Abstract
Heat shock factor Hsf in nonvertebrate animals and homologous heat shock factor Hsf1 in vertebrate animals are key transcriptional regulators of the stress protein response. Hsf/Hsf1 is constitutively present in cells but is, typically, only active during periods during which cells are experiencing a physical or chemical proteotoxic stress. It has become increasingly clear that regulation of Hsf/Hsf1 activity occurs at multiple levels: the oligomeric status of Hsf/Hsf1, its DNA-binding ability, posttranslational modification, transcriptional competence, nuclear/ subnuclear localization, as well as its interactions with regulatory cofactors or other transcription factors all appear to be carefully controlled. This review emphasizes work reported over the past several years suggesting that regulation at several of these levels is mediated by repressive interactions of Hsp90-containing multichaperone complexes and/or individual chaperones and Hsf/Hsf1.
INTRODUCTION
Heat shock transcription factors (Hsf) and regulation of the stress protein response were reviewed periodically before (eg, Wu 1995; Voellmy 1996; Morimoto 1998; Morano and Thiele 1999; Pirkkala et al 2001; Christians et al 2002; Holmberg et al 2002). The reader is referred to this earlier literature for aspects not discussed in detail herein. Although an attempt was made to at least introduce the many aspects of the subject area, this article is primarily intended to review the topic of regulation of Hsf activity by heat shock proteins and other chaperones and cochaperones. The reason for this focus is not only that this topic has been a major interest of the author's laboratory but also that much of what is now known has only been learned during the past several years and has not yet been properly reviewed. For many years, models of Hsf regulation by chaperones have been “in competition” with models invoking direct sensing of stress by the heat shock factor (HSF) molecule. The latter models are also reviewed herein, albeit from the perspective of one who favors a chaperone-based model. Finally, several additional factors that were shown to affect Hsf activity are discussed, especially those discovered only recently.
Nonvertebrate metazoans such as the fruit fly Drosophila melanogaster have a single Hsf. Vertebrates are capable of expressing 4 distinct but related Hsf species. Hsf1, Hsf2, and Hsf4 are found in mammalian cells, whereas Hsf3 appears to be an avian-specific factor. Depending on the context, the term “Hsf” is used herein to relate to heat shock factor generically or to a heat shock factor from a nonvertebrate, metazoan cell that expresses only a single type of factor.
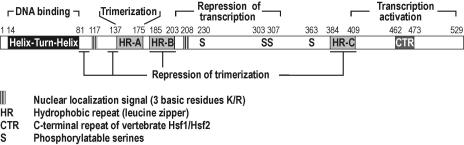
Sequence and functional features of a typical metazoan Hsf are outlined in Figure 1, using human Hsf1 as the example. A DNA-binding domain comprising a winged, helix-turn-helix motif (see Harrison et al 1994 for a crystal structure of the related yeast Hsf domain; Vuister et al 1994; see Schultheiss et al 1996 for a study on a related plant Hsf domain) is located near the amino terminus (Clos et al 1990). The domain is capable of interacting with the so-called heat shock element (HSE) sequences present in the promoters of heat shock protein (hsp) genes. Sequence comparison revealed that all Hsf types have a long, interrupted, hydrophobic repeat sequence (HR-A/B) situated adjacent to the DNA-binding domain. Oligomerization of Hsf requires the presence of at least a portion of this sequence (see Sorger and Nelson 1989; Clos et al 1990; see also Peteranderl and Nelson 1992). Most Hsf structures also include an additional hydrophobic repeat sequence (HR-C) in the carboxy-terminal third of their sequence. Studies on mammalian Hsf1 and Drosophila Hsf located transactivation domains near the carboxy ends of the respective factors (Green et al 1995; Shi et al 1995; Zuo et al 1995; Wisniewski et al 1996).
Fig 1.
Sequence features and functional properties of human Hsf1. Numbers refer to amino acid residues
There is convincing evidence that Hsf-Hsf1 is the key factor controlling stress-regulated gene expression in most vertebrate cells, excepting avian cells (see the comments below). Antibodies specific for Hsf1 supershift or inactivate essentially all stress-induced HSE DNA-binding activity (Baler et al 1993; Sarge et al 1993; Ali et al 1998). An hsf1−/− mouse, as well as a derived embryo fibroblast line, is deficient in heat induction of hsp gene expression (McMillan et al 1998; see also Zhang et al 2002). The defective stress protein response in the hsf1−/ − fibroblasts can be repaired by transfection of an Hsf1 expression construct (see, eg, Holmberg et al 2001). Drosophila expressing the temperature-sensitive Hsf4 mutation is incapable of heat-induced hsp gene expression (Jedlicka et al 1997).
Although it was recently proposed that Hsf1 and Hsf2 could coactivate hsp genes under certain conditions of stress (Mathew et al 2001) or that Hsf2 may coregulate Hsf1 (He et al 2003), the evidence supporting these roles for Hsf2 remains somewhat circumstantial. In contrast, it appears well established that Hsf2 is an important developmental regulator. Such a role was first suggested after the HSE DNA-binding activity of Hsf2 was found induced in hemin-treated K562 erythroleukemia cells (Sistonen et al 1992, 1994). Even though a subsequent study suggested that hemin-induced Hsp expression in K562 cells was mediated by Hsf1 rather than Hsf2 (Yoshima et al 1998), the notion that Hsf2 plays a developmental role has survived: several studies of mouse development, including studies using hsf2−/− mice, clearly implicated Hsf2 in developmental regulation (Rallu et al 1997; Eriksson et al 2000; Kallio et al 2002; Paslaru et al 2003; see also Wang et al 2003). Surprisingly, this regulation may not involve modulation of hsp gene expression (Rallu et al 1997; Wang et al 2003).
The carboxy half of Hsf4 lacks sequences present in other Hsf types (Nakai et al 1997). Interestingly, alternative splicing creates 2 Hsf4 proteins of slightly different sequence that have strikingly different functional properties (Tanabe et al 1999): when overexpressed, HsfF4a acts as an inhibitor of both constitutive and heat-induced Hsp expression (Nakai et al 1997; Tanabe et al 1999; Zhang et al 2001), whereas Hsf4b has properties of a transcriptional activator (Tanabe et al 1999). It is presently unknown whether endogenous Hsf4b significantly contributes to stress-induced HSF activity or whether endogenous Hsf4a modulates overall Hsf activity.
Avian cells but not mammalian cells express additional heat shock factor Hsf3 (Nakai and Morimoto 1993). The factor has a stress-induced HSE DNA-binding activity (Tanabe et al 1997). Hsf3 null cells are defective in stress-induced Hsp expression and thermotolerance to severe heat stress (Tanabe et al 1998). A recent study revealed that the avian (chicken) Hsf1 is only marginally capable of transactivating the major hsp genes but enables cells to tolerate moderately severe stress (Inouye et al 2003). Hence, in avian cells both Hsf1 and Hsf3 contribute to stress resistance. The independent functions of avian Hsf1 and Hsf3 appear to be combined in mammalian Hsf1.
ACTIVATION OF HSF1 IS A MULTISTEP PROCESS
Westwood and colleagues discovered that heat treatment of Drosophila cells induced homotrimerization of Hsf (Westwood et al 1991; Westwood and Wu 1993). Subsequent studies on other metazoan Hsf-Hsf1 revealed this to be a general feature of factor activation (eg, for observations on human Hsf1, see Baler et al 1993; Sarge et al 1993). Systematic mutagenesis experiments suggested that homotrimerization and acquisition of HSE DNA-binding activity are inseparable events (Zuo et al 1994). Thus, high-affinity binding of HSE sequences appears to depend on proper juxtaposition of multiple Hsf DNA-binding domains. HR-C and both ends of the HR-A/B region were shown to participate in repression of Hsf-Hsf1 oligomerization and DNA-binding activity in the unstressed cell (Rabindran et al 1993; Zuo et al 1994; Orosz et al 1996; Farkas et al 1998). These findings originally led to the hypothesis that intramolecular interactions between these repeat sequences may occur in the Hsf-Hsf1 polypeptide. Repression of Hsf-Hsf1 oligomerization could then be imagined to involve stabilization of the latter hydrophobic interactions by a repressor protein. Considering that direct evidence for such interactions was never obtained, the alternative explanation may now be preferable that trimerization of Hsf-Hsf1 is suppressed by interactions between the hydrophobic repeats and one or more repressor proteins. Mutations in the linker region between the DNA-binding domain and HR-A/B were also observed to deregulate factor trimerization (Orosz et al 1996; Liu and Thiele 1999).
Homotrimerization, leading to acquisition of HSE DNA-binding activity, is only the first of at least 2 steps involved in the process of activation of Hsf1 occurring in a stressed cell. This conclusion is supported by the identification of chemicals or conditions that induce HSE DNA-binding activity but not expression of hsp genes (Jurivich et al 1992; Bruce et al 1993). Furthermore, when Hsf1 is overexpressed from transfected genes, a large fraction of factor accumulates as DNA-binding trimers (Zuo et al 1995). These trimers possess only minimal transcription-enhancing activity. Thus, Hsf1 DNA-binding ability and transactivation competence are regulated independently.
Mutagenesis experiments suggested that Hsf1 transactivation competence is also controlled by a repression mechanism. Green et al (1995) examined Gal4 chimeras that included residues 201–310 or 221–330 and a transcription activation domain from human Hsf1 as the only Hsf-related sequences. These chimeras were capable of imparting heat regulation on a reporter gene. Some degree of heat regulation was even achieved when the Hsf1 transcription activation domain was replaced by Vp16 (Newton et al 1996). These experiments defined a “regulatory domain” approximately spanning residues 201– 330 of human Hsf1 (see also Zuo et al 1995). A single point mutation, a Lys298 to Ala substitution, compromised the regulatory domain in a Gal4-Hsf1 chimera, resulting in elevated transcriptional activity in the absence of a stress (Knauf et al 1996).
Unlike in human cells (see previous paragraph), oligomerization of human Hsf1 overexpressed from microinjected messenger RNA is stringently repressed in Xenopus leavis oocytes (Zuo et al 1994). Taking advantage of this robust regulation of exogenous Hsf1 in the Xenopus oocyte, experiments could be performed that provided further evidence for the independence of the mechanisms repressing Hsf1 oligomerization and transcriptional competence (Zuo et al 1995). Results showed that an Hsf1 mutant lacking the regulatory domain remained heat regulated for oligomerization–DNA-binding activity as well as transactivation ability. A mutant containing a disabled HR-C had HSE DNA-binding activity but lacked transactivation competence in the absence of a stress. A combination of the 2 mutations resulted in a mutant factor that was constitutively oligomeric and transactivation competent. A complementary observation was made in Drosophila: an Hsf mutant deleted in a sequence corresponding topologically to the Hsf1 regulatory domain (d259– 440) still oligomerized in a heat-regulated fashion (Orosz et al 1996).
MECHANISMS OF REGULATION OF HSF1-HSF ACTIVITY
Chaperone repression of Hsf-Hsf1 at multiple levels
In the early 1980s it dawned on many of us that the common denominator of most, if not all, conditions and chemicals known to induce Hsp expression was a potential for causing protein unfolding or accumulation of unfolded proteins, or both. Hence, an elevated level of unfolded proteins could be the proximate trigger of the stress protein response (eg, Kelley and Schlesinger 1978; Hightower 1980). The first aspect of this hypothesis, ie, that many inducers of Hsp expression cause accumulation of unfolded proteins, appears reasonable based on chemical and physical principles. It is further supported by a series of studies that led to the realization that a predominant mechanism of protein unfolding involves oxidation of nonprotein and protein thiols, resulting in glutathione-adducted and cross-linked proteins (Freeman et al 1995; Liu et al 1996; McDuffee et al 1997; Senisterra et al 1997; Zou et al 1998b). To test the second aspect of the hypothesis, ie, whether unfolded protein could activate Hsp expression, Xenopus oocytes were coinjected with an hsp70 promoter–driven reporter gene and either chemically denatured proteins or the same proteins in native form (Ananthan et al 1986). Reporter assays revealed that the denatured proteins, but not the native proteins, induced reporter activity. These observations were interpreted as evidence that denatured or nonnative proteins are the proximate trigger of activation of Hsf and, consequently, the stress protein response.
In the above microinjection experiments, chemically denatured proteins were generated by exposure to urea and subsequent carboxy-methylation using iodoacetic acid in the presence of dithiothreitol. Because reagents were carefully removed before microinjection, the microinjected, stably derived, reduced proteins were not expected to be capable of directly altering Hsf1 structure or of functioning as oxidants. Consequently, the results of the microinjection experiments led away from the idea that Hsf activation results from direct sensing of a stress by the Hsf. Instead, they suggested that the cell contains a factor that can sense an increased level of nonnative proteins and, somehow, translate this information to cause an appropriate level of Hsf activation. Because of their propensity for dynamic association with nonnative proteins, Hsps and other chaperones have an intrinsic capacity to function as sensors of levels of nonnative proteins. If an Hsp were a repressor of Hsf, factor activation would result from increased competition for the Hsp by proteins denatured during a stressful situation. Hsf would remain in an activated state until the level of nonnative proteins would have been reduced to the prestress level by refolding or degradation, or the level of “repressor Hsp” increased sufficiently to counteract competition by nonnative proteins. Note that this mechanism would include feedback components. The existence of such a component was postulated long ago by Lindquist (1980) and Didomenico et al (1982).
Discovery of the repressor Hsp proved to be more difficult and protracted than had been expected. For many years, Hsp70 was the preferred candidate for repressor. For reasons that can now be better appreciated, convincing evidence for Hsp70 as the key repressor of Hsf activation could not be obtained (Abravaya et al 1992; Baler et al 1992, 1996; Mosser et al 1993; Rabindran et al 1994). Several more recent studies finally began outlining the mechanisms that appear to keep Hsf-Hsf1 in an inactive state (Ali et al 1998; Zou et al 1998a; Bharadwaj et al 1999; Guo et al 2001; Marchler and Wu 2001; for a related study on yeast, see Duina et al 1998). These studies owed much to progress made in the understanding of the role of Hsp90 and cochaperones in steroid receptor regulation (Pratt and Toft 1997) and to in vitro experiments showing that Hsp90 and cochaperones can interact with Hsf1 (Nadeau et al 1993; Nair et al 1996). A role for Hsp90 in Hsf1 repression was also suggested by the finding that Hsp90-binding drugs of the benzoquinone ansamycin family, eg, herbimycin A and geldanamycin, activate (mammalian) Hsf1 (Hedge et al 1995; Zou et al 1998a).
Zou et al (1998a) developed a HeLa cell lysate system that was capable of recapitulating in vitro important aspects of Hsf1 regulation observed in vivo. In this system, Hsf1 oligomerization and DNA-binding activity could be induced by heat, geldanamycin, or addition of chemically denatured proteins. The system was used to determine whether immunodepletion of an Hsp, or another chaperone or cochaperone, could induce Hsf1 DNA-binding activity under nonstressful conditions. Depletion of Hsp90, but no other chaperone or cochaperone examined, produced Hsf1 DNA-binding activity. Back addition of purified Hsp90 prevented this effect. Ali et al (1998) found that “depletion” of Hsp90 by antibody injection induced Hsf1 DNA-binding activity in Xenopus oocytes. This induction could be prevented by injection of purified Hsp90, subsequent to Hsp90 antibody injection. Using the same system, Bharadwaj et al (1999) tested antibodies against other chaperones and cochaperones. These experiments revealed that p23 depletion also caused induction of HSE DNA-binding activity in the absence of a stress. Little importance should be accorded to the negative result that a similar effect of p23 depletion did not show in the HeLa lysate experiments discussed before. Several factors, including differences in the stability and dynamics of in vitro and in vivo interactions between Hsf1, Hsps, and other chaperones and cochaperones as well as differences in p23 antibodies used, may account for the difference in results observed. Hsp90 and p23 are components of the type of multichaperone complex that is part of the final steroid aporeceptor complex (Pratt and Toft 1997). Thus, Hsf1 oligomerization and DNA-binding activity appear to be repressed by an association of monomeric Hsf1 with a similar multichaperone complex that maintains steroid receptors in a latent form. Note that the latter multichaperone complex also includes an immunophilin. Perhaps, because of the redundancy of immunophilins, neither the HeLa in vitro system nor the Xenopus oocyte system identified an immunophilin as a corepressor. However, a genetic study found a Cyp40-like cyclophilin participating in the repression of yeast Hsf activation (Duina et al 1998).
On stabilization by in situ cross-linking, complexes containing Hsf1 and Hsp90 could be immunoprecipitated from unstressed HeLa cells. Heat treatment of the cells caused the majority of these complexes to disappear at a rate comparable with that at which Hsf1 trimerization occurred (Zou et al 1998a; Guo et al 2001). In these experiments, immunoprecipitations were carried out under stringent conditions such that only complexes cross-linked within the cells were detected. Hence, complexes that existed in the cells could be distinguished from associations that may have formed after cell lysis. A possible alternative interpretation of the data that Hsf1 may have relocalized during heat treatment and, as a consequence, was no longer accessible to cross-linker could be excluded (Guo et al 2001). To summarize, the available data suggest that repression of Hsf1 oligomerization– DNA-binding activity is relieved under conditions under which an association of Hsf1 polypeptide and an Hsp90-containing complex (the Hsp90-p23-immunophilin type complex discussed above) can no longer be maintained. Unbound Hsf1 polypeptides self-associate to form Hsf1 homotrimers. Additional support for this hypothesis was provided by Knowlton and Sun (2001) who showed that geldanamycin reduced the Hsf1-Hsp90 interaction in rat cardiac myocytes. Furthermore, Zhao et al (2002) observed that overexpression of Hsp90 substantially depressed heat induction of Hsf1 DNA-binding activity in mouse NIH3T3 cells.
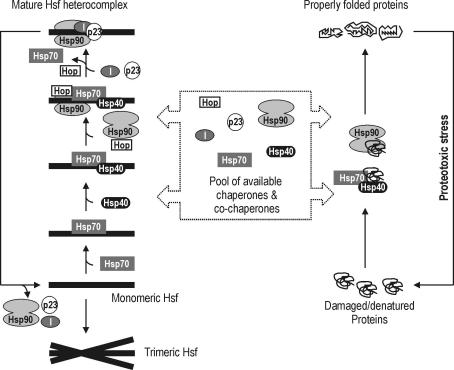
It is well known that an association of an Hsp90-p23-immunophilin complex with a substrate protein, such as a steroid receptor, is a dynamic end product of a pathway of chaperone interactions (Pratt and Toft 1997). These chaperone interactions involve initial binding of a substrate by Hsp70 and Hsp40, introduction of Hsp90 by Hsp70- and Hsp90-binding protein Hop, and replacement of the first Hsp90 complex with the final, “mature,” Hsp90-p23-immunophilin complex. Assuming that a similar assembly process occurs on Hsf1, one would predict that depletion of any of the proteins participating in the assembly process could result in induction of Hsf1 oligomerization and DNA-binding activity. Whether such an effect can be demonstrated may depend on the degree of depletion of a protein necessary for slowing the assembly process, the degree of depletion achievable in the experiment, functional redundancy of the protein, the rates of the assembly process and of dissociation of the interaction of Hsf and mature chaperone complex, etc. Conceivably, these factors may differ in vitro and in vivo as well as in cells of different origin and type. Neither in the HeLa lysate system nor in Xenopus oocytes did depletion of Hsp-c70, Hsp40, Hop, or Hip produce increased Hsf1 DNA-binding activity (Zou et al 1998a; Bharadwaj et al 1999). However, marked effects were observed in Drosophila cells, when proteins were depleted by specific RNAi (Marchler and Wu 2001). Depletion of Hsp70, Hsp40 (DroJ1) or Hsp90 enhanced HSF DNA-binding activity. Consistent with an involvement of these Hsps in a sequential assembly process, codepletion of Hsp70 and Hsp40, or of Hsp90 and Hsp40, had synergistic effects. Collectively, the above-discussed studies support a model in which inactive, nontrimeric Hsf-Hsf1 undergoes an assembly process similar to that described for steroid receptors to produce an inactive, mature heterocomplex that includes Hsp90, p23 and, presumably, an immunophilin. Intermediate complexes, as well as mature heterocomplex, are of a dynamic nature. When the cell is exposed to a stressful condition, protein unfolding increases, and the concentration of nonnative proteins rises. These nonnative proteins serve as substrate for Hsp chaperones, other chaperones, and cochaperones. Because of the competition between nonnative proteins and Hsf-Hsf1, the rate of the Hsf-Hsf1 heterocomplex assembly is reduced, resulting in a reduction or complete disappearance of the Hsf-Hsf1 heterocomplex. Released Hsf-Hsf1 self-assembles to form DNA-binding homotrimers. A view of this model is presented in Figure 2.
Fig 2.
Model showing assembly of mature Hsf heterocomplex. Most Hsf is believed to reside in this kind of complex in an unstressed cell. When the cell experiences a proteotoxic stress, cellular proteins unfold and compete with Hsf for chaperones and cochaperones. Because mature as well as intermediate Hsf heterocomplexes are dynamic assemblies, this competition will result in a reduction or disappearance of all Hsf heterocomplexes as well as inhibition of the assembly pathway. Unbound Hsf monomers will rapidly trimerize
As discussed before, trimeric, DNA-binding Hsf1 can be in a transcriptionally competent or incompetent state. This realization led to the hypothesis that Hsf1 may be subject to a second repression mechanism operating at the level of the trimeric factor. Hsp90-containing complexes, including Hsp90-p23-immunophilin complexes, had been observed to assemble on recombinant Hsf1 (Nair et al 1996). Presumably, recombinant Hsf1 was predominantly in the trimeric state in these experiments. To prove beyond a doubt that Hsp90-containing complexes could associate with trimeric Hsf1, assembly reactions were repeated on recombinant Hsf1 noncovalently bound to HSE DNA immobilized on beads (Guo et al 2001). The results were essentially the same as those of the earlier study. Antibodies against Hsp90, p23, and Fkbp52 were found to supershift HSE DNA-binding activity in electrophoretic mobility shift assays using extract from heat-treated Xenopus oocytes (Bharadwaj et al 1999). Hsf1 and Fkbp52 could be cross-linked in situ in heat-treated HeLa cells (Guo et al 2001). Additional experiments showed that Fkbp52 antibody coimmunoprecipitated Hsf1 DNA-binding activity from cells overexpressing Hsf1. Together, these in vitro and in vivo observations provided strong evidence that trimeric Hsf1 associates with an Hsp90-p23-Fkbp52 complex and raised the possibility that transcriptional activation of trimeric, DNA-binding HSF1 is repressed by this interaction. Note that Hsf1 and Fkbp52 could only be cross-linked in situ in heat-treated but not in untreated HeLa cells (Guo et al 2001). Additional experiments showed that Fkbp52 interacted much less well with an Hsf1 mutant impaired in its ability to trimerize than with wild-type Hsf1. Hence, the presence of Fkbp52 appears to be specific to trimeric Hsf1 complexes.
Experiments attempting to test the hypothesis that transcriptional activation of trimeric Hsf1 is repressed by an Hsp90-p23-Fkbp52 multichaperone complex made use of coimmunoprecipitation of Hsf1 and Fkbp52 for identification of trimeric Hsf1 complexes (Guo et al 2001). Advantage was also taken of the earlier finding that, in cells overexpressing Hsp1 (or LexA-Hsf1, a chimera containing the LexA DNA-binding domain instead of the Hsf1 HSE DNA-binding domain), a large fraction of factor accumulates as DNA-binding, but transcriptionally inert, trimers. A first type of experiment tested the prediction that, in cells overexpressing LexA-Hsf1, manipulations causing an increased demand for chaperones (chemical stress, overexpression of glucocorticoid receptor or misexpression of bovine serum albumin in the cytoplasm) should both activate LexA-Hsf1 as well as reduce the level of trimeric LexA-Hsf1 complexes. The predicted results were consistently observed. A second type of experiment critically examined the prediction that, if multichaperone complex repressed Hsf1 transcriptional activity, it needed to be capable of interacting with the Hsf1 regulatory domain. As discussed before, this domain had been genetically defined as the sequence through which Hsf1 transcriptional activity is repressed. Results revealed that mutants lacking portions of the regulatory domain were severely impaired or unable to associate with the multichaperone complex. Thus, both types of experiments supported the hypothesis that transcription competence of Hsf1 is repressed by an association of the trimeric factor with an Hsp90-p23-Fkbp52 multichaperone complex.
The work outlined above provided evidence that Hsf1 activation is repressed at successive stages by similar chaperone complexes. The regulatory domain is now known to be the Hsf1 sequence that is contacted when the multichaperone complex interacts with the trimeric factor. The target sequence(s) in nontrimeric Hsf1 has not yet been defined biochemically. However, the results from mutagenesis experiments (see above) indicate that portions of the hydrophobic repeat regions (HR-A/B and HR-C) and a linker sequence N-terminal of the HR-A/B sequence, but not the regulatory domain, are involved in repression of Hsf1 oligomerization. Hence, the prediction is that the Hsp90-containing complex interacts with the former sequence elements but not with the regulatory domain of nontrimeric Hsf1. One can only speculate, at this time, about the apparent need for 2 chaperone-based repression mechanisms operating in sequence. A plausible explanation may be derived from observations suggesting that Hsf1 oligomerization may be a practically irreversible reaction. A modest decrease in the availability of chaperones for binding to Hsf1 polypeptide, and the consequential fractional increase in the concentration of unbound Hsf1 polypeptide, may result in nearly quantitative oligomerization of the factor. Hence, a chaperone-based repression mechanism may not be capable of ensuring that Hsf1 oligomerization increases proportionally with the level of stress experienced by a cell. A second repression mechanism operating on trimeric Hsf1 may provide this proportionality because unbound and chaperone-bound trimeric Hsf1 may readily equilibrate. Based on this model, one would predict that, at an intermediate level of stress, inactive Hsf1 heterocomplexes should virtually disappear, and essentially all Hsf1 should assemble into homotrimers. Some of these trimers should then be bound by the Hsp90-p23-Fkbp52 complex, preventing acquisition of transcriptional competence. This prediction may not be far off the mark: Guo et al (2001) observed that essentially all Hsf1 in HeLa cells was trimeric after a 15 minute heat treatment at 43°C but that a small yet readily detectable fraction of Hsf1 could be cross-linked in situ to Hsp90 at that time. Moreover, Hsf1 could also be cross-linked to Fkbp52 after but not before the heat treatment. Conceivably, assembly of the Hsp90-containing chaperone complex on trimeric Hsf1 may serve other or additional purposes. For example, the bound chaperones could be imagined to maintain trimeric Hsf1 in a conformation competent for other protein interactions that may be required for full activation of the factor.
A recent study by Dai et al (2003) provided evidence for Chip as a new Hsf1 regulator. Chip is a cochaperone that binds to Hsp70 and Hsp90 through its tetratricopeptide repeats and attenuates the adenosine triphosphatase activity of Hsp70 (Ballinger et al 1999). Deletion of Chip was found to substantially reduce heat-induced or chemically induced Hsf1 activity, as assessed by Hsp70 Western blot or hsp70 reporter gene expression (Dai et al 2003). Overexpression of Chip activated Hsf1. The protein associated with Hsf1 in unstressed as well as stressed cells, suggesting that it interacts with both nontrimeric and trimeric Hsf1 complexes. It may be speculated that the role Chip plays in HSF1 regulation could be related to maintenance of proper rates of multichaperone complex assembly on both nontrimeric and trimeric Hsf1. Excessively rapid assembly may reduce responsiveness to stress, whereas excessively slow assembly may result in a constitutively active HSF1. The activating effect of Chip on Hsf1 may also be explained as a consequence of the known ability of the protein to remodel Hsp90-containing multichaperone complexes, resulting in the removal of p23 from these complexes (Connell et al 2001; reviewed by McDonough and Patterson 2003).
Although the above discussion was related to roles of chaperone complexes in repression of Hsf activation in the absence of a stress or in modulation of the activation process, it appears that chaperones are also involved in the deactivation of Hsf1 that occurs subsequent to a stressful event. Shi et al (1998) observed that Hsp70 and an Hsp40 (Hdj1) could interact with Hsf1 transcription activation domains. Overexpression of Hsp70 or Hdj1 inhibited transactivation by a Gal4-Hsf1 transcription activation domain chimera but not by a Gal4-Vp16 chimera.
Direct sensing of stress by Hsf-Hsf1
It can be imagined that other regulatory mechanisms could aid or enhance the effectiveness of the above-described chaperone-based mechanisms of repression of activation and of inactivation of Hsf1. Furthermore, it has not been demonstrated that virtually all Hsf-Hsf1 molecules are associated with chaperone complexes most of the time in the absence of a stress or during recovery from a stress. To date, all that can be extrapolated from effects of geldanamycin is that an important fraction of inactive Hsf1 is associated with and is repressed by Hsp90-containing complexes (Zou et al 1998a, 1998b; see also the genetic experiments of Duina et al 1998). Therefore, the possibility cannot be excluded at this time that a fraction of Hsf-Hsf1 molecules could be regulated by mechanisms that are entirely different from the previously discussed chaperone-based mechanisms.
The question, whether direct activation of Hsf by physical conditions or chemical stressors can occur, has been addressed repeatedly. Several studies concluded that Hsf-Hsf1 is capable of directly sensing heat and hydrogen peroxide stress (Goodson and Sarge 1995; Larson et al 1995; Zhong et al 1998). Experiments reported involved production of monomeric mammalian Hsf1 or purification of a mixture of monomeric and trimeric Drosophila Hsf and demonstration that exposure of these preparations to elevated temperature or oxidant increased Hsf-Hsf1 oligomerization and HSE DNA-binding activity. Although these in vitro experiments provided evidence that Hsf-Hsf1 is indeed capable of sensing certain types of stress, it remains unknown whether the findings can be extrapolated to Hsf-Hsf1 in cells. Furthermore, the studies did not offer a possible mechanism by which heat and hydrogen peroxide could alter Hsf-Hsf1 conformation such that a change in oligomerization state and DNA-binding activity is produced. Regarding the effects of heat, a mechanism based on temperature-induced conformational changes would appear to contradict results from earlier studies indicating that the temperature at which Hsf1 oligomerizes is determined by the cell but not the transcription factor (Clos et al 1993; Zuo et al 1995). In these studies, human Hsf1 was introduced into Drosophila cells or Xenopus oocytes. Human Hsf1 oligomerized at much lower temperatures in the heterologous cells than in human cells. Furthermore, mouse T lymphocytes were found to have a lower temperature threshold for activation of Hsf1 than other mouse cell types, and mouse motor neurons were found to have a higher threshold (Batulan et al 2003; Gothard et al 2003).
Recently, results were reported in support of the specific hypothesis that heat and hydrogen peroxide stress activate Hsf1 in vitro and in vivo through oxidation of 2 cysteine residues within the HSE DNA-binding domain, which cysteine residues then become engaged in redox-sensitive disulfide bonds (Ahn and Thiele 2003). The cysteine residues of mouse Hsf1 involved were Cys35 and Cys105 (corresponding to Cys36 and Cys103 in human Hsf1). Several experimental findings appear to suggest that oxidation of the mentioned Hsf1 cysteines does not provide an important, independent mechanism of stress regulation of the factor. A human Hsf1 lacking Cys36 was shown to be stringently heat regulated (Zuo et al 1994, 1995). The studies used chimeric transcription factor LexA-Hsf1 that contained the entire human Hsf1 sequence, except for the HSE DNA-binding domain (residues 1–78), which was replaced with the DNA-binding domain of bacterial repressor LexA (residues 1–87). When the chimera was expressed in Xenopus oocytes, its oligomerization was entirely dependent on heat shock. Transactivation assays in human cells and Xenopus oocytes, using an appropriate reporter gene containing LexA binding sites, revealed that transcriptional activity of the chimeric factor was similarly dependent on heat treatment. It is also noted that, because it is highly conserved, Cys36 could be expected to play a similar role in all Hsf species containing it. Deletion of the first 136 residues of Drosophila Hsf, including the conserved cysteine, was not found to abolish the ability of the factor to trimerize in response to heat stress (Rabindran et al 1993; Orosz et al 1996). Furthermore, the cysteine oxidation model was primarily concerned with regulation of Hsf1 oligomerization and DNA-binding activity and all but ignored the fact that Hsf1 activity is controlled at multiple levels. As discussed previously, transcriptional competence is regulated independently of factor oligomerization. Stress regulation of transcriptional activity is mediated through the so-called regulatory domain. The experiments that defined this domain used Gal4-human Hsf1 chimeras that included the regulatory domain (residues 201–330) and transcription activation domain (residues 431–529) sequences but no other Hsf1 sequences (Green et al 1995). The transcription competence of these chimeras was stringently heat regulated, even though they did not contain any cysteine residue. In summary, there is clear evidence that major mechanisms controlling Hsf1 oligomerization and transcriptional competence do not involve changes in the redox status of the cysteine residues in question. However, the literature discussed above did not address the possibility that oxidation of the 2 cysteines in the HSE DNA-binding domain of Hsf1 may specifically enhance the functionality of the DNA-binding domain. Thus, although HSE DNA-binding activity clearly can be induced in the absence of oxidation (Zou et al 1998a, 1998b), it may be enhanced by oxidation of the 2 cysteines. Depending on the precise redox status of the cell, such an enhancing effect may or may not be countermanded by the previously described inhibitory effect of oxidation on HSE DNA-binding activity (Jacquier-Sarlin and Polla 1996; Manalo and Liu 2001).
Notwithstanding the above comments, a redox mechanism is believed to play a key role in the activation of Hsf-Hsf1 and the stress protein response. Heat and many, perhaps most, chemical inducers of Hsf-Hsf1 activity cause extensive oxidation of nonprotein and protein thiols in the cell, resulting in the formation of adducted and cross-linked proteins (Freeman et al 1995; Liu et al 1996; McDuffee et al 1997; Senisterra et al 1997; Zou et al 1998b). These unfolded proteins are believed to compete with Hsf-Hsf1 for components of Hsp90-containing multichaperone complexes that sequentially repress Hsf-Hsf1 or other chaperones and cochaperones (or both) involved in the assembly of these multichaperone complexes. Hsf-Hsf1, deprived of associated chaperones and cochaperones, rapidly trimerizes and proceeds to acquire transcriptional competence.
Other aspects of Hsf-Hsf1 regulation
Hsf-Hsf1 has long been known to become hyperphosphorylated when cells experience a stress. The best available evidence suggests that phosphorylation of human Hsf1 at Ser303, Ser307, and Ser363 accelerates inactivation of the factor subsequent to a stressful event (for a review, see Holmberg et al 2002). How this phosphorylation relates to the aforementioned mechanism of deactivation of transcriptionally competent Hsf1 by Hsp70 and Hsp40 is unknown. Phosphorylation of Ser230 and Thr142 was reported to enhance Hsf1 activity (Holmberg et al 2001; Soncin et al 2003). Again, the mechanism(s) by which these phosphorylation events enhance Hsf1 activity remains to be elucidated. Several protein kinases, including Erk1/2, Gsk3, Jnk, CamkII, Rsk2, Ck2, and Pkc isoenzymes, were found to phosphorylate Hsf1 in vitro or in vivo on overexpression of the respective protein kinase. To date, it is not known which of these protein kinases actually phosphorylates Hsf1 in unmanipulated, stressed or unstressed cells.
Daxx was recently identified as an important cofactor of Hsf1 activation (Boellmann et al 2004). Although originally described as an enhancer of Fas-mediated apoptosis, Daxx has since been assigned several other functions, including repression of basal transcription (Michaelson 2000). Daxx is known to be concentrated in subnuclear structures called promyelocytic leukemia oncogenic domains but is released into the nucleoplasm during different types of stress. Heat induction of Hsp70 or of hsp70 promoter–directed reporter gene expression is markedly reduced on depletion of Daxx by RNAi or in a cell line lacking functional Daxx. Overexpressed Daxx weakly stimulates basal Hsf1 activity but dramatically activates overexpressed (trimeric) Hsf1. How Daxx enhances Hsf1 transcriptional activity is unknown at this time, but the protein interacts with trimeric Hsf1 and may compete with binding of the Hsp90-p23-Fkbp52 complex to trimeric HSF1. It is noted that an earlier study that examined cell lines lacking functional Daxx failed to reveal a reduced Hsf1 activity (Nefkens et al 2003). It is speculated that, under the conditions of mild stress used in the latter study, a Daxx effect could have been obscured by the known feedback regulation of the stress protein response.
Several other mechanisms were described that may prove to be important for Hsf1 regulation. Hong et al (2001) first reported that human Hsf1 is sumoylated at Lys298 in cells exposed to a stress. Hietakangas et al (2003) corroborated this finding but, contrary to claims of the original study, concluded that elimination of the sumoylation site affected neither the DNA-binding activity nor the transcriptional activity of Hsf1 (see also Knauf et al 1996). Hence, a regulatory role of this modification remains speculative. Hu and Mivechi (2003) reported that, when overexpressed, Ral-binding protein 1 (RalBP1) associates with Hsp90 complex-bound Hsf1 in the unstressed cell and downmodulates stress-induced Hsf1 activity. RalBP1 is known to be an effective transporter of glutathione-electrophile conjugates and xenobiotics (Awasthi et al 2003). Hence, the findings suggest the interesting possibility that there may exist a direct feedback mechanism to inhibit activation of Hsf1 and the stress protein response when sufficient RalBP1 is available for removing oxidation products. However, a more indirect mechanism can also be envisioned: when RalBP1 is overexpressed in a cell, export of proteotoxic products is accelerated. As a consequence, the cell may experience a reduced level of stress. Because RalBP1 is normally a membrane-associated protein, a demonstration that the protein also interacts with Hsf1 in the absence of overexpression would constitute an important additional piece of evidence in support of a direct feedback mechanism. Satyal et al (1998) discovered a 76-residue polypeptide containing 2 stretches of hydrophobic repeats. When overexpressed, this polypeptide, termed heat shock factor binding protein 1, interferes with stress induction of Hsf1 activity. It remains to be established whether this protein functions as a physiological regulator of Hsf1. Finally, in a number of cell types, Hsf1 accumulates in a few nuclear foci during stressful events (Sarge et al 1993). These granules, termed stress granules, were found to form on chromosome 9 heterochromatin of human cells (Jolly et al 2002). It was speculated that stress granules could represent sites of storage or buffering (or both) of active Hsf1. It would seem important to learn more about how this sequestration affects cellular Hsf activity.
CONCLUDING REMARKS
It appears that, thanks to the contributions of many laboratories, an initial understanding of mechanisms controlling Hsf-Hsf1 activity and the stress protein response may have been gained and a number of interesting avenues for follow-up studies were identified. It is abundantly clear, however, that our understanding is still very incomplete and our models are likely to be overly simplistic. Fortunately, there are good reasons to believe that interest in the regulation of the stress protein response will, if anything, grow in the future, not least because of the realization that a number of important diseases appear to be “chaperoning” diseases and that, therefore, a pharmacological approach that deliberately activates the response may potentially ameliorate or prevent these diseases. A deeper insight into the mechanisms that regulate Hsf-Hsf1 activity will likely be key to the development of such therapies.
The author wishes to apologize that because of the chosen focus and the space constraints, he was unable to comprehensively cite all work concerned with regulation of Hsf and the stress protein response.
Acknowledgments
The author thanks Frank Boellmann for artwork and Alexis Hall for critical reading of this review. The author's research on the regulation of hsp gene expression is supported by grant GM31125 from the National Institutes of Health.
REFERENCES
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein Hsp70 interacts with Hsf1, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153.0890-9369(1992)006<1153:THHSPH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ahn S-G, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503.0890-9369(2003)017<0516:RROMHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. Hsp90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949.0270-7306(1998)018<4949:HIWART>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan J, Goldberg AL, Voellmy R. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 1986;232:522–524. doi: 10.1126/science.3083508.0193-4511(1986)232<0522:APSAES>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Awasthi S, Singhal SS, Sharma R, Zimniak P, Awasthi YC. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RalBP1): a novel link between G-protein and tyrosine kinase signaling and drug resistance. Int J Cancer. 2003;106:635–646. doi: 10.1002/ijc.11260.0020-7136(2003)106<0635:TOGCAC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor Hsf1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486.0270-7306(1993)013<2486:AOHHSG>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: Hsp70 as a potential autoregulatory factor. J Cell Biol. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151.0021-9525(1992)117<1151:HSGRBN>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler R, Zou J, Voellmy R. Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones. 1996;1:33–39. doi: 10.1379/1466-1268(1996)001<0033:efaroh>2.3.co;2.1466-1268(1996)001<0033:EFAROH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of Chip, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535.0270-7306(1999)019<4535:IOCANT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batulan Z, Shinder GA, Minotti S, He BP, Doroudchi MM, Nalbantoglu J, Strong MJ, Durham HD. High threshold for induction of the stress response in motor neurons is associated with failure to activate Hsf1. J Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003.0270-6474(2003)023<5789:HTFIOT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj S, Ali A, Ovsenek N. Multiple components of the Hsp90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol Cell Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033.0270-7306(1999)019<8033:MCOTHC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boellmann F, Guettouche T, Guo J, Fenna M, Mnayer L, Voellmy R. Daxx interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci U S A. 2004;101:4100–4105. doi: 10.1073/pnas.0304768101.0027-8424(2004)101<4100:DIWHSF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce JL, Price BD, Coleman N, Calderwood SK. Oxidative injury rapidly activates the heat shock transcription factor but fails to increase levels of heat shock proteins. Cancer Res. 1993;53:12–15.0008-5472(1993)053<0012:OIRATH>2.0.CO;2 [PubMed] [Google Scholar]
- Christians ES, Yan LJ, Benjamin IJ. Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit Care Med. 2002;30:S43–S50.0090-3493(2002)030<S43:HSFAHS>2.0.CO;2 [PubMed] [Google Scholar]
- Clos J, Rabindran S, Wisniewski J, Wu C. Induction temperature of human heat shock factor is reprogrammed in a Drosophila cell environment. Nature. 1993;364:252–255. doi: 10.1038/364252a0.0028-0836(1993)364<0252:ITOHHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Clos J, Westwood JT, Becker PB, Wilson S, Lambert K, Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c.0092-8674(1990)063<1085:MCAEOA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hoehfeld J, Patterson C. Regulation of heat shock protein-mediated protein triage decisions by the co-chaperone Chip. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618.1465-7392(2001)003<0093:ROHSPP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dai Q, Zhang C, and Wu Y. et al. 2003 Chip activates Hsf1 and confers protection against apoptosis and cellular stress. EMBO J. 22:5446–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didomenico BJ, Bugaisky GE, Lindquist S. The heat shock response is self-regulated at both the transcriptional and post-transcriptional levels. Cell. 1982;31:593–603. doi: 10.1016/0092-8674(82)90315-4.0092-8674(1982)031<0593:THSRIS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a Cyp40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974.0021-9258(1998)273<18974:RFHAAC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Jokinen E, Sistonen L, Leppa S. Heat shock factor 2 is activated during mouse heart development. Int J Dev Biol. 2000;44:471–477.0214-6282(2000)044<0471:HSFIAD>2.0.CO;2 [PubMed] [Google Scholar]
- Farkas T, Kutskova YA, Zimarino V. Intramolecular repression of mouse heat shock factor 1. Mol Cell Biol. 1998;18:906–918. doi: 10.1128/mcb.18.2.906.0270-7306(1998)018<0906:IROMHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman ML, Borrelli MJ, Syed K, Senisterra G, Stafford DM, Lepock JR. Characterization of a signal generated by oxidation of protein thiols activates the heat shock transcription factor. J Cell Physiol. 1995;164:356–366. doi: 10.1002/jcp.1041640216.0021-9541(1995)164<0356:COASGB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Goodson ML, Sarge KD. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447.0021-9258(1995)270<2447:HDBOPH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gothard LQ, Ruffner ME, Woodward JG, Park-Sarge OK, Sarge KD. Lowered temperature set point for activation of the cellular stress response in T-lymphocytes. J Biol Chem. 2003;278:9322–9326. doi: 10.1074/jbc.M209412200.0021-9258(2003)278<9322:LTSPFA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Green M, Schuetz TJ, Sullivan EK, Kingston RE. A heat-shock-responsive domain of human Hsf1 that regulates transcription activation domain function. Mol Cell Biol. 1995;15:3354–3362. doi: 10.1128/mcb.15.6.3354.0270-7306(1995)015<3354:AHDOHH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200.0021-9258(2001)276<45791:EFAMOR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672.0193-4511(1994)263<0224:CSOTDB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- He H, Soncin F, and Grammatikakis N. et al. 2003 Elevated expression of heat shock factor 2a stimulates Hsf1-induced transcription during stress. J Biol Chem. 278:35465–35475. [DOI] [PubMed] [Google Scholar]
- Hedge RS, Zuo J, Voellmy R, Welch WJ. Short circuiting stress protein expression via a tyrosine kinase inhibitor, herbimycin A. J Cell Physiol. 1995;165:186–200. doi: 10.1002/jcp.1041650122.0021-9541(1995)165<0186:SCSPEV>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, and Jakobsson AM. et al. 2003 Phosphorylation of serine 303 is a prerequisite for the stress-inducible Sumo modification of heat shock factor 1. Mol Cell Biol. 23:2953–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower LE. Cultured animal cells exposed to amino acid analogues or puromycin rapidly synthesize several polypeptides. J Cell Physiol. 1980;102:407–427. doi: 10.1002/jcp.1041020315.0021-9541(1980)102<0407:CACETA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Holmberg CI, Hietakangas V, and Mikhailov A. et al. 2001 Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20:3800–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg CI, Tran SEF, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7.0376-5067(2002)027<0619:MPPSRO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson M, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced Sumo-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200.0021-9258(2001)276<40263:ROHSTF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hu Y, Mivechi NF. Hsf-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with Hsp90 in vivo. J Biol Chem. 2003;278:17299–17306. doi: 10.1074/jbc.M300788200.0021-9258(2003)278<17299:HIWRPI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Inouye S, Katsuki K, and Izu H. et al. 2003 Activation of heat shock genes is not necessary for protection by heat shock transcription factor 1 against cell death due to a single exposure to high temperatures. Mol Cell Biol. 23:5882–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier-Sarlin MR, Polla BS. Dual regulation of heat shock transcription factor (Hsf) activation and DNA-binding activity by H2O2: role of thioredoxin. Biochem J. 1996;318:187–193. doi: 10.1042/bj3180187.0264-6021(1996)318<0187:DROHST>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452.0261-4189(1997)016<2452:MFODHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, Morimoto RI, Vourc'h C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J Cell Biol. 2002;156:775–781. doi: 10.1083/jcb.200109018.0021-9525(2002)156<0775:IVBOAH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. Effect of sodium salicylate on the human heat shock response. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322.0193-4511(1992)255<1243:EOSSOT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kallio M, Chang Y, and Manuel M. et al. 2002 Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in Hsf2 null mice. EMBO J. 21:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley PM, Schlesinger MJ. The effect of amino acid analogues and heat shock on gene expression in chicken embryo fibroblasts. Cell. 1978;15:1277–1286. doi: 10.1016/0092-8674(78)90053-3.0092-8674(1978)015<1277:TEOAAA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Knauf U, Newton EM, Kyriakis J, Kingston RE. Repression of human heat shock factor activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782.0890-9369(1996)010<2782:ROHHSF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Knowlton AA, Sun L. Heat shock factor-1, steroid hormones, and the regulation of heat-shock protein expression in the heart. Am J Physiol Heart Circ Physiol. 2001;280:H455–H464. doi: 10.1152/ajpheart.2001.280.1.H455.0363-6135(2001)280<H455:HSFSHA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Larson JS, Schuetz TJ, Kingston RE. In vitro activation of purified human heat shock factor by heat. Biochemistry. 1995;34:1902–1911. doi: 10.1021/bi00006a011.0006-2960(1995)034<1902:IVAOPH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lepock JR, Frey HE, Ritchie KP. Protein denaturation in intact hepatocytes and isolated cellular organelles during heat shock. J Cell Biol. 1993;122:1267–1276. doi: 10.1083/jcb.122.6.1267.0021-9525(1993)122<1267:PDIIHA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol. 1980;77:463–479. doi: 10.1016/0012-1606(80)90488-1.0012-1606(1980)077<0463:VPOPSI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Liu H, Lightfoot R, Stevens JL. Activation of heat shock factor by alkylating agents is triggered by glutathione depletion and oxidation of protein thiols. J Biol Chem. 1996;271:4805–4812.0021-9258(1996)271<4805:AOHSFB>2.0.CO;2 [PubMed] [Google Scholar]
- Liu PCC, Thiele DJ. Modulation of human heat shock factor trimerization by the linker domain. J Biol Chem. 1999;274:17219–17225. doi: 10.1074/jbc.274.24.17219.0021-9258(1999)274<17219:MOHHSF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Manalo DJ, Liu AJ-C. Resolution, detection, and characterization of redox conformers of human Hsf1. J Biol Chem. 2001;276:23554–23561. doi: 10.1074/jbc.M011300200.0021-9258(2001)276<23554:RDACOR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Marchler G, Wu C. Modulation of Drosophila heat shock transcription factor activity by the molecular chaperone DroJ1. EMBO J. 2001;20:499–509. doi: 10.1093/emboj/20.3.499.0261-4189(2001)020<0499:MODHST>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Marthur SK, Jolly C, Fox SG, Kim S, Morimoto RI. Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001.0270-7306(2001)021<7163:SAAROH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C. Chip: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2.1466-1268(2003)008<0303:CALBTC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDuffee AT, Senisterra G, and Huntley S. et al. 1997 Proteins containing non-native disulfide bonds generated by oxidative stress can act as signals for the induction of the heat shock response. J Cell Physiol. 171:143–151. [DOI] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523.0021-9258(1998)273<7523:TDOHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Michaelson JS. The Daxx enigma. Apoptosis. 2000;5:217–220. doi: 10.1023/a:1009696227420.1360-8185(2000)005<0217:TDE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr. 1999;7:271–282.1052-2166(1999)007<0271:HSFFAR>2.0.CO;2 [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788.0890-9369(1998)012<3788:ROTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mosser DD, Duchaine J, Massie B. The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by Hsp70. Mol Cell Biol. 1993;13:5427–5438. doi: 10.1128/mcb.13.9.5427.0270-7306(1993)013<5427:TDAOTH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau K, Das A, Walsh CT. Hsp90 cochaperonins possess ATPase activity and bind heat shock transcription factors and peptidyl prolyl isomerases. J Biol Chem. 1993;268:1479–1487.0021-9258(1993)268<1479:HCPAAA>2.0.CO;2 [PubMed] [Google Scholar]
- Nair SC, Toran EJ, Rimerman RA, Hjermstad S, Smithgall TE, Smith DF. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, FES tyrosine kinase, heat shock transcription factor 1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2.1466-1268(1996)001<0237:APOMIC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Morimoto RI. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983.0270-7306(1993)013<1983:COANCH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. Hsf4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469.0270-7306(1997)017<0469:HANMOT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefkens I, Negorev DG, Ishov AM, Michaelson JS, Yeh ETH, Tanguay RM, Mueller WEG, Maul GG. Heat shock and Cd2+ exposure regulate Pml and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J Cell Sci. 2003;116:513–524. doi: 10.1242/jcs.00253.0021-9533(2003)116<0513:HSACER>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Newton EM, Knauf U, Green M, Kingston RE. The regulatory domain of human heat shock factor 1 is sufficient to sense stress. Mol Cell Biol. 1996;16:839–846. doi: 10.1128/mcb.16.3.839.0270-7306(1996)016<0839:TRDOHH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosz A, Wisniewski J, Wu C. Regulation of Drosophila heat shock factor trimerization: global sequence requirements and independence of nuclear localization. Mol Cell Biol. 1996;16:7018–7030. doi: 10.1128/mcb.16.12.7018.0270-7306(1996)016<7018:RODHSF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paslaru L, Morange M, Mezger V. Phenotypic characterization of mouse embryonic fibroblasts lacking heat shock factor 2. J Cell Mol Med. 2003;7:425–435. doi: 10.1111/j.1582-4934.2003.tb00245.x.1582-1838(2003)007<0425:PCOMEF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteranderl R, Nelson HC. Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled-coil. Biochemistry. 1992;31:12272–12276. doi: 10.1021/bi00163a042.0006-2960(1992)031<12272:TOTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev.0892-6638(2001)015<1118:ROTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303.0163-769X(1997)018<0306:SRIWHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783.0193-4511(1993)259<0230:ROHSFT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Wisniewski J, Li L, Li GC, Wu C. Interaction between heat shock factor and Hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol Cell Biol. 1994;14:6552–6560. doi: 10.1128/mcb.14.10.6552.0270-7306(1994)014<6552:IBHSFA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Loones M, Lallemand Y, Morimoto RI, Morange M, Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci USA. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392.0027-8424(1997)094<2392:FAROHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear translocation and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392.0270-7306(1993)013<1392:AOHSGT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by Hsbp1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962.0890-9369(1998)012<1962:NROTHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss J, Kunert O, Gase U, Scharf KD, Nover L, Rueterjans H. Solution structure of the DNA-binding domain of the tomato heat stress transcription factor Hsf24. Eur J Biochem. 1996;236:911–921. doi: 10.1111/j.1432-1033.1996.00911.x.0014-2956(1996)236<0911:SSOTDD>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Senisterra GA, Huntley SA, Escaravage M, Sekhar KR, Freeman ML, Borrelli M, Lepock JR. Destabilization of the Ca2+-ATPase of sarcoplasmic reticulum by thiol-specific, heat shock inducers results in thermal denaturation at 37 degrees C. Biochemistry. 1997;36:11002–11011. doi: 10.1021/bi9711590.0006-2960(1997)036<11002:DOTCOS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Shi Y, Kroeger PE, Morimoto RI. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol Cell Biol. 1995;15:4309–4318. doi: 10.1128/mcb.15.8.4309.0270-7306(1995)015<4309:TCTDOH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as Hsf1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654.0890-9369(1998)012<0654:MCAHTR>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Morimoto RI. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087.0270-7306(1994)014<2087:HHSFAA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104.0270-7306(1992)012<4104:AOHSFD>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncin F, Zhang X, Chu B, Wang X, Asea A, Stevenson MA, Sacks DB, Calderwood SK. Transcriptional activity and DNA binding of heat shock factor 1 involve phosphorylation on threonine 142 by Ck2. Biochem Biophys Res Commun. 2003;303:700–706. doi: 10.1016/s0006-291x(03)00398-x.0006-291X(2003)303<0700:TAADBO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sorger PK, Nelson HC. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1.0092-8674(1989)059<0807:TOAYTA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Kawazoe Y, Takeda S, Morimoto RI, Nagata K, Nakai A. Disruption of the hsf3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance. EMBO J. 1998;17:1750–1758. doi: 10.1093/emboj/17.6.1750.0261-4189(1998)017<1750:DOTHGR>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Nakai A, Kawazoe Y, Nagata K. Different thresholds in the responses of two heat shock transcription factors, Hsf1 and Hsf3. J Biol Chem. 1997;272:15389–15395. doi: 10.1074/jbc.272.24.15389.0021-9258(1997)272<15389:DTITRO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Sasai N, Nagata K, Liu XD, Liu PCC, Thiele DJ, Nakai A. The mammalian hsf4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845.0021-9258(1999)274<27845:TMHGGB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Voellmy R 1996 Sensing and responding to stress. In: Stress-inducible Cellular Responses, ed Feige U, Morimoto RI, Yahara I, Polla BS. Birkhaeuser Verlag, Basel, 121–137. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Kim SK, Wu C, Bax A. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix-turn-helix and Hnf-3/forkhead families of transcription factors. Biochemistry. 1994;33:10–16. doi: 10.1021/bi00167a002.0006-2960(1994)033<0010:NEFSBT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200.1061-2289(2003)036<0048:TDOTHS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0.0028-0836(1991)353<0822:SOACRO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Westwood JT, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481.0270-7306(1993)013<3481:AODHSF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J, Orosz A, Allada R, Wu C. The C-terminal region of Drosophila heat shock factor (Hsf) contains a constitutively functional transactivation domain. Nucleic Acids Res. 1996;24:367–374. doi: 10.1093/nar/24.2.367.0305-1048(1996)024<0367:TCRODH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301.1081-0706(1995)011<0441:HSTFSA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yoshima T, Yura T, Yanagi H. Heat shock factor 1 mediates hemin-induced hsp70 gene transcription in K562 erythroleukemia cells. J Biol Chem. 1998;273:25466–25471. doi: 10.1074/jbc.273.39.25466.0021-9258(1998)273<25466:HSFMHH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Frejtag W, Dai R, Mivechi NF. Heat shock factor-4 (Hsf-4a) is a repressor of Hsf1-mediated transcription. J Cell Biochem. 2001;82:692–703. doi: 10.1002/jcb.1191.0730-2312(2001)082<0692:HSFHIA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem. 2002;86:376–393. doi: 10.1002/jcb.10232.0730-2312(2002)086<0376:TDOHLT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zhao C, Hashiguchi K, Kondoh W, Du W, Hata J, Yamada T. Exogenous expression of heat shock protein 90 kDa retards the cell cycle and impairs the heat shock response. Exp Cell Res. 2002;275:200–214. doi: 10.1006/excr.2002.5501.0014-4827(2002)275<0200:EEOHSP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5.1097-2765(1998)002<0101:DSOHAO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor Hsf1 activation by Hsp90 (Hsp90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998a;94:471–480. doi: 10.1016/s0092-8674(00)81588-3.0092-8674(1998)094<0471:ROHSTF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zou J, Salminen WF, Roberts SM, Voellmy R. Correlation between glutathione oxidation and trimerization of heat shock factor 1, an early step in stress induction of the Hsp response. Cell Stress Chaperones. 1998b;3:130–141. doi: 10.1379/1466-1268(1998)003<0130:cbgoat>2.3.co;2.1466-1268(1998)003<0130:CBGOAT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557.0270-7306(1994)014<7557:AOTDAO>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319.0270-7306(1995)015<4319:MLOROH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]