Abstract
Transcription of mammalian heat shock genes can be regulated by heat shock factors (HSF) 1 and 2. Although it has been shown previously that these factors respond to distinct stimuli, a broad analysis of the induction and function of these factors in living cells has not been performed. In our study, we assayed binding of human HSF1 and HSF2 at the promoters of 32 genes identified through LocusLink as heat shock genes in response to elevated temperature and hemin-induced differentiation in human K562 erythroleukemic cells using the chromatin immunoprecipitation technique. We also measured the induced expression of these genes under these 2 conditions. We found that 17 of the 32 genes were transcriptionally induced during heat shock, and HSF1 binding was detected at 15 of the 17 promoters. Nearly all the genes induced by heat shock were also induced to a lesser degree during hemin treatment. However, some genes were induced significantly more during hemin treatment than during heat shock. A new finding is that HSF1 and HSF2 bind to the same targets, but HSF1 binding is activated more by heat than by hemin treatment, and HSF2 binding is only activated by hemin treatment and not by heat. This technology also identified previously unknown HSF1 binding sites near genes that were previously shown to be heat inducible that may contribute to gene-specific regulation.
INTRODUCTION
The mammalian heat shock response is a highly conserved process that is regulated by the heat shock transcription factor (HSF) family. These factors control the transcription of various heat shock genes that protect cellular proteins against environmental stresses such as elevated temperatures, amino acid analogs, and oxygen free radicals (Morimoto 1998). The heat shock response is also elicited in nonstress conditions, such as in early mouse development and during spermatogenesis (Sarge et al 1994; Rallu et al 1997). The heat shock response has also been elicited by treating K562 human erythroleukemic cells with hemin, a known proteasome inhibitor that also induces K562 cells to differentiate further down the erythroid lineage (Singh and Yu 1984; Mathew et al 1998). Two mammalian HSFs, HSF1 and HSF2, interact with specific sites in heat shock–induced genes during the stress or differentiation conditions and effect large increases in transcription of these target genes (Rabindran et al 1991; Sarge et al 1991, 1993; Morimoto 1998).
Previous studies have shown that mammalian HSF1 binds to the promoters and induces transcription of several genes during heat shock (Rabindran et al 1991; Sarge et al 1991, 1993). Fibroblasts from HSF1-deficient mice are unable to induce expression of known heat shock genes under thermal stress (McMillan et al 1998). This evidence demonstrates that HSF1 is necessary for directly inducing the transcription of several heat shock genes. In addition, HSF1 knockout mice exhibit prenatal lethality and other developmental defects, indicating that HSF1 is required for more than just the classical heat shock response (Xiao et al 1999).
Experiments aimed at characterizing the biological role of HSF2, specifically its contribution to the transcriptional regulation of heat shock genes under different conditions, have been less conclusive. One study provides biochemical evidence that HSF2 is inactive during the environmental stress response but acquires deoxyribonucleic acid (DNA)–binding activity during hemin-induced differentiation (Sistonen et al 1992). Furthermore, Yoshima et al (1998) showed by using a GAL4 fusion reporter construct that HSF1 mediates hsp70 transcription during hemin-induced differentiation and heat shock. Recently, Mathew et al (2001) showed that heat shock preferentially induces the DNA-binding activity of HSF1, whereas azetidine or the proteasome inhibitor MG312 coactivates both HSF1 and HSF2, and hemin preferentially induces HSF2. Two independent HSF2 mouse knockout experiments had different results, thus it is still unclear whether there is a unique requirement for HSF2 during development in vivo. Kallio et al (2002) reported that replacing exons 4 and 5 of HSF2 with a β-gal/G418 resistance cassette results in brain abnormalities, defective meiotic chromosome synapsis, and female subfertility. However, McMillan et al (2002) have shown by disrupting the first exon of HSF2 that this gene is not essential for embryonic development, fertility, or cognitive and psychomotor function in mice.
One limitation in previous studies of the HSF family is that binding activity has typically been assayed in vitro and only at the promoters of 1 or 2 target genes. To better understand the biological role of each of these transcription factors, we searched LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/) for genes similar to or annotated as heat shock genes and then measured the binding of HSF1 and HSF2 at the promoters of 32 genes in living K562 cells in response to elevated temperature and hemin-induced differentiation. In addition, we also measured the levels of transcript accumulation of these target genes under these 2 conditions. Of the 32 annotated heat shock genes, we were interested in which genes were induced by heat or hemin treatment. Furthermore, we wanted to know which of these genes bound HSF1 and HSF2 at their promoters and more importantly whether HSF1 and HSF2 bound to the same genes during heat shock and hemin treatment or whether they had different targets. Finally, we experimentally searched for additional HSF1 binding sites near known HSF1-regulated genes to see whether there was evidence of HSF1 binding outside of the promoter region.
MATERIALS AND METHODS
Cell culture
Human erythroleukemic K562 cells were grown in a spinner flask culture in Iscove's modified Dulbecco media supplemented with 10% fetal bovine serum (ATCC, Manassas, VA, USA). The culture was maintained at a density between 105 and 106 cells/mL at 37°C.
Complementary DNA preparation
We prepared messenger ribonucleic acid (mRNA) using the Micro-Fast track 2.0 kit (Invitrogen, Carlsbad, CA, USA) from duplicate samples (5 × 105 cells/sample) at 0, 20, and 60 minutes of 43°C heat shock, followed by 1, 2, and 4 hours of recovery at 37°C for the heat shock expression analysis. We used the same protocol for the hemin-induced expression analysis, except we took samples at 0, 6, 12, 24, 48, and 72 hours after addition of hemin to a final concentration of 30 μM in the culture medium. Then, we reverse transcribed the mRNA preparations by using a mix of random hexamers, poly-T first-strand synthesis primers, and Superscript reverse transcriptase (Invitrogen).
Chromatin immunoprecipitation
Adapted from the Farnham Lab (http://mcardle.oncology.wisc.edu/farnham/protocols/chips.html). We cross-linked the K562 cells by adding formaldehyde to a final concentration of 1% for 10 minutes. For the heat shock chromatin immunoprecipitation (ChIP), we cross-linked after 1 hour of heat shock. For the hemin treatment ChIP, we cross-linked 24 hours after addition of hemin. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M. Then, we collected 2 × 107 cells per IP and washed once with 1× phosphate-buffered saline (PBS). We resuspended the cells in lysis buffer (5 mM 1,4-piperazine-bis-(ethanesulphonic acid), pH 8.0, 85 mM KCl, 0.5% NP-40, Protease Inhibitor Cocktail [Roche, Indianapolis, IN, USA]) and incubated on ice for 10 minutes and then centrifuged to collect the crude nuclear preparation. We resuspended the crude nuclear preparation in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], Protease Inhibitor Cocktail) and sonicated full power 4 times for 30 seconds each on ice to produce an average DNA fragment size of 500 base pairs. We centrifuged the chromatin solution at 4°C for 10 minutes at full speed and then incubated with 20 μL of Staph A cells (Calbiochem, San Diego, CA, USA, catalog no. 507862) for 30 minutes to preclear the solution. Then, we immunoprecipitated with 10 μL of polyclonal antibody from Santa Cruz Biotech, Santa Cruz, CA, USA (aHSF2: sc-8062, aHSF1: sc-9144) and incubated overnight at 4°C. Then, we added 20 μL of Staph A cells for 30 minutes and centrifuged to collect the immunocomplexes. We washed the Staph A cells 2 times with RIPA buffer and 4 times with a LiCl wash buffer (100 mM Tris, 500 mM LiCl, 1% NP-40, 1% deoxycholate). Then, we resuspended the Staph A cells in 400 μL IP elution buffer (1% SDS, 0.1 M NaHCO3) and incubated at 67°C for 2 hours. We centrifuged the Staph A cells, saved the supernatant, and incubated at 67°C for another 4 hours to fully reverse the cross-links. Then, we extracted with phenol-chloroform and back extracted the organic phase once. We concentrated the DNA in the aqueous phase with a Microcon spin filter (Millipore), washed the DNA twice with Tris-EDTA (TE), and resuspended the DNA in a final volume of 50 μL TE.
Real-time polymerase chain reaction
We performed real-time polymerase chain reaction (PCR) to quantitate the amount of template containing a given amplicon defined by a primer pair (amplicon size range between 60 and 100 base pairs). Real-time reactions contained 3.5 mM MgCl2, 0.125 mM deoxynucleoside triphosphates, 0.5 μM forward primer, 0.5 μM reverse primer, 0.5× Sybr Green (Molecular Probes, Eugene, OR, USA), 1 U Stoffel fragment (Applied Biosystems, Foster City, CA, USA), and template DNA in a final volume of 20 μL. We measured product accumulation for 40 cycles on the Bio-Rad Icycler and calculated the threshold cycle for each reaction. The threshold cycle is the cycle at which the fluorescent signal reaches an arbitrarily set threshold near the middle of the log-linear phase of amplification.
Expression analysis
We designed amplicons to the complementary DNA (cDNA) sequence of each gene to indirectly measure the accumulation of each gene's transcript in both time course experiments. We measured the levels of β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in each cDNA preparation to normalize for any variation in absolute quantities of cDNA in each preparation. Then, we measured the relative fold-induction of each gene during heat shock and hemin induction by calculating the difference in threshold cycles of each time point and the zero time point. Fold-difference in the quantity of template was calculated as 2 raised to the power of the cycle number difference.
HSF1 and HSF2 binding analysis
To quantitate the enrichment of each promoter using ChIP with antibodies to HSF1 and HSF2, we again used Sybr Green–based real-time PCR. We designed each amplicon between −400 and 0 relative to the transcription start site of each gene. For each amplicon, we constructed a standard curve of threshold cycles from 50 ng, 5 ng, 500 pg, and 50 pg of mock-IP genomic DNA (DNA prepared with the ChIP protocol but omitting the IP step). Then, we calculated the threshold cycle from a PCR reaction that used ChIP-enriched DNA as template. We fit the experimental threshold cycle from an amplicon to the standard curve of that amplicon to calculate the quantity of template containing that amplicon in the ChIP-enriched sample. We also designed 3 negative-control amplicons to the promoters of β-actin, GAPDH, and histone H2A. The ratio of an experimental promoter quantity and the negative-control promoter quantities is equal to the fold-enrichment of a given promoter by ChIP.
RESULTS
Heat shock gene expression
To begin our study, we searched LocusLink at NCBI (http://www.ncbi.nlm.nih.gov/) to identify a large set of human genes that are either known heat shock proteins or family members of heat shock proteins based on annotation or sequence similarity. We also included 3 genes (IL8, TXN, and AKR1C2) previously shown to be induced by hemin in K562 cells (Leppa et al 1997; Lam et al 2000). We chose to use K562 cells because it is the cell culture model most widely used to describe HSF2 induction. Although a cultured cell line is an artificial system, previous studies show convincingly that K562 cells treated with hemin recapitulate some aspects of differentiation, and our ChIP protocol is currently feasible only in cultured cells. The goal of our first experiment was to determine how the transcript levels of these genes change in the K562 human erythroleukemic cell line during heat shock and during treatment with hemin, an agent that induces these cells to differentiate along the erythroid lineage and also has been shown to inhibit the proteasome. We designed PCR primers specific to each gene and performed real-time reverse transcriptase–polymerase chain reaction to measure the relative induction of these genes in K562 cells during heat shock and hemin treatment. We measured, in duplicate, the transcript levels at 6 points along a time course for heat shock and hemin treatment and normalized our mRNA samples to the levels of β-actin and GAPDH. The time points were determined from previously published studies on gene expression under these conditions (Singh and Yu 1984; Rabindran et al 1991; Sarge et al 1991, 1993; Mathew et al 1998).
Many of the same genes were induced under both conditions, but some genes were induced to a greater degree in one condition than the other (Fig 1). Most striking is the difference in expression of the hsp70-A6 gene under the 2 conditions. This gene is induced more than 500-fold during hemin treatment but less than 15-fold by heat shock. The hsp90-α gene is also induced more by hemin than by heat shock, whereas hsp70-A1A and DnaJ.C3 are induced more by heat shock than by hemin (Fig 2). The 3 genes IL8, TXN, and AKR1C2, shown previously to be induced by hemin treatment, were also induced by our hemin treatment. Furthermore, on hemin treatment, the cells became noticeably more red because of an increase in globin synthesis, thus indicating that hemin treatment was effective in inducing these cells to undergo a process similar to differentiation, as described by many others in the literature for K562 cells.
Fig 1.
Heat shock and hemin-induced gene expression patterns. The expression data for 32 genes during heat shock and hemin-induced differentiation show that many of the same genes are induced during heat shock and hemin treatment. Time points are listed at the top of the figure. The number of vertical bars in each box indicate the fold-induction of a given gene at that time point according to the key at the bottom of the figure
Fig 2.
Expression kinetics of 6 genes during heat shock and differentiation. Each panel displays the expression kinetics of a single gene during heat shock and hemin-induced differentiation. Fold-induction is shown in log scale on the y-axis for each panel. Time points for heat shock and differentiation are shown on the x-axis at the bottom of the graph
HSF1 and HSF2 transcription factor binding
To gain some understanding of how this class of genes is regulated by the 2 treatments of K562 cells, we studied the binding of HSF1 and HSF2 to the promoters of the genes and determined how binding correlated with expression. To this end, we used the ChIP protocol (see the Materials and Methods) with antibodies that recognize either HSF1 or HSF2 to determine their occupancy at the promoters of the 32 genes in intact K562 cells before and after treatment with heat shock or hemin.
To assay the enrichment achieved by the ChIP, we used real-time PCR to quantitate the overrepresentation of the genomic sequence upstream of each gene in the pool of ChIP-enriched DNA. We typically saw enrichments greater than 100-fold for promoters that were bound by HSF1 and enrichments up to 20-fold for HSF2 (likely because the HSF2 antibody was less efficient at the HSF2 IP). We interpret the variation we see in enrichment values between genes as a result of the distance that the PCR primers were designed to the actual binding site rather than as strength of binding at a given promoter. We repeated our ChIP experiments 3 times, and our results were very reproducible for the genes that we tested in each experiment (data not shown). Therefore, because the qualitative distinction between enriched and unenriched was clear and because we do not make arguments based on the relative enrichment between promoters, we calculated our ChIP enrichments for all 32 promoters from 1 experiment.
With only 2 exceptions, the ChIP experiments revealed that HSF1 occupancy at the promoter corresponds to the increased expression of these genes during heat shock (Fig 3A). Interestingly, DnaJ.C3 and DnaJ.B4 are clearly transcriptionally induced by heat shock, but we found no evidence of HSF1 or HSF2 binding near their predicted transcription start sites because there was no enrichment of their promoter fragments by ChIP. We designed 8 additional amplicons in the introns of both genes to test for binding of either factor at sites outside of the promoter, and we failed to detect any enrichment at any of these additional sites (data not shown). However, the possibility remains that binding might occur further upstream than the 500 base pairs we assayed.
Fig 3.
HSF1 and HSF2 binding during heat shock and differentiation. (A) The bars indicate the degree of HSF1 and HSF2 binding to each promoter. Binding was quantitated by the ChIP fold-enrichment of each gene's promoter, which is shown on the scale on the left of the y-axis. Solid bars are from the heat shock HSF1 ChIP, and hatched bars are from the heat shock HSF2 ChIP. Both ChIPs were done 1 hour after heat shock. The white diamonds indicate the fold-induced expression at the 2-hour recovery time point after heat shock. Fold-induced expression values are shown on the right of the y-axis. (B) Same as panel A, except binding was assayed by ChIP 24 hours after the addition of hemin, and the diamonds indicate the fold-induced expression at the 72-hour time point of hemin treatment. HSF, heat shock factor; ChIP, chromatin immunoprecipitation
We also studied the binding of these 2 transcription factors to the promoters of the 32 genes before and during treatment of K562 cells with hemin. In contrast to the results with heat shock, the expression of these genes after hemin treatment did not correspond as strongly with either HSF1 or HSF2 binding (Fig 3B). Four of the 7 genes whose transcripts increased the most after treatment with hemin were not bound to either factor according to the ChIP assay. This is not including the 3 marker genes, IL8, TXN, and AKR1C2, that are induced by hemin, but for which we neither expect nor detect binding.
We believe that it is not appropriate to use the PCR-derived enrichment values in our assays to compare binding strengths between different promoters, for reasons described above. Nevertheless, it is fair to estimate binding strengths for the same promoter under different conditions (ie, heat shock vs differentiation) because these measurements are performed with the same PCR amplicon. Consistent with previous in vitro studies (Sistonen et al 1992; Mathew et al 2001), the results from our ChIP assays show that HSF1 binds to the promoters of its target genes with a higher affinity during heat shock than during hemin treatment (Fig 4A), whereas HSF2 binds with a higher affinity during hemin treatment than during heat shock (Fig 4B). Very interestingly, for the 32 genes we studied, HSF1 and HSF2 bind to the same target genes, but their DNA binding is activated differently under different conditions.
Fig 4.
Reciprocal binding of HSF1 and HSF2 during heat shock and differentiation. Binding was quantitated in both panels by the ChIP fold-enrichment of each gene's promoter. Fold-enrichment is shown on the y-axis. (A) The white and black bars show HSF1 binding during heat shock and hemin treatment, respectively. (B) The white and black bars show HSF2 binding during heat shock and hemin treatment, respectively. ChIP, chromatin immunoprecipitation
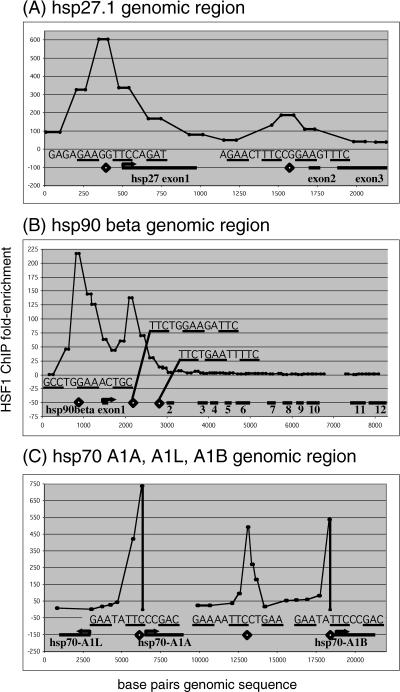
We further characterized HSF1 activity by assaying its localization throughout 3 genomic regions containing hsp27.1, hsp90-β, hsp70-A1A, hsp70-A1B, and hsp70-A1L during the heat shock response. We chose hsp27.1 and hsp90-β because they both had introns that contained sites similar to the consensus heat shock element (HSE). We chose the 20-kb genomic region containing 3 members of the hsp70 family to investigate whether there were long-range binding sites in this region that potentially contributed to the transcriptional regulation of these genes. To accomplish this, we used ChIP to enrich genomic binding sites of HSF1 and designed real-time PCR amplicons spaced evenly across the genomic sequence of the target genes to assay for enrichment of each amplicon. By using this approach, we identified a novel binding site for HSF1 located in the first intron of the hsp27 (HSPB1) gene (Fig 5A). We found that HSF1 binds at consensus HSE (TTCNNGAANNTTC) in this intron. In a similar experiment, we identified binding sites in the hsp90-β promoter and the first intron (Fig 5B). Interestingly, this experiment indicates that HSF1 binds to the first HSE in intron 1 but not to another HSE located further downstream. In addition, the strongest binding occurred at the nonconsensus HSE in the promoter of the hsp90-β gene. Finally, we measured HSF1 binding throughout the 20-kb genomic region that contains the hsp70-A1L, A1A, and A1B genes, 3 genes known to be involved in the heat shock response (Fig 5C). These experiments showed binding of HSF1 to the known sites in the promoters of each of these genes (Fig 5C). In addition, we also identified a new HSE that is bound by HSF1 during heat shock located 5 kb downstream from the hsp70-A1A gene and 5 kb upstream from the hsp70-A1B gene.
Fig 5.
HSF1 binding throughout the genomic regions of 5 heat shock genes. We assayed HSF1 binding at regularly spaced intervals throughout 3 genomic regions that contain 5 heat shock genes. Amplicons were designed to assay binding in the genomic regions of hsp27.1 (panel A), hsp90-β (panel B), and the region including hsp70-A1A, A1B, and A1L (panel C). White diamonds indicate HSE, and the sequence is shown for each. The enrichment of each amplicon is shown on the y-axis. HSF, heat shock factor; HSE, heat shock element
DISCUSSION
In this work, we studied the expression of and binding of HSF1 and HSF2 to 32 heat shock genes during thermal stress and hemin induction. Previous study has shown that HSF1 and HSF2 are induced differently during heat shock and hemin treatment, but these were primarily in vitro results that assayed binding at only 1 or 2 target genes. Our study extends the picture of HSF1 and HSF2 regulation of the heat shock response by assaying the binding in a cell culture system and to a much larger number of potential targets.
For the genes that are induced by both heat shock and hemin treatment, examination of the kinetics of transcript accumulation exposes differences in the degree of induction under one condition compared with the other. Most genes, exemplified by DnaJ.C3 and hsp70-A1A (Fig 2), are induced to a greater extent during heat shock than during hemin treatment. A few, such as Hsp90-α and hsp70-A6, show a higher induction during hemin than during heat shock (Fig 2). This difference is most striking for hsp70-A6, which is induced more than 500-fold during hemin treatment but only 12-fold during heat shock. These examples suggest that the response of cells to environmental stress and to differentiation requires unique biological activities from its collection of chaperone proteins.
Interestingly, although DnaJ.B4 and DnaJ.C3 are clearly transcriptionally induced during heat shock, we found no evidence that HSF1 or HSF2 bind at their predicted promoters. In subsequent experiments, we failed to detect binding in the first or second intron of both genes (data not shown). Therefore, although DnaJ.B4 and DnaJ.C3 are heat-inducible genes, our data suggest that they may not be regulated by HSF1 or HSF2, although the possibility exists that binding might occur upstream of the 500 base pairs we assayed.
We observed that certain genes are bound by both factors after hemin induction, raising the possibility that HSF1 and HSF2 work together to confer an appropriate response in some instances. For example, both hsp70-A1A and hsp70-A6 are transcriptionally induced after both heat shock and hemin induction and both show strong induction of HSF1 binding (but not HSF2 binding) after heat shock. After hemin induction, however, both HSF1 and HSF2 are bound to these 2 promoters (Figs 3 and 4). The behavior of these 2 promoters exemplifies 1 of the broad conclusions of this study that induction of genes that are regulated by HSF1 and HSF2 does not simply follow the extent of binding by HSF1 and HSF2. The data set presented here suggests that the contribution of HSF1 and HSF2 may vary widely in a gene-specific manner, although further experiments are needed to investigate the interactions of HSF1 and HSF2 with different regulatory inputs.
It has been proposed that HSF1 and HSF2 regulate different targets in addition to having separate roles in heat shock and differentiation (Mathew et al 1998; Morimoto 1998). As shown in Figure 4, these factors regulate the same set of genes in our study, but they may evoke different transcriptional responses by binding differentially under these 2 conditions. Our results are consistent with the hypothesis that HSF1 and HSF2 have distinct biological roles and may regulate the transcription of their target genes differently.
Assaying the binding of HSF1 along the entire stretch of 3 genomic regions containing 5 previously identified heat shock genes revealed additional HSF1-bound HSE outside of the promoter region in all 3 cases. In the hsp90-β gene, there is also an unbound HSE in the first intron. The bound HSE in the genomic region between the hsp70-A1A and hsp70-A1B genes is 5 kb from the start site of transcription of both genes. This is the first HSF1-bound HSE that has been located at a considerable distance from a transcription start site. These examples show that the regulation of transcription by HSF1 may involve binding at intronic sites and other long-range sites in addition to binding at the promoter of heat-inducible genes.
By assaying the binding of HSF1 and HSF2 at 32 stress-related genes in living cells, we have discovered new targets of these factors and have taken a first step toward understanding the global regulation of the human heat shock response by the heat shock transcription factor family.
Acknowledgments
We are grateful to members of the Myers lab for thoughtful discussion, specifically Shelley Force Aldred and Sara Hartman for help with the manuscript. We also thank John Murray for help with the manuscript and helpful discussion. We thank members of the Kingston lab, specifically Laura Corey for advice concerning heat shock protocols and help with cell culture. We also thank the Farnham lab for sharing their ChIP protocol and advice. This work was supported by the Stanford Genome Training Program (Training Grant NIH 5 T32 HG00044 to N.D.T. and W.C.C.)
REFERENCES
- Kallio M, Chang Y, and Manuel M. et al. 2002 Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 21:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Ronchini C, Norton J, Capobianco AJ, Bresnick EH. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by notch-1. J Biol Chem. 2000;275:19676–19684. doi: 10.1074/jbc.M002866200.0021-9258(2000)275<19676:SOEBNM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Leppa S, Pirkkala L, Chow SC, Eriksson JE, Sistonen L. Thioredoxin is transcriptionally induced upon activation of heat shock factor 2. J Biol Chem. 1997;28:30400–30404. doi: 10.1074/jbc.272.48.30400.0021-9258(1997)028<30400:TITIUA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Jolly C, Fox SG, Kim S, Morimoto RI. Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001.0270-7306(2001)021<7163:SAAROH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091.0270-7306(1998)018<5091:HSRAPD>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Christians E, and Forster M. et al. 2002 Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol. 22:8005–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523.0021-9258(1998)273<7523:TDOHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788.0890-9369(1998)012<3788:ROTHST>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci U S A. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906.0027-8424(1991)088<6906:MCAEOA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Loones MT, Lallemand Y, Morimoto RI, Morange M, Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc. Natl. Acad. Sci. U S A. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392.0027-8424(1997)094<2392:FAROHS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392.0270-7306(1993)013<1392:AOHSGT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol. Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334.0006-3363(1994)050<1334:EOHSFI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902.0890-9369(1991)005<1902:CACOTM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Singh MK, Yu J. Accumulation of a heat shock-like protein during differentiation of human erythroid cell line K562. Nature (Lond) 1984;309:631–633. doi: 10.1038/309631a0.0028-0836(1984)309<0631:AOAHSP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104.0270-7306(1992)012<4104:AOHSFD>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943.0261-4189(1999)018<5943:HIRFED>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshima T, Yura T, Yanagi H. Heat shock factor 1 mediates hemin-induced hsp70 gene transcription in K562 erythroleukemia cells. J Biol Chem. 1998;273:25466–25471. doi: 10.1074/jbc.273.39.25466.0021-9258(1998)273<25466:HSFMHH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]