ABSTRACT
Animal models for host-microbial interactions have proven valuable, yielding physiologically relevant data that may be otherwise difficult to obtain. Unfortunately, such models are lacking or nonexistent for many microbes. Here, we introduce organ agar, a straightforward method to enable the screening of large mutant libraries while avoiding physiological bottlenecks. We demonstrate that growth defects on organ agar were translatable to bacterial colonization deficiencies in a murine model. Specifically, we present a urinary tract infection agar model to interrogate an ordered library of Proteus mirabilis transposon mutants, with accurate prediction of bacterial genes critical for host colonization. Thus, we demonstrate the ability of ex vivo organ agar to reproduce in vivo deficiencies. Organ agar was also useful for identifying previously unknown links between biosynthetic genes and swarming motility. This work provides a readily adoptable technique that is economical and uses substantially fewer animals. We anticipate this method will be useful for a wide variety of microorganisms, both pathogenic and commensal, in a diverse range of model host species.
KEYWORDS: urinary tract infection, Proteus mirabilis, animal models, swarming
INTRODUCTION
Animal models have a longstanding track record in the study of microbial pathogens, including the fulfillment of Koch’s third postulate [that is, the cultured microorganism should cause disease when introduced into a healthy organism (1)]. Although an excellent in vivo model is indispensable for virulence studies, such models may be difficult to establish for fastidious or host-restricted pathogens. Furthermore, even when models exist, complex disease progression can confound experimental interpretation. For example, in the urinary tract, infecting microbes exhibit planktonic growth in urine, adhere or invade the bladder epithelium, ascend into the kidneys, and disseminate into the bloodstream (2, 3). Microbes compete with the host immune response and with established bladder microbiota (4, 5). Each of these challenges can manifest as a chokepoint, or physiological bottleneck, where only a limited number of microbes become established. These bottlenecks have a profound effect on experimental design, particularly in modern large-scale studies, where the number of mutants that can be feasibly studied without random loss is limited due to such founder effects (6 – 9).
Urinary tract infections (UTIs) are one of the most common infections worldwide, affecting most women at least once in their lifetime (10, 11). These infections are even more common in patients with urinary catheters, where a more diverse mix of bacterial species is likely to be the causative agent. In particular, Proteus mirabilis is especially problematic in patients with long-term (>30 days) indwelling urinary catheters (12 – 14). This species produces urease, which cleaves urea, abundantly present in urine, into ammonia and carbon dioxide. The resulting pH increase causes precipitation of calcium and magnesium ions naturally present in urine, to form struvite and hydroxyapatite crystals that can block catheter flow and directly damage urinary tract tissue via stone formation (urolithiasis) (15).
Previous studies on P. mirabilis virulence have relied on a well-established murine model of UTI (15). Most often, this model involves instillation of bacteria directly into the bladder via a catheter, which can either be removed to study UTI progression in the absence of a foreign body or fully pushed into the bladder to model long-term catheterization (16 – 18). In either case, P. mirabilis readily establishes infection and, similar to human UTI, may cause pyelonephritis and urolithiasis (15). In addition to challenging mice with one or two strains at a time, the mouse model has been used to screen pools of mutants from uropathogenic species of bacteria (6, 19).
We aimed to conduct similar large-scale studies to identify genes that contribute to UTI in mice. Toward this goal, we present data showing the experimental bottleneck for the total number of P. mirabilis mutants that can be tested in this model without stochastic loss is much narrower than anticipated. We recently built an ordered library containing single transposon insertions in 1,728 genes, representing 45% of predicted genes in P. mirabilis strain HI4320 (20). Based on the tight experimental bottleneck, we concluded that screening this library in a murine model would not be feasible. Thus, we devised a method to assay the ordered library on agar plates made by mixing organ homogenates with molten agar to create “organ agar.” In this study, we show that organ agar, as well as agar made from pooled human urine, identified genes that contribute to P. mirabilis fitness during experimental UTI in mice. Many of these genes were part of biosynthetic pathways, indicating that organ agar is especially useful for identifying nutritional availability in distinct organ sites. Excitingly, most (six of seven) of the mutants we selected for follow-up study were significantly impaired when cochallenged with the wild-type strain in mice. We anticipate that this economical model will open new avenues of research for many diverse pathogens.
RESULTS
The bottleneck effect for Proteus mirabilis in a murine model of UTI is much stronger than previously described
Our group and others have successfully enumerated genetic bottleneck effects observed in the traditional model of ascending UTI (6, 21, 22). Although relatively generous infection dynamics of P. mirabilis in mice have been estimated for a murine model where a catheter segment is entirely inserted into the bladder lumen (18, 19), we found that the bottleneck for P. mirabilis is narrow in the traditional ascending infection UTI model and would, therefore, be problematic for large screening experiments in vivo (Fig. 1). To measure the bottleneck, we used a spa47 mutant that we previously showed has no deleterious effect on bacterial recovery in our mouse model of ascending UTI (23). After testing different input ratios of spa47 and wild type in cochallenges, we determined that only very small groups of strains can be assembled to avoid stochastic loss of mutants. In urine, up to 100 strains could be tested with the ratio remaining stable throughout the infection, calculated as a log competitive index (CI) of zero (Fig. 1A). However, even at ratios as low as 1:8, we observed 1/5 mice displaying results outside of the ideal range, indicating stochastic loss of the mutant. Bladder samples became unacceptable at an even lower ratio of 1:59, where over half of mice fell outside the preferred range (Fig. 1B). Around 20% of mice with colonization in the urine and bladder had no detectable CFU burden in the kidneys (Table 1). This, in combination with the increased spread of data between biological replicates, caused cochallenge input ratios as low as 1:13 to be problematic (Fig. 1C). Collectively, we demonstrated that the bottleneck during murine UTI with P. mirabilis is severe. This is consistent with other studies using uropathogenic Escherichia coli (UPEC) (6, 21, 22) although the restriction for P. mirabilis is even narrower. For example, in the bladder at 24 h post-inoculation, a ratio of 1:500 exceeded the experimental capacity for testing individual E. coli mutants (6) while we found a ratio of 1:59 for P. mirabilis. This bottleneck, therefore, limits the feasibility of in vivo experimental screens with large pools of mutants. In response, we sought alternative approaches for identifying genes for further study.
Fig 1.
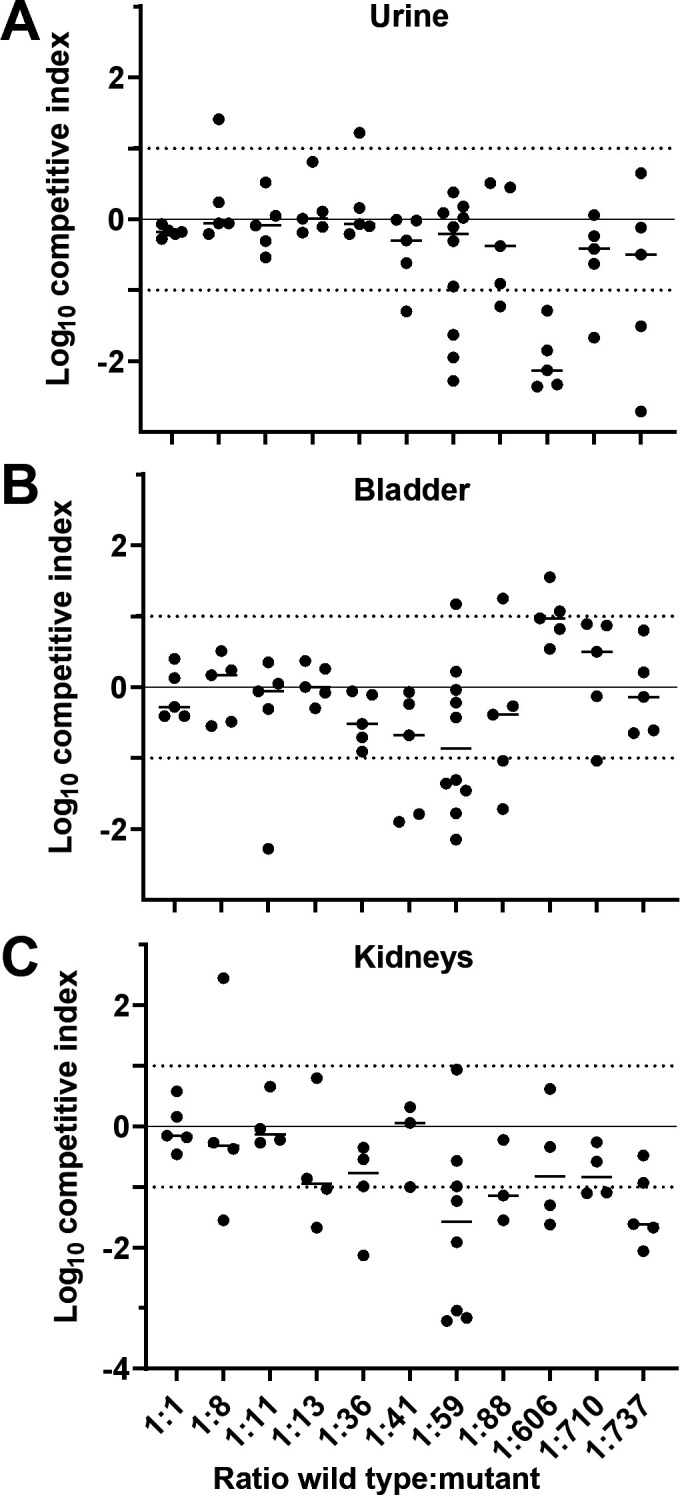
Determining the bottleneck of infection for P. mirabilis during UTI. (A–C) CBA/J mice were transurethrally inoculated with 107 CFU. The inoculum contained different ratios of wild-type HI4320 and marked mutant spa47 (kanR) that was previously determined to have no fitness defect (23). The measured ratios are indicated on the x-axis. At 24 h post-inoculation, (A) urine, (B) bladder, and (C) kidneys were harvested. Each sample was subjected to differential plating to enumerate the ratio of wild type to spa47 in the organ. A competitive index (CI) was calculated and is plotted on the y-axis. Each dot represents a single mouse (n = 5–10); bars indicate the median. A log CI of 0 indicates that the wild type and mutant were recovered in the same ratio as were introduced in the inoculum. Dotted lines at ±1 indicate the acceptable maximum variation for this experiment.
TABLE 1.
Recovery of bacteria from bottleneck experiments
| Number of mice with recovered bacteria | ||||
|---|---|---|---|---|
| Target ratio a | Actual ratio b | Urine | Bladder | Kidneys |
| 1:1 | 1:1 | 5/5 | 5/5 | 5/5 |
| 1:10 | 1:8 | 5/5 | 5/5 | 4/5 |
| 1:10 | 1:11 | 5/5 | 5/5 | 4/5 |
| 1:25 | 1:13 | 5/5 | 5/5 | 4/5 |
| 1:25 | 1:36 | 5/5 | 5/5 | 4/5 |
| 1:50 | 1:41 | 5/5 | 5/5 | 3/5 |
| 1:50 | 1:59 | 10/10 | 10/10 | 8/10 |
| 1:100 | 1:88 | 5/5 | 5/5 | 3/5 |
| 1:500 | 1:606 | 5/5 | 5/5 | 4/5 |
| 1:1000 | 1:737 | 5/5 | 5/5 | 5/5 |
| 1:1000 | 1:710 | 5/5 | 5/5 | 4/5 |
Intended input ratio of spa47 mutant to wild-type HI4320.
Measured input ratio of spa47 mutant to wild-type HI4320.
Development of organ and urine agar
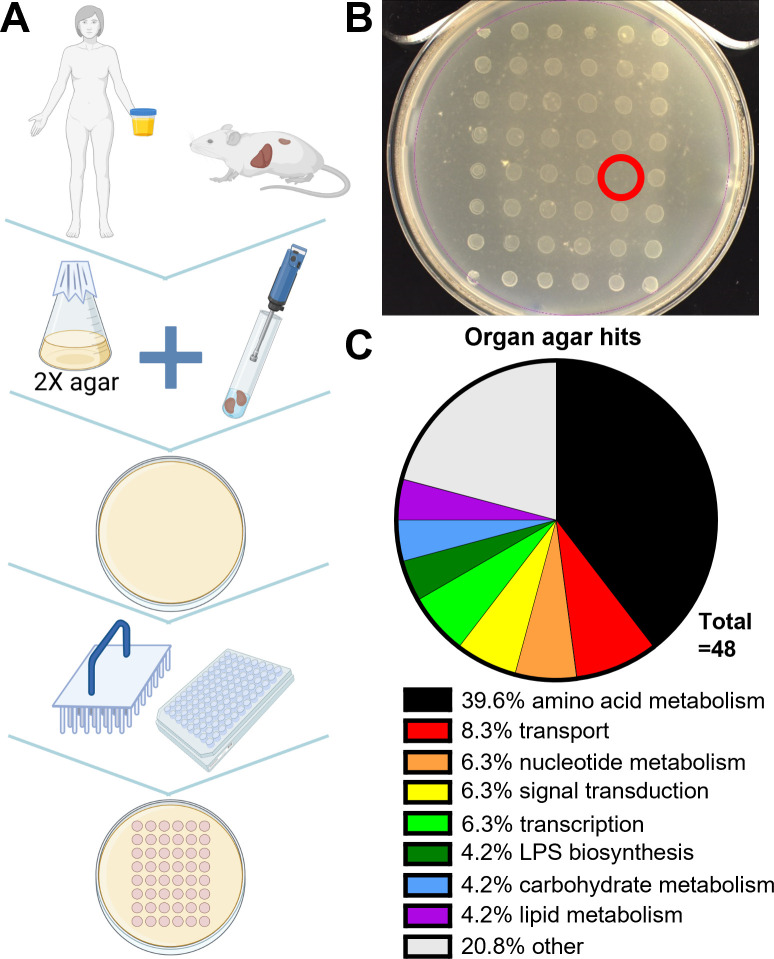
Our goal was to test the fitness of an ordered transposon library built for P. mirabilis strain HI4320 (20) in a physiologically relevant condition. Because of the severe bottleneck in mice, which would have required 70 mice to perform a single Tn-seq experiment using pools of 25 mutants from the 1,728-gene-ordered library, we devised a screen that did not require pooling of mutants. For the initial development and screen, we chose outbred male Swiss Webster mice because they are economical. Mouse bladders, kidneys, and spleens were aseptically collected to represent different stages of ascending UTI (cystitis, pyelonephritis, and bacteremia/urosepsis, respectively). Organs were homogenized in water to prevent salt-induced swarming motility by P. mirabilis (24, 25). These homogenates were either mixed 1:1 with autoclaved agar or further diluted before mixing with agar. All organs sustained growth of P. mirabilis; however, growth on spleens yielded tiny colonies at 24 h and colonies on bladder agar required at least 36 h of incubation to be readily visible. In contrast, the larger and denser kidneys could be diluted up to 1:10 and still sustain growth (Fig. S1). Uninoculated plates showed no outgrowth of microbiota during the duration of the experiment. We selected this 1:10 dilution for screening the ordered library on agar made from kidneys and switched spleens for larger livers to represent disseminated infection, which allowed us to screen 1,728 single gene insertions using organs pooled from only 15 mice.
Similarly, agar made from pooled human urine was adapted for P. mirabilis using two modifications. First, the agar was buffered to counteract growth-inhibitory increases in pH due to ammonia released from urease activity (Fig. S2A). Second, the agar concentration was increased to reduce swarming motility. This did not completely eliminate swarming but was sufficient for genetic screening (Fig. S2B). Interestingly, we observed swarming-deficient mutants in the transposon screen, suggesting that this modified urine agar could also be a useful tool for studying swarming motility (Fig. S3).
Screening of P. mirabilis ordered library on organ and urine agar
Mutants in the ordered library were stamped onto liver, kidney, and urine plates and incubated at 37°C, and growth was recorded after 24 and 48 h (Fig. 2; Fig. S3). A total of 48 mutants were qualitatively identified that had growth defects on one or more organ agars (Table 2). The largest category of hits was biosynthetic genes, particularly for amino acids (Fig. 2C). We found genes known to contribute to P. mirabilis fitness during UTI (i.e., high-affinity zinc uptake genes znuA and znuC) (26), as well as pathways known to be important (e.g., TCA cycle) (27). We also identified known contributors to UPEC-mediated UTI (e.g., purines and branched-chain amino acid biosynthesis) (6).
Fig 2.
Organ agar screen. (A) Schematic for generating organ agar and library screen. (B) 1:10 kidney agar with 48 stamped library mutants. One transposon hit (lack of growth) is circled in red. (C) Classification of organ agar hits obtained on one or more agars. Genes classified as “other” include singly-binned genes with predicted functions and hypothetical genes.
TABLE 2.
Organ agar hits
| Gene | Name | New locus tag | Plate a | Well a | OD600 | Organ b | Function | General function | PCR confirm c |
|---|---|---|---|---|---|---|---|---|---|
| PMI0003 | thrC | PMI_RS00015 | 216 | G4 | 0.862 | U | Threonine synthase | Amino acid metabolism | n/a |
| PMI0005 | PMI_RS00025 | 213 | F6 | 0.880 | U | Conserved hypothetical protein | n/a | ||
| PMI0006 | talB | PMI_RS00030 | 213 | G2 | 0.818 | UK | Transaldolase B | Carbohydrate metabolism | N |
| PMI0083 | nusB | PMI_RS00400 | 210 | C11 | 0.333 | K | N utilization substance protein B | Transcription machinery | n/a |
| PMI0200 | dksA | PMI_RS00965 | 211 | F9 | 0.877 | K | DnaK suppressor protein | rRNA transcription factor | n/a |
| PMI0205 | hemL | PMI_RS00985 | 219 | C2 | 0.500 | UK | Glutamate-1-semialdehyde 2,1-aminomutase | Amino acid metabolism | Y |
| PMI0206 | erpA | PMI_RS00990 | 211 | E9 | 0.576 | UK | Putative iron-sulfur protein | Y | |
| PMI0335 | proC | PMI_RS01600 | 202 | G2 | 0.644 | U | Pyrroline-5-carboxylate reductase | Amino acid metabolism | n/a |
| PMI0370 | proA | PMI_RS01770 | 208 | E11 | 0.727 | U | Gamma-glutamyl phosphate reductase | Amino acid metabolism | n/a |
| PMI0570 d | sucB | PMI_RS02805 | 207 | F6 | 0.863 | UKL | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex | TCA cycle e | Y |
| PMI0641 | sanA | PMI_RS03160 | 215 | C10 | 0.854 | K | Putative transport protein | Y | |
| PMI0711 | serC | PMI_RS03500 | 219 | C4 | 0.419 | UK | Phosphoserine aminotransferase | Amino acid metabolism | Y |
| PMI0765 | ompF/ nmpC | PMI_RS03760 | 218 | G6 | 0.716 | UK | Outer membrane porin | Signal transduction | Y |
| PMI1151 | znuC | PMI_RS05555 | 201 | G4 | 0.775 | KL | High-affinity zinc uptake system ATP-binding protein | Transport | Y |
| PMI1152 | znuA | PMI_RS05560 | 209 | C1 | 0.990 | K | High-affinity zinc uptake system substrate-binding protein | Transport | n/a |
| PMI1344 | trpD | PMI_RS06485 | 204 | B8 | 0.914 | K | Anthranilate synthase component (glutamine amidotransferase) | Amino acid metabolism | n/a |
| PMI1348 | trpA | PMI_RS06505 | 204 | C4 | 0.889 | UK | Tryptophan synthase alpha chain | Amino acid metabolism | n/a |
| PMI1545 | guaA | PMI_RS07520 | 201 | D12 | 0.807 | UKL | GMP synthase (glutamine-hydrolyzing) | Nucleotide metabolism | Y |
| PMI1562 | purC | PMI_RS07610 | 209 | G4 | 0.880 | U | Phosphoribosylaminoimidazole-succinocarboxamide synthetase (SAICAR synthetase) | Nucleotide metabolism | n/a |
| PMI1833 | cysW | PMI_RS09045 | 201 | A5 | 0.609 | K | Sulfate/thiosulfate ABC transporter, permease protein | Transport | Y |
| PMI1875 | purL | PMI_RS09255 | 201 | C11 | 0.737 | U | Phosphoribosylformylglycineamide synthetase | Nucleotide metabolism | n/a |
| PMI1912 | PMI_RS09435 | 207 | F3 | 0.737 | U | FtsK/SpoIIIE-family protein | Chromosome partitioning | n/a | |
| PMI1945 | ireA | PMI_RS09585 | 202 | A9 | 0.714 | U | Putative TonB-dependent ferric siderophore receptor | Transport | n/a |
| PMI2085 | leuB | PMI_RS10270 | 207 | C5 | 0.946 | U | 3-Isopropylmalate dehydrogenase | Amino acid metabolism | n/a |
| PMI2094 | speA | PMI_RS10315 | 201 | H1 | 0.571 | K | Biosynthetic arginine decarboxylase | Amino acid metabolism | n/a |
| PMI2288 | dapD | PMI_RS11300 | 210 | E12 | 0.746 | K | 2,3,4,5-Tetrahydropyridine-2-carboxylate N-succinyltransferase | Amino acid metabolism | n/a |
| PMI2309 | recB | PMI_RS11420 | 219 | B3 | 0.241 | UK | Exodeoxyribonuclease V beta chain | DNA repair | n/a |
| PMI2720 | PMI_RS13405 | 213 | D5 | 0.771 | U | BMC domain-containing protein | n/a | ||
| PMI2821 | argD | PMI_RS13935 | 210 | A4 | 0.720 | U | Acetylornithine/succinyldiaminopimelate aminotransferase | Amino acid metabolism | n/a |
| PMI2870 | PMI_RS14185 | 208 | F4 | 0.575 | UKL | Hypothetical protein | Y | ||
| PMI2930 | glpD/ glyD | PMI_RS14485 | 210 | D8 | 0.727 | K | Aerobic glycerol-3-phosphate dehydrogenase | Lipid metabolism | n/a |
| PMI3027 | aroB | PMI_RS14975 | 219 | A3 | 0.734 | U | 3-Dehydroquinate synthase | Amino acid metabolism | n/a |
| PMI3028 | PMI_RS14980 | 213 | F2 | 0.946 | U | SPOR domain-containing protein | Peptidoglycan binding | n/a | |
| PMI3175 | waaF/ rfaF | PMI_RS15700 | 219 | B2 | 0.402 | K | ADP-heptose--LPS heptosyltransferase II | Lipopolysaccharide biosynthesis | n/a |
| PMI3180 | envC | PMI_RS15725 | 211 | E2 | 0.564 | K | Putative exported peptidase/murein hydrolase activator EnvC | Cell division | Y |
| PMI3185 | cysE | PMI_RS15750 | 212 | G7 | 0.954 | UL | Serine acetyltransferase | Amino acid metabolism | n/a |
| PMI3210 | glpK | PMI_RS15875 | 214 | E2 | 0.958 | U | Glycerol kinase | Lipid metabolism | Y |
| PMI3236 | argE | PMI_RS16000 | 209 | F6 | 0.812 | U | Acetylornithine deacetylase | Amino acid metabolism | n/a |
| PMI3301 | ilvE | PMI_RS16410 | 207 | B4 | 1.016 | U | Branched-chain amino acid aminotransferase | Amino acid metabolism | Y |
| PMI3302 | ilvD | PMI_RS16415 | 207 | D2 | 0.913 | U | Dihydroxy-acid dehydratase | Amino acid metabolism | n/a |
| PMI3316 | wecA/ rfe | PMI_RS16480 | 219 | D6 | 0.351 | U | Undecaprenyl-phosphate alpha-N-acetylglucosaminyl 1-phosphate transferase | Lipopolysaccharide biosynthesis | Y |
| PMI3319 | rffD | PMI_RS16495 | 219 | C5 | 0.398 | K | UDP-N-acetyl-d-mannosamine dehydrogenase | Carbohydrate metabolism | n/a |
| PMI3384 | fklB | PMI_RS16845 | 213 | E11 | 0.588 | K | FkbP-type 22 kDa peptidyl-prolyl cis-trans isomerase | Chaperones and folding catalysts | N |
| PMI3431 | PMI_RS17100 | 213 | E9 | 0.974 | U | Two-component system sensor kinase | Signaling and cellular processes | N | |
| PMI3457 | argI | PMI_RS17230 | 204 | D1 | 0.911 | UL | Ornithine carbamoyltransferase chain I | Amino acid metabolism | Y |
| PMI3528 | metR | PMI_RS17535 | 201 | A2 | 0.728 | U | LysR-family transcriptional regulator | Amino acid metabolism | Y |
| PMI3538 | ubiB | PMI_RS17585 | 206 | A11 | 0.939 | U | Probable ubiquinone biosynthesis protein | Cofactor metabolism | N |
| PMIt068 | PMI_RS16125 | 219 | C1 | 0.490 | K | tRNA | Transcription machinery | n/a |
Location of transposon mutants in the ordered library.
Urine, Kidney, Liver.
The computationally predicted location of the transposon was tested by PCR for 20 mutants.
Seven PCR-confirmed mutants that were selected for follow-up studies are highlighted in bold.
sucB, primarily known as a component of the TCA cycle, is also involved in amino acid metabolism; it was included as a metabolic gene for purposes of categorization in Fig. 2.
Selection of candidates for further study
We selected 20 mutant hits for confirmation of the predicted transposon insertion using PCR. Consistent with our previous randomized testing of this library (20), 16 of 20 mutants were confirmed to have the predicted insertion (Table 2; Fig. S4). We selected seven genes to validate with targeted mutagenesis (Table 2, highlighted in yellow). We chose these specific genes to represent (i) the major functional categories from Fig. 2C (amino acid metabolism and purine biosynthesis) and (ii) a range of mutants with phenotypes on either a single or all three types of agar (urine, kidney, or liver).
Most organ agar mutants had nutritional deficiencies in vitro
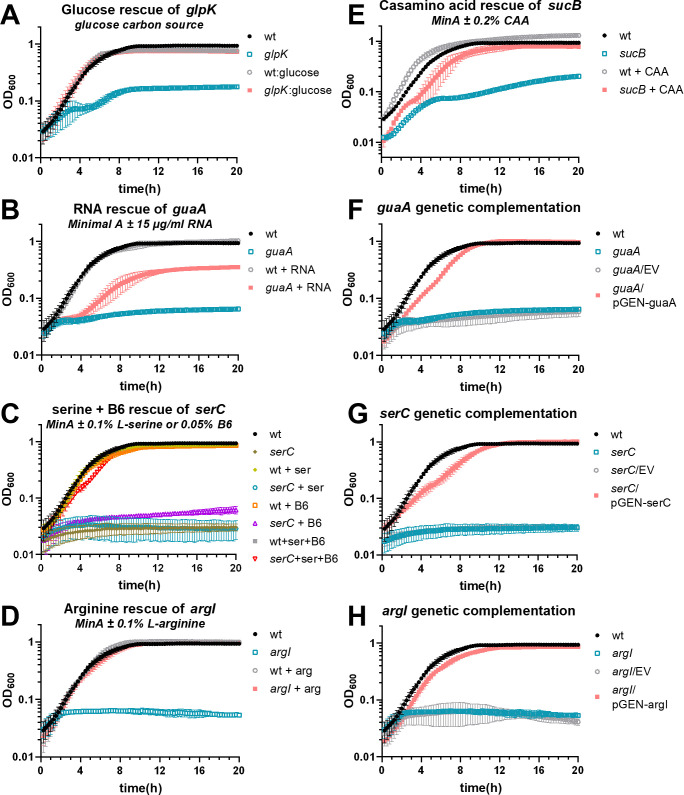
We examined the growth of all seven of the reconstructed mutants in a variety of media. Most exhibited robust growth in complex LB medium, as expected, because the transposon mutants were originally produced using this medium. The exceptions were sucB, which grew at a similar rate as wild type but saturated at a lower density, and serC, which grew more slowly but reached the same saturation as wild type (Fig. 3A; Fig. S5A). However, in the chemically defined minimal medium Minimal A, most mutants (5 of 7) had profoundly diminished growth (Fig. 3B; Fig. S4B). Only two, sanA and PMI2870, displayed normal growth. Based on the predicted function of each mutated gene, we supplemented Minimal A with appropriate substrates and rescued growth of all mutants. Thus, glpK mutant was rescued by swapping the carbon source from glycerol to glucose, argI was rescued by the addition of l-arginine, serC was rescued with a combination of l-serine and vitamin B6 (28), guaA was rescued by the addition of purified P. mirabilis RNA, and sucB was rescued by the addition of casamino acids (Fig. 4A through E).
Fig 3.
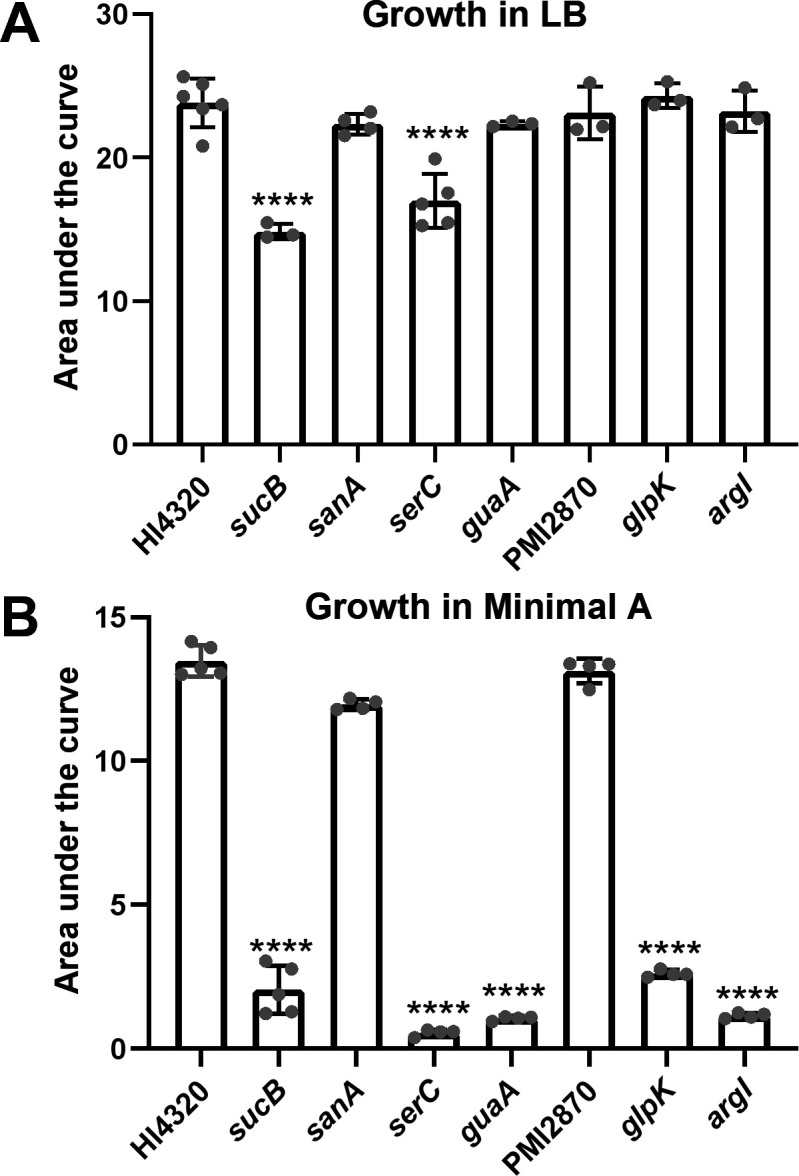
P. mirabilis wild type and mutant strains were assayed for growth in (A) LB (n = 3–6 independent experiments) or (B) chemically defined medium Minimal A (n = 4–5). Area under the curve was measured after culturing for 20 h in the indicated medium. Most mutants grew well in LB, which was the medium used for obtaining transposon mutants. However, most mutants had a deficiency in Minimal A. Error bars depict SD. Statistical significance was calculated vs wild type using one-way ANOVA with Dunnett’s multiple comparisons test.
Fig 4.
Chemical and genetic complementation is shown for a selection of mutants. For all growth curves, Minimal A medium with 0.2% glycerol as a carbon source was used as the base medium unless otherwise specified. (A–E) Chemical complementation growth curves. (A) Complementation of glpK was achieved by swapping the carbon source to 0.2% glucose. (F–H) Genetic complementation growth curves. MinA, Minimal A; B6, pyridoxine HCl; CAA, casamino acids; EV, empty vector (pGEN-MCS). n = 3–5 independent experiments for each condition. Error bars show SD.
We selected three of these mutants for genetic complementation (argI, serC, and guaA). The first two are standalone genes and were cloned as a single fragment with their native promoters. The last gene, guaA, follows guaB as part of an operon (guaBA). This gene was cloned with the gua promoter, omitting guaB. In all three cases, genetic complementation restored growth in Minimal A (Fig. 4F through H).
Some mutants had altered swarming motility
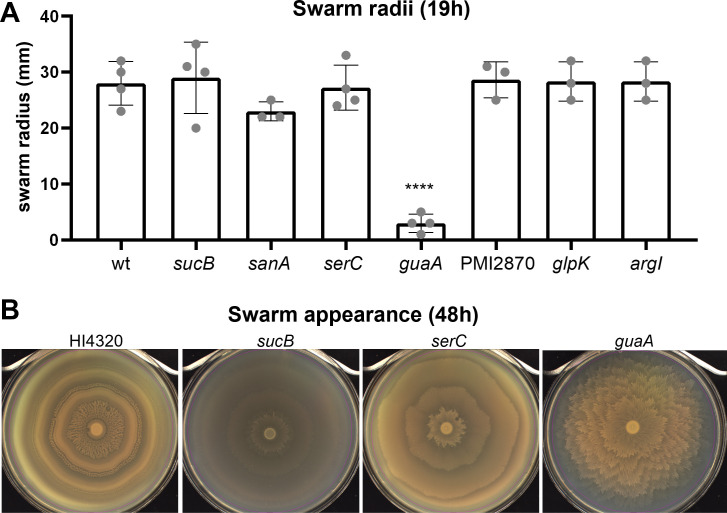
P. mirabilis readily colonizes surfaces via swarming motility, including catheters. Because genes involved in swarming often correlate with in vivo fitness defects (29, 30), we next assessed swarming motility by these mutants. When assayed for distance swarmed after 19 h of incubation, only the guaA mutant had a significant defect (P < 0.0001; Fig. 5A). However, guaA and two additional mutants, serC and sucB, exhibited swarming patterns strikingly distinct from the classic wild-type bullseye. These patterns were especially apparent with 48 h incubation time, allowing natural pigmentation to develop (Fig. 5B; Video S1). Swarming by all seven mutants after either 19 or 48 h incubation is shown in Fig. S6.
Fig 5.
P. mirabilis readily swarms in a bullseye pattern on LB agar containing 10 g/L NaCl. (A) Quantification of swarm radii after 19 h at 30°C, before wild-type swarms reached the edge of the agar surface. Error bars show SD. Statistical significance was calculated vs wild type using one-way ANOVA with Dunnett’s multiple comparisons test. (B) Photos of selected swarms after 48 h incubation at 30°C when swarming and consolidation rings are more visible. Three of 7 of the organ agar hits had strikingly different patterns compared with wild type.
Organ agar results were reproducible in another strain of mice
The initial organ agar screen was conducted with transposon mutants using outbred male Swiss Webster mice, selected for low cost and wide availability. We next examined agar made from our standard UTI model strain, female CBA/J mice, with the reconstructed mutants and found comparable results (Table 3). Notably, most phenotypes observed on diluted organs were also seen on agar made from undiluted homogenates, and we genetically complemented the growth defects for guaA, serC, and argI (Fig. S7A). Interestingly, complementation of argI led to a hyperaggressive swarming phenotype on urine agar (Fig. S7B), consistent with previous observations that arginine is a swarming cue (31, 32). The major exception was PMI2870, which had a profound defect on all three agar types in the initial screen but no observable defect in the rescreen using CBA/J mice. Because the original screen involved stamping of samples from frozen plates and the rescreen used fresh broth cultures, we reasoned the difference could have resulted from the number of bacteria deposited on the plate. Accordingly, there was a growth defect observed for the PMI2870 mutant on liver agar after 100-fold dilution (Fig. S7C).
TABLE 3.
Comparison of organ agar and in vivo mutant phenotypes
| Gene | Name | Organ a | CBA/J agar b | In vivo c | Function |
|---|---|---|---|---|---|
| PMI0570 | sucB | UKL | UKLBS | UBK | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex |
| PMI0641 | sanA | K | KS | US | Putative transport protein |
| PMI0711 | serC | UK | KS | UBKS | Phosphoserine aminotransferase |
| PMI1545 | guaA | UKL | UKLBS | UK | GMP synthase (glutamine-hydrolyzing) |
| PMI2870 | UKL | None d | UBKS | Hypothetical protein | |
| PMI3210 | glpK | U | None | US | Glycerol kinase |
| PMI3457 | argI | UL | ULB | None | Ornithine carbamoyltransferase chain I |
Transposon mutants on agar from Urine (human), Kidney, Liver.
Targetron mutants on agar from Urine (human), Kidney (0.1×), Liver (0.1×), Bladder, Spleen.
Targetron mutants in CBA/J mice from Urine, Kidney, Spleen.
PMI2870 defect was detected in dilution series (Fig. S7C).
Uropathogenic E. coli (UPEC) is the most common causative agent of uncomplicated UTI (3). To investigate whether our P. mirabilis findings extended to this species, we next tested whether UPEC mutants had similar deficiencies on organ agar. Homologous mutations in UPEC CFT073 were available for five of seven of the target genes. Interestingly, all five of these mutants grew on all organ agars (Fig. S7A). This is consistent with prior work where these mutants had no apparent defect in either human urine ex vivo or in murine bladders (6). Notably, UPEC often encodes redundancy that is not seen for P. mirabilis (27, 33 – 36).
Organ agar hits displayed defects in murine ascending UTI model
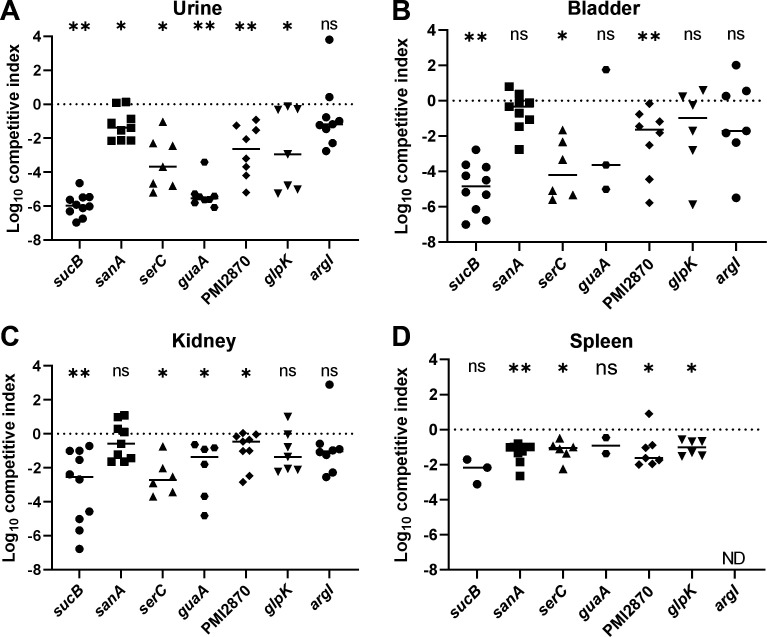
We were especially interested to see if poor growth of mutants on organ agar translated to deficiencies in establishing UTI. To this end, we transurethrally inoculated bladders of female CBA/J mice with a 1:1 mixture of wild-type P. mirabilis HI4320 and each mutant. After 7 days, urine was collected; then mice were sacrificed; and bladders, kidneys, and spleens were removed. In all cases, except argI, there was a statistically significant defect in colonization compared with wild type in one or more of the assayed sites (Fig. 6). In three cases (sucB, serC, and guaA), the mutant was almost completely outcompeted by wild type. A comparison of male Swiss Webster murine organ agar with transposon mutants, female CBA/J murine organ agar with targetron mutants, and in vivo female CBA/J UTI model fitness is shown in Table 3.
Fig 6.
Female CBA/J mice were transurethrally inoculated with a 1:1 mixture of HI4320 (wt) and the indicated mutant. After 7 days, bacteria were quantitatively cultured and competitive indices were calculated. Each symbol represents an individual mouse. Bars indicate median values and the dotted line at y = 0 represents no competitive advantage for either wild type or mutant. P values were assessed using the Wilcoxon signed rank test (*P < 0.05; **P < 0.01; ns, not significant). ND, not determined; a competitive index could not be determined because no bacteria were recovered.
The only mutant that was not statistically significantly outcompeted at any site in murine cochallenge was argI, which nevertheless was recovered from urine, bladder, and kidneys by a median of 1–2 log CFU less than wild type. Notably, the overall bacterial burden at 7 days post-inoculation was lower than usual in the argI cochallenge (Fig. S8). In summary, lack of growth on organ agar was a highly effective predictor of fitness loss in the urinary tract of mice.
DISCUSSION
Microbiologists have historically cultured bacteria using media that are reflective of their origins. For example, soil organisms would use soil medium and marine organisms would use seawater (37, 38). Organ-derived media have been used since the inception of bacteriological culture to study mammalian pathogens, sometimes as a method to distinguish related bacterial species (39), but not to our knowledge as a method to screen mutant libraries for host fitness.
P. mirabilis, like many other bacterial species, colonizes and infects multiple sites within the human body to cause disease. Specifically, Proteus can colonize the gut, skin wounds, and the urinary tract (40, 41). These unique host niches are studied using various in vivo models, each with their own limitations (16, 19, 42 – 44). Here, we demonstrated that the urinary tract has strict bottleneck effects in the murine model, requiring the use of many animals for testing mutant libraries. To address this, we present an alternative screening method by generating agar from whole organ samples, greatly reducing the number of mice required for experimentation while increasing feasibility of large screens in models exhibiting limitations.
We screened 1,728 individual P. mirabilis transposon mutants from an ordered transposon library, representing 45% of predicted genes in strain HI4320 (20), on human urine and murine kidney and liver agar. Stamping arrayed mutants onto agar only required the use of organs from 15 mice to screen this library. Using a calculated bottleneck of 25 mutants, a comparable Tn-seq experiment would have required 70 mice; thus, organ agar contributes to the conservation of animal lives. In addition to being more ethically and fiscally responsible, this alternative ex vivo model strongly correlated with colonization defects in vivo and, thus, serves as a proxy for more complex model systems. Indeed, six out of seven mutants had statistically significant defects in the urine, bladder, kidneys, or spleen of mice when examined in the traditional cochallenge model of ascending UTI (Fig. 6) and other validating hits were observed, such as zinc transporter genes znuA and znuC (26).
Our data show that organ agar is a physiologically relevant medium for testing colonization and virulence factors. However, certain variables may not be modeled as they are in vivo. For example, neutrophil recruitment plays an important role in Proteus uropathogenesis (45, 46). These non-resident immune cells would likely be in low numbers in the organs of naive mice, and those that are present in organ agar would not exhibit antimicrobial activities such as phagocytosis. Likewise, the architecture of different cell types is disrupted during homogenization. In the uroepithelium, for example, the surface-expressed residues used for bacterial adherence may be heterogeneously distributed throughout the agar. Deeper tissues such as the lamina and muscularis propria, which likely do not directly encounter the bacteria, are exposed in organ agar. Despite these potential pitfalls, organ agar detected fitness factors in functional categories including metabolism, transport, LPS biosynthesis, and even those with unpredicted function.
Most detected hits, 54.3%, fell into a metabolism-related category, suggesting that this method is particularly powerful for identifying the nutritional requirements of microorganisms in their respective host environments. Five of the seven mutants selected for further follow-up analysis were part of well-studied metabolic pathways. Although the lack of growth of most mutants in minimal medium was initially surprising, several mutants exhibiting no growth defects in LB or minimal medium were outcompeted in vivo, demonstrating the relevance and sensitivity of ex vivo organ agar for discovering new fitness factors beyond metabolism. These findings contrast with a UPEC Tn-seq study, where a much smaller percentage (8%) of detected fitness factors were identified as metabolic (6). Yet, tailored metabolism is increasingly recognized as an integral part of UPEC virulence (47). The E. coli accessory genome is remarkably broad (48), and organ agar would greatly facilitate screening a wide variety of UPEC strains for nutritional fitness.
Organ agar also uncovered new connections between nutrition and swarming motility. Swarming is the coordinated, flagella-dependent movement of bacteria across a solid surface. P. mirabilis swarming is particularly aggressive compared with other swarm-capable bacterial species and typically manifests as a characteristic bullseye pattern due to successive waves of swarming and consolidation (12). There is a long history establishing the dual importance of biosynthetic and nutrient acquisition pathways to both swarming motility and virulence or colonization fitness, including amino acid biosynthesis, glycolysis, TCA cycle, and peptide import (12). Three of the seven targeted mutants in this study displayed unusual swarm patterns, and all three of these contained mutations in biosynthetic genes (sucB, serC, and guaA). The guaA GMP synthase mutant is particularly interesting because of its striking swarming phenotype and a newly identified role for purine biosynthesis in both swarming and colonization of the urinary tract. Previous identification of purines in P. mirabilis pathogenesis focused on cyclic AMP, with swarming and virulence defects found for cyaA adenylate cyclase and crp cyclic AMP receptor protein mutants (34, 49). A different STM study also reported altered swarming for a serC mutant although the transposon insertion was upstream of the translational start site and that mutant was able to grow in minimal medium (50). Likewise, serine uptake and biosynthesis have both recently been shown to be important for P. mirabilis swarming motility and fitness in a catheterized mouse model of UTI (51, 52). Additionally, when the argI mutant was genetically complemented in trans, this strain gained an aggressive swarming phenotype, perhaps because extracellular arginine is a potent stimulus of swarming (32). Finally, we note that buffered urine agar mostly constrains P. mirabilis swarming but does not eliminate it. This modified urine agar would be a useful tool for screening for swarming-defective mutants. Thus, organ agar may facilitate further studies into the dynamics of swarming motility and its possible contribution to colonization of the urinary tract.
Organ agar reflects distinct niches; for example, the liver and kidney give different results. We speculate that use of particular host genetic backgrounds will also identify unique fitness determinates. For example, knockout mice may change the nutritional landscape for microorganisms. Organ agar could likewise be used to test age or sex differences. Similarly, urine agar could be made using samples from volunteers with specific diets or disease states.
Overall, we demonstrate that ex vivo organ agar is a reliable, sensitive method to predict fitness factors in model organisms that can be recapitulated in vivo. This is especially important for microorganisms that lack well-developed animal models. Additionally, organ agar can utilize animals that would normally be euthanized to maintain breeding colonies. Using different host models with unique genetic backgrounds, such as knockout mice, may yield surprising and exciting results and mitigate previously mentioned limitations such as immune interactions. We propose that additional refinements of the technique, such as improving plate clarity to allow quantitative densitometry measurements, will further expand the utility of organ agar. We hope that others will use this method to quickly perform large screens of other species and find similar success.
MATERIALS AND METHODS
Bacterial strains and plasmids
P. mirabilis strain HI4320 was isolated from the urine of an elderly female nursing home patient with a long-term (≥30 days) indwelling catheter (53, 54). This strain is well established as a model organism for P. mirabilis virulence studies and readily produces UTIs in mice (15, 16). The P. mirabilis HI4320 ordered library contains 1,728 transposon mutants, each insertion within a different open reading frame, and has been described elsewhere in detail (20). Uropathogenic Escherichia coli CFT073 was isolated from the blood and urine of a hospitalized patient with acute pyelonephritis and bacteremia (55). E. coli homologs of P. mirabilis organ agar hits were identified using BLAST, and selected E. coli CFT073 transposon mutants were pulled from a previously reported ordered library (6). Bacteria were routinely cultured at 37°C with aeration in lysogeny broth (LB; 10 g/L tryptone, 5 g/L yeast extract, 0.5 g/L NaCl) or on LB solidified with 1.5% agar. All strains and plasmids are listed in Table S1. All strains used in this study can be obtained by contacting the corresponding author.
Organ and urine agar
For initial development of organ agar, 5 male Swiss Webster mice (outbred, 6- to 7-week-old, Envigo) were humanely euthanized. To facilitate aseptic organ removal, skin and fur were removed from the abdominal region as previously described prior to opening the abdominal cavity (16). Organs (bladders, kidneys, and spleens) were removed, homogenized in 3 mL of water, pooled by organ type, and diluted in a range of ratios in sterile distilled water (Table 4). Three percent of agar was autoclaved, cooled to 55°C, and mixed 1:1 with the diluted organ homogenates. In general, larger organ masses could be diluted further and still support bacterial growth; for example, a 1:10 dilution of kidney homogenate was experimentally determined to sustain reliable growth of wild-type P. mirabilis HI4320 (Fig. S1). Because growth on spleen agar was relatively poor, for the subsequent ordered library screen, spleens were swapped for larger livers to represent disseminated infection. Urine agar was made with modifications from a previous protocol (24). Specifically, human urine was collected from seven healthy female volunteers, pooled, and filter sterilized. Aliquots of urine were stored at −20°C. Four percent of agar buffered with 500 mM HEPES, pH 6.8, was autoclaved, cooled to 55°C, and mixed 1:1 with pooled urine. Both organ and urine agar were precisely aliquoted (20 mL) to 100 mm Petri dishes and allowed to solidify and dry at room temperature overnight.
TABLE 4.
Preparation of organ agar, per plate
| Ratio a | Organs per plate (mL) | Homogenized organ (mL) | Sterile water (mL) | Molten 3% agar (mL) | Total volume (mL) |
|---|---|---|---|---|---|
| 2× b | 6.7 | 10 | 0 | 10 | 20 |
| 1× | 3.3 | 10 | 0 | 10 | 20 |
| 1:2 | 1.7 | 5 | 5 | 10 | 20 |
| 1:5 | 0.67 | 2 | 8 | 10 | 20 |
| 1:10 | 0.33 | 1 | 9 | 10 | 20 |
Ratio is determined by dilution of organ before mixing with agar.
For 2× agar, homogenize two organs per 3 mL water; otherwise prepare as for 1× agar.
Ordered library screen
For the library screen, 15 male Swiss Webster mice were euthanized as above. Livers and kidneys were collected, homogenized, and pooled and then diluted 1:10 with sterile distilled water. Diluted homogenates or pooled human urine were mixed with molten agar and aliquoted to Petri dishes as above. Twenty 96-well plates containing the frozen P. mirabilis ordered library were placed on dry ice. A sterile 48-pin stamper was used to transfer bacteria onto organ or urine agar plates, which were then incubated at 37°C for 24 or 48 h. Plates were visually assessed for decreased or absent growth in each spot. Interesting candidates were rescreened to confirm the phenotype. The genetic location of the transposon was confirmed for a selection of 20 of these mutants using PCR with transposon-specific primer CP-7 and a primer flanking the transposon insertion site (Table S2).
Construction of targeted mutants
Stable insertional mutations of selected genes were generated using the targetron method (23, 56). Briefly, a group II intron fragment was synthesized (eBlocks, Integrated DNA Technologies) to specifically target each gene using the ClosTron prediction algorithm (57). Reprogrammed intron fragments were cloned into pACD4K-CloxP using NEBuilder HiFi DNA Assembly master mix (New England Biolabs) with primers designed to amplify vector or intron templates and confirmed using Sanger DNA sequencing. Targetron-containing plasmids and a source for T7 polymerase, pAR1219 (58), were introduced into P. mirabilis HI4320 using electroporation and induced to jump into the specified genes. Insertional mutations in kanamycin-resistant mutants were confirmed using PCR. Targetron plasmids and mutants are listed in Table S1. All primers are listed in Table S2.
Growth curves and chemical complementation of mutants
Wild type and mutant P. mirabilis were cultured separately overnight in LB and then diluted 1:100 into target media for growth curve analysis using a Bioscreen C (Growth Curves USA). Readings were collected at an optical density of 600 nm (OD600) every 15 min for 24 h. Each experiment contained three technical replicates and was conducted at least three times. Minimal A is a minimal, chemically defined medium tailored for P. mirabilis and was used for most experiments (59). The carbon source in Minimal A was 0.2% glycerol unless otherwise specified. For chemical complementation with RNA, RNA purified from P. mirabilis HI4320 or isogenic mutants (RNeasy kit, Qiagen) was pooled together, quantified using a NanoDrop spectrophotometer, and then added to Minimal A at a final concentration of 15 µg/mL.
Genetic complementation of mutants
Complementation plasmid pGEN-MCS, chosen because it is low copy number and is maintained stably without antibiotic selection, was used for genetic complementation of selected mutants (60). Native P. mirabilis promoters were predicted using Softberry (61) and included all predicted DNA-binding protein sites. Coding sequences and promoters were amplified by PCR, cloned using the Gibson method (New England Biolabs), and constructs were confirmed by Sanger sequencing. Complementation plasmids are listed in Table S1, and primers used for cloning are shown in Table S2.
Swarming assays
Swarming motility was assayed on LB agar containing 10 g/L NaCl as previously described (62). Briefly, strains were cultured overnight in LB (LB 10 g/L NaCl), spotted onto swarm agar, and allowed to dry. Swarm plates were incubated at 30°C. Swarm radii were measured after incubation for 19 h, and swarms were photographed after incubation for 48 h using a Qcount (Advanced Instruments). Swarming experiments were independently repeated at least three times.
Mouse model of ascending UTI
Bacterial fitness during UTI was assessed using a well-established mouse model (17, 63, 64). Briefly, overnight cultures of P. mirabilis were diluted in LB to OD600 = 0.194 (~2 × 108 CFU/mL), and then wild type and mutant bacteria were mixed 1:1. Ten female CBA/J mice, aged 5–6 weeks (Jackson Laboratory), were transurethrally inoculated with 50 µL of this 1:1 mixture (107 CFU/mouse). At 7 days post-inoculation, urine was collected; mice were euthanized; and bladders, kidneys, and spleens were harvested. Organs were homogenized and plated to quantify CFU; mutants were distinguished from wild-type colonies using kanamycin. Competitive indices were calculated for each site by comparing the ratio of output wild type and mutant to the ratio of input bacteria (36). Statistical significance of competitive indices was calculated using the Wilcoxon signed rank test.
Mouse model bottleneck determination
Wild-type HI4320 and a kanamycin-resistant mutant that was previously shown to have no fitness defect in our mouse UTI model, spa47 (23), were mixed in different ratios to test the limitations of our mouse model. The OD600 was recorded to normalize strains prior to mixing and adjusted to target ratios. Aliquots of the input mixtures were taken as samples and plated to determine the actual ratio of mutant:wild type in the inoculum (Fig. 1). The output organ samples collected from CBA/J mice were homogenized and enumerated, via differential plating, to determine mutant and wild-type HI4320 CFU burden. A competitive index calculation was used to demonstrate the output ratio relative to the input ratio (6). A sample that was the same ratio at the beginning and end of the experiment would have log10 CI = 0. The acceptable range of error for these experiments was ±1 log (10-fold) CFU.
Organ agar for targeted mutants
Targetron mutants were tested on organ agar made from female CBA/J mice to directly compare with in vivo cochallenges in this standard UTI mouse model. Agar was made as above, but in addition to 1:10 dilutions of liver and kidneys, undiluted “1×” organs were used. For bladder and spleen agar, two organs were homogenized together in 3 mL of water to achieve a 2× concentration of each organ. Bacteria were cultured overnight in LB, adjusted to OD600 = 0.1 in LB, aliquoted into a 96-well plate, and stamped onto organ agar. Plates were incubated at 37°C, and growth was photographed at 24 and 48 h. Mutant PMI2870 was additionally screened on urine and 0.1× liver agar in a 1:10 dilution series to determine whether the amount of inoculum affected growth on organ agar.
Statistics and software
All graphs and charts were plotted and statistics calculated using GraphPad Prism 9. Error bars show SD. To calculate statistical significance for all bar graphs, one-way ANOVA with Dunnett’s multiple comparisons test was used. For mouse cochallenge experiments, statistical significance was calculated using the Wilcoxon signed rank test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
ACKNOWLEDGMENTS
We gratefully acknowledge Stephanie Himpslfor photography of swarm agar plates for the time-lapse images and Geoff Severin and Madison Fitzgerald for manuscript editing. Figure 2A was created using BioRender. Research reported in this publication was supported by National Institutes of Health award R01AI059722 (H.L.T.M. and M.M.P.) and F32AI147527 (A.E.S.).
Footnotes
This article is a direct contribution from Harry L. T. Mobley, a member of the Infection and Immunity Editorial Board, who arranged for and secured reviews by Sheryl Justice, The Ohio State University School of Medicine, and Ana Flores-Mireles, University of Notre Dame.
Contributor Information
Melanie M. Pearson, Email: mpears@umich.edu.
Andreas J. Bäumler, University of California, Davis, California, USA
ETHICS APPROVAL
Animal experiments were approved by the University of Michigan Medical School Institutional Animal Care and Use Committee, protocol number PRO00007111. During catheterization procedures, mice were anesthetized by intraperitoneal injection of ketamine/xylazine. Mice were euthanized by inhalant isoflurane anesthetic overdose prior to organ removal.
Collection of deidentified clean-catch human urine from healthy adult female volunteers was deemed to not involve human subjects by the University of Michigan Medical School Institutional Review Board (protocol HUM 4949), and therefore, this study required neither IRB exemption nor oversight.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00355-23.
Supplemental figures.
Bacterial strains, plasmids, and primers.
Side-by-side swarming comparison of wild type, serC, and guaA mutants after incubation at 30C from 2-46 h.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Falkow S. 2004. Molecular koch's postulates applied to bacterial pathogenicity--a personal recollection 15 years later. Nat Rev Microbiol 2:67–72. doi: 10.1038/nrmicro799 [DOI] [PubMed] [Google Scholar]
- 2. Subashchandrabose S, Mobley HLT. 2015. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr 3. doi: 10.1128/microbiolspec.UTI-0015-2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McLellan LK, Hunstad DA. 2016. Urinary tract infection: pathogenesis and outlook. Trends Mol Med 22:946–957. doi: 10.1016/j.molmed.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neugent ML, Hulyalkar NV, Nguyen VH, Zimmern PE, De Nisco NJ. 2020. Advances in understanding the human urinary microbiome and its potential role in urinary tract infection. mBio 11:e00218-20. doi: 10.1128/mBio.00218-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abraham SN, Miao Y. 2015. The nature of immune responses to urinary tract infections. Nat Rev Immunol 15:655–663. doi: 10.1038/nri3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shea AE, Marzoa J, Himpsl SD, Smith SN, Zhao L, Tran L, Mobley HLT. 2020. Escherichia coli CFT073 fitness factors during urinary tract infection: identification using an ordered transposon library. Appl Environ Microbiol 86:e00691-20. doi: 10.1128/AEM.00691-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warr AR, Hubbard TP, Munera D, Blondel CJ, Abel Zur Wiesch P, Abel S, Wang X, Davis BM, Waldor MK. 2019. Transposon-insertion sequencing screens unveil requirements for EHEC growth and intestinal colonization. PLoS Pathog 15:e1007652. doi: 10.1371/journal.ppat.1007652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hullahalli K, Waldor MK. 2021. Pathogen clonal expansion underlies multiorgan dissemination and organ-specific outcomes during murine systemic infection. Elife 10:e70910. doi: 10.7554/eLife.70910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abel S, Abel zur Wiesch P, Davis BM, Waldor MK, Hogan DA. 2015. Analysis of bottlenecks in experimental models of infection. PLoS Pathog 11:e1004823. doi: 10.1371/journal.ppat.1004823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 11. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schaffer JN, Pearson MM. 2015. Proteus mirabilis and urinary tract infections. Microbiol Spectr 3. doi: 10.1128/microbiolspec.UTI-0017-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pelling H, Nzakizwanayo J, Milo S, Denham EL, MacFarlane WM, Bock LJ, Sutton JM, Jones BV. 2019. Bacterial biofilm formation on indwelling urethral catheters. Lett Appl Microbiol 68:277–293. doi: 10.1111/lam.13144 [DOI] [PubMed] [Google Scholar]
- 14. Armbruster CE, Prenovost K, Mobley HLT, Mody L. 2017. How often do clinically diagnosed catheter-associated urinary tract infections in nursing homes meet standardized criteria? J Am Geriatr Soc 65:395–401. doi: 10.1111/jgs.14533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Armbruster CE, Mobley HLT, Pearson MM. 2018. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith SN. 2019. Independent transurethral urinary tract inoculation in a murine model of ascending infection with Proteus mirabilis. Methods Mol Biol 2021:159–172. doi: 10.1007/978-1-4939-9601-8_15 [DOI] [PubMed] [Google Scholar]
- 17. Smith SN. 2019. Cochallenge inoculation with Proteus mirabilis in a murine transurethral urinary tract model of ascending infection. Methods Mol Biol 2021:173–186. doi: 10.1007/978-1-4939-9601-8_16 [DOI] [PubMed] [Google Scholar]
- 18. Smith SN, Armbruster CE. 2019. Indwelling urinary catheter model of Proteus mirabilis infection. Methods Mol Biol 2021:187–200. doi: 10.1007/978-1-4939-9601-8_17 [DOI] [PubMed] [Google Scholar]
- 19. Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, Wu W, Mobley HLT, Baumler AJ. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13:e1006434. doi: 10.1371/journal.ppat.1006434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pearson MM, Pahil S, Forsyth VS, Shea AE, Mobley HLT. 2022. Construction of an ordered transposon library for uropathogenic Proteus mirabilis HI4320. Microbiol Spectr 10:e0314222. doi: 10.1128/spectrum.03142-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walters MS, Lane MC, Vigil PD, Smith SN, Walk ST, Mobley HLT, Miller JF. 2012. Kinetics of uropathogenic Escherichia coli metapopulation movement during urinary tract infection. mBio 3:e00303-11. doi: 10.1128/mBio.00303-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwartz DJ, Chen SL, Hultgren SJ, Seed PC. 2011. Population dynamics and niche distribution of uropathogenic Escherichia coli during acute and chronic urinary tract infection. Infect Immun 79:4250–4259. doi: 10.1128/IAI.05339-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pearson MM, Mobley HLT. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J Med Microbiol 56:1277–1283. doi: 10.1099/jmm.0.47314-0 [DOI] [PubMed] [Google Scholar]
- 24. Pearson MM. 2019. Culture methods for Proteus mirabilis. Methods Mol Biol 2021:5–13. doi: 10.1007/978-1-4939-9601-8_2 [DOI] [PubMed] [Google Scholar]
- 25. Naylor P. 1964. The effect of electrolytes or carbohydrates in sodium chloride deficient medium on formation of discrete colonies of proteus and the influence of these substances on growth in liquid culture. J. Appl. Bacteriol 27:422–431. doi: 10.1111/j.1365-2672.1964.tb05050.x [DOI] [Google Scholar]
- 26. Nielubowicz GR, Smith SN, Mobley HLT. 2010. Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infect Immun 78:2823–2833. doi: 10.1128/IAI.01220-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alteri CJ, Himpsl SD, Mobley HLT, Kubori T. 2015. Preferential use of central metabolism in vivo reveals a nutritional basis for polymicrobial infection. PLoS Pathog 11:e1004601. doi: 10.1371/journal.ppat.1004601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shimizu S, Dempsey WB. 1978. 3-hydroxypyruvate substitutes for pyridoxine in serC mutants of Escherichia coli K-12. J Bacteriol 134:944–949. doi: 10.1128/jb.134.3.944-949.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pearson MM, Rasko DA, Smith SN, Mobley HLT. 2010. Transcriptome of swarming Proteus mirabilis. Infect Immun 78:2834–2845. doi: 10.1128/IAI.01222-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allison C, Coleman N, Jones PL, Hughes C. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect Immun 60:4740–4746. doi: 10.1128/iai.60.11.4740-4746.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Armbruster CE, Hodges SA, Smith SN, Alteri CJ, Mobley HLT. 2014. Arginine promotes Proteus mirabilis motility and fitness by contributing to conservation of the proton gradient and proton motive force. Microbiologyopen 3:630–641. doi: 10.1002/mbo3.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Armbruster CE, Hodges SA, Mobley HLT. 2013. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess L-glutamine. J Bacteriol 195:1305–1319. doi: 10.1128/JB.02136-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Himpsl SD, Pearson MM, Arewång CJ, Nusca TD, Sherman DH, Mobley HLT. 2010. Proteobactin and a yersiniabactin-related siderophore mediate iron acquisition in Proteus mirabilis. Mol Microbiol 78:138–157. doi: 10.1111/j.1365-2958.2010.07317.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HLT. 2008. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol 57:1068–1078. doi: 10.1099/jmm.0.2008/002071-0 [DOI] [PubMed] [Google Scholar]
- 35. Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HLT. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190:4027–4037. doi: 10.1128/JB.01981-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia EC, Brumbaugh AR, Mobley HLT. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79:1225–1235. doi: 10.1128/IAI.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. GREENFIELD LJ, HINES JJ, BORAL LL. 1962. Polymorphism in a marine bacterium in relation to population growth. J Bacteriol 84:357–363. doi: 10.1128/jb.84.2.357-363.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stewart EJ. 2012. Growing unculturable bacteria. J Bacteriol 194:4151–4160. doi: 10.1128/JB.00345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huddleson IF. 1964. Enhancement of colonial growth of brucellae on beef liver agar. J Bacteriol 88:540. doi: 10.1128/jb.88.2.540-540.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuan L, Schaffer JN, Zouzias CD, Pearson MM. 2014. Characterization of 17 chaperone-usher fimbriae encoded by Proteus mirabilis reveals strong conservation. J Med Microbiol 63:911–922. doi: 10.1099/jmm.0.069971-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O’Hara CM, Brenner FW, Miller JM. 2000. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev 13:534–546. doi: 10.1128/CMR.13.4.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Armbruster CE, Forsyth VS, Johnson AO, Smith SN, White AN, Brauer AL, Learman BS, Zhao L, Wu W, Anderson MT, Bachman MA, Mobley HLT, Baumler AJ. 2019. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog 15:e1007653. doi: 10.1371/journal.ppat.1007653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seo SU, Kamada N, Muñoz-Planillo R, Kim YG, Kim D, Koizumi Y, Hasegawa M, Himpsl SD, Browne HP, Lawley TD, Mobley HLT, Inohara N, Núñez G. 2015. Distinct commensals induce interleukin-1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity 42:744–755. doi: 10.1016/j.immuni.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McManus AT, McLeod CG, Mason AD. 1982. Experimental Proteus mirabilis burn surface infection. Arch Surg 117:187–191. doi: 10.1001/archsurg.1982.01380260057010 [DOI] [PubMed] [Google Scholar]
- 45. Schaffer JN, Norsworthy AN, Sun TT, Pearson MM. 2016. Proteus mirabilis fimbriae- and urease-dependent clusters assemble in an extracellular niche to initiate bladder stone formation. Proc Natl Acad Sci U S A 113:4494–4499. doi: 10.1073/pnas.1601720113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Y, Kwon K, Tsitrin T, Bekele S, Sikorski P, Nelson KE, Pieper R, Weiss D. 2017. Characterization of early-phase neutrophil extracellular traps in urinary tract infections. PLoS Pathog 13:e1006151. doi: 10.1371/journal.ppat.1006151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan CCY, Lewis IA. 2022. Role of metabolism in uropathogenic Escherichia coli. Trends Microbiol 30:1174–1204. doi: 10.1016/j.tim.2022.06.003 [DOI] [PubMed] [Google Scholar]
- 48. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. [DOI] [PubMed] [Google Scholar]
- 49. Tsai YL, Chien HF, Huang KT, Lin WY, Liaw SJ. 2017. cAMP receptor protein regulates mouse colonization, motility, fimbria-mediated adhesion, and stress tolerance in uropathogenic Proteus mirabilis. Sci Rep 7:7282. doi: 10.1038/s41598-017-07304-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HLT. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: Identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect Immun 72:2922–2938. doi: 10.1128/IAI.72.5.2922-2938.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brauer AL, White AN, Learman BS, Johnson AO, Armbruster CE, D’Orazio SEF. 2019. D-serine degradation by Proteus mirabilis contributes to fitness during single-species and polymicrobial catheter-associated urinary tract infection. mSphere 4:e00020-19. doi: 10.1128/mSphere.00020-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brauer AL, Learman BS, Taddei SM, Deka N, Hunt BC, Armbruster CE. 2022. Preferential catabolism of L- vs D-serine by Proteus mirabilis contributes to pathogenesis and catheter-associated urinary tract infection. Mol Microbiol 118:125–144. doi: 10.1111/mmi.14968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warren JW, Tenney JH, Hoopes JM, Muncie HL, Anthony WC. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J Infect Dis 146:719–723. doi: 10.1093/infdis/146.6.719 [DOI] [PubMed] [Google Scholar]
- 54. Mobley HLT, Chippendale GR. 1990. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J Infect Dis 161:525–530. doi: 10.1093/infdis/161.3.525 [DOI] [PubMed] [Google Scholar]
- 55. Mobley HLT, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren J. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pearson MM, Himpsl SD, Mobley HLT. 2019. Insertional mutagenesis protocol for constructing single or sequential mutations. Methods Mol Biol 2021:61–76. doi: 10.1007/978-1-4939-9601-8_7 [DOI] [PubMed] [Google Scholar]
- 57. Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The clostron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018 [DOI] [PubMed] [Google Scholar]
- 58. Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 81:2035–2039. doi: 10.1073/pnas.81.7.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Belas R, Erskine D, Flaherty D. 1991. Transposon mutagenesis in Proteus mirabilis. J Bacteriol 173:6289–6293. doi: 10.1128/jb.173.19.6289-6293.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, N.Y. [Google Scholar]
- 62. Pearson MM. 2019. Methods for studying swarming and swimming motility. Methods Mol Biol 2021:15–25. doi: 10.1007/978-1-4939-9601-8_3 [DOI] [PubMed] [Google Scholar]
- 63. Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Edén C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. doi: 10.1128/iai.40.1.273-283.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Johnson DE, Lockatell CV, Hall-Craigs M, Mobley HLT, Warren J. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J Urol 138:632–635. doi: 10.1016/s0022-5347(17)43287-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures.
Bacterial strains, plasmids, and primers.
Side-by-side swarming comparison of wild type, serC, and guaA mutants after incubation at 30C from 2-46 h.