Abstract
Heat shock transcription factor (Hsf)–1 and Hsf2 are members of the heat shock factor (HSF) protein family involved in heat shock protein (hsp) gene regulation, a regulation that is critical for the ability of cells to survive exposure to stress conditions. Although the role of Hsf1 in binding and activating transcription of hsp gene promoters in response to cell stress is well established, how Hsf2 enhances stress-induced hsp expression is not understood. To gain an insight into the critical conserved features of the regulation and function of Hsf2, we have identified and characterized the Hsf2 protein from Xenopus laevis. We found that, similar to its human counterpart, Xenopus Hsf2 is sumoylated at lysine 82 and that, as it does in human Hsf2, the modification event of the small ubiquitin–related modifier 1 functions to increase the deoxyribonucleic acid–binding activity of this transcription factor in Xenopus. These results indicate that sumoylation is an evolutionarily conserved modification of Hsf2 proteins, supporting the position of this modification as a critical regulator of Hsf2 function.
INTRODUCTION
Heat shock transcription factor (Hsf)–2 is a member of the heat shock factor (HSF) family of transcription factors (Sarge et al 1991; Schuetz et al 1991). Another member of this family, Hsf1, has been well characterized as the transcriptional inducer of heat shock protein (hsp) gene expression in response to cell stress (Morano and Thiele 1999; Pirkkala et al 2001; Christians et al 2002). Overexpression of Hsf2 enhances stress-induced hsp gene transcription (He et al 2003), and Hsf2 is found in nuclear stress bodies with Hsf1 (Alastalo et al 2003), but precisely how Hsf2 is important for inducible hsp gene transcription is still a mystery. In light of the critical role of hsp gene expression in protecting cells from stress, it is essential to elucidate the mechanisms that control Hsf2 activity and the mechanism by which Hsf2 stimulates hsp gene expression.
We previously discovered that Hsf2 is covalently modified at lysine 82 by a protein called small ubiquitin–related modifier 1 (SUMO-1) and that this sumoylation functions to stimulate Hsf2 deoxyribonucleic acid (DNA) binding (Goodson et al 2001). SUMO-1 is a 97-amino protein with homology to ubiquitin whose conjugation to lysine residues of proteins appears to be involved in regulating functional properties of these proteins, including subcellular localization and protection against degradation (Hay 2001; Hochstrasser 2001; Muller et al 2001; Seeler and Dejean 2001; Pichler and Melchior 2002). A comparison of protein sequences flanking the lysine residues where SUMO-1 groups are attached to different proteins led to the identification of the sumoylation consensus sequence ΨKXE, where Ψ is a residue bearing a hydrophobic side chain (Hay 2001; Hochstrasser 2001; Muller et al 2001; Seeler and Dejean 2001; Pichler and Melchior 2002). The functionality of this consensus sequence has been validated by mutational analyses (Rodriguez et al 2001; Sampson et al 2001).
Only 1 HSF has been identified from Xenopus laevis and corresponds to the heat-inducible Hsf1 (Stump et al 1995). This Hsf1 is present in both pretranscriptionally active and transcriptionally active oocytes and is heat activatable (Ovsenek and Heikkila 1990). The Xenopus Hsf1 is present in oocytes as a nuclear protein, which gains DNA-binding ability on heat shock, suggesting that localization to the nucleus is not a part of the multistep process of Hsf1 activation (Mercier et al 1997).
As an avenue for increasing our understanding of Hsf2, we sought to determine whether Xenopus expressed an Hsf2 homologue, which thus far had only been identified in humans, mouse, and chicken (Sarge et al 1991; Schuetz et al 1991; Nakai and Morimoto 1993). Our reasoning was that identifying an Hsf2 homologue in a species widely divergent from humans would provide a wealth of important information including, (1) extending the boundary of when the HSF gene family split occurred during evolution, (2) giving us a system with which to determine how early Hsf2 sumoylation arose during evolution and whether its regulation and function are the same or different between widely divergent species, which (3) because Xenopus is a well-characterized model organism, could then be used for developmental and other studies to shed new light on Hsf2 sumoylation and the role of Hsf2 in the regulation of hsp gene expression.
We found that Xenopus does indeed have an Hsf2 homologue. It undergoes sumoylation at the same lysine position as does human Hsf2, and this modification stimulates the DNA-binding activity of Xenopus Hsf2 as it does in its human counterpart. These results demonstrate that the HSF gene family arose earlier than previously thought and suggest that Hsf2 sumoylation arose before the divergence of these evolutionary lines, indicating the importance of this modification as an integral mechanism of Hsf2 regulation.
MATERIALS AND METHODS
Identification of X laevis Hsf2
Candidate full-length X laevis Hsf2 expressed sequence tag (EST) clones were identified through BLAST search of the National Center for Biotechnology Information database using the human Hsf2 sequence. Two putative X laevis Hsf2 EST clones XL032d11 and XL059j06 (GenBank accession numbers BJ042603 and BJ058495) were obtained from Dr Naoto Ueno at the National Institute for Basic Biology (Okazaki, Japan). Subsequent sequencing of the EST clones revealed that the EST clone XL059j06 represented a partially spliced transcript by the identification of an intron still present in the EST. All experiments in this study were performed using the EST XL032d11, which represented a mature transcript based on the absence of any introns in the sequence.
Mutagenesis of Hsf2
To determine whether lysine 82 in the X laevis Hsf2 was the site of sumoylation, an Hsf2 mutant was generated using the QuickChange mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol. Mutation of lysine 82 to arginine was confirmed by DNA sequencing.
In vitro SUMO-1 modification assay
X laevis wild-type and mutant proteins were in vitro translated using the Promega TNT T7 Quick for polymerase chain reaction DNA rabbit reticulocyte and then subjected to in vitro SUMO-1 modification assay, essentially as described previously (Duprez et al 1999). Modification reactions were terminated using sodium dodecyl sulfate (SDS)-sample buffer containing β-mercaptoethanol and fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (10%) followed by gel fixation in 20% isopropanol: 10% acetic acid for 15 minutes. After fixation, gels were soaked in Amplify (Pharmacia, Piscataway, NJ, USA) according to the manufacturer's instructions and then dried and subjected to autoradiography.
Immunoprecipitation analysis
For immunoprecipitation, frozen X laevis tadpoles were lysed in 0.15 M Tris-HCl (pH 6.7), 5% SDS, and 30% glycerol in a tissue homogenizer. The lysate was then diluted 1:10 in phosphate-buffered saline containing 0.5% Nonidet P-40 containing complete protease inhibitor (Roche Molecular Biochemicals, Indianapolis, IN, USA). Subsequent immunoprecipitation analysis was performed using mouse Hsf2 polyclonal antibody (Sarge et al 1993), as described previously (Hong et al 2001). Immunoprecipitated proteins were then analyzed by SDS-PAGE and Western blot using anti–SUMO-1 monoclonal antibodies (21C7) (Matunis et al 1996).
Electrophoresis mobility shift assay
X laevis Hsf2 wild type and Hsf2 K82R were in vitro translated and sumoylated as described above. The translated proteins were then subjected to electrophoresis mobility shift assay. The binding reaction contained 20 μL of binding buffer (10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 1 mM ethylenediamine-tetraacetic acid [EDTA], 0.5 mM dithiothreitol, and 5% glycerol), 0.1 ng of 32P-end labeled– DNA probe, 0.5 μg of poly(dI-dC)poly(dI-dC), and 10 μg of bovine serum albumin and was incubated at 20°C for 10 minutes. Binding reactions were then subjected to electrophoresis on native 4% polyacrylamide gels in 0.5× Tris-borate–EDTA, and Hsf2 DNA-binding complexes were observed by autoradiography. The oligonucleotide probe contained 4 inverted repeats of the heat shock element consensus sequence 5′-nGAAn-3′.
RESULTS
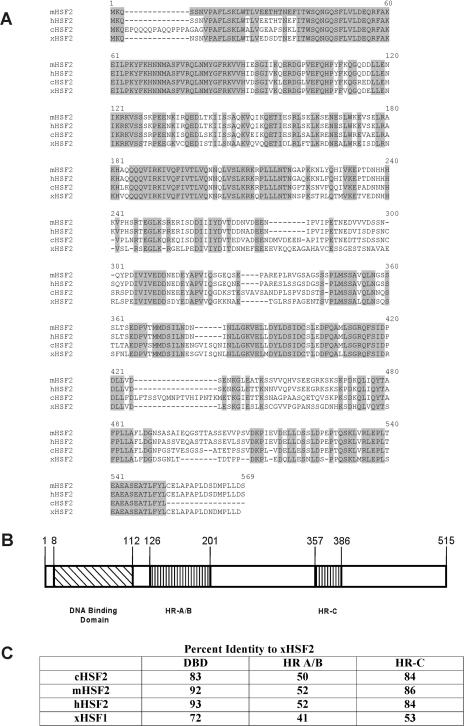
A BLAST search using the human Hsf2 protein sequence as a query revealed a match with an EST clone (GenBank accession number BJ042603) whose sequence is different from that of the known Xenopus Hsf1 protein. We obtained this clone and sequenced the regions not already present in the database to yield the nucleotide-protein sequence shown in Figure 1. The predicted open-reading frame predicts a polypeptide of 515 amino acids. An alignment of this Xenopus sequence with Hsf2 sequences from humans and mouse is shown in Figure 2A. Figure 2B shows the 3 major regions of homology in HSF proteins, which are the DNA-binding domain, trimerization domain containing the leucine zippers that interact between monomers to form trimers (HR-A, HR-B), and a more C-terminal region that also contains a leucine zipper motif (HR-C). A comparison of the percent identities in these 3 regions among Xenopus, mouse, and human Hsf2 proteins, as well as the Xenopus Hsf1 protein, is shown in Figure 2C. This analysis supports the contention that it is a member of the Hsf2 subfamily because it is much more distantly related to Hsf1.
Fig 1.
Nucleotide and amino acid sequence of Xenopus heat shock transcription factor–2. The putative polyadenylation signal is indicated by an underline
Fig 2.
Sequence comparisons of heat shock transcription factor–2 (Hsf2) proteins of different species. (A) Homology among Xenopus, mouse, chicken, and human Hsf2 proteins. Positions identical in all 3 proteins are shaded. (B) Three conserved functional domains of Hsf2 proteins. Indicated are the locations of the deoxyribonucleic acid (DNA)–binding domain, trimerization domain (HR-A, HR-B), and a C-terminal region that contains another leucine zipper motif (HR-C). (C) Percent identity comparison of DNA binding, trimerization (HR-A, HR-B), and C-terminal leucine zipper domains (HR-C) among Hsf2 proteins of different species as well as Xenopus heat shock transcription factor–1
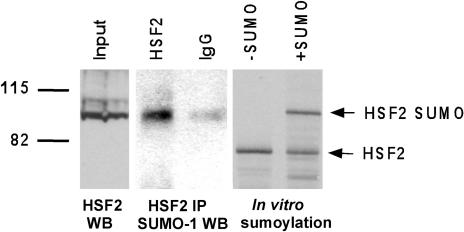
Our previous studies showed that human Hsf2 is covalently modified by the SUMO-1 protein at lysine 82. To test whether Hsf2 sumoylation is conserved in Xenopus, we immunoprecipitated Hsf2 from extracts of Xenopus tadpoles and then analyzed the immunoprecipitates by Western blot using anti–SUMO-1 antibodies. The results show a band of the expected size of sumoylated Xenopus Hsf2 (90 kDa), suggesting that Hsf2 in this species is indeed modified by SUMO-1 (Fig 3, middle panel). To strengthen the conclusion that this band indeed represents sumoylated Xenopus Hsf2, the products of an in vitro sumoylation reaction using in vitro–translated Xenopus Hsf2 protein were separated on the same SDS-PAGE gel, and the results (Fig 3, right panel) show that the size of the in vitro–sumoylated Xenopus Hsf2 does indeed match that of the immunoprecipitated band. To compare the size of these Hsf2-sumo products with that of the endogenous Xenopus Hsf2 protein, on this same SDS-PAGE gel we also separated a sample of the lysate used for the immunoprecipitation, which was then subjected to Western blot using Hsf2 antibodies. The results show that the predominant band of the endogenous Xenopus Hsf2 (Fig 3, left panel) comigrates with that of the sumoylated Hsf2 proteins in the middle and right panels, suggesting that a significant portion of the endogenous Xenopus protein is sumoylated.
Fig 3.
Xenopus heat shock transcription factor–2 (Hsf2) is sumoylated. Extracts of Xenopus tadpoles were subjected to immunoprecipitation using anti-Hsf2 antibodies or nonspecific immunoglobulin G (as a negative control) and then subjected to Western blot analysis using anti–SUMO-1 antibodies. To show that the mouse Hsf2 antibody detected Xenopus Hsf2, the input was subjected to Western blot using anti-mouse Hsf2. Size comparisons of the endogenous Xenopus Hsf2 were made to the in vitro–translated, sumoylated, and nonsumoylated forms of X.l. Hsf2. Direct size comparisons were made because all samples were run on the same SDS-PAGE gel and transferred to nitrocellulose. SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SUMO, ubiquitin–related modifier 1
Xenopus Hsf2 has a match to the SUMO modification consensus sequence at lysine 82 within its DNA-binding domain. The consensus SUMO-1 modification site comprises the sequence ΨKXE, where the lysine is the site of covalent attachment and Ψ indicates the requirement for a hydrophobic amino acid at this position (Rodriguez et al 2001; Sampson et al 2001). To test whether SUMO modification is occurring at this lysine, we performed in vitro sumoylation reactions on wild-type Xenopus Hsf2 and Xenopus Hsf2 in which lysine 82 was changed to a nonsumoylatable arginine. The results, shown in Figure 4, indicate that this mutation completely blocks sumoylation of the Hsf2 protein, indicating that this is indeed the site of sumoylation. Human Hsf2 is also sumoylated at lysine 82, indicating that the site of sumoylation has been strictly conserved between human and Xenopus Hsf2 proteins, strongly supporting the importance of sumoylation for Hsf2 function (Goodson et al 2001). A comparison of the Xenopus Hsf2 site with the sumoylation sites of other proteins is shown in Figure 4B (Sarge et al 1991; Schuetz et al 1991; Hay 2001; Hochstrasser 2001; Muller et al 2001; Seeler and Dejean 2001).
Fig 4.
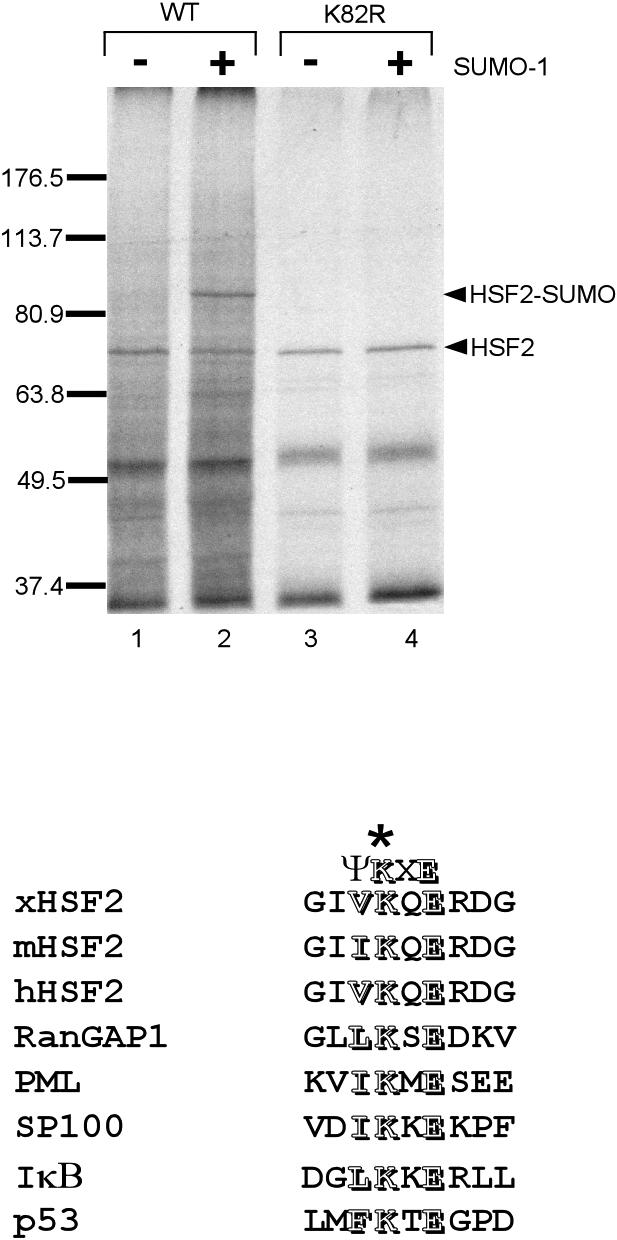
Xenopus heat shock transcription factor–2 (Hsf2) is sumoylated at lysine 82. (A) In vitro–translated, 35S-labeled wild-type, and K82R Xenopus Hsf2 were incubated in a reaction containing purified SUMO-1 and SUMO-1 E1 and E2 enzymes and an ATP-regenerating system. After the reaction, the protein samples were subjected to SDS-PAGE followed by autoradiography to observe nonsumoylated and sumoylated Hsf2 proteins. The positions of nonsumoylated and sumoylated Hsf2 are indicated by arrowheads. (B) Alignment showing conservation of sumoylation sites in Hsf2 proteins of different species and other known sumoylated proteins. Conserved residues of the SUMO-1 modification consensus sequence (ΨKXE) are indicated as outlined letters. ATP, adenosine triphosphate; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SUMO, ubiquitin–related modifier 1
We demonstrated previously that sumoylation stimulates the DNA-binding activity of human Hsf2 (Goodson et al 2001). To assess whether SUMO-1 modification has the same or a different regulatory role with respect to Xenopus Hsf2, we compared the DNA-binding activity of nonsumoylated Xenopus Hsf2 with that of Hsf2 we sumoylated using the reconstituted in vitro SUMO-1 modification reaction. The results reveal that SUMO-1 modification results in a significant increase in the DNA-binding ability of Xenopus Hsf2 (Fig 5). This increase in DNA binding was not observed when the K82R mutant was tested, indicating that sumoylation is required for this functional effect.
Fig 5.
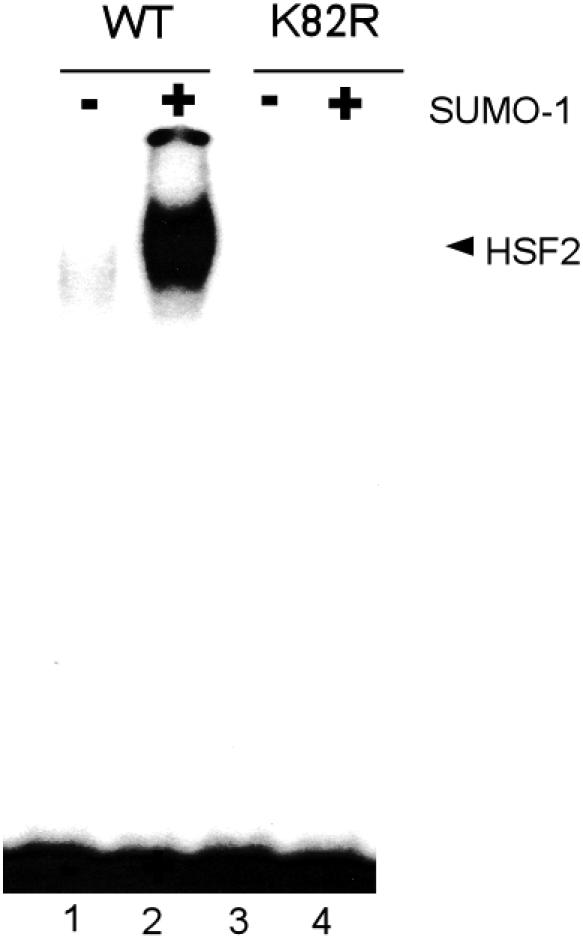
Sumoylation stimulates the deoxyribonucleic acid–binding activity of Xenopus heat shock transcription factor–2 (Hsf2). Nonsumoylated and sumoylated (using the in vitro sumoylation reaction) wild-type and nonsumoylatable K82R mutant Xenopus Hsf2 was subjected to gel shift assay using an heat shock element–containing probe. Input Hsf2 samples were aliquots of the Hsf2 samples shown in Figure 4
DISCUSSION
In this article we have identified and characterized the Hsf2 protein homologue in the African clawed toad X laevis. The close identity of this sequence with Hsf2 sequences of other species clearly places this Xenopus sequence in the Hsf2 subfamily of the HSF transcription factors. In addition, analysis of Xenopus ESTs suggests the existence of at least 2 other HSF family members in this species that are distinct from Hsf1 and Hsf2 and await characterization.
We have determined that, similar to its human counterpart, the Hsf2 protein in Xenopus is covalently linked with the SUMO-1 protein and that this occurs at the identical lysine residue in the Hsf2 proteins of both species, which is lysine 82. Our results also indicate that for both human and Xenopus Hsf2, sumoylation stimulates the DNA-binding activity of these transcription factors. The conservation of Hsf2 sumoylation at lysine 82 between such widely divergent species, and the conserved role of this modification in regulating Hsf2 DNA binding, underscores the importance of this modification event for Hsf2 function. Being able to compare Hsf2 proteins from these 2 species will be very useful for studies probing the mechanism(s) by which SUMO-1 modification regulates the DNA-binding and perhaps other activities of Hsf2 proteins. It also opens the door to comparative studies testing whether Hsf2 sumoylation may be developmentally regulated in mammals or amphibians (or both).
Acknowledgments
We would like to thank Rick McCann and Lisa Kelly for providing X laevis tadpoles and Hongyan Xing, Chad Wilkerson, and Hollie Skaggs for insightful discussions. This work was supported by NIH grant GM61053 and ACS grant MGO-98525 to K.D.S. and NIH grant HD36879 to O.-K.P.-S.
REFERENCES
- Alastalo TP, Hellesuo M, Sandqvist A, Hietakangas V, Kallio M, Sistonen L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J Cell Sci. 2003;116:3557–3570. doi: 10.1242/jcs.00671.0021-9533(2003)116<3557:FONSGI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Christians ES, Yan LJ, Benjamin IJ. Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit Care Med. 2002;30:S43–S50.0090-3493(2002)030<S43:HSFAHS>2.0.CO;2 [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, and Desterro JM. et al. 1999 SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 112:381–393. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–18518. doi: 10.1074/jbc.M008066200.0021-9258(2001)276<18513:SMRTDB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hay RT. Protein modification by SUMO. Trends Biochem Sci. 2001;26:332–333. doi: 10.1016/s0968-0004(01)01849-7.0376-5067(2001)026<0332:PMBS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- He H, Soncin F, and Grammatikakis N. et al. 2003 Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. Biol Chem. 278:35465–35475. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0.0092-8674(2001)107<0005:SFSNFB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD, Goodson M. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200.0021-9258(2001)276<40263:ROHSTF>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Matunis M, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Biol Chem. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457.0021-9258(1996)135<1457:ANUMMT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier PA, Foksa J, Ovsenek N, Westwood JT. Xenopus heat shock factor 1 is a nuclear protein before heat stress. J Biol Chem. 1997;272:14147–14151. doi: 10.1074/jbc.272.22.14147.0021-9258(1997)272<14147:XHSFIA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Exp. 1999;7:271–282.1052-2166(1999)007<0271:HSFFAR>2.0.CO;2 [PMC free article] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591.1471-0080(2001)002<0202:SUMC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nakai A, Morimoto RI. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983.0270-7306(1993)013<1983:COANCH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsenek N, Heikkila JJ. DNA sequence-specific binding activity of the heat-shock transcription factor is heat-inducible before the midblastula transition of early Xenopus development. Development. 1990;110:427–433. doi: 10.1242/dev.110.2.427.1011-6370(1990)110<0427:DSBAOT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pichler A, Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–387. doi: 10.1034/j.1600-0854.2002.30601.x.1398-9219(2002)003<0381:UMSANT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev.0892-6638(2001)015<1118:ROTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200.0021-9258(2001)276<12654:SCIVRB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200.0021-9258(2001)276<21664:TSUMSC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392.0270-7306(1993)013<1392:AOHSGT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902.0890-9369(1991)005<1902:CACOTM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911.0027-8424(1991)088<6911:IOACFH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758.0950-9232(2001)020<7243:SOBPAN>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Stump DG, Landsberger N, Wolffe AP. The cDNA encoding Xenopus laevis heat-shock factor 1 (XHSF1): nucleotide and deduced amino-acid sequences, and properties of the encoded protein. Gene. 1995;160:207–211. doi: 10.1016/0378-1119(95)00176-7.0378-1119(1995)160<0207:TCEXLH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]