Abstract
Aryl hydrocarbon receptor (AhR) is a transcription factor that is activated by the binding of xenobiotic and endogenous ligands. AhR interacts with heat shock protein (Hsp) 90 complexes and can be used as a functional substrate to detect chaperone-dependent processes. Yeast Hsp90 (hsp82) mutants that variably affected AhR signaling were identified using reporter gene assays. Some mutated alleles resided in the p23/adenosine triphosphate (ATP)–binding pocket of Hsp90, so the relationship between the cochaperone Sba1 (yeast p23) and adenosine triphosphatase (ATPase) activity was investigated. Deletion of the p23 gene in the hsp82G170D mutant background had a greater effect on AhR signaling than the individual mutations, suggesting that these 2 mutations have separate actions on AhR signaling. In contrast, p23 overexpression suppressed temperature sensitivity and AhR signaling defects in the hsp82G170D mutant strain, suggesting that there is a relationship between these 2 proteins. The mutated hsp82G170D protein lacked detectable ATPase activity and p23 binding in vitro, which may relate to the weakened AhR signaling observed in mutant cells. Sba1 (p23) suppressed Hsp82 ATPase activity in vitro. These studies implicate the p23 protein and the G170 region of Hsp90 as being important, but not essential, for AhR signaling. Our results are consistent with a model in which p23 inhibits Hsp90 ATPase activity, thereby stabilizing ATP-Hsp90-client protein complexes.
INTRODUCTION
Aryl hydrocarbon or dioxin receptor (AhR) binds numerous ligands, including the environmental contaminant 2,3,7,8-tetrachlorodibenzo(p)dioxin (TCDD) (Hankinson 1995; Denison and Heath-Pagliuso 1998). On ligand binding, AhR translocates to the nucleus and forms a heterodimeric transcription factor by associating with the aryl hydrocarbon nuclear translocator (Arnt) protein. The AhR/Arnt dimer binds to aryl hydrocarbon (dioxin) response elements (AHREs) on the deoxyribonucleic acid (DNA) and alters gene expression and cellular regulation. The AhR and Arnt proteins are members of the basic helix-loop-helix/Per-Arnt-Sim domain family of transcription factors (Gu et al 2000). Like some steroid hormone receptors, AhR is regulated by the 90-kDa heat shock protein (Hsp90) chaperone complex (Carver et al 1994; Chen and Perdew 1994; Coumailleau et al 1995; Whitelaw et al 1995). In contrast, Arnt does not bind ligand and does not associate with Hsp90 complexes.
Before ligand binding, AhR is retained in the cytoplasm in a complex with an Hsp90 dimer and other cochaperone proteins, including an immunophilin-like protein called the hepatitis B virus protein X–associated protein 2 (Xap2) (Ma and Whitlock 1997; Carver et al 1998; Meyer and Perdew 1999) and the p23 cochaperone (Caplan 1997; Pratt and Toft 1997; Pratt 1998). The Hsp90 complex is thought to localize AhR to the cytoplasm in the absence of ligand to protect AhR from degradation and to stabilize the high-affinity ligand-binding conformation of AhR (Pongratz et al 1992). The association of Xap2 with the Hsp90 complex reduces proteosomal degradation, influences cellular localization, and enhances signaling of AhR (Ma and Whitlock 1997; Meyer et al 1998; Meyer and Perdew 1999; Kazlauskas et al 2000, 2001). Two proteins with partial sequence similarity to Xap2, Cpr7 and Cns1, influence AhR signaling in a yeast model system (Miller 2002).
The role that the p23 cochaperone plays in Hsp90-dependent AhR signaling processes is presently under investigation. Studies in mammalian cells have demonstrated that overexpression of p23 differentially affects steroid hormone receptor–mediated signaling (Freeman et al 2000). The mammalian p23 cochaperone has a homologue, Sba1, in the budding yeast, Saccharomyces cerevisiae (Fang et al 1998). Studies in yeast have indicated that the p23 cochaperone functions in synthetic steroid hormone receptor signaling pathways (Caplan 1997). Deletion of the yeast SBA1 gene affected the levels of reporter gene activation mediated by steroid hormone receptor signaling pathways (Knoblauch and Garabedian 1999; Freeman et al 2000). More importantly, human p23 complemented the steroid hormone receptor signaling defects associated with the deletion of SBA1 in yeast (Freeman et al 1996; Knoblauch and Garabedian 1999). In mammalian cells, p23 exists as an abundant phosphorylated protein (Johnson et al 1994). It prevented protein aggregation in in vitro studies, suggesting a direct chaperone action on some protein substrates (Bose et al 1996; Freeman et al 1996). As a cochaperone, p23 is thought to interact indirectly with steroid hormone receptors through its association with Hsp90 (Nair et al 1996; Pratt and Toft 1997). p23 interacts with the N-terminal nucleotide-binding domain of Hsp90 in an adenosine triphosphate (ATP)–dependent manner, although residues in the C-terminal region are also required for p23 binding (Grenert et al 1997; Sullivan et al 1997; Chadli et al 2000). The ATP-dependent interaction of p23 with Hsp90 is disrupted by benzoquinone ansamycin antibiotics (geldanamycin, herbimycin A, and macbecins) that bind and displace ATP from the nucleotide-binding domain of Hsp90 (Whitesell et al 1994; Johnson and Toft 1995). Formation of glucocorticoid and progesterone receptor complexes with Hsp90 and p23 in vitro results in high ligand affinity, suggesting a positive role in signal transduction (Smith et al 1995; Dittmar et al 1997; Pratt and Toft 1997). A recent study suggested that p23 dissociates transcription factor complexes from DNA as a means of turning off gene expression (Freeman and Yamamoto 2002). Thus, p23 may play multiple roles in enhancing and inhibiting transcriptional responses.
The effects of p23 on AhR are not as well defined. In vitro studies have suggested that p23 acts to stabilize AhR-Hsp90 complexes (Kazlauskas et al 1999, 2001). The Hsp90-p23 complex facilitated the binding of AhR to importin α in vitro, suggesting a role in nuclear import as well (Kazlauskas et al 2001). Studies conducted in our laboratory demonstrated that deletion of SBA1 resulted in reduced levels of AhR-mediated expression of a reporter gene in yeast, and both SBA1 and human p23 expression restored AhR signaling in sba1 mutants (Cox and Miller 2002). The p23 cochaperone was not required, but was important for efficient AhR signaling in yeast.
Although several studies have identified the domains of AhR that interact with Hsp90 and other cochaperones, the domains of Hsp90 that are important for AhR function have not been studied extensively. Identification of Hsp90 mutations that alter AhR signaling might better define the functional domains of this chaperone. Consequently, we have examined AhR signaling in the presence of mutated hsp90 derivatives in this report. We also assessed the role of the p23 protein in temperature sensitivity and AhR signaling in cells that contained individual mutated hsp82 derivatives.
MATERIALS AND METHODS
Reagents
Reagent-grade chemicals were purchased from Fisher Scientific (Springfield, NJ, USA) and Sigma Chemical (St Louis, MO, USA) companies. All restriction enzymes used in this study were from New England Biolabs (Beverly, MA, USA). 5-Fluoroorotic acid (5-FOA) was purchased from Toronto Research Chemicals Inc. (North York, Ontario, Canada). The ansamycin antibiotic, geldanamycin, was purchased from Alexis Biochemicals (San Diego, CA, USA). The geldanamycin was stored as a 10-mM stock solution in dimethyl sulfoxide (DMSO) at −20°C. The β-naphthoflavone used as an AhR ligand in the lacZ assays was purchased from Acros Organics through Fisher Scientific and was of 99% purity.
Plasmids
Plasmid amplification was performed in the DH5α strain of Escherichia coli using standard techniques (Sambrook et al 1989). All HSP82 wild type and mutant expression plasmids (pKAT6, pHGPD/P82, pHGPD/T22I, pHGPD/A41V, pHGPD/G81S, pHGPD/T101I, pHGPD/G170D, pHGPD/G313S, and pHGPD/E381K) were kindly provided by Susan Lindquist (The Whitehead Institute, Boston, MA, USA) (Nathan and Lindquist 1995). The 2μ pKAT6 plasmid expresses HSC82 from a glyceraldehyde phosphate dehydrogenase (GPD) promoter and contains a URA3 selectable marker (Nathan and Lindquist 1995). The centromeric pHGPD vector expresses genes from a GPD promoter and contains a HIS3 selectable marker. The centromeric lacZ reporter plasmids pTXRE5-Z (Miller 1999) and pLXRE5-Z (Cox and Miller 2002) contain TRP1 and LEU2 selectable markers, respectively, and express a lacZ gene under control of 5 AHREs. The 2μ SBA1 expression vector, pBevy-GU-SBA1, expresses SBA1 from the GAL 1,10 promoter and contains a URA3 selectable marker (Cox and Miller 2002). The pBevy-GU parent vector was described previously (Miller et al 1998). The centromeric expression vector, pGAL-SBA1 (generously provided by Avrom Caplan, Mt Sinai Medical Center, New York, NY, USA), expresses SBA1 with an N-terminal tag of 6 consecutive histidines (Sba1(His6)) from a galactose-inducible (GAL 1) promoter and contains a URA3 selectable marker (Fang et al 1998). Sba1(His6) was capable of restoring AhR-mediated expression of a lacZ reporter gene to wild-type levels in the sba1 deletion strain, indicating that the tag has no effect on biological activity (see below for a description of the AhR-mediated reporter assay, data not shown). The bacterial expression vector, p23ET (generously provided by Avrom Caplan), is derived from pET15b (Novagen, Madison, WI, USA) and expresses an Sba1 derivative with an N-terminal tag of 6 consecutive histidines (Fang et al 1998). The 2μ human p23 expression vector, pBevy-GU-hp23, expresses human p23 from the GAL 1,10 promoter and contains a URA3 selectable marker (Cox and Miller 2002).
DNA manipulation
The HSP82 and hsp82G170D genes with a C-terminal tag of 6 consecutive histidines were constructed by polymerase chain reaction (PCR) using the Failsafe PCR amplification kit (Epicenter, Madison, WI, USA). The pHGPD/P82 or pHGPD/G170D plasmids were used as templates for the PCR. The forward primers consisted of either 5′-cgggatccatggctagtgaaactttt-3′ or 5′-ctccatggctagtgaaactttt-3′, depending on whether the product was intended for cloning into the yeast or bacterial expression vector. The reverse primer that introduced the His6 tag consisted of 5′-ctgagctcctaatgatgatgatgatgatgatctacctcttccatt-3′. After an initial denaturation at 95°C for 2 minutes, the reactions were cycled twice at a denaturing temperature of 95°C for 30 seconds, followed by an annealing temperature of 38°C for 30 seconds, and a 2-minute 30-second extension at 72°C. Thirty more PCR cycles were then performed using an annealing temperature of 50°C. The PCR products intended for cloning into the yeast expression vector were digested with BamHI and Sac I and subcloned into the unique BamHI and Sac I sites directly adjacent to the GPD promoter on the centromeric yeast expression vector pHGPD to create pHGPD-HSP82(His6) and pHGPD-G170D(His6). The His6-tagged fusions were expressed in the YCM82a (see below and Table 1) strain of yeast and supported normal growth and were similar to those of yeast expressing the nontagged counterparts (data not shown). The PCR products intended for cloning were digested with Nco I and Sac I and inserted into the unique Nco I and Sac I sites on pET28a(+) (Novagen) to create pET28a-HSP82(His6) and pET28a-G170D(His6).
Table 1.
Yeast strains used in this studya
The HSP82, hsp82G170D, and hsp82E381K genes were amplified by PCR (Platinum Taq High Fidelity, Invitrogen, Carlsbad, CA, USA) using pHGPD/P82, pHGPD/G170D, or pHGPD/E381K as templates and the primers 5′-ctccatggctagtgaaactttt-3′ and 5′-ctgagctcctaatctacctcttcca-3′. After an initial denaturation at 95°C for 2 minutes, the reactions were cycled twice at a denaturing temperature of 95°C for 30 seconds, followed by an annealing temperature of 40°C for 45 seconds, and a 3-minute extension at 68°C. Thirty more PCR cycles were then performed using an annealing temperature of 50°C. The PCR products were digested with Nco I and Sac I and ligated into the unique Nco I and Sac I sites on pET28a(+) to create pET28a-HSP82, pET28a-G170D, and pET28a-E381K, respectively.
Yeast strains
The genotypes and references for all strains used in this study are shown in Table 1. All yeast transformations were performed by a lithium acetate method (Gietz et al 1992). Transformants were selected for and propagated on synthetic minimal medium containing 2% glucose and lacking the appropriate amino acids or nucleosides (or both) needed to select for plasmid retention. The sugar in the synthetic minimal medium was switched to 2% galactose to express AHR and ARNT genes, which were under control of the bidirectional galactose-inducible promoter.
The YCM82a strain was constructed from the PCLD82a parent strain. PCLD82a was transformed with the lacZ reporter plasmid pTXRE5-Z. The AHR-GAL 1,10-ARNT expression construct was then integrated into chromosome III using a 2-step recombination method (Rothstein 1991) as described previously (Cox and Miller 2002). Integration of the AHR-ARNT expression construct into chromosome III was confirmed by Southern blot analysis (data not shown). The diploid strain YCM282a/α was constructed by mating YCM82a with PCLD82α. Note that the latter strain carries the HSC82 expression plasmid pKAT6, which expresses the Hsc82 protein (Table 1). YCM282a/α is the parent strain for all the temperature-sensitive hsp82 mutant strains used in the lacZ assays. To construct the wild-type HSP82 or temperature-sensitive strains, the pHGPD plasmid expressing wild-type HSP82 or hsp82 derivatives was transformed into YCM282a/α. Cells that lost pKAT6 were selected by growth for several generations in uracil-containing medium, followed by selection on 5-FOA. Temperature sensitivity was confirmed by a lack of growth at 37°C. These strains are named according to their Hsp90 status as YCM282/HSP82, YCM282/T22I, YCM282/A41V, YCM282/G81S, YCM282/G170D, YCM282/G313S, and YCM282/E381K in experiments shown below.
Sporulation and random spore analysis
The sba1/hsp82/hsc82 triple mutant (YMC5) was constructed by mating the sba1 deletion strain (YMC2α) with the YCM82a strain carrying the hsp82G170D expression vector (pHGPD/G170D). The resulting diploid strain was selected for growth on synthetic minimal glucose medium lacking leucine, adenine, histidine, and tryptophan. Sporulation was induced by growing the diploids in presporulation medium (0.8% yeast extract, 0.3% peptone, and 10% glucose) for 2 days, followed by approximately 4 weeks of growth in minimal sporulation medium (1% potassium acetate) containing the appropriate amino acids or nucleosides (or both). Spore formation was observed by light microscopy at 400-fold magnification. The sporulated cells were collected in tubes using centrifugation at 1300 × g for 5 minutes, washed once in distilled water, and digested for 6 minutes in 10 μL of zymolase solution (1.5 μL β-mercaptoethanol, 78.5 μL distilled water, and 20 μL zymolase [0.5 mg/mL final]). The reaction was diluted by the addition of 100 μL of distilled water, and the tetrads were disrupted by vigorously vortexing the mixture for 5 minutes. The cell suspension was plated onto yeast extract-peptone-dextrose medium containing adenine plates (YPAD plates) and grown for 2 days at room temperature. Colonies were replica plated on YPAD at 37°C and on synthetic minimal glucose medium containing 200 μg/mL G418 sulfate and lacking the appropriate amino acids or nucleosides (or both) at room temperature. We identified a clone that grew on G418 sulfate–containing medium at room temperature but did not grow on YPAD at 37°C. The temperature sensitivity indicated that the clone contained both the hsc82:LEU2 and hsp82:LEU2 deletions. Growth on G418 sulfate–containing medium indicated that the clone contained the sba1:Kanr deletion.
LacZ assay
The β-galactosidase (lacZ) assay was used to detect ligand-induced, AhR-mediated gene expression. The original assay protocol (Kippert 1995) was adapted to a 96-well plate format (Miller 1999). Cells from saturated cultures grown overnight in synthetic minimal glucose medium were diluted between 1:50 and 1:200 in 96-well plates containing synthetic minimal galactose medium with the appropriate amino acids or nucleosides (or both). β-Naphthoflavone was added to a final concentration of 1 μM with a final DMSO vehicle concentration of 1%. All other treatment doses presented were serial dilutions from the 1-μM β-naphthoflavone treatment and were performed in triplicate. The 96-well plates were incubated at the indicated temperatures for 18 hours. Negative controls performed along with the experimental treatments include cells with 1% DMSO or medium alone. Absorbance readings at 405 nm for the lacZ assays and cell densities were performed within the linear range of the microplate spectrophotometer. LacZ units were calculated as described previously (Miller 1999). All data presented are representative of at least 3 independent experiments of similar results.
Hsp82 immunoblot analysis
Yeast cells grown overnight in the appropriate medium to select for plasmid maintenance were harvested, washed once with deionized water, and resuspended in 0.5 mL of extraction buffer (0.3 M NH4SO4, 5% glycerol, 50 mM NaCl, 5 mM MgCl2, 1 mM ethylenediaminetetraacetic acid, 1 mM phenylmethane sulfonyl fluoride [PMSF], and protease inhibitor cocktail). The cells were broken in the presence of 0.4-mm glass beads by vigorous vortexing for 5 cycles of 60 seconds each, with intermittent cooling on ice. The cell extracts were clarified at 14 000 × g for 20 minutes at 4°C. Protein concentrations were measured using the BioRad protein assay (BioRad, Hercules, CA, USA) according to the manufacturer's instructions. The total protein extracts was resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a 0.45-μM nitrocellulose membrane (BioRad). The membrane was incubated with shaking in blocking buffer, which consisted of phosphate-buffered saline (PBS) with 0.05% Tween 20 and 5% nonfat dry milk for 1 hour at room temperature. The membrane was then incubated with the primary antibody Ab4-2, a rabbit anti-yeast Hsp90 serum, kindly provided by S. Lindquist. The antiserum was diluted 1:20 000 in blocking buffer and reacted for 1 hour with shaking at room temperature, followed by 6 washes for 5 minutes each in wash buffer (1× PBS, 0.05% Tween 20). The membrane was incubated with the secondary antibody (horseradish peroxidase–conjugated goat anti-rabbit IgG, BioRad) (diluted 1:100 000 in blocking buffer) for 1 hour with shaking at room temperature. The membrane was washed 6 times for 5 minutes each in wash buffer, followed by chemiluminescent detection using the SuperSignal West Chemiluminescent Detection/Optimization Kit (Pierce, Rockford, IL, USA) according to the manufacturer's instructions.
Bacterial expression and preparation of bacterial cell lysates
E coli BL21(DE3) (Novagen) containing the bacterial expression vectors was grown overnight at 37°C, followed by 2 hours of growth in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside at 37°C to induce expression of the desired protein. When the temperature-sensitive hsp82G170D mutant was expressed, the cells were grown in the presence of 1% glucose before induction. Bacterial cell lysates were prepared in extraction buffer (buffer A) for the His6-tagged proteins for use in the adenosine triphosphatase (ATPase) assays (see below). Buffer A consists of 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1% Triton X-100, 1 mM PMSF, 1 mM β-mercaptoethanol, and protease inhibitor cocktail. The extraction buffer (buffer B) for nontagged proteins used in the in vitro interaction experiments (see below) consisted of 20 mM Tris-HCl (pH 7.4), 50 mM KCl, 5 mM MgCl2, 0.01% nonidet P-40, 1 mM PMSF, 1 mM β-mercaptoethanol, and protease inhibitor cocktail. Harvested cells were washed once in extraction buffer and then resuspended in 4 mL of extraction buffer for every 100 mL of cell culture harvested. The resuspended cells were incubated at 30°C in the presence of 100 μg/mL lysozyme for 20 minutes and disrupted by sonication using a Vibra Cell Sonicator (model VC-375, Sonics & Materials Inc., Danbury, CT, USA). The sonicator was used with a microtip probe, set at a power level of 5 at 50% duty for 4 bursts of 10 seconds each, with samples placed on ice in the intermittent periods. The crude cell lysates were clarified by centrifugation at 16 000 × g for 20 minutes.
Affinity isolation of His6-tagged proteins
Bacterial cell lysates containing His6-tagged proteins were prepared as described above. His6-tagged proteins were purified using a cobalt metal-affinity resin (Talon, Clontech, Palo Alto, CA, USA). Approximately 3 mL of suspended resin (capacity = 3 mg His6-tagged protein/mL of suspended resin) was equilibrated with extraction buffer A and then incubated with 20 mL of His6-tagged proteins at 4°C for 20 minutes. The resin was clarified by centrifugation at 700 × g for 2 minutes, washed once in 20 mL of extraction buffer A, and once in 20 mL of a stringent wash buffer (extraction buffer A containing 5% glycerol and 5 mM imidazole). The resin was transferred to a 2-mL gravity flow column and washed with 20 mL of stringent wash buffer. Elution buffer (buffer A lacking Triton X-100 and containing 5% glycerol and 150 mM imidazole) was then applied to the column, and eluted proteins were collected in 1-mL fractions. Typically, most of the protein eluted in the first 5 fractions. Protein concentrations were determined using the BioRad protein assay. The fractions were pooled, dialyzed extensively against 20 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol (DTT), and then concentrated in a Centricon YM-50 centrifugal filter device (Millipore Corporation, Bedford, MA, USA).
ATPase activity assays
The ATPase activity of Hsp82 was measured in vitro using the EnzChek phosphate assay kit (Molecular Probes, Eugene, OR, USA). The phosphate assay is based on the conversion of 2-amino-6-mercapto-7-methylpurine riboside to ribose-1-phosphate and 2-amino-6-mercapto-7-methylpurine by purine nucleoside phosphorylase, which results in a change in absorption at 360 nm (Webb 1992). The typical ATPase assay contained 1–5 μM Hsp82 or 1 μM GroEL (generously provided by Sam Landry and Frank Shewmaker, Tulane University, New Orleans, LA, USA), 1 mM ATP, and either buffer C (20 mM Tris-HCl [pH 7.4], 10 mM KCl, 6 mM MgCl2, and 1 mM DTT) or buffer D (20 mM Tris-HCl [pH 7.4], 100 mM potassium acetate, 10 mM magnesium acetate, 5% glycerol, and 1 mM DTT) in a final volume of 150 μL. The reactions also contained 0.01% Triton X-100 where indicated. The reactions were incubated at the indicated temperature for 60–90 minutes and then diluted 1:10 into a 1-mL phosphate assay as outlined by the manufacturer's instructions. A standard curve was generated using the inorganic phosphate solution provided by the manufacturer. The linear range of the phosphate assay is between 5 and 150 μM inorganic phosphate. Given the slow nature of the Hsp90 ATPase activity, all phosphate assays containing 1 μM Hsp82 were supplemented with 5 μM inorganic phosphate to bring all readings within the linear range of the assay. Reactions containing 60 μM geldanamycin were performed in parallel with other reactions. ATPase activity measured in the presence of geldanamycin (60 μM) was considered to represent nonspecific ATPase activity (background) and was subtracted from the total activity to obtain the specific Hsp90 ATPase activity. Specific Hsp90 ATPase activity was typically 70–75% of the total ATPase activity measured.
In vitro interaction experiments
Hsp82, hsp82G170D, and hsp82E381K proteins were expressed in bacteria as described above. One hundred microliters of bacterial cell lysate containing Hsp82 or its derivatives was incubated with 20 μg of affinity-isolated Sba1(His6) for 30 minutes at 30°C. The assays were supplemented with 1 mM ATPγS and 5 μM geldanamycin where indicated. The mixture was then incubated with 50 μL of suspended Talon Metal-Affinity Resin preequilibrated in extraction buffer B for 20 minutes at 4°C. The resin was clarified at 700 × g for 2 minutes at 4°C, followed by 2 washes in buffer B containing 5 mM imidazole. Proteins remaining on the resin were removed with 50 μL of 2-fold concentrated SDS-PAGE sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 0.7 M β-mercaptoethanol, and 0.01% bromophenol blue) and denatured at ∼95°C for 3 minutes. Thirty-five microliters of each sample was run on a 7.5% SDS-PAGE gel, fixed, and visualized by Coomassie blue staining.
RESULTS
A functional AhR-mediated reporter assay constructed in yeast
We previously developed a reporter gene assay to detect the regulatory factors that modulate AhR-mediated gene expression in yeast (Miller 1999; Cox and Miller 2002). In brief, the system consists of a genomic copy of a human AHR-ARNT expression construct driven by a bidirectional GAL 1,10 promoter integrated into chromosome III and a β-galactosidase reporter plasmid driven by 5 AHREs (Fig 1). Cells grown in galactose medium in the presence of β-naphthoflavone (an AhR ligand) expressed the lacZ reporter gene, and its protein product was detected enzymatically. The β-galactosidase activity provided a quantitative indicator of AhR signaling status in the yeast cells.
Fig 1.
Components of the AhR-mediated lacZ reporter assay. The key elements of the β-galactosidase (lacZ) reporter plasmids (pLXRE5-Z and pTXRE5-Z) and the AHR-GAL 1,10 promoter-ARNT expression construct in chromosome III are shown. The reporter plasmids are centromeric (low copy number) and contain either leucine or tryptophan biosynthetic pathway genes as selectable markers, respectively. AhR/Arnt-mediated expression of the lacZ reporter gene is driven by 5 upstream AHREs. AhR, aryl hydrocarbon receptor; Arnt, aryl hydrocarbon nuclear translocator; AHRE, aryl hydrocarbon response elements
Yeast Hsp90 (hsp82) mutants are defective in AhR-mediated signaling
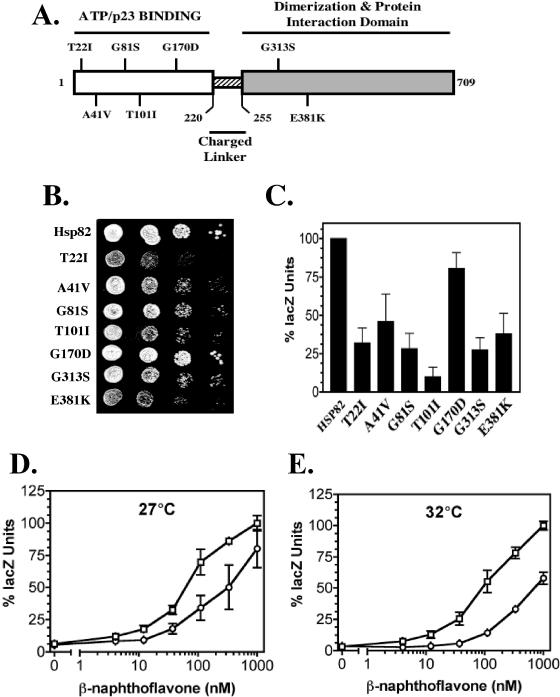
We tested yeast Hsp90 (hsp82) derivatives with point mutations (T22I, A41V, G81S, T101I, G170D, G313S, and E381K) for specific effects on AhR signaling (Table 1). Figure 2A shows the positions of the mutations in relation to various structural and functional features of the Hsp90 protein. Several of these mutations (T22I, A41V, G81S, T101I, and G170D) are within the ATP/p23-binding site of Hsp90 and could potentially affect p23 interactions and ATPase function (Fig 2A). We constructed wild type (YCM282/Hsp82) and mutant strains (YCM282/T22I, YCM282/A41V, YCM282/G81S, YCM282/T101I, YCM282/G170D, YCM282/G313S, and YCM282/E381K) that contained the functional human AHR-ARNT expression construct and the lacZ reporter plasmid (Fig 1). These strains were constructed from the YCM282a/α (Table 1) parent strain, which lacks the endogenous HSP82 and HSC82 genes. Although deletion of both Hsp90 genes is normally lethal, YCM282a/α retains viability by virtue of plasmid-borne Hsp90 genes. These cells were assayed for AhR-mediated expression of the lacZ reporter gene at the permissive temperature of 27°C (the ambient temperature of our shaking incubator). All the mutant strains were significantly defective in AhR signaling at β-naphthoflavone concentrations of 1 μM, with the exception of cells containing the hsp82G170D derivative (Fig 2B). The hsp82T101I mutant displayed an AhR signaling phenotype that was reduced to ∼10% of the levels observed in the parent strain, and the other derivatives were reduced to ∼30% signaling capacity. Several of the mutant strains grew more slowly than wild-type cells at permissive temperatures (22–27°C, Fig 2C), suggesting that cell cycle or other growth defects were present. The growth defects were not correlated with the degree of impaired AhR signaling, suggesting differential effects of the hsp82 mutations on cellular substrates other than AhR. The hsp82 mutant strains, with the exception of YCM282/G170D, required high ligand concentrations (1 μM) to detect AhR signaling. Thus, the apparent potency and efficacy of the AhR ligand is altered in the mutant strains. AhR signaling in the YCM282/G170D mutant was consistently reduced to ∼70% of wild-type levels at lower ligand concentrations (12–333 nM) at 27°C (Fig 2D). The AhR signaling defect in YCM282/G170D was more severe at 32°C (Fig 2E), although the cells still exhibited wild-type growth rates (not shown) at this permissive temperature. Thus, AhR-specific effects were observed at growth-permissive temperatures in the hsp82G170D mutant.
Fig 2.
Effects of temperature-sensitive hsp82 point mutants on AhR signaling. (A) A diagram of the Hsp82 protein showing the relative positions of the temperature-sensitive point mutations is shown (white = N-terminal nucleotide-binding site; striped = charged linker region; shaded = C-terminal dimerization and interaction domains). (B) Wild-type HSP82 and hsp82 mutant strains (see Materials and Methods) were grown at 27°C with a range of β-naphthoflavone concentrations and assayed for AhR-mediated expression of the lacZ reporter gene. The data were plotted as a percentage of the lacZ activity measured in the YCM282/Hsp82 strain at 1000 nM β-naphthoflavone, and relative signaling at this dose is shown. AhR signaling in all mutant strains except the hsp82G170D strain was significantly different from that in the wild-type HSP82 strain (P ≤ 0.001). (C) Serial dilutions of wild type and hsp82 mutant strains plated at 27°C showing growth rate differences. (D, E) The wild-type HSP82 strain (open squares) and the hsp82G170D temperature-sensitive mutant strain (open circles) were grown at 27°C (D) and 32°C (E) in the presence of the indicated concentrations of β-naphthoflavone and assayed for AhR-mediated signaling. The data were plotted as a percentage of the lacZ activity measured in the YCM282/Hsp82 strain at 1000 nM β-naphthoflavone. AHR-mediated signaling in the hsp82G170D strain was consistently and significantly different from the HSP82 wild-type strain at β-naphthoflavone concentrations between 12 and 333 nM at 27°C (P ≤ 0.05) and between 12 and 1000 nM at 32°C (P ≤ 0.05). AhR, aryl hydrocarbon receptor; Hsp. heat shock protein
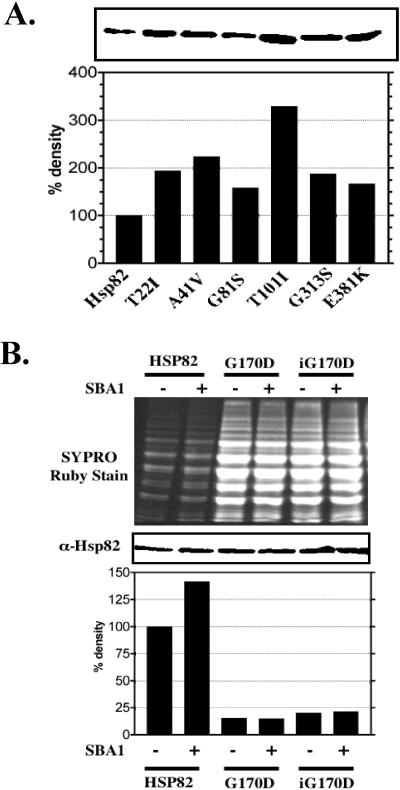
Immunoblot analysis of extracts from the wild type (YCM282/Hsp82) and mutant strains grown at the permissive temperature (27°C) was performed to determine whether differences in hsp82 levels could explain the AhR signaling defects. All the mutated hsp82 proteins, with the exception of the hsp82G170D derivative, were present at about the same or higher levels than the wild-type Hsp82 protein (Fig 3A). The enhanced levels of the hsp82 proteins per se were not responsible for reduced AhR-mediated signaling in the mutant strains because expression of excess wild-type HSP82 from a plasmid did not inhibit AhR signaling (data not shown). In contrast to the other mutated proteins, the hsp82G170D protein (data not shown) was not detectable in immunoblots under the conditions shown in Figure 3A.
Fig 3.
Hsp82 protein levels in wild type and mutant strains. (A) Extracts from wild-type and temperature-sensitive hsp82 strains were compared by immunoblotting using rabbit anti-yeast Hsp90 (Ab-4) serum. The bands were quantified by densitometry and normalized to the strain expressing wild-type Hsp82. (B) YCM282/Hsp82 (HSP82), YCM282/G170D (G170D), and YMCiG170D (iG170D) strains with or without an SBA1 expression vector were compared by immunoblotting using the rabbit anti-yeast Hsp90 (Ab-4) serum. The top panel shows the SYPRO ruby dye–stained SDS-PAGE gel containing 2 μg of resolved protein extract from the wild-type strain (HSP82) and 20 μg from the hsp82G170D strains (G170D and iG170D). The middle panel illustrates the relative immunoreactivity corresponding to the amount of Hsp82 or hsp82G170D protein in extracts shown in the top panel. The bands in the immunoblot (middle panel) were measured by densitometry and normalized to reflect differences in protein loading (bottom panel). Hsp, heat shock protein; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis
The hsp82G170D protein was detectable by immunoblot analysis when the total cellular protein extract from the YCM282/G170D and YMCiG170D strains was loaded at a 10-fold higher concentration (20 μg) than that from the wild type YCM282/HSP82 strain (2 μg, Fig 3B). When corrected for protein loading, hsp82G170D protein levels in both the YCM282/G170D and YMCiG170D strains were approximately 10-fold lower than the levels of the wild-type Hsp82 protein in YCM282/Hsp82 (Fig 3B). The reduced hsp82G170D protein levels were not due to reduced solubility because insoluble pelleted material displayed a similar reduction in hsp82G170D protein concentration (data not shown). It is not likely that the hsp82G170D point mutation results in reduced immunoreactivity because the antiserum was made using a small peptide from the distal C-terminal region of Hsp82. Thus, it is likely that the low levels of hsp82G170D protein were due to inefficient expression or instability in yeast. Expression of SBA1 (yeast p23) in both the YCM282/G170D and YMCiG170D strains had no effect on the hsp82G170D protein levels (Fig 3B). The levels of hsp82G170D protein from the YMCiG170D strain were slightly higher than in the YCM282/G170D strain. The nonpermissive growth temperature for the YCM282/G170D strain was approximately 35°C (varied by 1–2°C), and the nonpermissive temperature for the strain expressing a genomic copy of hsp82G170D (YMCiG170D) was approximately 2°C higher than its counterpart. The small differences in Hsp82G170D protein levels between these 2 mutants may account for the small differences in temperature sensitivity.
p23 (Sba1) suppresses AhR signaling defects and temperature sensitivity associated with the hsp82G170D allele
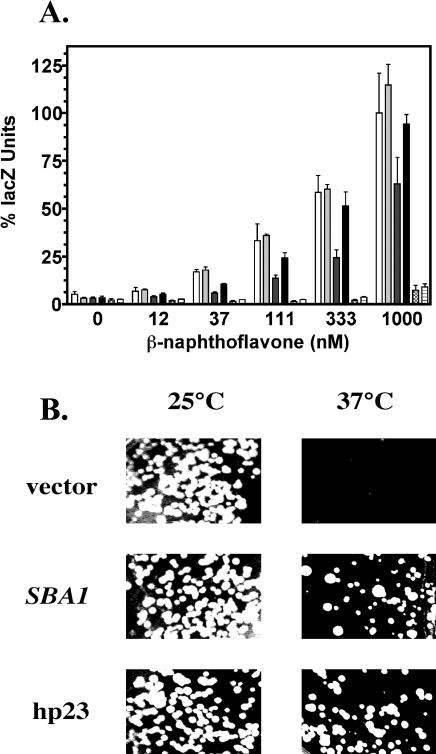
Given that several of the mutations are within the ATP/p23-binding site of Hsp90, we reasoned that overexpression of p23 might suppress the AhR signaling defects and temperature sensitivity associated with these mutations. To test this possibility, we assayed the wild-type HSP82 (YCM282/Hsp82) and mutant strains for AhR-mediated expression of the lacZ reporter gene in the presence or absence of a high–copy number SBA1 expression vector. SBA1 overexpression specifically suppressed the AhR signaling defects associated with the hsp82G170D mutation but did not affect signaling in the other hsp82 mutants described above. SBA1 overexpression in the YCM282/G170D strain restored AhR signaling to wild-type levels at 32°C (Fig 4A) and at 27°C (data not shown). SBA1 overexpression had no positive or negative effects on AhR signaling in the wild-type YCM282/Hsp82 strain or in the YCM282/E381K strain that contained a mutation outside the ATP/p23-binding region of Hsp82 (Figs 2A and 4A). This result suggests that p23's enhancement of signaling in the YCM282/G170D strain is a receptor-specific effect and implicates the G170 region of Hsp82 in the regulation of AhR function. The G170D mutation had no effect on β-galactosidase expression from a separate galactose-inducible lacZ reporter plasmid (data not shown), demonstrating that there is no effect on general transcription, translation, or β-galactosidase protein stability in this strain. This control experiment helps rule out the presence of potential nonspecific factors that might affect general features of the galactose-regulated reporter gene system. We also observed that overexpressed Sba1 and human p23 proteins enhanced AhR signaling in the strain containing an integrated copy of hsp82G170D (YMCiG170D, data not shown).
Fig 4.
SBA1 suppression of AhR signaling defects and temperature sensitivity associated with the G170D mutation. (A) The HSP82 strain with (light gray bars) or without (white bars) the SBA1 expression vector, the hsp82G170D strain with (black bars) or without (dark gray bars) the SBA1 expression vector (pGal-SBA1), and the hsp82E381K strain with (horizontal lined bars) or without (hatched bars) the SBA1 expression vector (pGal-SBA1) were compared for AhR signaling. All strains carried the pTXRE5-Z reporter vector and the genomic AHR-GAL1,10-ARNT expression construct and were assayed at 32°C. (B) The YMCiG170D derivatives with the parent vector (pBevy-GU), SBA1 expression vector (pBevy-GU-SBA1), or human p23 expression vector (pBevy-GU-p23) were grown on synthetic minimal galactose medium for 3 days at the indicated temperatures. AhR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon nuclear translocator
We next questioned whether SBA1 overexpression could suppress the temperature sensitivity associated with the Hsp82 mutations. The YMCiG170D strain was tested for growth at permissive and nonpermissive temperatures in the presence or absence of an SBA1 expression vector, a human p23 expression vector, or the parent null expression vector. Both SBA1 and human p23 overexpression suppressed the temperature sensitivity of the YMCiG170D strain (Fig 4B). All the cells tested were able to grow at 25°C (permissive temperature), but when the cells were grown at 37°C (the typical nonpermissive temperature for this mutant), only the YMCiG170D cells expressing excess SBA1 or human p23 from plasmids were able to grow. Expression of SBA1 and human p23 enhanced the nonpermissive temperature of the YMCiG170D strain within a narrow window of ∼2°C. When the experiment was performed using synthetic glucose medium rather than galactose medium, the suppression of temperature sensitivity in cells carrying the SBA1 and human p23 expression vectors was abolished (data not shown). This result indicated that the suppression of temperature sensitivity in the YMCiG170D strain required the expression of either the SBA1 or human p23 genes because both were expressed under control of a galactose-inducible promoter.
The hsp82G170D protein lacks Sba1 binding and ATPase activity in vitro
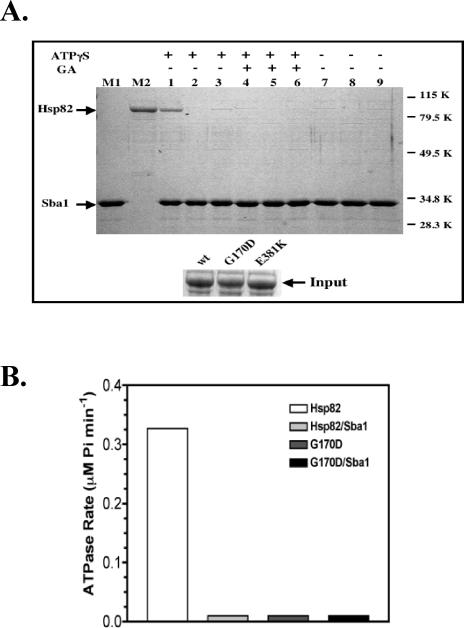
Given that SBA1 expression suppressed the hsp82G170D-mediated AhR signaling phenotype, we questioned whether hsp82G170D binding to Sba1 was normal. The Hsp82, hsp82G170D, and hsp82E381K proteins were assessed for their ability to bind purified Sba1(His6) using an in vitro coprecipitation assay. During culture, we noted that E coli that expressed the hsp82G170D protein grew slowly relative to cells that expressed wild-type Hsp82 and hsp82E381K proteins. This growth delay could be indicative of some toxicity of the hsp82G170D protein. The inset at the bottom of Figure 5A shows that the Hsp82, hsp82G170D, and hsp82E381K proteins were expressed at equal levels and were equally soluble in the extracts. We observed an interaction between wild-type Hsp82 and Sba1(His6) in the presence of 1 mM ATPγS (Fig 5A; lane 1), and the specificity of the interaction was confirmed by disruption in the presence of geldanamycin (lane 4). An interaction between the hsp82G170D protein and Sba1(His6) was not observed in the presence of ATPγS (lane 2), whereas a very weak interaction was observed between Sba1(His6) and the hsp82E381K protein (lane 3). The original preparations of the hsp82G170D protein were made from bacteria grown at 37°C and may have inactivated this derivative. Therefore, we repeated the experiment using protein extracts from bacteria that were grown at 27°C (data not shown) and obtained the same results as in Figure 5A. Thus, the purified hsp82G170D protein was unable to bind p23 in vitro.
Fig 5.
Sba1(His6) binding and ATPase activity of Hsp82 proteins. (A) Bacterial extracts (100 μL) containing wild type or mutated Hsp82 proteins were incubated with 20 μg of purified Sba1(His6) at 30°C for 30 minutes. p23 complexes were isolated using 50 μL of suspended cobalt metal-affinity resin, and a portion was analyzed by SDS-PAGE. The reactions contained wild-type Hsp82 (lanes 1, 4, and 7), hsp82 G170D (lanes 2, 5, and 8), or hsp82 E381K (lanes 3, 6, and 9). The binding assays in lanes 1–3 were supplemented with 1 mM ATPγS, and lanes 4–6 were supplemented with 1 mM ATPγS and 5 μM geldanamycin (GA). The assays in lanes 7–9 lacked both ATPγS and geldanamycin. Lane M1 contains purified Sba1(His6) and lane M2 contains purified Hsp82(His6) as markers. Molecular weight markers (kDa) are shown on the right. The inset at the bottom shows that equivalent amounts of Hsp82, hsp82 G170D, and hsp82 E381K proteins were added into the coprecipitation experiment. (B) Purified Hsp82(His6) and G170D(His6) (1 μM) were incubated at 30°C in the presence of 1 mM ATP with or without 2.5 μM purified Sba1(His6), and ATPase activity was assessed as described in the Materials and Methods. ATPase activity was only detectable in assays containing wild-type Hsp82 (white bar). A small positive value was used to mark the position of samples with undetectable ATPase activity for graphical purposes. ATPase, adenosine triphosphatase; Hsp, heat shock protein; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; ATP, adenosine triphosphate
The G170D point mutation resides within the p23- and ATP-binding domains of Hsp82. After finding that the hsp82G170D point mutation disrupted p23 binding, we questioned whether the hsp82G170D protein had ATPase activity. No ATPase activity was detected from the hsp82G170D protein at 30°C, which is a permissive temperature (Fig 5B; dark gray bar). Addition of a 2.5-fold molar excess of purified Sba1 did not restore ATPase activity to the hsp82G170D protein (Fig 5B; black bar). The ATPase rate of the wild-type Hsp82 protein averaged 0.32 μM Pi min−1 μM−1 protein (Fig 5B; white bar) and at 37°C averaged 0.94 μM Pi min−1 μM−1 protein. Interestingly, the presence of a 2.5-fold molar excess of Sba1 strongly inhibited ATPase activity of the Hsp82 protein (Fig 5B; light gray bars).
Sba1 inhibits the wild-type Hsp82 ATPase activity in vitro
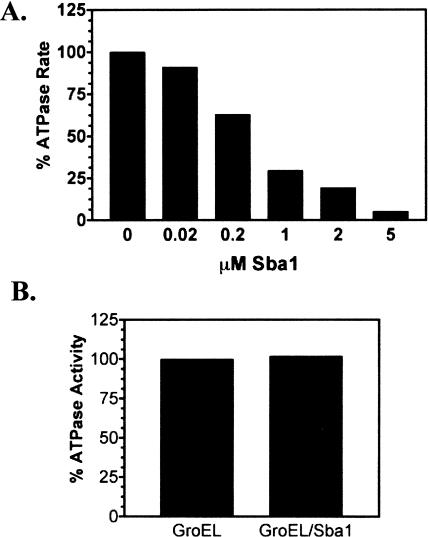
Young and Hartl (2000) reported that Sba1 had no effect on the ATPase activity of the wild-type Hsp82 protein in vitro. Given the discrepancies between the findings of Young and Hartl and our finding that Sba1 inhibited the Hsp82 ATPase activity (Fig 5B), we investigated this effect in more detail. The wild-type Hsp82 ATPase was assayed at 37°C, and Sba1 inhibited the enzyme in a concentration-dependent manner (Fig 6A). At equimolar concentrations of Hsp82 and Sba1 (1 μM), the Hsp82 ATPase activity was reduced by approximately 75% and was undetectable in the presence of a 5-fold molar excess of Sba1 (Fig 6A). ATPase activity experiments were repeated in a high salt buffer (buffer D) similar to that used by Young and Hartl (2000), and Sba1-dependent inhibition was observed. The Hsp82 ATPase activity was unaffected by addition of a mock bacterial cell extract lacking Sba1 (data not shown), which demonstrates that inhibition required Sba1. In contrast to Hsp82, addition of a 5-fold molar excess of Sba1 had no effect on the ATPase activity of the bacterial chaperone GroEL (Fig 6B). Therefore, the p23 inhibition observed was specific for the Hsp82 ATPase function.
Fig 6.
Sba1 inhibition of Hsp82 ATPase activity in vitro. (A) Purified Hsp82(His6) was incubated at 37°C in 1 mM ATP and different Sba1(His6) concentrations. After 90 minutes, the inorganic phosphate concentration (Pi released) specifically by the Hsp82 ATPase was measured. The data were plotted as a percentage of the ATPase activity relative to that measured in the absence of Sba1 after subtraction of the geldanamycin (60 μM)-inhibited background activity. (B) Purified GroEL (1 μM) was incubated at 37°C in 1 mM ATP with or without 5 μM Sba1(His6). After 90 minutes, the inorganic phosphate concentration (Pi released) was measured. The data were normalized to the ATPase activity measured for GroEL in the absence of Sba1 and plotted. ATPase, adenosine triphosphatase; ATP, adenosine triphosphate; Hsp, heat shock protein
Detection of significant ATPase activity from 1 μM Hsp82 in our assay system required the presence of a very low concentration of nonionic detergent. In the presence of 0.01% Triton X-100 detergent, the geldanamycin-sensitive (specific) Hsp90 ATPase activity was 70–75% of the total activity measured. In the absence of Triton X-100, the background (nonspecific) ATPase activity was constant, and Hsp90-specific activity was greatly reduced to near background levels (data not shown). The ATPase activity of the Hsp82(His6) protein that was purified in the presence of Triton X-100, dialyzed in buffer lacking Triton X-100, and assayed in the absence of Triton X-100 was equivalent to the activity of Hsp82(His6) protein assayed in the presence of Triton X-100. Perhaps, residual detergent remained associated with Hsp82 after dialysis and was sufficient to produce the positive effect in the ATPase assays. Therefore, the effects of detergents on Hsp90 ATPase activity might have gone unnoticed by those who have purified these chaperone proteins by various methods. We also noted that at higher Hsp90 concentrations at which there was detectable ATPase activity, the activity was enhanced (2-fold) by detergent (data not shown).
SBA1 deletion and the hsp82G170D mutation produce an additive inhibition of AhR signaling
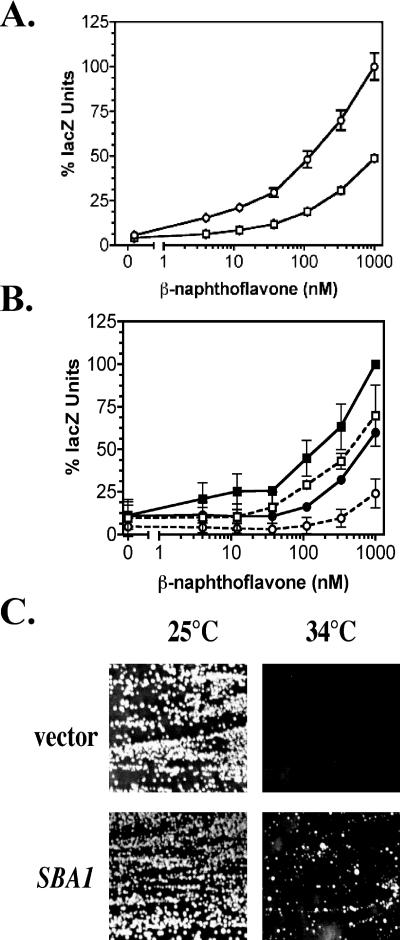
We next questioned whether a lack of SBA1 would have an effect on the hsp82G170D mutant strain. By mating and random spore analysis, we constructed an sba1 deletion strain that carried an expression vector that provided the sole source of Hsp90 (YMC5, Table 1). This strain also carried the pTXRE5-Z reporter plasmid and AHR-ARNT expression construct (Fig 1A). After replacing the wild-type HSC82 expression vector with the wild-type HSP82 expression vector (YMC5/Hsp82) or the hsp82G170D expression vector (YMC5/G170D), the cells were assayed for AhR-mediated signaling at lower and higher permissive temperatures. We found that new nonisogenic YMC5 strain grew slower and could not be compared directly to signaling of its parents, so relative signaling differences (normalized as percentages) were used for purposes of comparison. The AhR signaling defect in the parental sba1 deletion strain is shown in Figure 7A, and the effect of hsp82G170D mutation is shown in Figure 2E. Deletion of sba1 reduced AhR signaling by ∼50% as compared with the wild-type strain (YMC1). The lack of Sba1 (p23) protein in conjunction with the G170D mutation produced an additive effect in the double-mutant strain (Fig 7B, dashed line with open circles), as indicated by a dramatic shift in the dose-response curve and approximately an 80% decline in signaling at the highest ligand concentration. This result suggests that the 2 mutations have distinct actions on AhR signaling. Hsp82 immunoblot analysis of the YMC5/Hsp82 and YMC5/G170D strains revealed an analogous relationship as seen with the YCM282/Hsp82 and YCM282/G170D strains, respectively (Fig 3B). Thus, sba1 deletion had no effect on the hsp82G170D protein levels, ruling out this possibility as a mechanism for reduced AhR signaling in the double-mutant strain.
Fig 7.
AhR signaling defects and temperature sensitivity in the hsp82G170D, sba1 double-mutant strain. (A) The parent strain (YMC1) (open circles) and the sba1 deletion strain (YMC2) (open squares) carrying the pLXRE5-Z reporter vector were assayed for AhR-mediated signaling in the presence of the indicated concentrations of β-naphthoflavone. The data were plotted as a percentage of the lacZ activity relative to that measured in the parent YMC1 strain at 1000 nM β-naphthoflavone. The lacZ activity in the YMC2 strain was significantly different (P < 0.05) from that of the parent strain at all ligand concentrations tested. (B) The YMC5/Hsp82 strain (squares) and the YMC5/G170D temperature-sensitive mutant strain (circles) were grown at 32°C and assayed for AhR-mediated signaling. Solid lines and symbols indicate strains that carry an SBA1 expression vector, and dashed lines with open symbols indicate sba1 null strains. All strains carry the pTXRE5-Z reporter plasmid and the AHR-GAL 1,10 promoter-ARNT expression construct integrated on chromosome III shown in Figure 1. The data were plotted as a percentage of the lacZ activity relative to that measured in the YMC5/Hsp82 strain at 1000 nM β-naphthoflavone. (C) The YMC5/G170D cells with the parent vector or SBA1 expression vector were grown on galactose-containing medium for 4 days at the indicated temperatures. AhR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon nuclear translocator
We again detected a small difference between the nonpermissive temperatures for the YMC5/G170D and YCM282/G170D strains. SBA1 overexpression suppressed the temperature sensitivity in the YMC5/G170D strain (Fig 7C) and cells grew slowly at 34°C relative to the permissive temperature (25°C). This result suggested that some chaperone substrates other than AhR were relatively sensitive to this combination of mutations.
DISCUSSION
The Hsp90 chaperone complex is required for the proper functioning of a specific subset of cellular proteins with diverse cellular functions. Among the proteins known to require the Hsp90 complex for proper functioning are specific steroid hormone receptors and other transcription factors (eg, AhR), several serine-threonine kinases, and the oncogenic tyrosine kinase pp60v-src (Carver et al 1994; Nathan and Lindquist 1995; Nair et al 1996; Nathan et al 1997; Pratt and Toft 1997). AhR signaling is a complex process that requires numerous cellular factors. It is likely that Hsp90 cochaperones such as Xap2 and p23 are important factors in AhR signaling (Ma and Whitlock 1997; Pratt and Toft 1997; Meyer et al 1998; Pratt 1998; Kazlauskas et al 1999, 2000; Meyer and Perdew 1999; Cox and Miller 2002; Miller 2002). The studies presented here provide insight into the relationships among AhR signaling, Hsp90 ATPase activity, and Sba1 (p23) function.
Nathan and Lindquist (1995) developed a panel of yeast Hsp90 (hsp82) temperature-sensitive mutations for the study of Hsp90 target protein interactions. Yeast expressing the temperature-sensitive hsp82 derivatives as the sole source of Hsp90 were not only temperature sensitive but were also defective in glucocorticoid receptor signaling and pp60v-src activation (Nathan and Lindquist 1995). We placed these mutated hsp82 derivatives into our yeast reporter system (Fig 1) and demonstrated that they were defective in AhR signaling, albeit to varying degrees (Fig 2). These results suggest that a general defect in Hsp82 chaperone function exists among these derivatives. The hsp82G170D mutation was the only classical temperature-sensitive derivative that provided for wild-type growth up to the nonpermissive temperature of ∼35°C. Nathan and Lindquist (1995) reported that the hsp82G170D mutant was not defective in glucocorticoid receptor signaling at 27°C, and that signaling declined as temperature was increased further. We observed specific signaling defects at lower ligand concentrations in our AhR system at 27°C for the hsp82G170D mutant (Fig 2D). This difference may reflect the sensitivities of the 2 systems or a divergence in the mechanisms of Hsp90 regulation for the 2 receptors. The previous study showed that glucocorticoid receptor levels were unchanged in the different Hsp82 backgrounds, but the Hsp82 protein levels and functional activities other than signaling and temperature sensitivity were not reported. We demonstrated that all the hsp82 proteins, except for hsp82G170D, were present at approximately equivalent or higher levels when compared with the wild-type Hsp82 protein in yeast (Fig 3A). The elevated expression levels of some mutated hsp82 derivatives might reflect a greater half-life or a compensatory mechanism to counteract a weakened functional activity. The only true temperature-sensitive protein, hsp82G170D, was present at a 10-fold lower level than the wild-type Hsp82 protein (Fig 3B). This result suggests that a 10-fold reduction in Hsp82 protein levels may have a modest effect on the levels of AhR signaling because the YCM282/G170D strain functions relatively well at 27°C (Fig 2D). However, we favor the idea that rather than reduced protein levels, it is the reduced activity of the hsp82G170D protein that accounts for reduced AhR signaling. The observation that overexpression of Sba1 (p23) suppresses the hsp82G170D mutation (Fig 4) without altering hsp82G170D protein expression (Fig 3B) supports this notion. There may be rapid recycling of the receptor-chaperone complexes in this mutant, and perhaps, only a small amount of active AhR complex is needed to achieve efficient signaling. The latter idea is in agreement with results of Hestermann et al (2000), which suggest that only 1–2% of the total cellular AhR is needed to mediate efficient ligand-directed signaling in vertebrate cells.
We previously demonstrated that either Sba1 (yeast p23) or human p23 is required for efficient AhR signaling in yeast (Fig 7A, Cox and Miller [2002]). p23 is thought to act indirectly on steroid hormone receptors through its ATP-dependent interaction with the N-terminal nucleotide-binding site on Hsp90 (Nair et al 1996; Pratt and Toft 1997). We showed here that overexpression of Sba1 (p23) protein suppressed the AhR signaling defect and temperature sensitivity associated with the hsp82G170D mutation (Fig 4). This finding further implicates p23 as a facilitator of AhR signaling. The hsp82G170D mutation is within the ATP/p23-binding site on Hsp90, and it is associated with a loss of ATPase activity and p23 binding in vitro (Fig 5). Thus, these findings suggest that the mechanism by which p23 supports normal AhR signaling in yeast occurs, at least in part, through p23's interaction with the nucleotide-binding site on Hsp90. In contrast to hsp82G170D, the hsp82E381K derivative weakly interacted with p23; however, this mutation was not rescued by overexpression of p23 in cells. The E381K mutation lies outside the ATPase domain, which could explain why it is unaffected by excess p23.
Human p23 genetically complements the loss of SBA1 in yeast with regard to AhR and steroid hormone receptor signaling defects (Knoblauch and Garabedian 1999; Freeman et al 2000; Cox and Miller 2002). This functional equivalence of the yeast and human proteins is further supported by the result showing that human p23 overexpression suppressed the temperature sensitivity in the hsp82G170D mutant (Fig 4B). This result also suggests that human p23 supports other Hsp90-dependent cellular processes in yeast apart from its role in AhR signaling because AhR is a foreign protein in this system and has no effect on the yeast cell cycle.
The finding that p23 (Sba1) suppresses the hsp82G170D mutant phenotypes suggests that the basis for the defect is a disruption of the p23/Hsp90 interaction. An interaction between immobilized Sba1(His6) and the hsp82G170D protein was not detected in a previous study that used yeast cell lysates (Fang et al 1998). This negative result might have been confounded by the low hsp82G170D protein levels in yeast (Fig 3B) and competition from untagged Sba1 in the lysate. However, we too were unable to detect an interaction between tagged Sba1 and hsp82G170D using purified proteins in vitro under conditions in which purified wild-type Hsp82 and Sba1 proteins interacted well (Fig 5A). It is possible that purified hsp82G170D protein is in an inactive form, and for this reason, p23 interactions are not detected. However, the SBA1 overexpression experiments in which hsp82G170D effects were suppressed argue in favor of Sba1-mediated restoration of function in vivo. It is possible that the in vitro methods used to detect interactions in our study were simply not sensitive enough to detect a low-affinity p23-hsp82G170D interaction. In support of this idea, an analogous mutated human Hsp90 derivative (G182D) was shown to bind human p23 in vitro, although it was a very low-affinity interaction that was considered to be insignificant (Grenert et al 1997). The hsp82G170D protein that we purified from bacteria lacked ATPase activity. Thus, it is likely that the hsp82G170D protein either is severely impaired for ATPase function or is in an inactive form when produced in E coli, whereas the wild-type Hsp82 is functional. Taken literally, the lack of p23 binding and ATPase activity might suggest that these factors are not required for Hsp90-dependent processes, given that yeast expressing hsp82G170D as their sole source of Hsp90 display wild-type growth characteristics and moderate residual AhR and glucocorticoid receptor signaling activity at 27°C (Fig 2, Nathan and Lindquist [1995] ). However, others have suggested that ATP hydrolysis is required for Hsp90 function, and drugs (eg, geldanamycin) that disrupt this activity are cytotoxic (Obermann et al 1998; Grenert et al 1999). Thus, function of the Hsp90 ATPase domain appears to be critical for biological activity and cell viability.
Sba1 (p23) interacts with the nucleotide-binding site on Hsp90 in an ATP-dependent manner (Fig 5A) (Nair et al 1996; reviewed by Pratt and Toft 1997). Thus, it is plausible that p23 could affect the rate of ATP hydrolysis by Hsp90. One study showed that Sba1 did not affect the rate of ATP hydrolysis by Hsp90 (Young and Hartl 2000). Another study showed that human p23 inhibits the ATPase activity of human Hsp90β but only in the presence of a client protein (McLaughlin et al 2002). These results differed from our finding that Sba1, in the absence of a client protein, inhibited the ATPase activity of wild-type Hsp82 in vitro (Figs 5B and 6A). Thus, studies from 3 laboratories suggest different results with respect to the role of Sba1/p23 on Hsp90 ATPase function. If a minor protein contaminant in our purified protein preparations acted as an Hsp90 client protein, it might help explain the discrepancies between our results and the other studies (Figs 5B and 6A). However, our protein preparations appeared relatively pure by SDS-PAGE analysis. Based on our results, we conclude that Sba1 (p23) reduces Hsp90 ATPase activity, and this may explain the reported p23-mediated stabilization of Hsp90-client protein complexes.
Small amounts of nonionic detergents were reported to stabilize the interaction of human p23 with human Hsp90 in vitro (Sullivan et al 1997, 2002). Thus, the presence of low levels of Triton X-100 may have stabilized the interaction of Sba1 with Hsp82 in our study, thereby favoring ATPase inhibition. However, the fact that Sba1 still inhibited the ATPase activity in the absence of detergent at higher Hsp82 concentrations (data not shown) suggests that this is not the case. The presence of 0.01% Triton X-100 enhanced detection of Hsp82 ATPase activity when assayed at a concentration of 1 μM protein but was not needed to detect ATPase activity at higher (5 μM) concentrations. Factors such as protein adhesion to the plastic surface of reaction tubes may have come into play when we did not detect Hsp82 ATPase activity at lower protein concentrations in the absence of detergent. Detergent also enhanced the amount of ATPase activity detected at higher (5 μM) Hsp82 concentrations, so it appears to have a general facilitative effect. In addition it is possible that the detergent prevented the association of inhibitors that might have copurified with Hsp82. However, the presence of detergent only enhanced Hsp90-specific ATPase activity (geldanamycin-sensitive activity) and not other ATPases.
In the final series of experiments (Fig 7), we compared AhR signaling in a new strain that contained an sba1 deletion mutation and the hsp82G170D mutation. The combined action of these mutations produced an approximately additive reduction of ∼80% or more in the AhR reporter assays relative to the control strain. This result suggests that Sba1 and G170 regions of Hsp90 have different actions on AhR signaling. In contrast, the ability of excess Sba1 (p23) to suppress the hsp82G170D mutation (Fig 4) argues that the 2 mutations are related. We propose that the 2 chaperones may be cooperating in AhR signaling and suggest that the role of p23 is to regulate or “chaperone” the G170/ATPase region of Hsp90. Thus, in the absence of p23, the G170D mutation enhances AhR signaling defects and temperature sensitivity, whereas extragenic copies of SBA1 (p23) suppress the G170D mutation. If this idea is correct, then p23 is probably facilitating AhR signaling through an indirect mechanism that involves its action on Hsp90.
The p23 and Hsp90 mutations described here affect AhR signaling through mechanisms that remain unknown at present. The potential mechanisms of these chaperone mutations may include a reduction in AhR levels and changes in ligand-binding affinity. We have not been able to test these possibilities because of the very low levels of AhR expressed in this model system, which are required for proper ligand-mediated regulation of AhR in yeast (Miller 1999; Miller 2002). In addition, the insolubility of radiolabeled TCDD in this yeast system has been problematic for conducting binding studies. Regardless of potential effects on AhR levels or ligand affinity, our data indicate that the modulation of AhR signaling by p23 occurs through the interaction with the G170/ATP-binding domain of Hsp90. The data presented here are in general agreement with other in vitro studies, suggesting that the interaction of p23 with Hsp90 stabilizes the Hsp90-client protein interaction (Smith et al 1995; Dittmar et al 1997; Pratt and Toft 1997; Kazlauskas et al 1999). Data from in vitro studies suggest that p23 does not affect ligand binding by AhR (Kazlauskas et al 2001). In contrast, p23 has a dramatic effect on ligand affinity of some steroid hormone receptors (Hutchison et al 1995). Whether p23 stabilization of Hsp90-AhR complexes has a positive effect on AhR signaling by altering ligand affinity deserves further study, and testing this idea will require development of a new or modified experimental system other than our current yeast strains. If p23 and other chaperones enhance AhR signaling by a mechanism that does not involve enhanced ligand affinity, it will entail a mechanism that is distinct from that described for specific steroid hormone receptors.
Acknowledgments
We thank Avrom Caplan and Susan Lindquist for providing reagents, strains, and plasmids. We thank Sam Landry and Frank Shewmaker for providing the purified GroEL protein used in the ATPase assays, as well as for helpful discussions regarding the ATPase assays. This study was supported by NIH grant ES 09055, the Tulane-Xavier Center for Bioenvironmental Research, Tulane Cancer Center, and the Cancer Association of Greater New Orleans.
REFERENCES
- Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715.0193-4511(1996)274<1715:CFOHP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Caplan AJ. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrinol Metab. 1997;8:271–275. doi: 10.1016/s1043-2760(97)00079-9.1043-2760(1997)008<0271:YMCATM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Carver LA, Jackiw V, Bradfield CA. The 90-kDa heat shock protein is essential for Ah receptor signaling in a yeast expression system. J Biol Chem. 1994;269:30109–30112.0021-9258(1994)269<30109:TKHSPI>2.0.CO;2 [PubMed] [Google Scholar]
- Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem. 1998;273:33580–33587. doi: 10.1074/jbc.273.50.33580.0021-9258(1998)273<33580:COTARP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, Toft DO. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci U S A. 2000;97:12524–12529. doi: 10.1073/pnas.220430297.0027-8424(2000)097<12524:DANDPU>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Perdew GH. Subunit composition of the heteromeric cytosolic aryl hydrocarbon receptor complex. J Biol Chem. 1994;269:27554–27558.0021-9258(1994)269<27554:SCOTHC>2.0.CO;2 [PubMed] [Google Scholar]
- Coumailleau P, Poellinger L, Gustafsson JA, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291.0021-9258(1995)270<25291:DOAMDO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cox MB, Miller CA. The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol Lett. 2002;129:13–21. doi: 10.1016/s0378-4274(01)00465-9.0378-4274(2002)129<0013:TPCFDR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull Environ Contam Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973.0007-4861(1998)061<0557:TARARO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dittmar KD, Demady DR, Stancato LF, Krishna P, Pratt WB. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor.hsp90 heterocomplexes formed by hsp90.p60.hsp70. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213.0021-9258(1997)272<21213:FOTGRB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727.0270-7306(1998)018<3727:SEAYHC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Felts SJ, Toft DO, Yamamoto KR. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 2000;14:422–434.0890-9369(2000)014<0422:TPMCAA>2.0.CO;2 [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718.0193-4511(1996)274<1718:MCMCAO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051.0193-4511(2002)296<2232:DOTRCB>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425.0305-1048(1992)020<1425:IMFHET>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525.0021-9258(1999)274<17525:TIOABA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Grenert JP, Sullivan WP, and Fadden P. et al. 1997 The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 272:23843–23850. [DOI] [PubMed] [Google Scholar]
- Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519.0362-1642(2000)040<0519:TPSSOE>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515.0362-1642(1995)035<0307:TAHRC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hestermann EV, Stegeman JJ, Hahn ME. Relative contributions of affinity and intrinsic efficacy to aryl hydrocarbon receptor ligand potency. Toxicol Appl Pharmacol. 2000;168:160–172. doi: 10.1006/taap.2000.9026.0041-008X(2000)168<0160:RCOAAI>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hutchison KA, Stancato LF, Owens-Grillo JK, Johnson JL, Krishna P, Toft DO, Pratt WB. The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J Biol Chem. 1995;270:18841–18847. doi: 10.1074/jbc.270.32.18841.0021-9258(1995)270<18841:TKAPIR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Johnson JL, Beito TG, Krco CJ, Toft DO. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956.0270-7306(1994)014<1956:COANKP>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Toft DO. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513.0888-8809(1995)009<0670:BOPAHD>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J Biol Chem. 1999;274:13519–13524. doi: 10.1074/jbc.274.19.13519.0021-9258(1999)274<13519:ETTCPR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kazlauskas A, Poellinger L, Pongratz I. The immunophilin-like protein XAP2 regulates ubiquitination and subcellular localization of the dioxin receptor. J Biol Chem. 2000;275:41317–41324. doi: 10.1074/jbc.M007765200.0021-9258(2000)275<41317:TIPXRU>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kazlauskas A, Sundstrom S, Poellinger L, Pongratz I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol Cell Biol. 2001;21:2594–2607. doi: 10.1128/MCB.21.7.2594-2607.2001.0270-7306(2001)021<2594:THCCRI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x.0378-1097(1995)128<0201:ARPPFA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Knoblauch R, Garabedian MJ. Role for Hsp90-associated cochaperone p23 in estrogen receptor signal transduction. Mol Cell Biol. 1999;19:3748–3759. doi: 10.1128/mcb.19.5.3748.0270-7306(1999)019<3748:RFHCPI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Whitlock JP Jr.. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1997;272:8878–8884.0021-9258(1997)272<8878:ANCPTI>2.0.CO;2 [PubMed] [Google Scholar]
- McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245.0022-2836(2002)315<0787:SOTWAA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Meyer BK, Perdew GH. Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry. 1999;38:8907–8917. doi: 10.1021/bi982223w.0006-2960(1999)038<8907:COTACC>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol. 1998;18:978–988. doi: 10.1128/mcb.18.2.978.0270-7306(1998)018<0978:HBVXPI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA. A human aryl hydrocarbon receptor signaling pathway constructed in yeast displays additive responses to ligand mixtures. Toxicol Appl Pharmacol. 1999;160:297–303. doi: 10.1006/taap.1999.8769.0041-008X(1999)160<0297:AHAHRS>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Miller CA. Two tetratricopeptide repeat proteins facilitate human aryl hydrocarbon receptor signalling in yeast. Cell Signal. 2002;14:615–623. doi: 10.1016/s0898-6568(02)00002-5.0898-6568(2002)014<0615:TTRPFH>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Miller CA, Martinat MA, Hyman LE. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3577–3583. doi: 10.1093/nar/26.15.3577.0305-1048(1998)026<3577:AOAHRC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SC, Toran EJ, Rimerman RA, Hjermstad S, Smithgall TE, Smith DF. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2.1466-1268(1996)001<0237:APOMIC>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917.0270-7306(1995)015<3917:MAOHFI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci U S A. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949.0027-8424(1997)094<12949:IVFOTS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901.0021-9525(1998)143<0901:IVFOHI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267:13728–13734.0021-9258(1992)267<13728:DROTKH>2.0.CO;2 [PubMed] [Google Scholar]
- Pratt WB. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252.0037-9727(1998)217<0420:THCSII>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrinol Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5.0076-6879(1991)194<0281:TDRAAR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, and Maniatis T 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- Smith DF, Whitesell L, Nair SC, Chen S, Prapapanich V, Rimerman RA. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804.0270-7306(1995)015<6804:PRSAFA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan WP, Owen BA, Toft DO. The influence of ATP and p23 on the conformation of hsp90. J Biol Chem. 2002;24:24. doi: 10.1074/jbc.M207754200.0021-9258(2002)024<0024:TIOAAP>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri ES, Litwack G, Toft D. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007.0021-9258(1997)272<8007:NATFSO>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884.0027-8424(1992)089<4884:ACSAFI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw ML, McGuire J, Picard D, Gustafsson JA, Poellinger L. Heat shock protein hsp90 regulates dioxin receptor function in vivo. Proc Natl Acad Sci U S A. 1995;92:4437–4441. doi: 10.1073/pnas.92.10.4437.0027-8424(1995)092<4437:HSPHRD>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324.0027-8424(1994)091<8324:IOHSPH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930.0261-4189(2000)019<5930:PRBHIA>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]