ABSTRACT
Different fungal species of the Pleosporaceae family infect rice, causing similar symptoms. Reference genomic sequences are useful tools to study the evolution of these species and to develop accurate molecular diagnostic tools. Here, we report the complete genome sequences of Bipolaris bicolor, Curvularia hawaiiensis, Curvularia spicifera, and Exserohilum rostratum.
KEYWORDS: phytopathology, rice disease, fungi, population genetics, diagnostic tools, genotypic identification, molecular detection, rice brown spot, genomics, pathogenesis
ANNOUNCEMENT
Rice is a worldwide crop widely consumed in Asia and Africa. Many diseases affect rice production, with an estimated annual loss of 20% in yield (1). Different fungal species infect rice (2), some of which are not sufficiently well studied, in particular, fungi responsible for the rice brown spot diseases, Bipolaris bicolor, Curvularia hawaiiensis, Curvularia spicifera, and Exserohilum rostratum (3, 4). In the perspective of developing molecular diagnostic markers, the availability of their complete genomes is essential (5). In 2018, rice seeds and leaves with typical brown spot disease symptoms were collected in rice fields in Burkina Faso, France, Mali, and Pakistan. They were incubated for 5 to 7 days on wet filter paper at 25°C with a 12-h photoperiod. One conidium was isolated from each sample, giving rise to monospore isolates. Genomic DNAs extracted from these isolates were used to amplify glyceraldehyde-3-phosphate dehydrogenase and translation elongation factor 1 alpha (TEF1-a) genes using the primers GPD1/GPD2 (6) and TEF1-983/TEF1-2218 (7), respectively. These PCR products were sequenced, and the alignments of these sequences with sequences in databases showed a 100% identity to reference sequences from B. bicolor, C. hawaiiensis, C. spicifera, and E. rostratum, respectively, for the strains ML9021, PK9021, FR9030, and BF9006. Sequences were deposited in NCBI GenBank under accession numbers OP554702, OP554633, OP473603, OP473604, OP473605, OP554576, OP554691, and OP473583. For whole-genome sequencing, each isolate was cultured for 7 days at 25°C (12 h photoperiod) using the Corn Meat Agar (18 g/L) medium previously covered with a sterile cellophane disc. DNA was extracted using approximately 60 mg of fresh mycelium using the cetyl trimethyl ammonium bromide method (8). The genomic library was prepared using the TruSeq Nano DNA library preparation kit from Illumina. Pair-end sequencing (2 × 150 pb) was performed with an Illumina NovaSeq 6000, with an average depth of 50×. The demultiplexing and the production of the fastq files were performed using the bcl2fastq software v2.20.0.422. The quality of sequencing reads was determined using the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and FastQ Screen (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/) especially for contaminant detection. Assembly was performed with ABySS software v2.2.1 (9) for different kmer values, and genome completeness was evaluated using BUSCO v5.4.3 with the Pleosporales lineage data set (6,641 genes) (10).
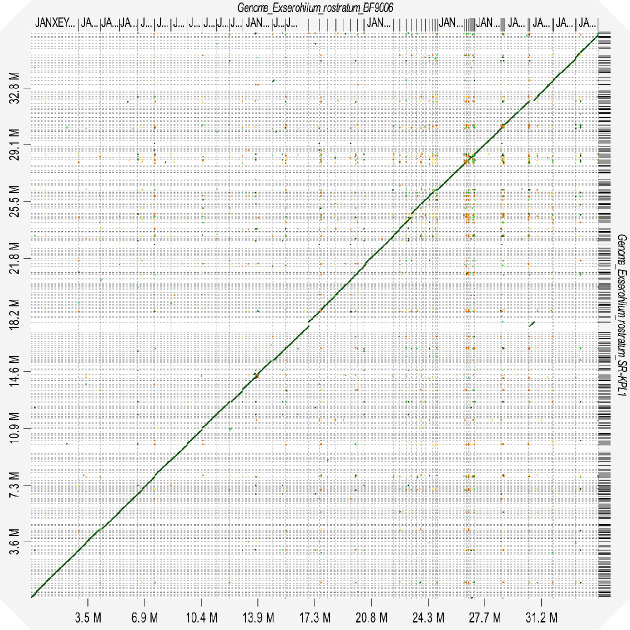
Total genome lengths ranged from 30 to 35 Mb, the number of scaffolds ranged from 99 to 412, and GC content ranged from 49.14 to 51.17% (Table 1). The BUSCO evaluation showed that the assembly recovered over 93% of the expected single-copy genes in the Pleosporales order. Compared to the E. rostratum genome of strain SR-KPL1 isolated from rice in India (11), strain BF9006 described in this study has fewer scaffolds (99 versus 769) and a larger N50 value (340,895 versus 122,824 bp) attesting data quality. The average nucleotide identity (ANI) analysis of the two complete genomes performed using pyani v0.2.11 (12) showed a similarity of 98.32%. Visualization of the alignment of the two genomes (Fig. 1) using D-GENIES v1.5.0 (13) corroborates the high ANI values.
TABLE 1.
General features of the complete genome sequences of Bi. bicolor, C. hawaiiensis, C. spicifera, and E. rostratum isolated from rice
| Sequencing | Assembly | BUSCO analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Coverage depth (x) | No. of scaffolds | Size (bp) | GC % | N50 (bp) | % of complete | % of complete and single copy | % of complete and duplicated | % of fragmented | % of missing |
| B. bicolor strain ML9021 | 62.9 | 218 | 35,093,885 | 49.14 | 965,787 | 95.45 | 95.38 | 0.07 | 0.42 | 4.12 |
| C. hawaiiensis strain PK9021 | 205.5 | 412 | 30,043,159 | 51.17 | 179,076 | 94.28 | 92.82 | 1.46 | 0.62 | 5.10 |
| C. spicifera strain FR9030 | 69.4 | 171 | 31,738,811 | 50.98 | 745,506 | 94.85 | 94.64 | 0.21 | 0.39 | 4.80 |
| E. rostratum strain BF9006 | 74 | 99 | 34,670,870 | 50.70 | 1,163,278 | 94.60 | 94.40 | 0.21 | 0.45 | 4.94 |
Fig 1.
Dot plot analysis for genome comparison of E. rostratum strains BF9006 and SR-KPL1 using D-GENIES (13). BF9006 as a target is on the x-axis, and SR-KPL1 as a query is on the y-axis. Genomic alignment regions are presented as four-colored lines, corresponding to different similarity values (Yellow: <25% similarity, Orange: 25–50% similarity, Green: 50–75% similarity, and Dark Green: >75%). The long stretches of dark green lines in the diagonal indicate high nucleotide similarity between the two strains of E. rostratum.
ACKNOWLEDGMENTS
This work was financially supported by the RICE CRP and the Islamic Development Bank.
We thank Dr. Sajid Ali (The University of Agriculture Peshawar, Pakistan) for providing us with the rice sample from Pakistan. We also thank the Montpellier GenomiX (MGX) platform for sequencing our genomes.
Contributor Information
Kouka Hilaire Kaboré, Email: koukas2@yahoo.fr.
Jason E. Stajich, University of California, Riverside, Riverside, California, USA
DATA AVAILABILITY
Whole-genome sequences were deposited in NCBI/GenBank under accession numbers JAODYD000000000, JAODYC000000000, JAODYB000000000, and JANXEY000000000. The raw data were deposited in the NCBI Sequence Read Archive (SRA) under SRA accession number SRR23346300, SRR23346299, SRR23346298, and SRR22883102.
REFERENCES
- 1. Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A. 2019. The global burden of pathogens and pests on major food crops. 3. Nat Ecol Evol 3:430–439. doi: 10.1038/s41559-018-0793-y [DOI] [PubMed] [Google Scholar]
- 2. Barro M, Kassankogno AI, Wonni I, Sérémé D, Somda I, Kaboré HK, Béna G, Brugidou C, Tharreau D, Tollenaere C. 2021. Spatiotemporal survey of multiple rice diseases in irrigated areas compared to rainfed lowlands in the Western Burkina Faso. Plant Dis 105:3889–3899. doi: 10.1094/PDIS-03-21-0579-RE [DOI] [PubMed] [Google Scholar]
- 3. Kaboré KH, Kassankogno AI, Adreit H, Milazzo J, Guillou S, Blondin L, Chopin L, Ravel S, Charriat F, Barro M, Tollenaere C, Lebrun M-H, Tharreau D. 2022. Genetic diversity and structure of Bipolaris oryzae and Exserohilum rostratum populations causing brown spot of rice in Burkina Faso based on genotyping-by-sequencing. Front Plant Sci 13:1022348. doi: 10.3389/fpls.2022.1022348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaboré KH, Diagne D, Milazzo J, Adreit H, Lebrun M-H, Tharreau D. 2022. First report of rice brown spot caused by Exserohilum rostratum in Mali. Plant Dis:DIS03210662PDN. doi: 10.1094/PDIS-03-21-0662-PDN [DOI] [PubMed] [Google Scholar]
- 5. Lang JM, Hamilton JP, Diaz MGQ, Van Sluys MA, Burgos MRG, Vera Cruz CM, Buell CR, Tisserat NA, Leach JE. 2010. Genomics-based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis 94:311–319. doi: 10.1094/PDIS-94-3-0311 [DOI] [PubMed] [Google Scholar]
- 6. Berbee ML, Pirseyedi M, Hubbard S. 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91:964–977. doi: 10.1080/00275514.1999.12061106 [DOI] [Google Scholar]
- 7. Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98. doi: 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- 8. Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical bulletin 19:11–15. [Google Scholar]
- 9. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38:4647–4654. doi: 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pramesh D, Prasanna KMK, Kirana KM, Mahesh HB, Pushpa HD, Bharath K, Buella P, Saddamhusen A, Chidanadappa E, Sharanabasava H. 2022. Data from "Genomics of rice fungal pathogen Setosphaeria rostrata (Syn. Exserohilum rostratum). GenBank. https://www.ncbi.nlm.nih.gov/genome/104827?genome_assembly_id=1882813.
- 12. Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- 13. Cabanettes F, Klopp C. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6:e4958. doi: 10.7717/peerj.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome sequences were deposited in NCBI/GenBank under accession numbers JAODYD000000000, JAODYC000000000, JAODYB000000000, and JANXEY000000000. The raw data were deposited in the NCBI Sequence Read Archive (SRA) under SRA accession number SRR23346300, SRR23346299, SRR23346298, and SRR22883102.