were inoculated into 250-ml Erlenmeyer flasks, each containing 100 ml of GMS medium, GMS−Zn, GMS+NaNO3, or GMS+NH4NO3. All flasks were incubated at 29°C on a rotatory shaker (150 rpm). Mycelia and filtrates from duplicate flasks were collected and analyzed as described above. All studies were conducted with multiple flasks and were repeated at least once.
Analysis of the AflR protein in fungal cell extracts.
Preparation of cell protein extracts and Western blotting analysis of AflR were conducted as described by Liu et al. (20). The relative AflR levels in each sample on the filter membrane of a Western blot were determined by densitometry (SI personal densitometer; Molecular Dynamics Inc.).
Enzymatic assay of O-MTase.
O-MTase activity for conversion of ST to O-methyl-ST was determined as described by Keller et al. (17).
Generation of riboprobes.
To construct a plasmid for the generation of an aflR-specific cRNA probe, aflR cDNA from A. flavus (4) was digested with SmaI-BssHII, and the resulting 185-bp fragment was subcloned into the SmaI-digested pGEM-7Zf(+) vector. The plasmid was linearized with BamHI and transcribed in vitro by use of T7 RNA polymerase with [α-32P]rCTP (800 Ci/mmol; Dupont) as a 272-nucleotide RNA riboprobe. This probe was used to protect a 189-nucleotide fragment corresponding to the zinc finger motif in the putative AflR protein. To construct a plasmid for the generation of omtA-specific cRNA, omtA DNA from A. parasiticus (35) was digested with EcoRI-XbaI, and the resulting 1,754-bp fragment was subcloned into the EcoRI-XbaI-digested pGEM-11Zf(+) vector. The plasmid was linearized with NcoI and transcribed as described above to generate a 178-nucleotide riboprobe. This probe was used to protect a 150-nucleotide fragment corresponding to a highly conserved region of the omtA gene in which differences in only 2 nucleotides are present between A. parasiticus and A. flavus (35). The sizes of the riboprobes and their protected fragments were confirmed on a denaturing sequencing gel along with a known DNA sequence. An 18S rRNA riboprobe generated from plasmid pT7 18S RNA (Ambion) was used as an internal control.
Isolation and analysis of fungal RNA.
Mycelia were collected by filtration through cheesecloth and quickly frozen in liquid nitrogen. Total RNA was isolated and purified by a single-step method (7) with RNA STAT-60 isolation reagents (TEL-TEST “B” Inc., Friendswood, Tex.). An RNase protection assay (RPA) was performed as described by Kaestner et al. (16) with a slight modification. Total RNA samples were hybridized with 5 × 105 cpm of 32P-labeled aflR cRNA or omtA cRNA probes in a solution containing 80% formamide, 100 mM Tris-HCl (pH 7.4), 2.5 N NaCl, and 10 mM EDTA at 45°C for 16 to 18 h. Each hybridized sample was digested with RNase A at 37°C for 60 min, treated with proteinase K at 37°C for 20 min, extracted with phenol-chloroform, and precipitated with isopropanol at −20°C. After centrifugation, the pellets were resuspended in sequencing gel loading buffer (27). Protected fragments were resolved on a 6% polyacrylamide–8 M urea denaturing gel. The gel was autoradiographed on Kodak BIOMAX MS film at −70°C for 40 h (aflR) or 16 h (omtA). The hybridized mRNA levels for aflR and omtA were estimated by densitometry.
RESULTS
Effect of temperature on AflR expression.
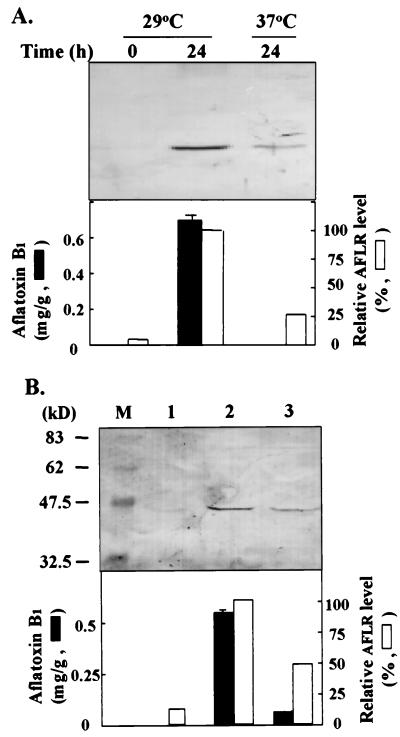
We grew A. parasiticus NRRL 2999 in PMS medium at 29°C and then transferred it to GMS medium and incubated it at either 29 or 37°C. Neither AFB1 nor AflR was detected in the 29°C culture in PMS medium (Fig. 1A). However, after transfer to GMS medium, AFB1 production occurred at 29°C but not at 37°C. Western blot analysis with anti-AflR antibodies showed that the level of AflR in the 37°C GMS medium culture was four times lower than that in the 29°C GMS medium culture.
FIG. 1.
Effects of temperature (A) and agitation (B) on AflR (AFLR) expression. (A) A. parasiticus NRRL 2999 was first grown in PMS medium at 29°C for 64 h and then transferred to fresh GMS medium and grown at either 29 or 37°C for another 24 h. Duplicate fungal mycelial samples were harvested at time zero and after being transferred. Fungal extracts were prepared, subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (50 μg of protein/lane), and tested by Western blot analysis with anti-AflR antibodies. AflR levels in cultures grown under different conditions, as measured by Western blotting, are shown in the top panel. The accumulation of AFB1 (milligrams of AFB1 per gram of mycelial wet weight) in culture filtrates and the relative AflR levels in Western blots are shown in the bottom panel. The intensity of the AflR signal for the 29°C GMS medium culture was assigned as 100%. (B) A. parasiticus NRRL 2999 was first grown in PMS medium for 64 h with shaking. The harvested mycelia were then (i) transferred to PMS medium with continued shaking for 24 h (lane 1), (ii) transferred to GMS medium with continued shaking for 24 h (lane 2), or (iii) transferred to GMS medium with 12 h of shaking and 12 h of no shaking (lane 3). The intensity of the AflR signal for the 24-h shaking GMS medium culture was assigned as 100%. Lane M represents protein markers.
Effect of agitation on AflR accumulation.
A. parasiticus NRRL 2999 was grown in PMS medium with shaking, and the harvested mycelia were (i) transferred to fresh PMS medium and grown for 24 h with shaking; (ii) transferred to GMS medium and grown for 24 h with shaking; and (iii) transferred to GMS medium and grown for 12 h with shaking and then for another 12 h without shaking. More mycelia were obtained from the cultures agitated for 24 h than from those agitated for 12 h (4 versus 2.8 g). No AFB1 was detected in 24-h shaking PMS medium cultures (Fig. 1B), even though trace amounts of AflR were found. The AFB1 concentration in the GMS medium cultures with 12 h of shaking and 12 h of static incubation (66 μg/g of mycelial wet weight) was eight times lower than that in the 24-h shaking GMS medium cultures (550 μg/g). Western blot analysis revealed that the amount of AflR in the GMS medium cultures with 12 h of shaking and 12 h of static incubation was half that in the 24-h shaking GMS medium cultures.
Effect of nutritional factors on AflR accumulation.
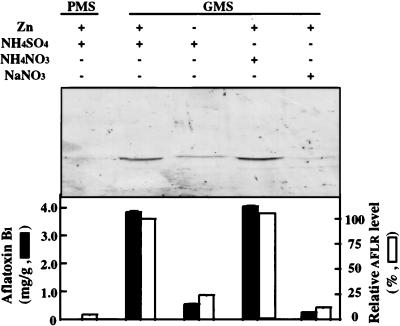
We used batch fermentation to determine the effects of nutritional factors, such as trace metals, zinc, and nitrogen source, on the accumulation of AflR in A. parasiticus. PMS medium, GMS medium, GMS−Zn, GMS+NH4NO3, and GMS+NaNO3 were used as the culture media. The average wet weights of the mycelial mass harvested from these media were 2.0, 2.7, 1.5, 2.8, and 2.4 g/100 ml of medium, respectively. AFB1 formation in GMS−Zn and GMS+NaNO3 cultures was reduced 85 and 93%, respectively (Fig. 2), compared to that in GMS medium. Western blot analysis revealed that AflR levels in GMS−Zn and GMS+NaNO3 cultures were 25 and 12% that in GMS medium, respectively. However, the AFB1 concentration and the AflR level in GMS+NH4NO3 cultures were slightly higher (<5%) than those in GMS medium.
FIG. 2.
Effect of nutritional factors on AflR (AFLR) expression. A. parasiticus NRRL 2999 was grown in various media (PMS, GMS, GMS−Zn, GMS+NH4NO3, and GMS+NaNO3) for 70 h. Duplicate samples in each group were collected, subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (50 μg of protein/lane), and tested by Western blot analysis with anti-AflR antibodies. Results of the Western blot analysis for AflR are shown in the top panel. The accumulation of AFB1 and relative AflR levels in Western blots are shown in the bottom panel. The intensity of the AflR signal for the GMS medium culture was considered 100%.
Kinetic expression of the aflR and omtA genes in A. parasiticus.
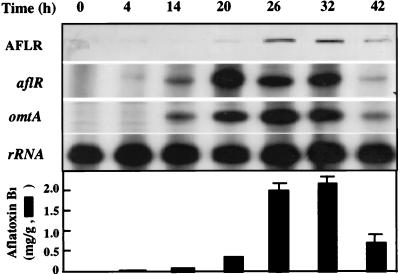
The nutritional shift assay was used to study the kinetic expression of the aflR regulatory gene and the omtA structural gene. AFB1 was not detected in PMS medium, but the concentration of the toxin increased with time after the shift to GMS medium (Fig. 3). The maximum accumulation of AFB1 occurred 32 h after the shift and then declined gradually. aflR mRNA appeared 4 h after the shift to GMS medium, peaked at approximately 20 h, and decreased dramatically after 32 h. The AflR protein was clearly detected at 20 h, reached its maximum at about 26 to 32 h, and then decreased. omtA mRNA was clearly detected at 14 h, peaked at 26 h, and decreased thereafter. In all of these analyses, the accumulation of aflR transcripts correlated with omtA expression and AFB1 formation.
FIG. 3.
Kinetics of aflR and omtA expression in A. parasiticus following a nutritional shift. A. parasiticus NRRL 3386 was first grown in PMS medium for 48 h and then transferred to GMS medium and grown for another 4 to 42 h under the same incubation conditions. Fungal extracts and RNA of duplicate samples were isolated before transfer (0 h) and at the indicated times following transfer. Fungal extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (60 μg of protein/lane) and then Western blot analysis with anti-AflR (AFLR) antibodies. RNA samples at 25 and 8 μg were subjected to RPA with either aflR cRNA and an 18S rRNA riboprobe or omtA cRNA and an 18S rRNA riboprobe as the probes, respectively. The results are shown in the top panel. The accumulation of AFB1 is shown in the bottom panel.
Expression of aflR and omtA in selected species in Aspergillus section Flavi.
Cell protein extracts and total RNA samples obtained from several fungal species, including aflatoxigenic A. parasiticus and A. flavus and nonaflatoxigenic A. flavus, A. sojae, and A. oryzae, were subjected to Western blot analysis and RPAs, respectively. aflR mRNA and AflR were found in all eight strains grown in GMS medium for 24 h (Fig. 4). However, omtA transcripts and O-MTase activity were detected only in the aflatoxigenic strains A. parasiticus NRRL 3386 and A. flavus NRRL 3357. We confirmed, by Southern hybridization, that omtA was present in all tested strains (data not shown).
FIG. 4.
Expression of aflR and omtA in selected nonaflatoxigenic species. Eight fungal species were first grown in PMS medium for 64 h and then distributed to GMS medium and grown for another 24 h. Levels of AflR (AFLR) and of aflR and omtA transcripts were determined as described in the legend to Fig. 3. Lanes 1′ and 1, A. parasiticus NRRL 3386; lane 2, A. sojae NRRL 5596; lanes 3′ and 3, A. flavus NRRL 3357; lane 4, A. flavus SRRC 2111; lane 5, A. flavus NRRL 6341; lane 6, A. flavus NRRL 482; lane 7, A. oryzae NRRL 451; lane 8, A. oryzae NRRL 1911. Samples in lanes 1′ and 3′ were collected at transfer; others were collected after being transferred to GMS medium and grown for 24 h. The results are shown in the top panel. The accumulation of AFB1 in the culture filtrate and O-MTase activity are shown in the bottom panel.
Regulated expression of the aflR and omtA genes in A. oryzae.
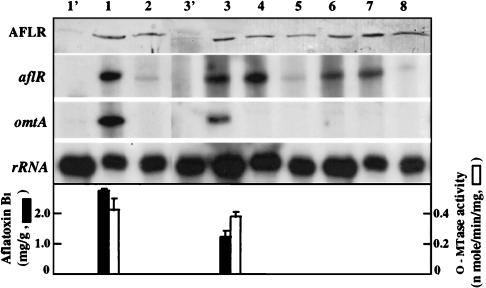
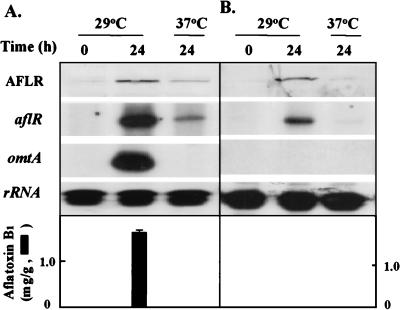
We used a nutritional shift assay to investigate the regulated expression of aflR and omtA in A. oryzae NRRL 451 and A. parasiticus NRRL 3386. Both aflR and omtA genes in A. parasiticus grown at 29°C in GMS medium were transcribed, and AFB1 accumulated (Fig. 5A). Neither AFB1 nor omtA transcripts were detected in cultures after a transfer to 37°C, even though low levels of aflR transcripts and AflR were found. The expression of the aflR gene in A. oryzae was regulated by carbon source and temperature, as it was in A. parasiticus (Fig. 5B). AflR was almost undetectable in A. oryzae grown in PMS medium; AflR accumulated in the 29°C GMS medium culture but not in the 37°C culture. However, despite the presence of AflR, neither omtA mRNA nor AFB1 was detected in A. oryzae cultures.
FIG. 5.
Regulated expression of aflR and omtA in A. parasiticus (A) and A. oryzae (B). A. parasiticus NRRL 3386 (A) and A. oryzae NRRL 451 (B) grown at 29°C in PMS medium for 48 h were transferred to GMS medium and grown at 29 or 37°C for another 24 h. Fungal mycelia were harvested at the time of transfer (0 h) and after transfer (24 h). Western blot analysis and RPA were conducted as described in the legend to Fig. 3. The results are shown in the top panels. The accumulation of AFB1 is shown in the bottom panels.
Expression of AflR in various fungal species.
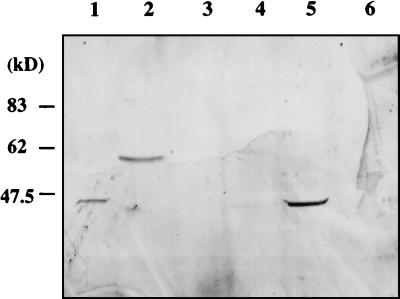
We grew six additional fungal species in SLS medium for 70 h by using the batch fermentation approach to determine if AflR or related proteins were present. Anti-AflR antibodies recognized a single band in extracts obtained from species in Aspergillus section Flavi, including A. nomius and nonaflatoxigenic A. flavus and A. sojae (Fig. 6). The size of the detected protein from A. nomius was approximately 58 kDa, which is larger than the expected size of AflR (47 kDa). No AflR was detected in the protein preparations from A. nidulans, A. versicolor, and C. sativus. The appearance of AflR in non-aflatoxin-producing species was not limited to aflatoxin-supporting culture media, such as GMS and SLS media, or dependent on culture conditions.
FIG. 6.
Western blot analysis of AflR in fungal extracts of various species. Various fungal species were grown in SLS medium at 29°C for 70 h. Protein extracts from these cultures were prepared, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (60 μg of protein/lane), and tested by Western blot analysis with anti-AflR antibodies. Lane 1, A. flavus NRRL 5565; lane 2, A. nomius NRRL 3161; lane 3, A. versicolor SRRC 111; lane 4, A. nidulans FGSC 26; lane 5, A. sojae NRRL 6271; lane 6, C. sativus ATCC 24081. Molecular masses for the marker proteins are indicated.
DISCUSSION
Although intensive studies of the factors affecting aflatoxin formation have been carried out in the last three decades, the mechanism that regulates aflatoxin formation remains poorly understood. The availability of readily utilizable carbohydrates, levels of certain intermediates in primary metabolism, and the cytoplasmic anabolic reduction charge ratio of NADPH to NADP are all important (24, 29). More recently, most of the genes involved in the biosynthesis of aflatoxin were identified, and evidence for a complex interaction between environmental and nutritional factors and the genetic characteristics of the fungi was obtained (21). In the present study, we found that the expression of aflR was regulated by environmental and nutritional factors that have long been known to affect aflatoxin formation.
Our results suggest that a high temperature (i.e., 37°C) suppresses the formation of aflatoxin through the down regulation of AflR formation. Temperatures above 35°C are known to be unsuitable for aflatoxin formation in A. flavus (28). Feng et al. (9) suggested that the failure of aflatoxin production in A. parasiticus at 37°C is due to the inhibition of transcription of certain aflatoxin biosynthetic genes. Our results also indicate that AflR repression at a nonpermissive temperature may be responsible for the inhibition of transcription of structural genes, including omtA. However, as shown in Fig. 1A and 5A, the presence of low levels of AflR at 37°C cannot induce aflatoxin production and omtA transcription. Chang et al. (6) found that a transformant of A. parasiticus containing an extra copy of the aflR gene did not overcome the inhibitory effect of a high temperature on aflatoxin formation. Thus, although our data suggest that a high temperature suppresses aflatoxin formation by down regulating AflR, it is likely that, in addition to AflR, another factor(s) may also play a critical role in the temperature-induced regulation of aflatoxin biosynthesis.
Our results suggest that nitrate negatively regulates the expression of AflR, leading to the suppression of aflatoxin production. Previous studies showed that nitrate exerted a suppressive effect on aflatoxin biosynthesis (15) and inhibited the transcription of the aflR gene (6). We have demonstrated that there is a direct correlation among nitrate, AflR level, and aflatoxin formation. Chang et al. (6) demonstrated that a transformant of A. parasiticus containing an additional copy of the aflR gene restored the nitrate-induced repression of the transcription of aflR and several pathway genes. Likewise, Flaherty and Payne (10) showed that the overexpression of aflR activated all related pathway genes that were inhibited by nitrate, although it did not alleviate nitrate inhibition on aflatoxin production. Thus, the mechanism of nitrate repression of aflatoxin production is more complex than previously envisaged and is not solely dependent on aflR.
Zinc is essential for aflatoxin biosynthesis (23, 31), and we found that the failure of aflatoxin production in Zn-deficient medium was also due to the repression of AflR formation. While Zn serves as a prosthetic group of many enzymes (33) and affects both the primary metabolism and the secondary metabolism of many fungi (30), Johnston (13) found that zinc is required for the structural integrity and DNA-binding activity of the zinc finger motif of the GAL4 regulatory protein.
AflR was found in all of the nonaflatoxigenic strains in Aspergillus section Flavi examined; however, omtA was not expressed, even though it was present in all of the strains. Thus, AflR in the nonaflatoxigenic aspergilli is insufficient to activate omtA, and the presence of AflR in these strains is not correlated with aflatoxin formation. The role of AflR in the nonaflatoxigenic aspergilli is not clear. It is possible that a repressor in nonaflatoxigenic strains interacts with AflR to suppress aflatoxin formation. Some fungal regulatory proteins interact with a repressor(s) to regulate gene expression (14). For example, in the absence of galactose, the GAL80 protein binds to the acidic C-terminal end of the GAL4 protein, preventing it from transcriptionally activating the expression of several genes responsible for the conversion of galactose (22). Because of the similarity of the C-terminal regions of AflR and GAL4 (6), they may be regulated by a similar mechanism.
No AflR homolog was found in ST-producing strains when polyclonal antibodies raised against the recombinant AflR protein were used (Fig. 6). However, Brown et al. (3) identified an aflR gene homolog in the ST gene cluster in A. nidulans. Because the putative A. nidulans aflaR gene product is only 33% identical to AflR in amino acid sequence (36), it is likely that they do not have similar epitopes for antibody recognition. On the other hand, due to the different growth characteristics of various species, we cannot rule out the possibility that growing conditions and time points for harvesting the mycelia of examined isolates were not suitable for accumulating an AflR homolog.
Solution hybridization-RPA was developed for efficiently detecting trace amounts of aflR mRNA. Because only a specific nucleotide transition (C in A. flavus and A. oryzae and T in A. parasiticus and A. sojae at position 102) occurs in the zinc finger motif (5) in these four species, riboprobes complementary to the motif and its downstream region were generated and successfully used to detect aflR mRNA. However, due to the slight mismatch of riboprobes, aflR mRNA levels in various isolates could not be quantitatively compared with each other. This mismatch also led to the inconsistency between aflR mRNA levels and AflR protein amounts in some examined isolates (Fig. 4). Likewise, riboprobes complementary to the omtA gene of A. parasiticus did not hybridize perfectly with omtA transcripts from other species. In addition to the absence of omtA mRNA determined by RPA, Western blot analysis (data not shown) and methyltransferase activity assays confirmed the lack of the omtA gene product, ST O-MTase, in the nonaflatoxigenic isolates.
In conclusion, our data indicate that environmental and nutritional factors exert their effect on aflatoxin formation through the presence of AflR. This report is the first to show that the regulated expression of aflR also occurs in nonaflatoxigenic species, including A. oryzae, which is widely used in the food industry. An understanding of the regulation of AflR formation and the role of this protein in aflatoxigenic and nonaflatoxigenic strains could lead to novel approaches to reduce aflatoxin levels in food supplies. The immunochemical method and riboprobes that we have developed could be used in the identification of these regulatory mechanisms.
ACKNOWLEDGMENTS
This work was supported by grant NC-129 from the College of Agricultural and Life Sciences, University of Wisconsin—Madison, and a cooperative research agreement with the U.S. Department of Agriculture Agricultural Research Service (58-6435-1-116).
We thank Deepak Bhatnagar, U.S. Department of Agriculture, for supplying crude AFLR protein and valuable discussions and Barbara Cochrane for help in preparing the manuscript.
Footnotes
Contribution 314 from the Environmental Toxicology Center of the University of Wisconsin—Madison.
REFERENCES
- 1.Abdollahi A, Buchanan R L. Regulation of aflatoxin biosynthesis: characterization of glucose as an apparent inducer of aflatoxin production. J Food Sci. 1981;46:143–146. [Google Scholar]
- 2.Bennett J W, Bhatnagar D, Chang P-K. The molecular genetics of aflatoxin biosynthesis. In: Powell K A, et al., editors. The genus Aspergillus. New York, N.Y: Plenum Publishing Corp.; 1994. pp. 51–58. [Google Scholar]
- 3.Brown D W, Yu J-H, Kelkar H S, Fernandes M, Nesbitt T C, Keller N P, Adams T H, Leonard T J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang P-K, Cary W J, Bhatnagar D, Cleveland T E, Bennett J W, Linz J E, Woloshuk C P, Payne G A. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang P-K, Bhatnagar D, Cleveland T E, Bennett J W. Sequence variability in homologs of the aflatoxin pathway gene aflR distinguishes species in section Flavi. Appl Environ Microbiol. 1995;61:40–43. doi: 10.1128/aem.61.1.40-43.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang P-K, Ehrlich K C, Yu J, Bhatnagar D, Cleveland T E. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl Environ Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Chu F S, Fan T S L, Zhang G S, Xu Y C, Faust S, McMahon P L. Improved enzyme-linked immunosorbent assay for aflatoxin B1 in agricultural commodities. J Assoc Off Anal Chem. 1987;70:854–857. [PubMed] [Google Scholar]
- 9.Feng G H, Chu F S, Leonard T J. Molecular cloning of genes related to aflatoxin biosynthesis by differential screening. Appl Environ Microbiol. 1992;58:455–460. doi: 10.1128/aem.58.2.455-460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty J E, Payne G A. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microbiol. 1997;63:3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S K, Maggom K K, Venkitasubramanian T A. Effect of zinc on tricarboxylic acid cycle intermediates and enzymes in relation to aflatoxin biosynthesis. J Gen Microbiol. 1977;99:43–48. doi: 10.1099/00221287-99-1-43. [DOI] [PubMed] [Google Scholar]
- 12.Hamid A B, Smith J E. Degradation of aflatoxin by Aspergillus flavus. J Gen Microbiol. 1987;133:2023–2029. doi: 10.1099/00221287-133-8-2023. [DOI] [PubMed] [Google Scholar]
- 13.Johnston M. Genetic evidence that zinc is an essential cofactor in the DNA binding domain of GAL4 protein. Nature (London) 1987;328:353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- 14.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kachholz T, Demain A L. Nitrate repression of averufin and aflatoxin biosynthesis. J Nat Prod. 1983;46:499–506. [Google Scholar]
- 16.Kaestner K H, Ntambi J M, Kelley T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: a second differentially expressed gene encoding stearoyl-coenzyme A desaturase. J Biol Chem. 1989;264:14755–14761. [PubMed] [Google Scholar]
- 17.Keller N P, Dischinger H C, Cleveland T E, Ullah A H J. Purification of a 40-kilodalton methyltransferase active in the aflatoxin biosynthetic pathway. Appl Environ Microbiol. 1993;59:479–484. doi: 10.1128/aem.59.2.479-484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klich M A, Yu J, Chang P-K, Mullaney E J, Bhatnagar D, Cleveland T E. Hybridization of genes involved in aflatoxin biosynthesis to DNA of aflatoxigenic and non-aflatoxigenic Aspergilli. Appl Microbiol Biotechnol. 1995;44:439–443. doi: 10.1007/BF00169941. [DOI] [PubMed] [Google Scholar]
- 19.Klich M A, Montalbano B, Ehrlich K. Northern analysis of aflatoxin biosynthesis genes in Aspergillus parasiticus and Aspergillus sojae. Appl Microbiol Biotechnol. 1997;47:246–249. [Google Scholar]
- 20.Liu B H, Brewer J F, Flaherty J E, Payne G A, Bhatnagar D, Chu F S. Immunochemical identification of AFLR, a regulatory protein involved in aflatoxin biosynthesis. Food Agric Immunol. 1998;9:289–298. [Google Scholar]
- 21.Luchese R H, Harrigan W F. Biosynthesis of aflatoxin—the role of nutritional factors. J Bacteriol. 1993;74:5–14. doi: 10.1111/j.1365-2672.1993.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 22.Ma J, Ptashne M. The carboxyl-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 23.Marsh P B, Simpson M E, Trucksess M W. Effects of trace metals on the production of aflatoxins by Aspergillus parasiticus. Appl Microbiol. 1975;30:52–57. doi: 10.1128/am.30.1.52-57.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niehaus W G, Dilts R P. Purification and characterization of mannitol dehydrogenase from Aspergillus parasiticus. J Bacteriol. 1982;151:243–250. doi: 10.1128/jb.151.1.243-250.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne G A, Nystrom G J, Bhatnagar D, Cleveland T E, Woloshuk C P. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roebuck B D, Maxuitenko Y Y. Biochemical mechanisms and biological implications of the toxicity of aflatoxins as related to aflatoxin carcinogenesis. In: Eaton D L, Groopman J D, editors. The toxicology of aflatoxins: human health, veterinary, and agricultural significance. San Diego, Calif: Academic Press, Inc.; 1994. pp. 27–41. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schindler A F, Palmer J G, Eisenberg W V. Aflatoxin production by Aspergillus flavus as related to various temperatures. Appl Microbiol. 1967;15:1006–1009. doi: 10.1128/am.15.5.1006-1009.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shantha T, Murthy V S. Influence of tricarboxylic acid cycle intermediates and related metabolites on the biosynthesis of aflatoxin by resting cells of Aspergillus flavus fungi. Appl Environ Microbiol. 1981;42:758–761. doi: 10.1128/aem.42.5.758-761.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinberg E D. Biosynthesis of microbial metabolites—regulation by mineral elements and temperature. In: Krumphanzl V, Sikyta B, Vanek Z, editors. Overproduction of microbial products. London, England: Academic Press, Inc.; 1982. pp. 181–194. [Google Scholar]
- 31.Wlostowski T, Bernacka B, Pepinski W. The relationship of mycelial zinc to aflatoxin production by Aspergillus parasiticus. Acta Microbiol Pol. 1989;38:37–43. [Google Scholar]
- 32.Woloshuk C P, Foutz K R, Brewer J F, Bhatnagar D, Cleveland T E, Payne G A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F Y H, Wu C W. The role of zinc in DNA and RNA polymerases. In: Sigel H, editor. Zinc and its role in biology and nutrition. New York, N.Y: Marcel Dekker, Inc.; 1983. pp. 157–192. [Google Scholar]
- 34.Yu J, Cary J W, Bhatnagar D, Cleveland T E, Keller N P, Chu F S. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3564–3571. doi: 10.1128/aem.59.11.3564-3571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Chang P-K, Payne G A, Cary J W, Bhatnagar D, Cleveland T E. Comparison of the omtA genes encoding O-methyltransferase involved in aflatoxin biosynthesis from Aspergillus parasiticus and Aspergillus flavus. Gene. 1995;163:121–125. doi: 10.1016/0378-1119(95)00397-o. [DOI] [PubMed] [Google Scholar]
- 36.Yu J-H, Butchko R A E, Fernandes M, Keller N P, Leonard T J, Adams T H. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]