Abstract
Bias introduced by the simultaneous amplification of specific genes from complex mixtures of templates remains poorly understood. To explore potential causes and the extent of bias in PCR amplification of 16S ribosomal DNAs (rDNAs), genomic DNAs of two closely and one distantly related bacterial species were mixed and amplified with universal, degenerate primers. Quantification and comparison of template and product ratios showed that there was considerable and reproducible overamplification of specific templates. Variability between replicates also contributed to the observed bias but in a comparatively minor way. Based on these initial observations, template dosage and differences in binding energies of permutations of the degenerate, universal primers were tested as two likely causes of this template-specific bias by using 16S rDNA templates modified by site-directed mutagenesis. When mixtures of mutagenized templates containing AT- and GC-rich priming sites were used, templates containing the GC-rich permutation amplified with higher efficiency, indicating that different primer binding energies may to a large extent be responsible for overamplification. In contrast, gene copy number was found to be an unlikely cause of the observed bias. Similarly, amplification from DNA extracted from a natural community to which different amounts of genomic DNA of a single bacterial species were added did not affect relative product ratios. Bias was reduced considerably by using high template concentrations, by performing fewer cycles, and by mixing replicate reaction preparations.
The PCR has become an invaluable tool because of the speed and simplicity with which specific DNA segments can be amplified from a background of complex genomes (3, 5). In studies of molecular evolution (29) and microbial ecology (26) this property has facilitated the characterization of both single genes and families of related genes in single or multiple species. This is generally done by designing degenerate primers which target conserved regions of homologous genes, thereby accelerating the detection, amplification, and, ultimately, sequence analysis of the genes under study.
One of the most innovative applications of the PCR has been the cataloging of bacterial and archaeal species richness in the environment. Mixtures of 16S rRNA genes amplified from natural communities are considered representative of the native organisms from which they originated. This approach has revealed the existence of numerous uncultured microorganisms because it circumvents bias introduced by traditional culture-based methods (7), which typically detect only a fraction (<1%) of the total bacteria present in an environment (2). The protocols involve extraction of nucleic acids from an environmental sample, PCR amplification of the 16S rRNA genes with universal, degenerate primers, and separation of amplified products by cloning or by denaturing gradient gel electrophoresis (DGGE) (15). Subsequently, clones or bands on DGGE gels can be used in sequencing and in analyzing phylogenetic diversity. Since in most cases the ultimate goal is to obtain a picture of microbial community composition that is not affected by selective cultivation, the protocols include the implicit assumption that PCR amplification proceeds without major bias; that is, numerically important organisms in the environment are expected to be represented by dominant clones in libraries or by strong bands on DGGE gels.
The following two major classes of processes may skew template-to-product ratios based on theoretical modeling of PCR: (i) PCR selection and (ii) PCR drift (29). The first class comprises all mechanisms which inherently favor the amplification of certain templates due to properties of the genes, of their flanking sequences, or of the overall genome. Potentially important contributors to PCR selection among these mechanisms are preferential denaturation due to overall low GC content, higher binding efficiency of GC-rich permutations of degenerate primers, differential accessibility of rRNA genes within genomes, and correlation between amplification probabilities and gene copy numbers within genomes. The second type of bias is assumed to be caused by stochastic variation in the early cycles of the reaction (when amplification still proceeds largely from the genomic templates), and its outcome should therefore not be reproducible in replicate PCR amplifications. Bias in amplification from mixtures of 16S ribosomal DNAs (rDNAs) has only recently begun to be explored experimentally (4, 6, 21, 27).
In the most extensive study to date on bias in amplification of 16S rDNAs, Suzuki and Giovannoni (27) demonstrated that the importance of different bias-causing mechanisms may change over the course of an amplification. These authors used combinations of different primers to amplify pairs of PCR products. Under the conditions used, primer pairs with high amplification efficiency resulted in reactions entering the plateau phase (i.e., products arriving at saturation concentrations [10−7 M]) (23). Since templates which reach saturation concentrations essentially stop amplifying while others are still increasing (23), a kinetic bias towards 1:1 product ratios independent of the starting template concentrations was observed (27). However, primer pairs with lower amplification efficiency resulted in product concentrations below the saturation concentrations, and depending on the template pair, either the expected product ratio or bias was observed, for which no explanation could be given (27). Similarly, in an attempt to evaluate the effect of 16S rRNA gene copy number and genome size, Farrelly et al. (6) noted bias in amplifications from template pairs which could not be explained.
In the present study we investigated the potential extent, causes, and minimization of bias in PCR amplification from mixtures of 16S rDNA templates. Amplification of full-length genes with commonly used universal, degenerate primers was used to mimic realistic conditions in molecular diversity studies. In all experiments, bias due to template saturation (27) was avoided by adjusting reaction parameters so that the plateau phase was not reached. Initially, mixtures of genomic DNAs of different species were used to determine the relative contribution of PCR selection and PCR drift to bias in template-to-product ratios. The effect of varying the ratios of rDNA templates in reaction mixtures (gene dosage) and the effect of different AT-GC contents of the degenerate primers were investigated as potential major causes of PCR selection. In addition, the effect of the relative amount of a specific template in a complex mixture (genome dosage) on amplification efficiency was tested by adding different amounts of genomic DNA of one species to nucleic acids extracted from a natural community. Based on these experiments, we investigated alterations of reaction protocols that may reduce bias in amplification of multitemplate mixtures.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Vibrio fischeri ES1114 and Vibrio anguillarum 775 were generous gifts from Edward Ruby (University of Hawaii). Cells were grown at room temperature in SWT medium containing (per liter) 5 g of Bacto Tryptone (Difco), 3 g of yeast extract (Difco), 3 ml of glycerol, 700 ml of seawater, and 300 ml of distilled water. Escherichia coli INVαF′ was purchased from Invitrogen and was grown in Luria-Bertani broth (22).
Nucleic acid extraction.
DNAs from both Vibrio strains and from E. coli were extracted and purified by the method of Jarrell et al. (8), with slight modifications (17). Purified DNA from Bacillus subtilis RL202 was a generous gift from Len Duncan (Harvard University). Community nucleic acids were extracted from a coastal microbial community (Woods Hole, Mass.). Cells from 20 liters of prefiltered (pore size, 1 μm) water were concentrated with a 0.22-μm-pore-size Micro Culture Capsule filter (Gelman Sciences) in September 1994 provided by Meredith Hullar (Harvard University). Cells were lysed by incubation with sodium dodecyl sulfate and proteinase K freeze-boiling cycles as described previously (16) followed by standard phenol-chloroform extraction (22).
PCR primers.
Primers 27F and 1492R (Table 1) were used for amplification from genomic DNA and from PCR products in experiments performed to determine the causes and extent of bias; these primers are frequently used in molecular diversity studies because they result in a nearly full-length 16S rDNA product and are considered universal for the domain Bacteria and for the domains Archaea and Bacteria, respectively (11). Each primer contains a single degeneracy, which is between A and C at position 19 (E. coli numbering) in primer 27F and between T and C at position 1497 in primer 1492R (Table 1).
TABLE 1.
Designations and targets of amplification primers and hybridization probes
| Primer or probe | Sequencea | Positionsb | Target |
|---|---|---|---|
| Primers | |||
| 27F | AGAGTTTGATC(C/A)TGGCTCAG | 8–27 | (Eu)bacterial 16S rDNA |
| 27F(A) | AGAGTTTGATC A TGGCTCAG | 8–27 | (Eu)bacterial 16S rDNA containing T at position 19 |
| 27F(C) | AGAGTTTGATC C TGGCTCAG | 8–27 | (Eu)bacterial 16S rDNA containing G at position 19 |
| 1492R | TACGG(C/T)TACCTTGTTACGACTT | 1492–1513 | (Eu)bacterial 16S rDNA |
| 1492R(T) | TACGG T TACCTTGTTACGACTT | 1492–1513 | (Eu)bacterial 16S rDNA containing T at position 1497 |
| 1492R(C) | TACGG C TACCTTGTTACGACTT | 1492–1513 | (Eu)bacterial 16S rDNA containing C at position 1497 |
| Probes | |||
| Bsu | CGCGGGTCCATCTGTAAGTG | 219–238 | B. subtilis 16S rDNA |
| Van | CCTAGGCATATCCTGACGCG | 219–238 | V. anguillarum 16S rDNA |
| Vfi | CCTGGGCTAATCTTAGCGCG | 219–238 | V. fischeri 16S rDNA |
| Eco | CTTTACTCCCTTCCTCCCCG | 443–462 | E. coli mutagenized 16S rDNA with C [Eco(GC)] or A/T [Eco(AT)] in priming regions |
| EcoM | CTTTACTGGGAAGCTCCCCG | 443–462 | E. coli mutagenized 16S rDNA with A/T in priming region and nucleotides 450 to 455 exchanged [Eco(AT)m] |
| Eubc | GCTGCCTCCCGTAGGAGT | 338–355 | (Eu)bacterial 16S rDNA |
The nucleotides in boldface type are degenerate nucleotides in amplification primers and their permutations.
E. coli numbering.
Probe Eub is identical to S-D-Bact-0338-a-A-18 (1).
PCR templates.
Amplifications with primers 27F and 1492R were conducted with the following template mixtures: (i) three bacterial genomic DNAs, (ii) purified PCR products of different mutagenized E. coli 16S rDNAs, and (iii) V. anguillarum DNA and nucleic acids extracted from the aquatic community.
(i) Genomic DNAs.
In experiments performed to determine PCR drift and selection and reduction of bias, the template used consisted of a mixture of equal amounts (as determined by spectrophotometry) of total genomic DNAs of B. subtilis, V. anguillarum, and V. fischeri.
(ii) Mutagenized E. coli 16S rDNAs.
The effects of primer degeneracies and gene dosage were determined with pairwise mixtures of three mutagenized E. coli 16S rDNA templates, Eco(GC), Eco(AT), and Eco(AT)m. Eco(GC) and Eco(AT) differed only at the single degenerate position in each of the priming sites for 27F and 1492R (Table 1). Eco(AT)m differed from Eco(AT) and from Eco(GC) as follows: six nucleotides in the middle of the molecule were altered by site-directed mutagenesis. Eco(GC) and Eco(AT)m templates were mixed in equal amounts in the primer degeneracy experiments, and Eco(AT) and Eco(AT)m were mixed at 1:1, 1:5, 1:10, and 1:20 ratios in the gene dosage experiments.
The three mutagenized templates were generated as follows. Nondegenerate versions of primers 27F and 1492R (Table 1) were used in two combinations, 27F(A)-1492R(T) and 27F(C)-1492R(C). The resulting PCR products, Eco(AT) and Eco(GC), were cloned. Subsequently, in a cloned Eco(AT) 16S rDNA fragment, nucleotides 450 to 455 were changed to their complements by using a PCR site-directed mutagenesis protocol (7a). First, a reaction was carried out with a mutagenesis primer (TTAACTTTACTGGGAAGCTCCCCGCTGA; positions 439 to 466) and primer 27F(A) (Table 1), which created a 458-bp product. This product was gel purified and used as a primer in a second reaction together with primer 1492R(T) (Table 1). The mutagenized 16S rDNA was cloned and is referred to below as Eco(AT)m.
The effects of primer degeneracies and gene dosage were tested by using PCR products amplified from the Eco(GC), Eco(AT), and Eco(AT)m clones as templates. These products were generated with primers M13 reverse and M13(−40), which resulted in 16S rRNA gene fragments flanked by roughly 200 bp of plasmid-derived sequence, allowing purification of amplification products from templates before blotting and quantitative analysis. In all cases, templates were generated from clones in which the 16S rDNAs had the same orientation to avoid any potential influence of different flanking sequences on primer hybridization during the PCR. PCR products were quantified by comparison with standards on an agarose gel by using the Eagle Eye gel imaging and quantification system (Stratagene).
(iii) V. anguillarum and community DNA.
The influence of the relative amount of a specific template in a complex mixture on product distribution was tested with a mixture of V. anguillarum DNA and nucleic acids extracted from a natural community. Both types of nucleic acids were quantified spectrophotometrically, and the template mixture was generated by adding V. anguillarum DNA to final concentrations of 10, 1, and 0.1%.
PCR conditions.
All reactions were performed with a Twin Block System and a Power Block I System thermal cycler (Ericomp). The reaction volume was either 100 or 25 μl, and each reaction mixture contained 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 1% Triton X-100), each deoxynucleoside triphosphate at a concentration of 200 μM, 2.0 mM MgCl2, 5% acetamide (in reactions in which genomic DNA was the template), each primer at a concentration of 100 pM, and 0.025 U of Taq polymerase (Promega) per μl. Acetamide was included in the reaction mixtures containing genomic DNAs because it has been reported to increase denaturation of templates with high GC contents during the PCR temperature cycles (21). For replicate PCR amplifications, aliquots were taken from a single master mixture. The template concentration used was 0.1 ng of total genomic DNA per μl or 5 pg of purified PCR product per μl in 25-cycle amplifications and 5 ng of total genomic DNA per μl in 5- and 10-cycle amplifications.
All amplifications started with an initial denaturation step consisting of 94°C for 3 min; this was followed by cycles consisting of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C. To avoid bias associated with product saturation (27), the amounts of product accumulated after different numbers of cycles with each of the different template combinations were determined by spectrophotometry and by liquid scintillation counting of incorporated 32P-labeled dCTP (12). This showed that after 25 cycles the products were still being produced exponentially (data not shown). Thus, the number of cycles used in the experiments designed to identify the extent and possible mechanisms of bias was 25. In other experiments, the numbers of cycles were decreased to 5 and 10 in order to determine the effect of fewer cycles on product bias. To avoid false priming of the genomic templates at low temperatures (3, 5), a type of hot-start PCR was used. In each amplification tube, a lower reservoir containing water, buffer, and enzyme was created by sealing it off with 50 μl of molten Paraplast wax. After solidification of the wax, the rest of the reagents were added to the tube and sealed with an additional 50 μl of molten wax. This wax had a melting point of 56°C and floated to the top of the liquid during the initial denaturation step of the amplification.
Oligonucleotide probe design, labeling, and determination of Td and specificity.
Specific oligonucleotide probes for the different bacterial species were designed based on an alignment obtained from the Ribosomal Database Project (RDP) (13). For differentiation of the Vibrio species and B. subtilis, a 20-nucleotide stretch was identified (positions 219 to 238 [E. coli numbering]) which had the same GC content (60%) (Table 1). Probes Eco and EcoM were designed to differentiate the native E. coli 16S rDNA template from mutagenized versions (Table 1).
The midpoint dissociation temperatures (Tds) of oligonucleotides were determined experimentally to optimize the relationship between signal strength and specificity of the probes as described by Raskin et al. (20), with modifications (18). Each nucleic acid type was blotted in duplicate with a Minifold I dot blotter (Schleicher & Schuell) onto Zetaprobe nylon membranes (Bio-Rad) by using the alkaline denaturation method performed according to the instructions supplied. The oligonucleotide probes were labeled with polynucleotide kinase (Gibco BRL) so that they contained 5 × 106 cpm/pmol and were purified with NenSorb 20 cartridges (Du Pont NEN). Hybridizations were performed at 30°C overnight in the recommended buffer (Zetaprobe) by using the specific probes. Subsequently, the membranes were washed twice for 15 min at the same temperature. Individual dots were then cut out and washed in 2 ml of wash buffer in 7-ml scintillation vials which had been prewarmed in water baths at temperatures ranging from 20 to 65°C at 2 to 5°C intervals. After 10 min the membranes were removed, and the amounts of radioactivity in the wash solutions and on the membranes were determined by liquid scintillation counting. The Tds were calculated by dividing the counts remaining on each membrane by the total counts for each temperature point. The resulting values were then plotted as percentages of probe washed off versus temperature, and the 50% value was considered the Td.
Probe specificity was determined (i) by hybridizing the Van, Vfi, and Bsu probes with a blot containing both genomic DNAs and PCR-generated 16S rDNAs of the three species and (ii) by hybridizing the Eco and EcoM probes with a blot containing PCR products of native and mutagenized E. coli 16S rDNAs. The blots were hybridized and washed by using the conditions specified above except that the 15-min specific wash was at the Td only. Specific labeling and background were determined by exposing membranes on Reflection NEF-496 (Du Pont) X-ray film and by quantification of the radioactivity by using a Fujix BAS200 phosphorimager and BAS2000 Image File Manager 2.1 analysis software.
Quantitative dot blot hybridizations.
Quantitative dot blot hybridizations were carried out to determine 16S rDNA template and product ratios. Membranes were blotted with template and/or PCR product combinations and hybridized with specific probes as follows: (i) for PCR drift and PCR selection tests, three identical blots containing a mixture of three genomic templates (V. anguillarum, V. fischeri, and B. subtilis), five individual replicate amplifications (PCR 1 to 5), and a mixture of 10 replicate amplifications (PCR mixture) hybridized with probes Bsu, Van, and Vfi; (ii) for primer degeneracy tests, two identical blots containing a 1:1 mixture of Eco(GC) and Eco(AT)m mutagenized E. coli 16S rDNA templates and five replicate amplifications after 15, 25, and 35 cycles hybridized with probes Eco and EcoM; (iii) for gene dosage tests, two identical blots containing five replicate amplifications from 1:1, 1:5, 1:10, and 1:20 template mixtures of Eco(AT) and Eco(AT)m mutagenized E. coli 16S rDNA fragments hybridized with probes Eco and EcoM; (iv) for genome dosage tests, two identical blots containing three replicate amplifications from nucleic acids extracted from a natural community to which V. anguillarum DNA had been added at concentrations corresponding to 0.1, 1, and 10% of the total amount hybridized with probes Van and Eub; and (v) for a test of reduced cycle numbers, three identical blots containing the original three-genomic-template mixture (V. anguillarum, V. fischeri, and B. subtilis) and mixtures of 10 replicate amplifications after 5 and 10 cycles hybridized with probes Bsu, Van, and Vfi.
Before blotting, PCR products were purified from templates on 0.8% agarose gels (Qiaquick; Qiagen). In experiments in which 10 replicate PCR amplifications were mixed, the products of the reactions were concentrated individually by using Microcon 100 filtration devices (Amicon), purified, and eluted from 0.8% agarose gels. Then, after their concentrations were determined by spectrophotometry, subsamples of each of the 10 PCR amplifications containing equal amounts of DNA were mixed and blotted (PCR mixture).
In the experiments performed to explore PCR drift and PCR selection, three membranes containing nine replicate dots of each of the three classes of nucleic acids (template mix, PCR 1 to 5, and PCR mixture) were blotted. In the experiments designed to test the influence of reduced cycle numbers, three identical membranes containing eight replicate dots of the template mixture and the product mixture from 10 replicate PCR carried out for 5 and 10 cycles were blotted. This resulted in standard deviations for the replicate dots that were less than 5% of the mean for all samples spotted with nine and eight replicates. In all other experiments, two membranes were blotted with three replicate dots per class of nucleic acid. For 37 of the 76 samples in which three replicates were used the standard deviations were less than 5% of the mean. For most of the rest of the samples the standard deviations were less than 10%; the only exceptions were two samples which had standard deviations of 12 and 16%.
All hybridizations were carried out as described above, using a 10-fold molar excess of probe over target and 1 ml of hybridization buffer per dot. Bound probe was quantified by phosphorimaging, and average signals were calculated for each specific template or PCR product in the different classes of nucleic acids (e.g., B. subtilis in template mixture, in PCR mixture, or in replicate PCR). Subsequently, pairwise ratios of the averages (e.g., B. subtilis signal over V. fischeri signal in PCR mixure) were determined. To facilitate interpretation, the PCR product signals were normalized to the template mixture signal by calculating a constant factor for each template pair. For example, the ratio of 1.1 obtained from the hybridization intensities of the genomic mixture with the B. subtilis-V. anguillarum pair was multiplied by 0.91 to normalize it to 1.0. Subsequently, all other ratios (PCR mixture, PCR 1 to 5) determined for this species pair were multiplied by the same factor. In gene and genome dosage experiments in which PCR product ratios were compared to one another, signal ratios were corrected for differences in specific activities of the hybridization probes. Standard deviations of the ratios were calculated from all possible combinations of denominators and numerators in a given experiment and generally were less than 10% of the average.
Nucleic acid sequencing.
The 16S rRNA genes of all three species were sequenced partially to ensure sequence identity between all of the strains used in this study and the strains represented in the RDP. Three clones of each species were sequenced by using primer 519R (11), which covers two of the most variable regions in the 16S rRNA molecule. Furthermore, sequences in the 1492R priming region of the two Vibrio species were determined since they were not available in the databases. A 16S rDNA fragment was amplified by PCR as specified above, except that primer 1525R (11) was used in place of 1492R. The resulting product was purified and cloned into the PCR II vector (Invitrogen, San Diego, Calif.). Three clones of each of the Vibrio species were sequenced by using primers 1406F and 1525R (11).
All of the clones resulting from the in vitro mutagenesis experiments were checked by sequencing for the correct, expected sequence by using M13 primers or internal 16S rDNA primers (11).
RESULTS
Td and specificity of oligonucleotides.
The experimentally determined Tds for oligonucleotide probes Bsu, Van, Vfi, Eco, and EcoM were 52.5, 53.0, 46.2, 49.0, and 44.1°C, respectively (data not shown). These temperatures were used in the high-temperature wash step of the quantitative dot blot hybridizations.
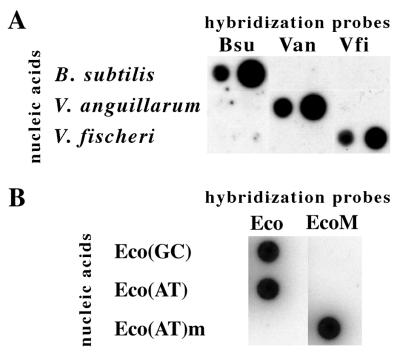
Under the conditions used, all of the probes reacted specifically with their target molecules (Fig. 1). No background was observed with probe-target mismatches with either the different genomic DNAs or the PCR. X-ray films remained completely clear after 5-h exposures. Likewise, quantification by phosphorimaging yielded background values only for the exposure times used for the quantitative analysis (data not shown).
FIG. 1.
Dot blot analyses showing the specificity of the oligonucleotide probes for their targets. (A) Genomic DNAs (left dots) and PCR-amplified 16S rDNAs (right dots) of B. subtilis, V. fischeri, and V. anguillarum were blotted together on three replicate membranes and hybridized with the specific probes Bsu, Van, and Vfi, respectively. (B) PCR-amplified 16S rDNAs of mutagenized plasmids Eco(GC), Eco(AT), and Eco(AT)m were blotted on two separate membranes and hybridized with the specific probes Eco and EcoM, respectively. The electronic image was taken from X-ray film exposed for 5 h.
Quantification of PCR bias.
The underlying rationale of the initial experiments was that if bias in PCR amplifications is due to stochastic fluctuations (PCR drift), it would not be reproducible in replicate reactions, whereas if it is a property of the templates (PCR selection), the same pattern of bias would be observed in individual amplifications. When signal ratios for the different species pairs were compared, the ratios of the genomic templates never corresponded to the ratios of the PCR products (Table 2). If all three templates had been amplified with the same efficiency, all of the ratios should have been similar to the ratios of the genomic DNAs. The error due to between-dot variability in the hybridizations was small because a large number of replicates were blotted for all treatments. The largest bias was observed for B. subtilis 16S rDNA, which was amplified with much higher efficiency than the DNAs of the two Vibrio species. When the two Vibrio templates were compared, V. anguillarum DNA was amplified less than V. fischeri DNA and overall was the least-well-represented DNA (Table 2). A detailed comparison also revealed variation among the ratios of the individual PCR products (Table 2). Most values remained within ±0.3 unit of the PCR mixture. However, some extreme cases occurred, as illustrated by the B. subtilis-V. fischeri pair in PCR 5, which differed 1.2-fold.
TABLE 2.
Comparison of 16S rDNA gene template and PCR product ratios in simultaneous PCR amplifications of three bacterial genomes with degenerate primersa
| Prepn | Species ratiosb
|
||
|---|---|---|---|
| B. subtilis/ V. fischeri | B. subtilis/ V. anguillarum | V. fischeri/ V. anguillarum | |
| Genomic mixture | 1.0 ± 0.06 | 1.0 ± 0.08 | 1.0 ± 0.07 |
| PCR mixture | 2.3 ± 0.18 | 3.2 ± 0.29 | 1.4 ± 0.13 |
| PCR 1 | 2.3 ± 0.12 | 3.7 ± 0.18 | 1.6 ± 0.09 |
| PCR 2 | 2.2 ± 0.13 | 3.2 ± 0.21 | 1.4 ± 0.09 |
| PCR 3 | 2.5 ± 0.17 | 3.2 ± 0.31 | 1.3 ± 0.12 |
| PCR 4 | 2.0 ± 0.15 | 3.5 ± 0.35 | 1.8 ± 0.15 |
| PCR 5 | 3.5 ± 0.25 | 3.5 ± 0.30 | 1.0 ± 0.08 |
The data were generated by dot blotting and hybridization with species-specific probes of equal amounts of template DNAs from the three species (genomic mixture), a mixture of 10 replicate PCR amplifications (25 cycles) performed with the genomic mixture (PCR mixture), and a subsample of 5 of the 10 replicate PCR amplifications (PCR 1 to 5).
Means ± standard deviations were calculated from data for nine replicate dots for each of the samples which was hybridized with the specific probes and quantified by phosphorimaging. Subsequently, the PCR product signals were normalized to the genomic mixture signal by using a constant factor for each species pair (see text).
The GC content of the priming region had a significant effect on the efficiency of amplification of the templates studied. In 1:1 mixtures of a pair of mutagenized E. coli 16S rDNAs [Eco(GC) and Eco(AT)m], the template with the GC-rich permutation in the priming site was consistently amplified better than the AT-rich permutation (Table 3). This unequal effectiveness of amplification increased with cycle number (Table 3); the average product ratios were 1.4, 1.7, and 2.2 after 15, 25, and 35 cycles, respectively.
TABLE 3.
Effect of primer degeneracies on PCR product ratiosa
| Prepn | Eco(GC)/Eco(AT)m ratiob
|
||
|---|---|---|---|
| 15 cycles | 25 cycles | 35 cycles | |
| Template mixture | 1.0 ± 0.04 | 1.0 ± 0.04 | 1.0 ± 0.04 |
| PCR 1 | 1.4 ± 0.16 | 1.5 ± 0.06 | 2.3 ± 0.10 |
| PCR 2 | 1.4 ± 0.14 | 1.8 ± 0.11 | 2.3 ± 0.07 |
| PCR 3 | 1.3 ± 0.11 | 1.7 ± 0.07 | 2.5 ± 0.20 |
| PCR 4 | 1.4 ± 0.26 | 1.9 ± 0.07 | 1.9 ± 0.14 |
| PCR 5 | 1.3 ± 0.16 | 1.8 ± 0.09 | 2.0 ± 0.17 |
The data were generated by dot blot hybridization by using the template-specific probes Eco and EcoM and mixtures containing equal amounts of templates Eco(GC) and Eco(AT)m. PCR products were amplified from the same template mixtures for 15, 25, and 35 cycles.
Means ± standard deviations were calculated from data for three replicate dots for each of the samples which was hybridized with the specific probes and quantified by phosphorimaging.
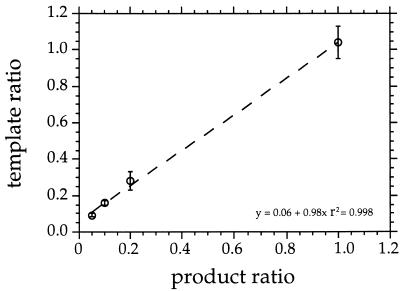
Gene dosage alone had no discernible effect on product ratios (Fig. 2). Two mutagenized E. coli 16S rDNA templates, Eco(AT) and Eco(AT)m, which differed only in the six nucleotides recognized by probes Eco and EcoM, respectively, were mixed at ratios of 1:1, 1:5, 1:10, and 1:20. Least-squares linear regression analysis of the average product ratios for three replicate amplifications per ratio indicated that amplification was proportional to template representation (r2 = 0.998) (Fig. 2).
FIG. 2.
Effect of gene dosage as determined by amplification with different template ratios of Eco(AT) and Eco(AT)m and quantification of product ratios by quantitative dot blotting with probes Eco and EcoM. Regression analysis showed that the relationship between template and product ratios was linear. The vertical bars indicate standard deviations.
Genome dosage also did not influence product ratios to a large and consistent extent under the conditions used (Table 4). V. anguillarum genomic DNA was added at concentrations equivalent to 10, 1, and 0.1% of the total DNA to DNA extracted from a natural microbial community and was amplified with the universal primers. The product ratios determined with the specific probe Van and the universal (eu)bacterial probe Eub (Table 1) (1) indicated that proportional amplification occurred (Table 4). The values for the product ratios at the 10% dilution were approximately 10 times higher than the values at the 1% dilution (Table 4). However, the coefficient of variation of the product ratios for the three replicate amplifications increased from 3% at the 10% dilution to 12% at the 1% dilution, indicating that there was a lower level of reproducibility at the lower template concentration. Ratios for the 0.1% dilution could not be determined because there was not enough V. anguillarum product.
TABLE 4.
Reproducibility of PCR amplification of a single template in a complex mixture of nucleic acids from a natural communitya
| Prepn |
V. anguillarum/(eu)bacterial 16S rDNA ratiob
|
||
|---|---|---|---|
| 10% V. anguillarum | 1% V. anguillarum | 0.1% V. anguillarum | |
| PCR 1 | 0.156 ± 0.008 | 0.017 ± 0.001 | NDc |
| PCR 2 | 0.147 ± 0.011 | 0.020 ± 0.000 | ND |
| PCR 3 | 0.153 ± 0.005 | 0.016 ± 0.001 | ND |
The data were generated by dot blot hybridization by using the V. anguillarum-specific probe Van219 and the universal (eu)bacterial probe S-D-Bact-0338-a-A-18. PCR products were amplified from nucleic acids extracted from a natural community to which known amounts of V. anguillarum DNA were added.
Means ± standard deviations were calculated from data for three replicate dots for each of the samples which was hybridized with the specific probes and quantified by phosphorimaging.
ND, not detected (below detection limit).
To test the hypothesis that accumulation of bias is largely template inherent and additive with every cycle, the effect of decreasing the number of cycles on PCR product ratios was examined. The same three-species mixture was used in these experiments. While the same pattern of overamplification was observed, the differences between template ratios and PCR mixture ratios were considerably smaller in all cases (Table 5). Indeed, the product ratio approached the template ratio when the numbers of cycles were 10 and 5 (Table 5).
TABLE 5.
Effect of lower number of cycles of the PCR on skewing of product ratiosa
| Prepn | Species ratiosb
|
||
|---|---|---|---|
| B. subtilis/ V. fischeri | B. subtilis/ V. anguillarum | V. fischeri/ V. anguillarum | |
| Genomic mixture | 1.0 ± 0.05 | 1.0 ± 0.06 | 1.0 ± 0.05 |
| PCR mixture, 10 cycles | 1.7 ± 0.10 | 2.2 ± 0.10 | 1.3 ± 0.04 |
| PCR mixture, 5 cycles | 1.3 ± 0.15 | 1.5 ± 0.14 | 1.1 ± 0.05 |
The data were generated by dot blotting and hybridization with species-specific probes containing equal amounts of template DNA from the three species (genomic mixture) and a mixture of 10 replicate PCR amplifications (5 and 10 cycles).
Means ± standard deviations were calculated from data for eight replicate dots for each of the samples which was hybridized with the specific probes and quantified by phosphorimaging.
DISCUSSION
The results of this study indicate that PCR product ratios can be significantly biased in standard amplifications of mixed templates (Table 2). Under the experimental conditions used, mechanisms summarized under PCR drift appeared to cause little bias, but occasional extremes occurred (e.g., PCR 5 in Table 2). PCR selection emerged as the force driving unequal amplification of templates, and different binding energies of degenerate primers were a major contributor (Table 3). Considerable bias was observed even though the effects of PCR selection may have been counterbalanced to a large extent by kinetic bias as observed by Suzuki and Giovannoni (27) (i.e., progressive reduction in the amplification efficiency of specific products). Overall, the results suggest that product distributions are reproducible despite being biased in an a priori unpredictable fashion. In addition, the effects of PCR selection can be reduced by performing short-cycle PCR amplifications with high template concentrations (Table 5).
PCR drift.
The observed deviations could be caused by (i) true PCR drift rooted in the reaction mechanism and (ii) errors perceived as PCR drift but really introduced by the experimenter. A low template concentration in the early cycles may lead to stochastic fluctuation in the interactions of PCR reagents, especially primer annealing to the genomic template. In the experiments presented here, PCR drift may actually have been minimized because templates were added at relatively high starting concentrations (Table 2). However, in other investigations performed to test the effect of low template concentrations, product distribution and yield exhibited very low reproducibility, and some specific products were missing from some replicate amplifications (4, 9, 14). Perceived PCR drift may stem from pipetting errors between replicates, from variations in the thermal profiles of different wells, or from unequal ramping temperatures in thermal cyclers, which may affect templates differentially. While the first possibility was minimized by using master mixtures, we have no means of differentiating the second possibility from true PCR drift. However, independent of the causes, the data emphasize the danger of using a single PCR amplification for analysis of microbial communities by cloning or DGGE because the variation between replicates is unpredictable and can be large (Table 2).
PCR selection.
PCR selection may be caused to a large extent by differences in the GC content at degenerate positions in the primer target sites in the 16S rDNAs. This was indicated by the occurrence of bias in the product pairs of the three species and of the E. coli 16S rDNAs, which were mutagenized to differ essentially only in the amplification sites (Tables 2 and 3). The consistent overamplification of the B. subtilis template may also have been largely due to higher primer affinity for the priming region due to higher GC content. Inspection of the sequences in the RDP database and partial sequencing indicated that at both degenerate positions of the two primers B. subtilis has a G, whereas the two Vibrio species have an A or T (Fig. 3). This sequence variation is reflected in the widely used 16S rDNA amplification primers 27F and 1492R (11), each of which contains a single degeneracy (between A and C and between T and C, respectively) (Table 1; Fig. 3). Because both G and C form a triple hydrogen bond, the melting temperatures of the GC-rich permutations of both primers are theoretically about 2°C higher than the AT-rich permutation. Thus, at each annealing step a greater proportion of the templates containing GC complements in the priming region should hybridize to their matched primers. The alternative explanation for the observed continuous buildup of bias (Tables 2 and 3) is that AT-containing primers are more effective than GC-containing primers in forming mismatched hybrids. However, due to the much lower thermal stability of mismatches, this possibility appears less likely. The unexplained bias observed in other studies (6, 27) may also have been due to primer degeneracy effects, but interpretation of the data is hampered by a lack of sequence information in the databases for the templates used.
FIG. 3.
Alignment of the sequences of universal amplification primers 27F and 1492R and their target regions on the 16S rRNA genes of B. subtilis, V. fischeri, and V. anguillarum. The two primers each contain a single degeneracy (between C and T and between C and A, respectively). In the B. subtilis gene both priming sites contain a G at the degenerate site, which most likely results in a higher melting temperature for the primer-target duplex than the melting temperature for the two Vibrio genes, which contain an A and a T at the two positions.
PCR bias due to gene and genome dosage effects was not detected (Fig. 2, Table 4). Farrelly et al. (6) have suggested that such dosage effects cause bias (6) and have argued that 16S rDNAs from species with higher rrn operon numbers should be amplified better than 16S rDNAs from species with lower rrn operon numbers. Indeed, a similar explanation for the overamplification of the B. subtilis 16S rDNA from the mixture containing three species (Table 2) could not be ruled out a priori. Total genomic DNAs were mixed at a ratio of 1:1:1, and both operon number and genome size could have skewed the product distribution in favor of B. subtilis. This species has 10 rrn operons and a genome size of 4,165 kb (6); in contrast, V. fischeri has only 8 rrn operons (10) and a slightly larger genome (4,550 kb) (24) (no rrn operon data are available for V. anguillarum). However, a strong effect of gene copy number or genome copy number in amplification was not supported by our experiments. Regression analysis of the products amplified with the different template ratios gave no indication of deviation from a linear relationship (Fig. 2). Similarly, different amounts of the V. anguillarum genome in a complex community resulted in no major or consistent amplification bias (Table 4). Whether equally high levels of reproducibility occur with templates present at much lower levels, such as V. anguillarum added at a concentration equivalent to 0.1% of the total community DNA concentration, could not be tested by the quantitative hybridization approach used here.
Two lines of evidence point to the existence of additional factors that cause PCR selection in addition to primer degeneracies. First, when 25 cycles were used, the average B. subtilis/V. fischeri ratio was 2.3 (Table 2), whereas the average Eco(GC)/Eco(AT)m ratio was only 1.7 (Table 3), suggesting that primer degeneracies accounted for only about one-half of the overamplification. Second, there was also bias with the closely related Vibrio species (Table 2). Both of these species have the same sequence in the priming sites (Fig. 3), and their 16S rDNAs are 93.9% identical, yet the V. fischeri template was consistently amplified better (Tables 2 and 5). Although it is impossible to determine a definitive cause, a number of additional factors may have contributed to the bias observed. Sequence regions immediately adjacent to the priming sites may have influenced the hybridization efficiency of the primers, as suggested by Td studies of universal oligonucleotide probes with different templates (30). Single strands of 16S rDNA are potentially prone to secondary-structure formation during product extension, which may cause the polymerase to fall off. Furthermore, differences in the GC contents of the 16S rDNA templates or the whole genomes may lead to differential denaturation of templates. However, in the case of the vibrios, the GC contents are only slightly different; V. fischeri genomes contain 39 to 41% GC, whereas V. anguillarum genomes contain 43 to 44% GC. If overall differences in GC content, as well as the specific priming sites, are a cause of product bias, this problem may be exaggerated in natural samples, in which the differences between genomes typically far exceed the 5% maximum difference between the two Vibrio species examined.
Reduction of PCR bias.
The effects of PCR bias were decreased by (i) mixing several replicate PCR amplifications and (ii) reducing the numbers of cycles. The five replicate amplifications which were assayed individually showed good agreement with the PCR mixture, which was a composite of 10 replicate amplifications (Table 2). Thus, as suggested by Chandler et al. (4), pooling replicates may be an effective way to decrease variation in the amplification process; this is especially true for those templates which are present at low initial concentration in the sample. Reduction of the number of cycles had the most dramatic effect (Table 5). Overamplification of the Bacillus template was reduced to ratios of 1.7 and 2.2 with 10 cycles and decreased to ratios of 1.3 and 1.5 with 5 cycles (Table 5). The bias between the two Vibrio species was also reduced, and the ratio was close to the original template ratio (Table 5).
Recommendations and conclusions.
The following recommendations for limiting bias in PCR amplifications emerged from the data presented above. First, whenever possible, degeneracies should be avoided when universal primers are designed. Second, to increase reproducibility between replicates, amplifications should be carried out by using high template concentrations. Third, to minimize PCR drift, several replicate PCR amplifications should be combined. Fourth, to diminish PCR selection, the smallest number of cycles should be used. Since cloning utilizes only a very small amount of DNA, 10 cycles or even 5 cycles may be enough if a high template concentration (>500 ng of genomic DNA per tube) is used. In a PCR which started with 500 ng of template, after 10 cycles a discrete band on a standard agarose gel was easily detectable with a subsample as small as 5 μl (unpublished observations). Alternatively, combining several replicates should yield enough product to be analyzed by electrophoretic methods, such as DGGE.
Overall, the results indicate that PCR analysis is a method with relatively high precision but potentially low accuracy; that is, product distributions are reproducible, but template-inherent factors may lead to significant deviations from template distributions. These results support the validity of quantitative PCR approaches, in which internal standards are added, but show the limitations of multitemplate amplifications. Even in the simple three-species community tested, relatively large PCR selection was observed. How PCR selection will skew amplifications from natural communities with potentially thousands of species (28) and with templates that may be even more prone to overamplification cannot be predicted at this time. In addition, estimates of cell numbers based on amounts of products are skewed by the highly variable operon numbers in different species (6, 10). This emphasizes the fact that quantitative interpretation of PCR-based results should still be viewed with caution. In the future, it will be important to explore PCR bias further to arrive at measures which result in confidence in product distributions in molecular diversity studies of natural communities. Currently, quantitative oligonucleotide probing (18, 19, 25) and in situ hybridization (2) still provide ecologically more meaningful measures of the relative importance of specific microorganisms.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation and the Office of Naval Research to C.M.C.
We thank Christopher Harbison for help with sequencing, Charles Harvey and Dennis McLaughlin for help with statistics, Stephen Giovannoni, Daniel Distel, and Christian Luschnig for critical discussions, and Edward Ruby and Len Duncan for providing the strains used in this study.
REFERENCES
- 1.Alm W E, Oerther D, Larsen N, Stahl D A, Raskin L. The oligonucleotide database project. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnheim N, Erlich H. Polymerase chain reaction strategy. Annu Rev Biochem. 1992;61:131–156. doi: 10.1146/annurev.bi.61.070192.001023. [DOI] [PubMed] [Google Scholar]
- 4.Chandler D P, Fredrickson J K, Brockman F J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–483. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 5.Erlich H A, Gelfand D, Sninsky J J. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 6.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 7a.Hektor, H. Personal communication.
- 8.Jarrell K F, Faguy D, Herbert A M, Kalmkoff M L. A general method of isolating high molecular weight DNA from methanogenic archaea (archaebacteria) Can J Microbiol. 1991;38:65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- 9.Karrer E E, Lincoln J E, Hogenhout S, Bennet A B, Bostock R M, Martineau B, Lucas W J, Gilchrist D G, Alexander D. In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA. 1995;92:3814–3818. doi: 10.1073/pnas.92.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerkhof L, Speck M. Ribosomal RNA gene dosage in marine bacteria. Mol Mar Biol Biotechnol. 1997;6:260–267. [PubMed] [Google Scholar]
- 11.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 12.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutter G, Boynton K A. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 1995;23:1411–1418. doi: 10.1093/nar/23.8.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norrman B, Zweifel U L, Hopkinson C S, Fry B. Production and utilization of dissolved organic carbon during an experimental diatom bloom. Limnol Oceanogr. 1995;40:898–907. [Google Scholar]
- 17.Polz M F, Odintsova E V, Cavanaugh C M. Phylogenetic relationships of the filamentous sulfur bacterium Thiothrix ramosa based on 16S rRNA sequence analysis. Int J Syst Bacteriol. 1996;46:94–97. doi: 10.1099/00207713-46-1-94. [DOI] [PubMed] [Google Scholar]
- 18.Polz M F, Cavanaugh C M. A simple method for the quantification of uncultured microorganisms in the environment based on in vitro transcription of 16S rRNA. Appl Environ Microbiol. 1997;63:1028–1033. doi: 10.1128/aem.63.3.1028-1033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sardelli A D. Plateu effect—understanding PCR limitations. Amplifications. 1993;9:1–5. [Google Scholar]
- 24.Splinter P L, Rott M A. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Physical and genetic map of Vibrio fischeri MJ1, abstr. H-212; p. 636. [Google Scholar]
- 25.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffan R J, Atlas R M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M, Giovannoni S J. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner A, Blackstone N, Cartwright P, Dick M, Misof B, Snow P, Wagner G P, Bartels J, Murtha M, Pendleton J. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst Biol. 1994;43:250–261. [Google Scholar]
- 30.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]