Abstract
Two complementary deoxyribonucleic acid (cDNA) clones encoding 2 different 70-kDa heat shock proteins (HSPs) were isolated from the prawn Macrobrachium rosenbergii. The cDNA clones were 2448 and 2173 bp in length and contained 1950- and 1734-bp open reading frames (ORFs), respectively. The ORFs encoded 649– and 577–amino acid polypeptides, which were named Mar-HSC70 and Mar-HSP70, respectively, according to the sequence identities with other known HSC70s and HSP70s and based on their inducibility in response to heat shock stress (at 35°C). Genomic DNA sequence analysis revealed no introns in either gene. The major structural differences between the 2 proteins were a 60–amino acid segment and a 14–amino acid segment present in the N-terminal and C-terminal, respectively, of Mar-HSC70 that were not found in Mar-HSP70. Northern blotting and semiquantitative reverse transcription–polymerase chain reaction analyses indicated that the Mar-HSP70 gene was expressed under heat shock (35°C) stress in a non–tissue-specific manner. In contrast, Mar-HSC70 messenger ribonucleic acid was constitutively expressed in every tissue except muscle, and its expression in response to heat shock (at 35°C) changed only in muscle.
INTRODUCTION
Heat shock proteins (HSPs), found in all eukaryotic organisms, play essential roles in protein metabolism, folding, translocation, and refolding of denatured proteins under both stress and non-stress conditions (Blackman and Meselson 1986; Deshaies et al 1988; Nelson et al 1992; Rubin et al 1993). HSPs themselves and their genes are highly conserved throughout evolution and are found universally from bacteria to humans (Gupta and Golding 1993; Hartl 1996; Morimoto et al 1997). The various HSP families, including HSP100, HSP90, HSP70, HSP60, and small HSPs, are grouped according to their molecular mass (Georgopoulos and Welch 1993).
The HSP70s constitute one of the most highly conserved protein and gene families and are the most widely studied stress proteins (Boorstein et al 1994). Their expression is regulated by environmental and physiological stresses and nonstressful conditions such as cell growth, development, and pathophysiological conditions (Santacruz et al 1997; Morimoto 1998). Members of one HSP70 subfamily are expressed at extremely low levels under normal conditions and increase significantly in response to stressors, such as temperature shifts, heavy metals, hypoxia, and infectious microorganisms (Shinnick 1991; Baler et al 1993; Hartl 1996). Others (HSP70 cognates, also known as HSC70s) are constitutively expressed under normal conditions and change relatively little on exposure to stress (Park et al 2001; Kregel 2002). Although expression patterns of HSP70 and HSC70 are quite different, they share common structural features. They consist of 2 domains: the 44-kDa N-terminal adenosine triphosphatase–binding domain and a 30-kDa C-terminal substrate-binding domain (Dang and Lee 1989).
Several HSP70 and HSC70 genes from aquatic organisms, including reef coral (Tom et al 1999), oyster (Boutet et al 2003), and fishes (Molina et al 2000; Ali et al 2003), had been identified. However, there are only a little data on HSP70 and HSC70 of crustaceans. To our knowledge, it had been reported that expression of HSP70 was transiently induced by heavy metals (Hg and Cu) in a tissue-specific manner in freshwater prawn Macrobrachium malcolmsonii (Yamuna et al 2000); using homologous complementary deoxyribonucleic acid (cDNA) probes, levels of HSP70 messenger ribonucleic acid (mRNA) in the lobster Homarus americanus was characterized during hypo- and hyperosmotic stress (Spees et al 2002); and a heat-inducible HSP70 was identified in the deep-sea vent shrimp Rimicaris exoculata (Ravaux et al 2003). Cheng et al cloned the HSP70 gene in barnacle larvae and characterized its expression under hypoxic conditions, which was the first report on the HSP70 gene sequence in crustaceans (Cheng et al 2003). In prawns, HSP70 and HSC70 gene structure has not been reported to date.
The giant freshwater prawn, Macrobrachium rosenbergii, is a commercially important organism. In 1999, aquaculture of the species in 37 countries exceeded 130 000 tons of prawns per annum. Research has shown that M rosenbergii can exist in water that fluctuates between 15°C and 35°C, but that optimum growth occurs, when the animals are reared in a thermal environment of 27°C to 29°C (Smith 1981). Temperature is thus a limiting factor in the distribution of this species and thus restricts the locations where farming is successful. Thermal acclimation can involve all life cycle stages and can result in physiological compensatory temperature responses that may permit thermal niche expansion. Thus, an insight into the molecular adaptations and changes in response to temperature is important for understanding the biology, distribution, and ability of M rosenbergii to adapt to different thermal regimens across various geographic regions. Recently, Herrera et al (1998) demonstrated that the critical maxima and minima of M rosenbergii are significantly affected by acclimation temperature and that this species shows a high degree of adaptability to various thermal regimes.
In this study, we identified 2 cDNAs (2448 and 2173 bp) encoding Mar-HSC70 and Mar-HSP70 in M rosenbergii. Transcription of both genes was compared in several tissues in response to heat shock treatment. The results demonstrated that expression of Mar-HSC70 was not tissue specific with the exception of muscle and changed relatively little on exposure to heat shock. In contrast, expression of Mar-HSP70 was found to respond to heat shock (at 35°C) and expression also was not tissue specific. This is the first report describing the cloning and characterization of HSP70 and HSC70 mRNA in prawns.
MATERIALS AND METHODS
Animals
Adult prawns, M rosenbergii (7.2 to 12.0 g), of both sexes, were obtained from a market in Hangzhou, China, and were reared in a 15-L glass aquarium with circulating freshwater at 25°C. Prawns were acclimated for at least 2 weeks before experimentation and were fed twice daily.
Stress treatments
In the cold-treatment group, 20 prawns were transferred from 25°C to 15°C or 20°C for 2 hours, whereas for heat treatment, 20 prawns were transferred from 25°C to 30°C or 35°C for 2 hours. The 10 prawns remaining at 25°C served as controls. After the treatments, tissues including thoracic ganglia, hepatopancreas, ovary, gill, gut, and muscle were removed, snap-frozen in liquid nitrogen, and stored at −80°C.
Nucleic acid preparation
All tissues were homogenized in Trizol Reagent (Invitrogen), and total RNA was prepared according to the manufacturer's instructions. Genomic DNA for polymerase chain reaction (PCR) analysis was obtained using the DNeasy Tissue Kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer's instructions.
Reverse transcription–polymerase chain reaction
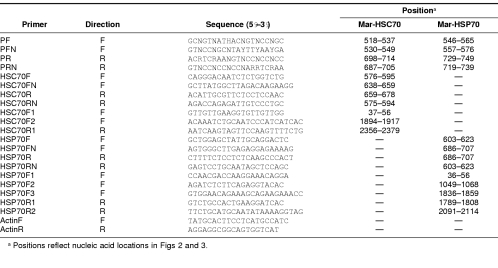
First-strand cDNA was synthesized from 2 μg of total RNA from hepatopancreas and thoracic glands obtained from the prawns treated at 35°C, using the Super-script™/First-strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. The first-strand cDNAs were used as the template, and cDNA fragments encoding the 2 HSP70s were amplified by 2 rounds of PCR amplification. Oligonucleotide primers for the 2 HSP70s were designed based on the highly conserved amino acid sequences corresponding to positions 125–146 and 197–220 of Escherichia coli K12 (AAC74362), Mozambique tilapia (CAA04673), Mediterranean fruit fly (P91902), cow (P19120), and humans (NP_002146). In the first PCR, amplification was primed by pairs of degenerate oligonucleotides (PF and PR, Fig 1; Table 1), and the following program was used: 94°C for 4 minutes, followed by 40 cycles of 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 30 seconds. In the second PCR, the first-round PCR products were used as templates, and amplification was primed by pairs of nested primers (PFN and PRN, Fig 1; Table 1) using the same program as in the first PCR but with the addition of a final polymerization step at 72°C for 10 minutes. The PCR products were subcloned into the PCR® 2.1-TOPO® vector using the TOPO™ TA cloning kit (Invitrogen) for sequencing analysis.
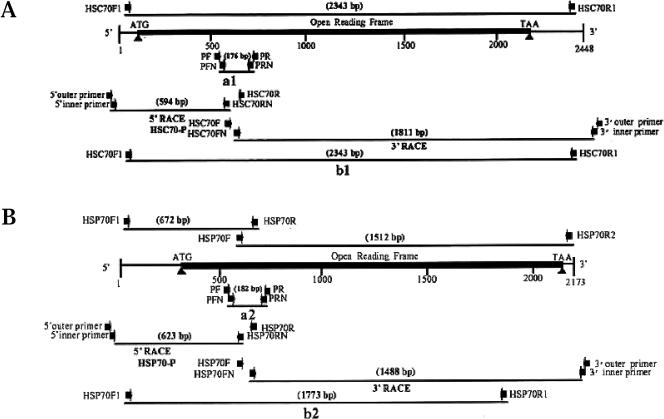
Fig 1.
Schematic diagram of Mar-HSC70 (A) and Mar-HSP70 (B) complementary deoxyribonucleic acids (cDNAs) showing locations of primers for polymerase chain reaction (PCR) and strategy of the gene cloning. Arrowheads represent primers, and lines under them indicate cDNA fragments amplified with the primers by PCR, 5′–rapid amplification of cDNA ends (RACE), and 3′-RACE
Table 1.
Nucleotide sequences and positions of primers used in polymerase chain reaction
3′ and 5′ rapid amplification of cDNA ends
The full-length Mar-HSC70 and Mar-HSP70 cDNAs were obtained by using the rapid amplification of cDNA ends (RACE) method. The cDNAs for 5′ RACE were synthesized from thoracic ganglia and hepatopancreas total RNAs using the FirstChoice™ RLM-RACE Kit (Ambion, USA) according to the manufacturers' protocol. The gene-specific primers (HSC70R, HSC70RN, HSP70R, and HSP70RN, Fig 1; Table 1) for 5′-RACE were designed based on the nucleotide sequences of the Mar-HSC70 and Mar-HSP70 cDNA fragments amplified by reverse transcription (RT)–PCR (a1 and a2, Fig 1). cDNA fragments encoding the 5′-regions of Mar-HSC70 and Mar-HSP70 were amplified by 2 rounds of PCR. The first round of PCR was performed using gene-specific primers (HSC70R and HSP70R, Fig 1; Table 1) and the 5′-RACE adapter outer primer included in the RACE kit. The programs for PCR amplification were 94°C for 5 minutes, followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 62°C, 35 seconds at 72°C, and a final step of 72°C for 10 minutes for Mar-HSC70 and 94°C for 5 minutes, followed by 38 cycles of 30 seconds at 94°C, 30 seconds at 54°C, 30 seconds at 72°C, and a final step of 72°C for 10 minutes for Mar-HSP70. In the nested PCR, the first-round PCR products were used as templates, and each amplification was primed by nested gene-specific primers (HSC70RN and HSP70RN, Fig 1; Table 1) and the 5′-RACE adapter inner primer included in the 5′-RACE kit. The programs for nested PCR amplification were as described for the first PCR. The nested PCR products were subcloned and sequenced.
The cDNAs for 3′-RACE were synthesized from ganglia and hepatopancreas total RNAs using FirstChoice™ RLM-RACE Kit (Ambion, Austin, TX, USA) according to the manufacturers' protocol and used as templates for PCR. The first round of PCR was primed by gene-specific primers (HSC70F and HSP70F, Fig 1; Table 1) and the 3′-RACE adapter outer primer provided in the kit. The programs for PCR amplification were 94°C for 5 minutes, followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 60°C, 90 seconds at 72°C, and a final polymerization step at 72°C for 10 minutes. The nested PCR was performed using nested gene-specific primers (HSC70FN and HSP70FN, Fig 1; Table 1) and the 3′-RACE adapter inner primer provided in the kit. The programs for nested PCR amplification were as described above. The nested PCR products were subcloned and sequenced.
Cloning of Mar-HSC70 and Mar-HSC70 genes from genomic DNA
To determine whether the identified Mar-HSC70 and Mar-HSP70 genes contained introns, genomic DNAs for Mar-HSC70 and Mar-HSP70 were amplified by PCR, and their nucleotide sequences were analyzed. For Mar-HSC70 intron analysis, the isolated genomic DNA (100 ng) was used as a template for PCR to amplify a genomic DNA fragment. The amplification was primed by pairs of gene-specific primers (HSC70F1 and HSC70R1, Fig 1A) and a program consisting of 3 steps: denaturation at 94°C for 5 minutes; 38 cycles of 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 4 minutes; and a final polymerization step at 72°C for 10 minutes. For Mar-HSP70 intron analysis, 2 genomic DNA fragments were amplified by pairs of gene-specific primers (HSP70F1 and HSP70R, HSP70F and HSP70R2, Fig 1B) using the program described above. The DNA fragments were then subcloned and sequenced.
Sequencing analysis
Purified recombinant plasmids were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA), diluted 1:1 in halfBD™ Sequencing Reagent (GenPak, Stony Brook, NY, USA), according to the manufacturer's instructions. The DNA was sequenced from both strands on an ABI PRISM 377 Genetic Analyzer (Applied Biosystems). Sequences were aligned using Vector NTI version 9.0.
Northern blot analysis
Each set of total RNA was extracted from various tissues (thoracic ganglia, hepatopancreas, ovary, gill, gut, and muscle) using the Trizol Reagent described above. Total RNA was quantified on a spectrophotometer at 260 nm. Aliquots containing 10 μg of total RNA corresponding to each tissue were mixed with glyoxal and dimethyl sulfoxide and heated at 50°C for 30 minutes and then placed on ice for more than 1 minute. The treated RNAs were then separated through a 1% agarose (Seakem) gel. The intensities of ethidium bromide–stained ribosomal ribonucleic acid bands were examined visually to ensure that equal amounts of total RNA were loaded onto the gels. The RNA was then transferred to a nylon membrane (Hybond-N, Amersham Pharamacia Biotech, Uppsala, Sweden) and ultraviolet cross-linked. The 5′-RACE products of cDNA fragments HSC70-P (594 bp, bases 1–594 in 5′ untranslated region of Mar-HSC70 cDNA, in Fig 1A) and HSP70-P (623 bp, bases 1–623 in untranslated region of Mar-HSP70 cDNA, in Fig 1B) were amplified and purified using QIAquick PCR Purification Kit (Qiagen). At the same time, the 3′-end region probes for both genes were amplified by pairs of primers HSC70F2-HSC70R1 for Mar-HSC70 and HSP70F3-HSP70R2 for Mar-HSP70 and also used in the northern blotting analysis. The fragments were labeled with digoxigenin–deoxyuridine triphosphate at 37°C overnight using the DIG High Prime Labeling Kit, and hybridization was performed overnight at 50°C using the DIG Easy Hyb system (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. The blots were washed at room temperature, twice at low stringency (2× standard saline citrate [SSC], 0.01% sodium dodecyl sulfate [SDS]) for 5 minutes and then at 55°C twice at high stringency (0.1× SSC, 0.1% SDS) for 15 minutes. After washing, hybridized probes were detected by alkaline phosphatase– conjugated anti-DIG antibody and the CSPD chemiluminescent detection system.
Semiquantitative RT-PCR
Because Northern blotting analysis could not detect expression of mRNA at very low amounts, we further performed semiquantitative RT-PCR experiments to measure expression levels of muscle Mar-HSC70, thoracic ganglia, and hepatopancreas Mar-HSP70 mRNA at each temperature point. Total RNA extraction and RT were described as above. The cDNA fragments were amplified using primers HSC70F2-HSC70R1 for the 486-bp Mar-HSC70 fragment and HSP70F2-HSP70R1 for the 760-bp Mar-HSP70 fragment. As an internal control, a 198-bp fragment of the constitutively expressed β-actin gene (AF221096) was amplified using primers actinF and actinR (Table 1). Ten microliter aliquots of PCR reaction were collected after 26, 28, 30, 32, and 34 cycles to determine the linage range of the reaction. Thirty cycles for Mar-HSC70, 32 cycles for Mar-HSP70, and 26 cycles for β-actin were chosen, and the same program was used in these reactions: 94°C for 5 minutes, followed by indicated cycles of 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 30 seconds, and a final extension step at 72°C for 10 minutes.
Nucleotide sequence accession numbers
The nucleotide sequences of the cDNA-encoding Mar-HSC70 and Mar-HSP70 were submitted to GenBank under accession numbers AY466445 and AY466497, respectively.
RESULTS
Identification of Mar-HSC70 and Mar-HSP70 genes
To design PCR primers for HSC70 or HSP70 amplification (or both), we aligned amino acid sequences corresponding to both inducible and constitutive forms of HSP70 from E coli K12, Mozambique tilapia, Mediterranean fruit fly, cow, and humans. The conserved regions from both HSC70 and HSP70 were selected for initial PCR primer design (primers PF, PR and their nested primers PFN and PRN, Fig 1; Table 1). The cDNAs were synthesized from total RNA isolated from thoracic ganglia and hepatopancreas of heat shocked shrimp (treated at 35°C) using RT. After 2 rounds of PCR amplification, cDNA fragments (a1 and a2 in Fig 1) were subcloned and sequenced by analyzing 5 independent clones for thoracic ganglia and hepatopancreas. Two fragments, a 176-bp (a1) fragment from thoracic ganglia and a 182-bp fragment from the hepatopancreas (a2), which exhibited sequence variations, were selected for designing the gene-specific primers (HSC70F, FN, R, RN and HSP70F, FN, R, and RN in Fig 1) for 3′- and 5′-RACE. To confirm the orientation of the 3 DNA fragments (fragments a1, a2 and their 3′-RACE and 5′-RACE fragments, respectively), PCR was performed to amplify cDNA fragments that spanned those regions (b1 and b2, Fig 1). Sequence analysis of fragments b1 and b2 indicated that the 3 fragments (5′-, 3′-RACE and a1 for Mar-HSC70, and a2 for Mar-HSP70) overlapped.
The full-length cDNA sequence from thoracic ganglia, based on 3′- and 5′-RACE, was 2448 bp in length and contained an open reading frame (ORF) of 1950 bp, which encoded a 649–amino acid protein (Mar-HSC70) with a calculated molecular mass of 70 904 Da. HSP70 signatures-1, -2, and -3, the adenosine triphosphate (ATP)-AGT–binding site, and nuclear localization signal sequences were identified (Fig 2). The Mar-HSC70 gene included a 94-bp 5′–untranslated region (UTR) and a 404-bp 3′-UTR that contained a polyadenylation signal (AATAAA) (Fig 2). The Mar-HSP70 cDNA sequence determined from hepatopancreas by 3′- and 5′-RACE was 2173 bp in length and contained an ORF of 1734 bp, which encoded a 577–amino acid protein with a calculated molecular mass of 63 731 Da. HSP70 signatures-2, and -3, the ATP-AGT–binding site, and nuclear localization signal sequences were identified (Fig 3), although the HSP70 signature-1 was not found. The Mar-HSP70 gene contained a 302-bp 5′-UTR, a 137-bp 3′-UTR, and a polyadenylation signal (AATAAA) (Fig 3).
Fig 2.
Nucleotide sequence and deduced amino acid sequence of Mar-HSC70. Deduced amino acid sequences of Mar-HSC70 are shown in the single letter representation below the respective codons. The asterisks indicate stop codons in the complementary deoxyribonucleic acid (cDNA). The consensus polyadenylation signals AATAAA is double-underlined. HSP70 signatures-1, -2, and -3, the adenosine triphosphate-guanosine triphosphate (ATP-GTP)–binding site, and the nuclear localization signal sequences are underlined
Fig 3.
Nucleotide sequence and deduced amino acid sequence of Mar-HSP70. Deduced amino acid sequences of Mar-HSP70 are shown in the single letter representation below the respective codons. The asterisks indicate stop codons in the complementary deoxyribonucleic acid (cDNA). The consensus polyadenylation signals AATAAA is double-underlined. HSP70 signatures-2 and -3, the adenosine triphosphate-guanosine triphosphate (ATP-GTP)–binding site, and the nuclear localization signal sequences are underlined
The similarity between amino acid sequences of HSC70s and HSP70s in different species
The similarity between HSP70 and HSC70 amino acid sequences in different species suggests similar molecular functions, and thus HSP70 and HSC70 are considered separate subfamilies of the HSP70 family. Greater similarity is commonly observed between members of the 2 subfamilies from different species than between HSP70 and HSC70 from the same species. Comparison of the Mar-HSC70 amino acid sequence with HSC70s of other species (oyster, zebrafish, house mouse, insect, humans) revealed 79.8–86.5% sequence identity. Slightly lower identities (68.2–81.0%) were observed between Mar-HSC70 and Mar-HSP70s from these species. On the other hand, comparison of the Mar-HSP70 amino acid sequence with HSP70s and HSC70s from the above species revealed 61.2–68.5% and 67.4–68.8% identity, respectively.
The alignment analysis showed that the major differences between the amino acid sequences of Mar-HSP70 and Mar-HSC70 (Fig 4) were deletions in the N-terminal (60 amino acids) and C-terminal (14 amino acid residues). The deleted N-terminal segment usually contains 1 of 3 HSP70 family signatures (IDLGTTYS), and thus Mar-HSP70 contains only 2 of these signatures. We confirmed the absence of the N-terminal segment from Mar-HSP70 by sequencing 3 hepatopancreas HSP70 cDNA clones and 3 genomic DNA clones. All 6 clones lacked the 60–amino acid segment.
Fig 4.
Cluster W formatted alignments of representative HSC70 and HSP70 amino acid sequences. Identical residues among all HSC70s and HSP70s are highlighted by light gray boxes, and identical HSC70s and HSP70s residues within each are indicated by the open boxes
Analysis of genomic DNA structure of Mar-HSP70 and Mar-HSC70
A 2343-bp genomic DNA fragment was amplified by using the gene-specific primers, HSC70F1 and HSC70R1, and sequenced. The sequence and length of the genomic DNA fragment were identical to those of the cDNA for Mar-HSC70 (Fig 1A). Two genomic DNA fragments (672 bp and 1512 bp) were amplified for Mar-HSP70 by using the gene-specific primers (HSP70F1 and HSP70R, and HSP70F and HSP70R2) and sequenced. The sequences and lengths of these genomic DNA fragments were also identical to those from the Mar-HSP70 cDNA (Fig 1B). These results suggested that neither gene contained introns.
Stress-induced expression of Mar-HSC70 and Mar-HSP70 genes
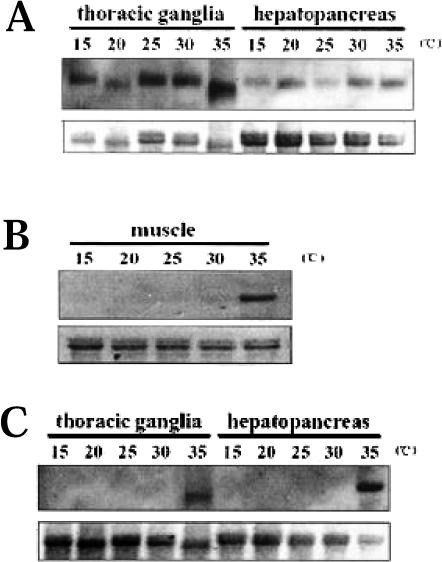
Northern blot analysis of total RNA was used to determine whether expression of Mar-HSC70 and Mar-HSP70 genes increased under heat and cold shock. A cDNA fragment produced by 5′-RACE (HSC70-P, 594 bp, in Fig 1A) as described above was used as a probe for detecting Mar-HSC70 gene expression. The probe hybridized to a ≈2.4-kb transcript in both thoracic ganglia and hepatopancreas at all tested temperatures: 15, 20, 25, 30, and 35°C (Fig 5A). However, expression of Mar-HSC70 was low at 25°C but increased significantly in muscle after exposure to 35°C (Fig 5B). Another cDNA fragment (HSP70-P, 623 bp, in Fig 1B) produced by 5′-RACE was used as probe for Mar-HSP70 in Northern blot analysis. Significant accumulation of a ≈2.1-kb mRNA was observed in thoracic ganglia and hepatopancreas only at 35°C heat shock but not at 15°, 20°, 25°, and 30°C (Fig 5C). The expression patterns of the 2 genes were also confirmed to be similar by using 3′-end probes of Mar-HSC70 and Mar-HSP70 (data not shown).
Fig 5.
Expression of Mar-HSC70 and Mar-HSP70 genes in response to heat shock. Total ribonucleic acids (RNAs) (10 μg) extracted from thoracic ganglia (A), muscle (B), and hepatopancreas (C) at different temperatures (15°, 20°, 25°, 30°, and 35°C) were analyzed by Northern blotting. Gene transcripts were detected by hybridization to a 594-bp (A and B) and 623-bp (C) 5′–untranslated region (UTR) fragment produced by 5′–rapid amplification of cDNA ends (RACE) for Mar-HSC70 and Mar-HSP70, respectively. The ethidium bromide–stained 28S/18S ribosomal ribonucleic acid (rRNA) bands were used as controls for loading variation. The figure was prepared from X-ray film using an HP 3500c Scanjet
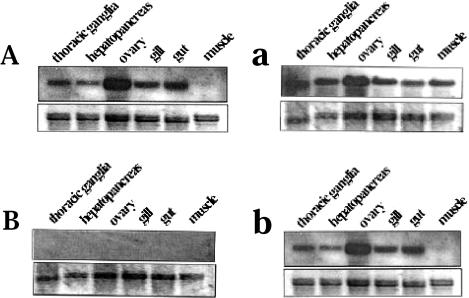
Tissue-specific expression of Mar-HSC70 and Mar-HSP70 genes
To characterize the tissue-specific expression of Mar-HSC70, the HSC70-P probe (Fig 1A) described above was used to detect HSC70 gene expression in thoracic ganglia, hepatopancreas, ovary, gill, gut, and muscle. Mar-HSC70 mRNA expression was observed in all test tissues except muscle at 25°C (Fig 6A), and the expression levels remained unchanged or increased only slightly in most tissues after a 35°C heat shock treatment (Fig 6a). In muscle, a heat shock of 35°C significantly increased the expression of Mar-HSC70 (Fig 5B). In contrast, Mar-HSP70 gene expression in thoracic ganglia, hepatopancreas, ovary, gill, gut, and muscle was not observed under control conditions (25°C) using the HSP70-P probe (Fig 6B). However, Mar-HSP70 gene expression was induced in all tissues on exposure to the 35°C heat shock for 2 hours (Fig 6b). We confirmed these observations using probes targeting the 3′ end of Mar-HSC70 and Mar-HSP70 (data not shown). The data clearly suggest that Mar-HSC70 is noninducible in most tissues except muscle and Mar-HSP70 is inducible under heat shock stress.
Fig 6.
Northern blot analyses of tissue-specific expression of Mar-HSC70 (A) and Mar-HSP70 (B) genes. Total RNAs (10 μg) extracted from thoracic ganglia, hepatopancreas, ovary, gill, gut, and muscle at different temperatures (25°C, A and B; 35°C, a and b) were analyzed. Both gene transcripts genes were detected by hybridization to a 594-bp and 623-bp 5′–untranslated region (UTR) fragment produced by 5′–rapid amplification of cDNA ends (RACE) for Mar-HSC70 and Mar-HSP70, respectively. The ethidium bromide–stained 28S/18S rRNA bands were used as a controls for loading variation. The figure was prepared from X-ray film using an HP 3500c Scanjet
DISCUSSION
Although HSC70 and HSP70 genes have been previously identified in many species, this study is the first to report isolation and sequencing of both HSC70 and HSP70 in prawns. In this article, 2 genes encoding Mar-HSC70 and Mar-HSP70, were identified and characterized from the prawn M rosenbergii. Based on amino acid sequence identities with other known HSC70s and HSP70s, the 2 genes were identified as members of the HSP gene family, which encoded HSP70 family proteins. One of the genes was not expressed under normal conditions but was inducible by heat shock (35°C) and was thus named Mar-HSP70. The second gene was expressed constitutively and was not induced by heat shock stress in most tissues and was thus named Mar-HSC70 (HSP70 cognate).
The major structural differences between the 2 proteins (Mar-HSC70 and Mar-HSP70) were found to be a 60–amino acid segment and a 14–amino acid segment present in the N-terminal and C-terminal, respectively, in Mar-HSC70 that were not found in Mar-HSP70, the accuracy of which has been further confirmed. However, the functional and evolutionary significance of this deletion is unknown. The 14–amino acid segment absent from the C-terminal of Mar-HSP70 was also found to be absent from the C-terminal of inducible HSP70s from other species. This segment contains a glycine-alanine-proline (GAP) repeat that may play an important role in the structure and function of the HSC70 subfamily. A review of previously reported sequences found that HSC70s contain at least 1 GAP sequence in this region. For example, HSC70s from the midges Chironomus tentans (AAN14525) and C yoshimatsui (AAN14526) contain 2 GAP sequences (Karouna-Renier et al 2003), mouse HSC70 (AAB18391) (Hunt et al 1999) contain 1 GAP, and oyster HSC70 (CAC83684) (Boutet et al 2003) contains 3.
An ATP-guanosine triphosphate–binding site motif was found at positions 131–138 in Mar-HSC70 (AEAFLGST) and 71–78 in Mar-HSP70 (AEAYLGKT). Likewise, putative bipartite nuclear localization signals (KKDPSENKRALRRL and RALRRLRTACERAKRTL) were found at positions 246–263 and 258–274 in Mar-HSC70 and positions 180–197 and 192–208 (QRKYKKDLTTNKRAIRRL and RAIRRLRTACERAKRTL) in Mar-HSP70. The 2 signals, which were characterized by an abundance of the basic amino acids lysine and arginine, are needed for the selective translocation of HSP70s into the nucleus (Knowlton and Salfity 1996). Although the C-terminal end appears to be less conserved, the last 4 amino acids, EEVD, are identical in all species. This feature was previously shown to be characteristic of cytosolic-nuclear HSP70s and HSC70s (Boorstein et al 1994). Three HSP70 signatures were identified in Mar-HSC70 at positions 8– 15 (GIDLGTTY), 198–206 (DLGGGTFD), and 334–48 (IVLVGGSTRIPKIQK). However, only 2 HSP70 signatures were found in Mar-HSP70 at positions 133–140 (DLGGGTFD) and 268–282 (IVLVGGSTRIPKIQK). To our knowledge, this is the first report of an inducible HSP70 in which signature-1 was deleted. We speculated that N-terminal deletion in Mar-HSP70 may be of great significance in evolution.
In the HSP70 family, introns are generally found only in constitutively expressed genes but not found in the inducible forms (Yost and Lindquist 1986). It was suggested that the lack of introns in inducible HSP70s allows rapid transcription and accumulation on stress, the nuclear export signal being probably provided by the mRNA secondary and tertiary structure (Huang et al 1999). However, in our study, genomic DNA sequence analysis revealed a lack of introns in both genes. Although previous studies have reported introns in inducible HSP70s (Muller et al 1992; Stefani and Gomes 1995), to our knowledge this is the first report of a constitutively expressed HSC70 that contained no intron.
Significant accumulation of Mar-HSC70 mRNA was observed in every test tissue except muscle under normal conditions. Mar-HSC70 mRNA was particularly enriched in the ovary of prawns. Similar enriched patterns of HSC70 transcripts have been reported in zebrafish and Drosophila during embryogenesis (Perkins et al 1990; Santacruz et al 1997). Therefore, the tissue-specific enrichment of Mar-HSC70 transcripts in prawns may signify a critical role for this stress protein during reproductive events.
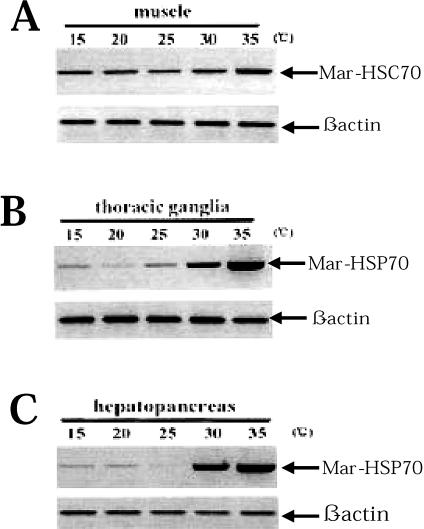
Northern blot analysis indicated that Mar-HSP70 mRNA expression in M rosenbergii was induced by 35°C heat shock, but expression was not observed at 15°, 20°, 25°, or 30°C. Semiquantitative RT-PCR results showed that expression of the Mar-HSP70 gene remained at low levels at 15, 20, 25, or 30°C in thoracic ganglia and hepatopancreas; whereas Mar-HSP70 expression was slightly upregulated exposed to 30°C, significantly increased to 35°C (Fig 7 B,C). The results obtained by semiquantitative RT-PCR demonstrated that that mRNA of Mar-HSP70 are expressed at low levels at 15°, 20°, 25°, or 30°C. These levels were lower than the detection limit of Northern blotting. In contrast, significant expression of Mar-HSC70 mRNA was detected in every test tissue except muscle, and, with the exception of muscle, no significant response to heat stress was observed. Mar-HSC70 mRNA expression was not detected by Northern blotting in the muscle of prawns cultured at 15°, 20°, or 25°C. Using RT-PCR, however, we detected the expression of the Mar-HSC70 gene at low levels in muscle at 15°, 20°, or 25°C (Fig 7A). Similarly, Boone and Vijayan (2002) observed significantly lower baseline levels of HSC70 protein in skeletal muscle than in liver, heart, gill, or white muscle of rainbow trout. They attributed these lower levels of HSC70 to slower protein turnover in skeletal muscle compared with the other tissues. M rosenbergii may also exhibit slower turnover of HSC70 protein in muscle than other tissues, which would maintain baseline levels of HSC70 protein without the need for high baseline levels of gene induction. Further investigation of HSC70 protein levels in M rosenbergii tissues is required to confirm this theory. In contrast to other tissues, exposure to a 30°C heat shock induced a slight increase in Mar-HSC70 expression in muscle, and exposure to 35°C heat shock for 2 hours induced expression of the mRNA to an even greater extent. Significantly higher induction of HSC70 mRNA has also been observed in muscle from carp in response to heat shock as compared with other tissues (Ali et al 2003). Under these conditions, an increase in HSC70 in addition to HSP70 may be required to protect against muscle damage.
Fig 7.
Expression analysis of Mar-HSC70 (A) and Mar-HSP70 (B and C) in response to heat shock by semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) experiments. RT-PCR amplification products for Mar-HSC70 (486 bp) fragment, Mar-HSP70 (760 bp), and the constitutive control β-actin (198 bp) from total ribonucleic acid (RNA) (1 μg) extracted from muscle, thoracic ganglia, and hepatopancreas at different temperatures (15°, 20°, 25°, 30°, and 35°C) are shown
No Mar-HSC70 and Mar-HSP70 mRNAs were observed in the muscle and in the hepatopancreas of prawns after cold shock (15°C and 20°C). Similar HSC70 and HSP70 expression patterns after cold shock have been reported in the flesh fly (Rinehart et al 2000). Furthermore, previous studies demonstrated that a recovery period is required before HSP70 expression and HSC70 upregulation are observed, indicating that these genes are not involved in the immediate response to a cold shock that leads to rapid cold hardening (Chen and Denlinger 1987; Rinehart et al 2000). At the same time, these results suggest that some members of the HSP70 family function in a fundamentally different manner during heat and cold shock stress.
Identification and characterization of 2 genes encoding Mar-HSC70 and Mar-HSP70 may provide valuable information for farming of the giant freshwater, M rosenbergii. Mar-HSC70, a non–heat-inducible form of the HSP70 family, may play a very important role in growth and development of M rosenbergii. The synthesis of Mar-HSP70 is slightly increased after exposure to 30°C, significantly increased to 35°C (Figs 5C and 7 B,C), suggesting that the appearance of severe cellular damage occurred at 35°C, at the same time, high levels of expressed Mar-HSP70 allowed their survival during and after thermal stress by refolding and repairing the denatured proteins. In addition, we will use Mar-HSP70 as a useful probe to investigate the relationship between expression of the Mar-HSP70 gene and other stressors (heavy metals, bacteria, and salt water), which we will expect to expand M rosenbergii as a model organism for biomonitoring in the environment.
Acknowledgments
The authors thank Mr Zhong-Min Dai and Jin-Shu Yang of the College of Life Sciences, Zhejiang University for their valuable advice and assistance with the data analysis. This work was supported by the National Natural Sciences Foundation of China (30225034 and 30371097).
REFERENCES
- Ali KS, Dorgai L, Abraham M, Hermesz E. Tissue- and stressor-specific differential expression of two hsc70 genes in carp. Biochem Biophys Res Commun. 2003;307:503–509. doi: 10.1016/s0006-291x(03)01206-3.0006-291X(2003)307<0503:TASDEO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486.0270-7306(1993)013<2486:AOHHSG>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman RK, Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986;188:499–515. doi: 10.1016/s0022-2836(86)80001-8.0022-2836(1986)188<0499:INSCUT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Boone AN, Vijayan MM. Constitutive heat shock protein 70 (HSC70) expression in rainbow trout hepatocytes: effects of heat shock and heavy metal exposure. Comp Biochem Physiol C. 2002;132:223–233. doi: 10.1016/s1532-0456(02)00066-2.1367-8280(2002)132<0223:CHSPHE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490.0022-2844(1994)038<0001:MEOTHM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Boutet I, Tanguy A, Moraga D. Organization and nucleotide sequence of the European flat oyster Ostrea edulis heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes. Aquat Toxicol. 2003;65:221–225. doi: 10.1016/s0166-445x(03)00137-1.0166-445X(2003)065<0221:OANSOT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Chen CP, Denlinger DL. Cold shock injury and rapid cold hardening in the flesh fly Sarcophaga crassipalpis. Physiol Zool. 1987;60:297–304.0031-935X(1987)060<0297:CSIARC>2.0.CO;2 [Google Scholar]
- Cheng SH, So CH, Chan PK, Cheng CW, Wu RS. Cloning of the HSP70 gene in barnacle larvae and its expression under hypoxic conditions. Mar Pollut Bull. 2003;46:665–671. doi: 10.1016/S0025-326X(03)00059-6.0025-326X(2003)046<0665:COTHGI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dang CV, Lee WM. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989;264:18019–18023.0021-9258(1989)264<18019:NANTSO>2.0.CO;2 [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0.0028-0836(1988)332<0800:ASOSPF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Welch WJ. Role of the major heat shock proteins as molecular chaperones. Ann Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125.0743-4634(1993)009<0601:ROTMHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743.0022-2844(1993)037<0573:EOHGAI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0.0028-0836(1996)381<0571:MCICPF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Herrera FD, Uribe ES, Ramirez LFB, Mora AG. Critical thermal maxima and minima of Macrobrachium rosenbergii (Decapoda: Palaemonidae) J Therm Biol. 1998;23:381–385.0306-4565(1998)023<0381:CTMAMO>2.0.CO;2 [Google Scholar]
- Huang Y, Wimler KM, Carmichael GG. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642.0261-4189(1999)018<1642:IMTEMA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CR, Parsian AJ, Goswami PC, Kozak CA. Characterization and expression of the mouse Hsc70 gene. Biochim Biophys Acta. 1999;1444:315–325. doi: 10.1016/s0167-4781(98)00285-1.0006-3002(1999)1444<0315:CAEOTM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Karouna-Renier NK, Yang W-J, Rao K. Cloning and characterization of a 70 kDa heat shock cognate gene (HSC70) from two species of Chironomus. Insect Mol Biol. 2003;12:19–26. doi: 10.1046/j.1365-2583.2003.00383.x.0962-1075(2003)012<0019:CACOAK>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Knowlton AA, Salfity M. Nuclear localization and the heat shock proteins. J Biosci. 1996;21:123–132.0250-5991(1996)021<0123:NLATHS>2.0.CO;2 [Google Scholar]
- Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001.8750-7587(2002)092<2177:HSPMFI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Molina A, Biemar F, and Muller F. et al. 2000 Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Lett. 474:5–10. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788.0890-9369(1998)012<3788:ROTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Kline MP, Bimston DN, Cotto JJ. The heat-shock response: regulation and function of heat-shock proteins and molecular chaperones. Essays Biochem. 1997;32:17–29.0071-1365(1997)032<0017:THRRAF>2.0.CO;2 [PubMed] [Google Scholar]
- Muller FW, Igloi GL, Beck CF. Structure of a gene encoding heat-shock protein HSP70 from the unicellular alga Chlamydomonas reinhardtii. Gene. 1992;111:165–173. doi: 10.1016/0378-1119(92)90684-h.0378-1119(1992)111<0165:SOAGEH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kD heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i.0092-8674(1992)071<0097:TTMAKH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Park JH, Lee JJ, and Yoon S. et al. 2001 Genomic cloning of the Hsc71 gene in the hermaphroditic teleost Rivulus marmoratus and analysis of its expression in skeletal muscle: identification of a novel muscle-preferred regulatory element. Nucleic Acids Res. 29:3041–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Doctor JS, Zhang K, Stinson L, Perrimon N, Craig EA. Molecular and developmental characterization of the heat shock cognate 4 gene of Drosophila melanogaster. Mol Cell Biol. 1990;10:3232–3238. doi: 10.1128/mcb.10.6.3232.0270-7306(1990)010<3232:MADCOT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravaux J, Gaill F, Le Bris N, Sarradin PM, Jollivet D, Shillito B. Heat-shock response and temperature resistance in the deep-sea vent shrimp Rimicaris exoculata. J Exp Biol. 2003;206:2345–2354. doi: 10.1242/jeb.00419.0022-0949(2003)206<2345:HRATRI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rinehart JP, Yocum GD, Denlinger DL. Developmental upregulation of inducible hsp70 transcripts, but not the cognate form, during pupal diapause in the flesh fly, Sarcophaga crassipalpis. Insect Biochem Mol Biol. 2000;30:515–521. doi: 10.1016/s0965-1748(00)00021-7.0965-1748(2000)030<0515:DUOIHT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rubin DM, Mehta AD, Zhu J, Shoham S, Chen X, Wells QR, Palter KB. Genomic structure and sequence analysis of Drosophila melanogaster HSC70 genes. Gene. 1993;128:155–163. doi: 10.1016/0378-1119(93)90558-k.0378-1119(1993)128<0155:GSASAO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Santacruz H, Vriz S, Angelier N. Molecular characterization of a heat shock cognate cDNA of zebrafish, hsc70, and developmental expression of the corresponding transcripts. Dev Genet. 1997;21:223–233. doi: 10.1002/(SICI)1520-6408(1997)21:3<223::AID-DVG5>3.0.CO;2-9.0192-253X(1997)021<0223:MCOAHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Shinnick TM. Heat shock proteins as antigens of bacterial and parasitic pathogens. Curr Top Microbiol Immunol. 1991;167:145–160. doi: 10.1007/978-3-642-75875-1_9.0070-217X(1991)167<0145:HSPAAO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Smith KC 1981 A Layman's Guide to Geothermal Aquaculture. Oregon Institute of Technology, Klamath Falls, OR. [Google Scholar]
- Spees JL, Chang SA, Snyder MJ, Chang ES. Osmotic induction of stress-responsive gene expression in the lobster Homarus americanus. Biol Bull. 2002;203:331–337. doi: 10.2307/1543575.0006-3185(2002)203<0331:OIOSGE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Stefani RM, Gomes SL. A unique intron-containing hsp70 gene induced by heat shock and during sporulation in the aquatic fungus Blastocladiella emersonii. Gene. 1995;152:19–26. doi: 10.1016/0378-1119(95)00645-m.0378-1119(1995)152<0019:AUIHGI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tom M, Douek J, Yankelevich I, Bosch TC, Rinkevich B. Molecular characterization of the first heat shock protein 70 from a reef coral. Biochem Biophys Res Commun. 1999;262:103–108. doi: 10.1006/bbrc.1999.1165.0006-291X(1999)262<0103:MCOTFH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yamuna A, Kabila V, Geraldine P. Expression of heat shock protein 70 in freshwater prawn Macrobrachium malcolmsonii (H. Miline Edwards) following exposure to Hg and Cu. Indian J Exp Biol. 2000;38:921–925.0019-5189(2000)038<0921:EOHSPI>2.0.CO;2 [PubMed] [Google Scholar]
- Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–93. doi: 10.1016/0092-8674(86)90382-x.0092-8674(1986)045<0185:RSIIBH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]