Abstract
Protein engineering holds immense promise in shaping the future of biomedicine and biotechnology. This Review focuses on our ongoing development of Mutexa, a computational ecosystem designed to enable “intelligent protein engineering”. In this vision, researchers will seamlessly acquire sequences of protein variants with desired functions as biocatalysts, therapeutic peptides, and diagnostic proteins through a finely-tuned computational machine, akin to Amazon Alexa’s role as a versatile virtual assistant. The technical foundation of Mutexa has been established through the development of a database that combines and relates enzyme structures and their respective functions (e.g., IntEnzyDB), workflow software packages that enable high-throughput protein modeling (e.g., EnzyHTP and LassoHTP), and scoring functions that map the sequence-structure–function relationship of proteins (e.g., EnzyKR and DeepLasso). We will showcase the applications of these tools in benchmarking the convergence conditions of enzyme functional descriptors across mutants, investigating protein electrostatics and cavity distributions in SAM-dependent methyltransferases, and understanding the role of nonelectrostatic dynamic effects in enzyme catalysis. Finally, we will conclude by addressing the future steps and fundamental challenges in our endeavor to develop new Mutexa applications that assist the identification of beneficial mutants in protein engineering.
1. Introduction
Protein engineering refers to the process of optimizing protein sequences for enhanced physical (e.g., thermal stability, solubility, and complex stoichiometry), chemical (e.g., reactivity, substrate specificity, selectivity, and substrate scope), biological, and pharmaceutical functions. Typical strategies in protein engineering include directed evolution,1−4 gene shuffling/recombination,5,6 site-directed mutagenesis,7,8 and protein truncation and fusion.9,10 Enabled by protein engineering, researchers can create enzymes to accelerate low-efficiency11−14 or even new-to-nature reactions,15,16 develop peptides with targeted therapeutic effects,17,18 innovate diagnostic tools for early stage cancer detection,19−21 and advance our understanding of fundamental life processes.22,23
A “holy grail” challenge in protein engineering is the effective identification of desired protein variants within a mutational landscape.24,25 This difficulty results from the combinatorial explosion associated with sequence mutation. Sampling mutations across only a dozen amino acid sites creates an astronomical number of variants. Despite advances in screening strategies for protein engineering, the success rate for identifying beneficial mutants is around 1% or lower.26−33De novo design of new functional proteins provides a promising alternative, but the hit rate to identify successful designs among all design candidates is similar to the chance of experimental discovery.34−37 The time-consuming, labor-intensive, and expensive process of experimental screening is largely unavoidable.
To reduce the size of mutant libraries for functional sreening, computational approaches have been augmented with protein engineering.25,38−40 These methods, such as bioinformatics,24,41 classical molecular simulations,42,43 quantum chemistry,44−47 and data-driven modeling,22,48−52 span a wide breadth of computational subfields. Each modeling strategy has a specific strength. Bioinformatics reveals the evolutionary coupling and patterns behind function-encoding sequence regions; classical molecular simulations elucidate the dynamics and conformational ensembles that constitute effective protein–protein/ligand interactions or enzyme catalysis; quantum chemistry informs the variation of electronic structure that underlies enzymatic reactions and covalent inhibition; and data-driven modeling predicts the formal, nonlinear relationships between sequence, structure, and function. Each of the aforementioned computational methods has associated strengths and weaknesses with respect to accuracy, efficiency, resolution, and reproducibility. The combination of these computational approaches promises to establish an integrative strategy that we call “intelligent protein engineering”. Similar to human intelligence that leverages both physical insight and statistical observation to guide decision making, intelligent protein engineering refers to a platform that employs physics-based molecular simulations and data-driven modeling to generate, discover, predict, and design new protein variants with enhanced chemical, mechanical, thermal, and pharmaceutical properties. Intelligent protein engineering aims to guide experimental discovery of desired protein mutants by effectively shrinking the number of mutations that have to be screened. In turn, intelligent protein design will save extensive experimental resources in the pursuit of identifying functional protein variants.
With a long-term goal to create a platform that enables intelligent protein engineering, our lab has been building a computational ecosystem called Mutexa (Scheme 1). Mutexa is short for “Alexa for mutants”, and we believe that how people engineer proteins in the future should mirror how Amazon Alexa is used today—when researchers want to create protein sequence variants with desired functions, they will simply consult our comprehensive computational platform. Mutexa integrates high-throughput computation, bioinformatics, quantum chemistry, multiscale simulations, and data-driven modeling to identify protein mutants that can enhance functions including enzyme catalysis, peptide therapeutics, and disease biomarker detection.23 Over the past three years, we have been establishing the technical foundation of Mutexa by developing 1) a database that integrates enzyme structure and function data (IntEnzyDB53,54), 2) software tools for high-throughput construction and modeling of enzymes (EnzyHTP55,56) and lasso peptides (LassoHTP57), and 3) scoring functions to predict the impact of mutations on substrate-positioning dynamics,23,58 enzymatic kinetic resolution (EnzyKR59), and peptide antimicrobial activity (DeepLasso60). The database, workflow software, and scoring functions will be discussed in detail in Sections 2, 3, and 4, respectively. In addition, we will briefly introduce applications of these tools including determining the convergence criteria for computing enzyme functional descriptors,61 investigating the distribution of protein electrostatics and cavity geometries for SAM-dependent methyltransferases,62 and understanding the role of nonelectrostatic dynamic effects in mediating enzyme catalysis.63 Finally, we will conclude by addressing the next steps and challenges in building new Mutexa applications for functional protein engineering.
Scheme 1. Overview of Mutexa, a Computational Ecosystem for Protein Engineering.
Mutexa consists of three components, including a database that integrates the structural and functional information of proteins; workflow software that allows automatic, high-throughput modeling for proteins; and a scoring function that describes sequence-structure-function relationship of proteins. Combining these three basic components, new applications for predictive modeling are being developed into Mutexa, including tools that enable enzyme engineering for non-native substrates or new-to-nature reactions, peptide engineering for antimicrobial uses, and binder protein engineering for disease biomarker recognition.
2. IntEnzyDB: An Integrated Structure–Function Enzymology Database
Building an integrated database that merges related enzyme sequence, structure, and function data in one place is essential for developing accurate physical methods and holistic data-driven models for enzyme engineering. However, data collection, cleaning, and joining present three major challenges. Data collection is often impeded by different design (e.g., relational, object-oriented, or hybrid), storage hierarchy, query mechanism, and API protocols of various existing databases. Data cleaning is tricky because existing data entries involve missing or inaccurate mutational spot labels and experimental conditions, as well as manual typos and rounding errors. Data joining between enzyme structure and function data is challenging due to inconsistent keys—enzyme kinetics databases typically store data entries using EC number and often lack PDB IDs, creating challenges for one-to-one mapping to structural databases.
To address these challenges, we developed an integrated structure-kinetics enzymology database, IntEnzyDB, for facile data-driven modeling and machine learning.53,54 The database merges related enzyme sequence, structure, and function data in one place to address the challenges associated with the collection, cleaning, and joining of enzymology data. In contrast to object-oriented databases that store enzyme records in separate data files,64 IntEnzyDB employs a relational database architecture with a flattened data structure. This approach enhances scalability and enables the integration of additional enzyme function data, such as folding stability and solubility, into the database. A similar database architecture has been employed by Fleischmann et al. to build IntEnz, an integrated enzymology database for nomenclature and classification of enzyme-catalyzed reactions.65
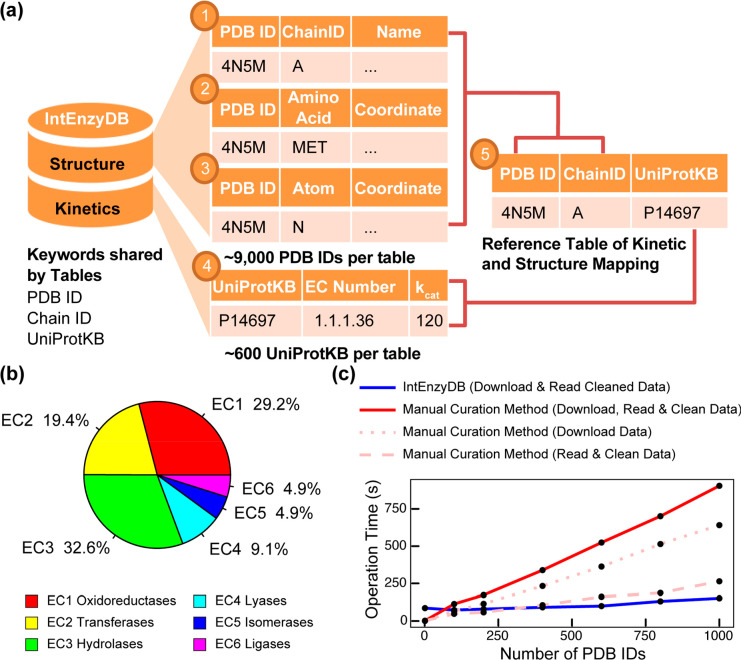
To store kinetics and structure information, IntEnzyDB implements five data tables (Figure 1a). We curated three tables to store cleaned enzyme structure data derived from RCSB PDB,64 including a chain table that contains general protein structure information (e.g., nomenclature, organism, resolution, etc.), an amino acid table that contains amino acid attributes, properties, and physiochemical parameters, and an atomic structure table that contains the atom types and Cartesian coordinates. We curated one table for kinetics data derived from BRENDA,66 Sabio-RK,67 ProtaBank,68 and Design2 Data.69 The table contains information of enzyme kinetic assays such as EC number, substrate, mutation, temperature, turnover number, Michaelis constant, and so on. We adopted a protocol to eliminate the data entries with no annotation of experimental conditions. If multiple kinetic values were measured for the same enzyme–substrate system under an identical experimental condition by different research groups, these values were averaged out and stored in IntEnzyDB. Finally, we curated one reference table to achieve one-to-one mapping between enzyme kinetics and PDB based on foreign keys PDB ID, Chain ID, and UniProtKB. Using IntEnzyDB, we constructed a data table containing 4243 kcat/KM values, which represent enzyme catalytic efficiency for variants with single amino acid substitutions. The data set includes 691 wild-type (WT) enzymes, 2592 enzyme mutants, and 943 substrates. Of the stored kcat/KM values, 29.2% pertain to oxidoreductases (EC 1), 19.4% to transferases (EC 2), 32.6% to hydrolases (EC 3), 9.1% to ligases (EC 4), 4.9% to isomerases (EC 5), and 4.9% to lyases (EC 6) (Figure 1b).
Figure 1.
Architecture, kinetics data statistics, and performance benchmark for IntEnzyDB. (a) The database architecture consists of five tables, including three tables for enzyme structure information (chain-level, amino acid-level, and atom-level), one table for kinetics, and a reference table that includes foreign keys from the structure and kinetics tables. The mapping of the tables is established using the PDB ID, Chain ID, and UniProtKB keys. (b) The distribution of kinetics data for six enzyme commission classes. (c) The comparison of operation time between IntEnzyDB and manual curation methods. The operation time for downloading, reading, and cleaning data is measured for processing 1, 100, 200, 400, 600, 800, and 1000 PDB IDs, with data downloading and reading/cleaning indicated by dotted and dashed lines, respectively. The total operation time for the manual curation method is shown by the red solid line. All operation times are reported in seconds. Adapted with permission from ref (53). Copyright 2022 American Chemical Society.
To assess the efficiency of retrieving enzyme structure data using IntEnzyDB, we compared it against a manual curation strategy (Figure 1c). Unlike the manual approach, IntEnzyDB allows the user to filter and download precleaned and tabulated structural data directly using SQL queries. Our results indicate that for processing 200 enzymes, IntEnzyDB is approximately two times faster than the manual curation approach (80 s vs 173 s), and for 1000 enzymes, it is around six times faster (151 s vs 905 s). The results indicate that the operating time using IntEnzyDB is nearly independent of data size, which is particularly beneficial when handling a large amount of structural data (i.e., thousands or more). The flattened data structure of IntEnzyDB likely accounts for its high data processing efficiency. By loading all data entries at once, IntEnzyDB outperforms the traditional approach, where data tables and files are accessed serially in CPU. While processing smaller amounts of data (e.g., for one enzyme structure), IntEnzyDB may take longer (86 s vs 1.9 s) than the manual approach. However, IntEnzyDB can save a substantial amount of time when managing large amounts of data by avoiding repeatedly opening and reading files.
Although only ∼10 min are saved when operating on the 1000 structures in the benchmark (Figure 1), time savings are expected to proportionally increase with the number of data entries. We expect that more quality enzyme structure and function data will be collected and stored in the future. As such, IntEnzyDB provides an efficient solution for extracting enzyme structural features for statistical analysis or machine learning. The high quality structure and function data stored in IntEnzyDB also serve as a benchmark for the systematic assessment and development of new molecular modeling methods used for enzyme engineering. As the next steps for developing IntEnzyDB, we will further expand the mapped structure-kinetics data table by using predicted structures and active site annotations. Text mining strategies will be implemented to enable more comprehensive data validation and collection. We will incorporate new types of enzyme property data to IntEnzyDB, including stability, solubility, expressibility, and even molecular modeling data derived from high-throughput simulations.56 The incorporation of a diverse range of quality data from molecular level to macroscopic scale has the potential to enhance the learning efficiency, predictive accuracy, and generalizability of the models.
3. Software Tools That Enable High-Throughput Molecular Simulations of Proteins
3.1. EnzyHTP: A High-Throughput Computational Platform for Enzyme Modeling
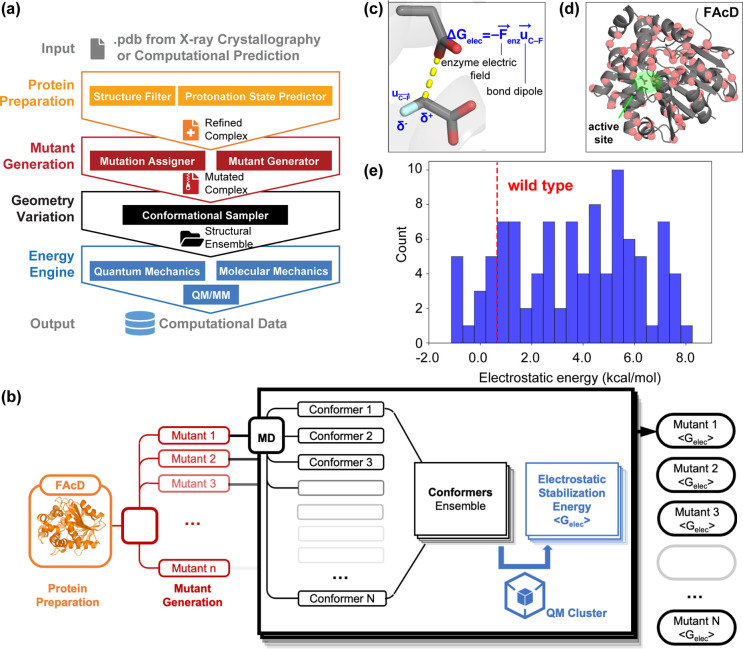
Different types of computational theories and methods, including quantum mechanics (QM), molecular mechanics (MM), and multiscale QM/MM modeling, have been extensively employed in protein engineering to guide the selection of function-enhancing enzyme mutants for late-stage functionalization,70 polymer upcycling,71,72 degradation of environmental pollutants,73,74 and treatment of food allergies.75,46 To maximize the potential of molecular simulations in biocatalyst development,76−79 it is essential to perform enzyme modeling in an automatic and high-throughput fashion. To address this challenge, we developed a computational platform, EnzyHTP, to automate the entire life cycle of enzyme modeling in a high-throughput manner. EnzyHTP has four levels of operation arranged in a top-down hierarchy (Figure 2a). The four levels are protein preparation, mutant generation, geometry variation, and energy engine. Each level was implemented as an independent Python module. The protein preparation module emphasizes constructing computational models for enzyme structures obtained from X-ray crystallography experimental data or computational predictions. The mutant generation module is responsible for generating novel enzyme variants based on a common enzyme sequence and scaffold by altering an existing amino acid’s side chain type and conformation. The geometry variation module samples enzyme conformation and substrate reaction coordinates using external molecular dynamics or Monte Carlo software packages. The energy engine makes use of QM, MM, or multiscale QM/MM calculations using quantum chemistry toolboxes. In particular, the QM treatment of enzyme active sites and reacting species is critical to elucidating the catalytic mechanisms of enzymes and predicting the impact of mutations on enzyme catalysis.
Figure 2.
Design framework and application of EnzyHTP. (a) The workflow of high-throughput enzyme modeling. The workflow comprises four levels of operation, namely protein preparation, mutant generation, geometry variation, and energy engine. The input to this framework is the enzyme structure, and the output is computational modeling data. (b) Application of EnzyHTP to compute the electrostatic stabilization energy values (i.e., <Gelec>) for 100 fluoroacetate dehalogenase (FAcD) mutants. For each mutant, the workflow automatically conducts 1 ns molecular dynamics simulations, 100 single point quantum mechanics calculations, dipole moment analysis, and output an averaged <Gelec> value. (c) Definition of electrostatic stabilization energy, which is computed by the dot product between the enzyme interior electric field and the dipole moment of the breaking C–F bond. (d) Spatial distribution of 100 single mutation spots on FAcD. (e) The distribution of Gelec values for 100 FAcD mutants, where the red dashed line indicates the Gelec value for the WT FAcD. Adapted with permission from ref (56). Copyright 2022 American Chemical Society.
To demonstrate the high-throughput capability of EnzyHTP, we employed the software to investigate the impact of single mutations on the interior enzyme electrostatics for 100 fluoroacetate dehalogenase (FAcD) mutants (Figure 2b). The model enzyme, Rhodopseudomonas palustris FAcD, hydrolyzes the C–F bond of fluoroacetate (FAc) via an SN2 mechanism (Figure 2c).80−84 The cleavage of the C–F bond contributes to the rate-determining step. The enzyme electric field accelerates the reaction by stabilizing the dipole moment along the breaking C–F bond.85 The electrostatic effect is quantified using electrostatic stabilization energy (i.e., Gelec), which is computed by the dot product between the electric field and the C–F bond dipole (Figure 2c). Using EnzyHTP, we created a Python workflow to compute Gelec values for 100 FAcD variants with random single amino acid substitution. The workflow first generates 100 variants using the mutant generation module based on a curated FAcD crystal structure (Figure 2b). The mutation spots are distributed over the entire FAcD enzyme scaffold (Figure 2d), with a spatial proximity to the active site ranging from 7 to 32 Å. The workflow performs an MD simulation for each variant and then samples 100 conformers from a 1 ns MD production run. The structure involves a restrained prereaction complex in which the residue Asp110 is aligned with the substrate C–F bond for a potential SN2 attack. A short propagation time is used for the MD simulations to ensure that the sampled enzyme conformers resemble the crystal structure. Third, the workflow computes the ensemble average of Gelec values (denoted by <Gelec>) using 100 conformational snapshots extracted from a 1 ns MD trajectory. The bond dipole is computed using a single-point QM calculation (HF/6-31G(d)) that consists of the substrate and Asp110, followed by wave function-based localized molecular orbital (LMO) analysis using Multiwfn. The electronic field strength of a mutant is computed based on the RESP charges of enzyme atoms using Coulomb’s law. Solvent molecules and counterions are excluded. Using the workflow, we completed the computation of <Gelec> values for 100 FAcD variants in 7 h with 10 GPUs (NVIDIA V100 SMX2) and 160 CPUs (Xeon Gold 6248). In contrast, performing this process manually for 100 enzyme variants would take several weeks due to tedious processes of mutant structure curation and file preparation, in addition to the computational runtime.
Figure 2e displays the distribution of <Gelec> values for 100 FAcD variants. The computed <Gelec> values exhibit a range of −1.1 to 8.2 kcal/mol. Comparing to the reference <Gelec> value of the WT FAcD (i.e., 0.5 kcal/mol), a small proportion of mutations (∼10%) cause a reduction in the < Gelec> value, indicating the formation of a more favorable electrostatic environment that can between stabilize the developing C–F dipole in the FAcD mutant compared to the WT FAcD. However, the majority of mutations (∼90%) have the opposite effect, which are likely to reduce or even abolish the catalytic effect. Despite an enhanced enzyme electric field strength for breaking the C–F bond, the 10% mutations are not necessarily the actual beneficial mutations due to the impact of mutation on other untested aspects, such as stability, solubility, expressibility, and so on. Our work on developing EnzyHTP software sets the basis for in silico high-throughput enzyme screening that identifies beneficial enzyme variants, which can accelerate the development cycle of new biocatalysts that catalyze non-native substrates or new-to-nature reactions. EnzyHTP will facilitate the comprehension of enzyme catalytic mechanisms across numerous enzymes within a protein family. EnzyHTP can also assist in generating computational data for our database IntEnzyDB, which in turn contributes to enhancing our statistical understanding and machine learning capabilities. Inspired by the code base and architecture of EnzyHTP, we are developing more high-throughput software suites to address specific challenges of automatic molecular modeling in protein engineering. For one, we developed a tool for automatic construction and modeling of lasso peptides. This will be discussed in Section 3.2.
3.2. LassoHTP: A High-Throughput Tool for Lasso Peptide Structure Construction and Modeling
Lasso peptides are a class of ribosomally synthesized and post-translationally modified natural products. They were first discovered in 199186 and have been increasingly reported as candidates for new antibiotics,87−90 enzyme inhibitors,88,91 and receptor antagonists,86 (e.g., microcin J2591,92). Lasso peptides involve a 1-rotaxane topology93,94 with a macrolactam ring held in position by sterically bulky residues above and below the ring. The ring in the lasso peptide is formed by an isopeptide bond between the N-terminal α-amino group and the carboxylate group of an aspartate or glutamate. Bioinformatic analyses estimate that the lasso peptides with a known structure and function occupy ∼10% of all possible lasso peptides that exist in nature. To accelerate the discovery of functional lasso peptides, computational tools capable of predicting the structures and functions of uncharacterized lasso peptides can aid in prioritizing pharmaceutically valuable candidates for experimental evaluation. However, due to the distinct topology of lasso peptides, computational tools that were designed for structural prediction of globular proteins (e.g., AlphaFold295) or cyclic peptides96 fail to predict the structure of lasso peptides with high fidelity.
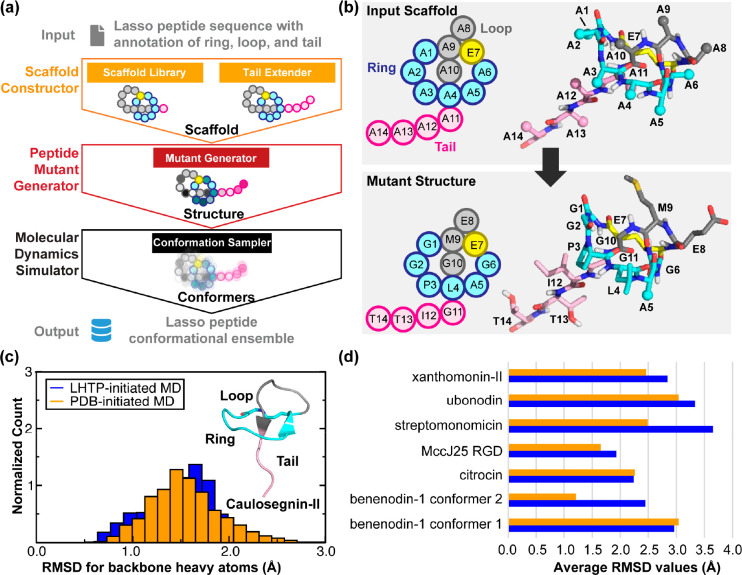
To address this challenge, we developed LassoHTP as a tool for high-throughput lasso peptide structure prediction and conformational sampling. LassoHTP converts a user-defined lasso peptide sequence (with annotation of ring, loop, and tail) into a three-dimensional structure and a conformational ensemble using three software modules, including a scaffold constructor, a mutant generator, and an MD simulator (Figure 3a). The scaffold constructor is responsible for generating a poly alanine lasso peptide scaffold based on a structural library and tail extender function, while the mutant generator module mutates this scaffold to produce a lasso peptide structure that corresponds to the user-defined sequence or sequences resulting from mutagenesis (Figure 3b). Finally, the MD simulator uses the AMBER software package97 to parametrize the resulting lasso peptide structure and conduct MD simulations to output a conformational ensemble. The modular architecture of LassoHTP ensures its flexibility and versatility, similar to that of EnzyHTP.56 Each module can be independently operated for building, modifying, or modeling a lasso peptide, and the three modules can be sequentially executed as part of an automatic workflow to convert user-defined lasso peptide sequences into conformational ensembles.
Figure 3.
Design framework and application of LassoHTP. (a) A schematic of LassoHTP’s workflow, which involves three modules: scaffold constructor, peptide mutant generator, and MD simulator, to transform a user-input sequence into a conformational ensemble. (b) Application of the mutant generator module to convert the poly alanine lasso peptide scaffold into the lasso peptide that is consistent with the user-input sequence. Sequence shown is for xanthomonin-II105 (PDB ID: 2MFV). (c) Distribution of root-mean-square deviation (RMSD) for LassoHTP-initiated and PDB-initiated MD conformational ensemble for caulosegnin-II.98 (d) Average RMSD values of LassoHTP (LHTP)-initiated (colored in blue) and PDB-initiated (colored in orange) MD ensembles for eight lasso peptides involved in the benchmark. The structures of the lasso peptides were determined mostly by NMR except for caulosegnin-II by X-ray crystallography (PDB ID: 5D9E). For (c) and (d), the RMSD was calculated using backbone atoms (i.e., Cα, N, C, and O) with reference to the reference crystal and NMR structure. Adapted with permission from ref (57). Copyright 2023 American Chemical Society.
To test LassoHTP, we employed the software to predict conformational ensembles for different types of lasso peptides (called LHTP-initiated MD) and then benchmarked their consistency against the MD ensembles initiated from the corresponding crystal- or NMR-structures (called PDB-initiated MD, Figure 3c and 3d). The first test case is the WT caulosegnin-II98 (PDB ID: 5D9E), as a crystal structure (resolution: 0.86 Å) exists for this peptide. For both LHTP-initiated and PDB-initiated MD ensembles, trajectories were simulated using identical force field parameters and the ensembles were constructed by evenly taking 1000 snapshots from a 100 ns MD trajectory. The RMSD value calculated from the LHTP-initiated ensemble (1.48 Å) closely align with that from the PDB-initiated ensemble (1.55 Å).
Furthermore, we tested LassoHTP using seven lasso peptides whose structures have been determined by NMR, including benenodin-1 conformer 1,99 benenodin-1 conformer 2,99 citrocin,100 the RGD variant of microcin J25,101 streptomonomicin,102 ubonodin,103,104 and xanthomonin-II105 (Figure 3d). They involve a wide range of structural constructs. The first structural model of each peptide’s NMR-resolved structural ensemble was used to initiate the MD simulation and as a reference structure for RMSD calculations in both LHTP- and PDB-initiated MD ensembles. The two ensembles are reasonably consistent: the difference of the RMSD values between the two ensembles ranges from ∼0.0 Å for benenodin-1 conformer 1 and citrocin to ∼1.2 Å for streptomonomicin and benenodin-1 conformer 2, with the average being 0.48 Å. The consistency between the two ensembles were also validated using principal component analysis (PCA). The benchmark shows that LassoHTP can generate reasonable lasso peptide structures and conformational ensembles from sequence. As such, LassoHTP provides a platform to build modules for high-throughput functional predictions including binding affinity to drug target, thermostability against harsh conditions, and permeability across membrane transport proteins.
Nonetheless, we should note some technical limitations that we would like to address in LassoHTP. For one, the isopeptide bonds with a cis-configuration, which populate with high abundance in benenodin-1,106 have not been constructed in the scaffold library. Additionally, enhanced sampling methods have yet to be used for navigating the conformational space of lasso peptides. Both aspects are expected to be addressed in the next version of LassoHTP.
3.3. ARMer: A Python Library for Adaptive Resource Allocation of Molecular Modeling Workflows on High-Performance Computing Clusters
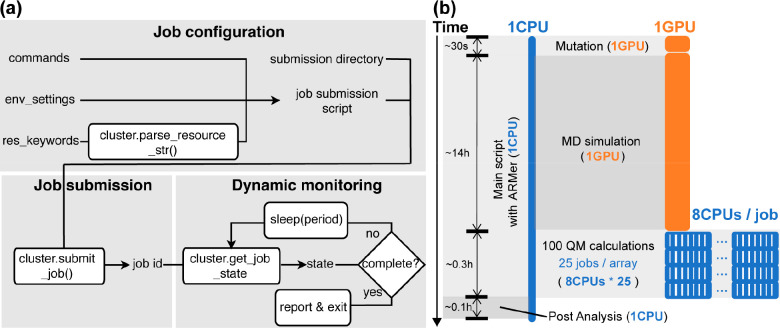
High-throughput computation has emerged as a new paradigm to facilitate mechanistic study,107 catalyst screening,108 functional material design,109,110 drug discovery,111,112 and enzyme modeling.56 Our lab has developed EnzyHTP56 and LassoHTP57 as open-access software packages to enable the high-throughput modeling of enzymes and lasso peptides, respectively. High-throughput computation needs to allocate different types of computing resources (e.g., CPU, GPU, etc.) for multiple subtasks in high-performance computing (HPC) clusters. Resource allocation in the workflow to minimize resources and overall runtime remains a technical challenge in the computational community. To address this challenge, we developed a new Python library, Adaptive Resource Manager (ARMer), to dynamically request computing resources based on the need of a specific modeling subtask in the workflow.55 Using commands implemented in the ARMer library, a Python “workflow script” is prepared that runs on a single-CPU thread to configure, submit, and monitor molecular simulation jobs for a high-throughput workflow in HPC clusters. This is in sharp contrast to the traditional resource allocation scheme where a fixed amount of computing resources is requested for all types of molecular modeling tasks.
The ARMer Python library contains a Job class that defines variables and functions associated with each job’s configuration, submission, and dynamic monitoring of completion (Figure 4a). ARMer also contains an HPC class that supports the Job class with variables and functions to mediate external input/output in a local HPC cluster where ARMer is deployed. In the Job class, a job object is instantiated based on information provided by the user through the arguments: commands, cluster, env_settings, and res_keywords. With the Job object created, a script for the required tasks can be generated and then submitted by the submit() method (Figure 4a). A job ID is added to the object by the function. By tracing the job ID, the “workflow script” can monitor the status of a job object in the queue, and mediate the status by killing, holding, or releasing the job. The “workflow script” will dynamically detect the timing of the job completion by retrieving error or completion messages from the output file. Notably, dynamic monitoring of job completion status is critical to a high-throughput modeling workflow because multiple types of simulation subtasks must be sequentially operated in the process. In the case of high-throughput enzyme modeling, after submitting an MD sampling task, the “workflow script” must put the rest of the subtasks on hold and wait for the conformational ensemble to generate before submission of the subsequent QM calculations.
Figure 4.
Framework and application of adaptive resource manager (i.e., ARMer), a Python library used for adaptive computing resource allocation on high-performance computing cluster. (a) Variables and functions used by ARMer for configuration, submission, and dynamic monitoring of computational tasks. The variables and functions are encapsulated in a Job class. They can be called by a user to prepare a Python script that enables the construction of a high-throughput molecular modeling workflow with effective allocation of computing resources (e.g., CPU and GPU). (b) An exemplary application of ARMer to construct a workflow for high-throughput modeling of fluoroacetate dehalogenase (FAcD) mutants. In the workflow, a Python script that runs on a single-CPU thread leverages functions and variables from the Job class to manage the modeling subtasks (i.e., mutation, molecular dynamics, and quantum mechanics simulations) by configuring, submitting, and monitoring new job scripts. The MD job requests 1 GPU (in orange) and each QM job requests 8 CPUs (in blue). To submit and run individual QM calculations in parallel, a job array with a size of 25 is employed. The type of modeling subtasks, time usage, and resource cost are noted in the figure. Adapted with permission from ref (55). Copyright 2023 American Chemical Society.
We tested the resource and time consumption of the high-throughput molecular modeling workflow enabled by adaptive resource allocation on our local HPC at Vanderbilt’s advanced computing center for research and education (ACCRE). A single-CPU job was submitted to execute a “workflow script” that employs built-in commands from the ARMer library to manage computing resources for molecular simulation tasks involved in the high-throughput modeling of FAcD mutants (Figure 4b). Compared to traditional allocation strategy that directly execute subtasks using a fixed amount of allocated CPU or GPU nodes, this Python script configures resource-demanding subtasks (i.e., needing >1 CPU or ≥1 GPU) in a new job script and then submits the job to the queue (i.e., setting ifcluster = “True” in the code). This job was configured with a 96-h wall-clock running time to oversee the entire workflow.
For the MD simulation task, the workflow script configures shell commands in a job script to request GPU resource, set environment variables, and conduct MD modeling using AMBER. The workflow script then submits the job and regularly monitors the completion status of the job. After receiving the signal of completion, the workflow script will continue operating the QM calculation subtasks in the workflow. Due to the independence of individual QM tasks, the workflow script can submit multiple QM jobs (8 CPU for each QM job) simultaneously to the job array so that they can run in parallel up to the size limit of job array (i.e., 25 jobs) in local HPC cluster (Figure 4b). New jobs are submitted once the “workflow script” detects open slots on the array. With an array size of 25 jobs, one would expect an approximate time acceleration by a factor of 25 if no major time is spent on job queueing. As such, the ARMer library makes it possible to adaptively allocate computing resources to effectively accomplish a high-throughput molecular modeling workflow. This is different from the traditional resource allocation strategy in which one relies on the initially requested/assigned GPU or CPU nodes for the entire computational workflow.
4. Scoring Functions That Describe Sequence-Structure–Function Relationships for Protein Engineering
4.1. A Molecular Dynamics-Derived Descriptor for Enzyme Catalysis
To guide predictive protein engineering, physical descriptors have been identified that correlate with enzyme catalytic efficiency, including enzyme electrostatics in ketosteroid isomerase,113 Kemp eliminase,114,115 methyltransferase,116 and P450 enzymes;117 and binding affinity in endoglucanases and cellobiohydrolases.118−120 Protein dynamics have been proposed as a critical factor to favor substrate positioning,121−128 control reaction dynamics,129−134 regulate dynamic network for thermal activation,135 and tune protein thermal capacity.136 However, the molecular dynamics-derived descriptors that represent the quantitative impact of protein dynamics on catalysis remain largely unexplored. Here, we used statistical modeling with PCA to identify molecular dynamics-derived descriptors that guide the search of enzyme variants that accommodate non-native substrates with optimal substrate-positioning dynamics.
We used lactonase SsoPox as a model system (Figure 5a), which catalyzes the hydrolysis of 3-oxo-CX acyl-homoserine lactone (X = 10 or 12).137−142 The WT SsoPox is most reactive toward the C10 substrate, while the W263T mutant for the C12 substrate (Figure 5b).138 This enzyme system was chosen primarily because kinetic turnover numbers have been characterized for both C10 and C12 substrates combined with the same set of SsoPox variants (i.e., WT, W263F, W263T, W263I, and W263 V). This allows us to identify physical descriptors that inform distinct substrate-positioning behaviors of the same enzyme scaffold toward different substrates.
Figure 5.
A molecular dynamics-derived descriptor for representing the impact of mutation on enzyme catalysis. (a) The crystal structure for the model enzyme used in the study: lactonase SsoPox (PDB ID: 2VC7). Flexible loops are colored in pink. Substrate binding pocket is indicated by an orange oval. W263 is the spot in which mutations have been performed to investigate the role of mutation on enzyme kinetics. (b) The reaction activation free energies (ΔG‡) for 3-oxo-CX acyl-homoserine lactone substrates (X = 10 or 12, colored in black and blue, respectively) combined with different enzyme variants (WT, W263F, W263T, W263I, and W263 V). ΔG‡ value is converted from the turnover rate using Eyring’s equation. (c) The PCA loading plot for the descriptors tested in the study. The descriptor is ranked based on its contribution in principal components (from major to minor): 1. SASAsub/SASApkt; 2. SASApkt; 3. RMSDpro; 4. dloop1–3; 5. RMSDpkt_sub; 6. RMSDsub; 7. RMSDpkt; 8. d99–229; 9. d258-sub; 10. EFall; 11. SASAsub; 12. d97-sub; 13. EFnoion; 14. d223–256; 15. dtail-loop8. The percentage in each axis label indicates the contribution of the principal component to the total variation. (d) Distribution of ΔG‡ values versus SASA ratio (i.e., SASAsub/SASApkt) in enzyme variants across C10 and C12 substrates. The red dashed lines indicate the optimal point of ΔG‡ and the black dashed lines are the linear fitting of data points on each side of the optimal point. Adapted with permission from ref (58). Copyright 2022 American Chemical Society.
Using molecular dynamics trajectories for each substrate-enzyme variant complex, we calculated 15 molecular features that are associated with the structural and dynamics characteristics. The descriptors fall into four groups: 1) solvent accessible surface area (SASA); 2) electric field; 3) root-mean-square deviation; and 4) functionally important substrate-residue, residue–residue, and loop–loop distances. We utilized a PCA loading plot to rank the importance of these descriptors–a higher importance rank indicates that the descriptor contains more information to predict the change of experimental activation free energies (Figure 5c). The PCA analysis identified the SASA ratio of substrate to active-site pocket (i.e., SASAsub/SASApkt) as the top predictor for the mutation effect on activation free energy. This descriptor has been defined to be the substrate-positioning index (SPI) in our later study.63 The form of SPI is similar to the definition of hydrophobicity index.143
We further investigated the distribution of ΔG‡ values versus SPI for C10 and C12 substrates combined with different enzyme variants. The distribution conforms to a two-segment, piecewise linear correlation plot with a volcano shape (Figure 5d). This quantitative relationship is very similar to the Sabatier principle observed for cellobiohydrolases by Jeppe et al.118−120 The SPI value ranges from 1.67 to 1.96, and the activation free energy reaches the minimum (∼16.5 kcal·mol–1) under an optimal SPI value. For the C10 substrate, WT SsoPox, which is most favorable, has an SPI of 1.85. In contrast, for C12 substrate, the SPI value for WT drifts to 1.96. The ΔG‡ reaches the minimum value of 16.4 kcal·mol–1 in W263T, where the reaction turnover number for C12 is comparable to the native reaction for C10 in the WT enzyme (16.6 kcal·mol–1). The shift of the SPI value upon mutation is dominated by the size variation of the active-site pocket. As such, the optimal SPI value shown in Figure 5a likely reflects the desired enzyme cavity that best accommodates a substrate to achieve efficient catalysis. Replacing the native substrate C10 with C12 leads to an increase of substrate size, which is beyond the accommodation capacity of the WT enzyme but presents a good fit in the W263T variant that has a larger active-site pocket. The results show that SPI can be employed as a predictive descriptor to guide the search for optimal enzyme mutants for catalyzing non-native substrates. To achieve efficient hydrolysis, a non-native substrate-bound enzyme variant needs to involve a similar range of SPI value to the native substrate-bound WT enzyme.
4.2. Deep Learning Models for Protein Function Prediction
In this section, we introduce two deep learning models that our group recently developed for engineering of enantioselective biocatalysts (i.e., EnzyKR59) and antimicrobial peptides (i.e., DeepLasso60). EnzyKR was developed to predict the enantiomeric outcome of kinetic resolution reactions catalyzed by hydrolases. DeepLasso was built to predict the antimicrobial activity of ubonodin variants.
Hydrolases, such as lipases, esterases, and dehalogenases, have been widely employed for kinetic resolution in synthetic reactions in the chemical and pharmaceutical industries.144−147 Despite the development of empirical,148 statistical,149 machine learning,150 and deep learning models,151,152 the “generalist” models that can predict enantioselectivity across a broad spectrum of hydrolase scaffolds, mechanisms, and substrate types remain undeveloped.153 To address this challenge, our group developed a deep learning model, EnzyKR, to predict the activation free energy of a hydrolase-substrate enantiomer complex. The difference of predicted free activation energies between enantiomers (i.e., ΔΔG‡ = ΔGR‡ – ΔGS‡) informs outcome of hydrolytic kinetic resolution. The training and test data include a total of 224 hydrolase-substrate complexes curated from 13 enzyme commission subclasses under the category of hydrolases, which are curated from our integrated enzyme structure-kinetic database IntEnzyDB.53
The model consists of a classifier that distinguishes reactive hydrolase-enantiomer complexes from unreactive binding poses, and a regressor predicts the hydrolytic activation free energy (i.e., ΔG‡) for the reactive complex. The classifier employs convolutional and graph neural networks to separately encode three types of input: enzyme sequences, substrate SMILES strings, and the distance maps for the hydrolase-substrate complex (Figure 6a). The regressor of EnzyKR takes input from both the classifier embedding and substrate-enzyme interaction maps (a stacked form of atomic distance map). Notably, the atomic distance map differentiates substrate chirality, allowing the model to effectively learn the enantiomeric preference of hydrolases. EnzyKR exhibits a decent prediction accuracy with a Pearson R of 0.91, Spearman R of 0.86, and a mean absolute error (MAE) of 0.8 kcal/mol on the training set (204 data points). EnzyKR also achieves a Pearson R of 0.66, Spearman R of 0.70, and MAE of 1.5 kcal/mol on the test set (20 data points held out from the training set). For both training and test sets, the value of Spearman R is close in range to that of Pearson R. This indicates that EnzyKR balances the regression of target values or ranking without overfitting.
Figure 6.
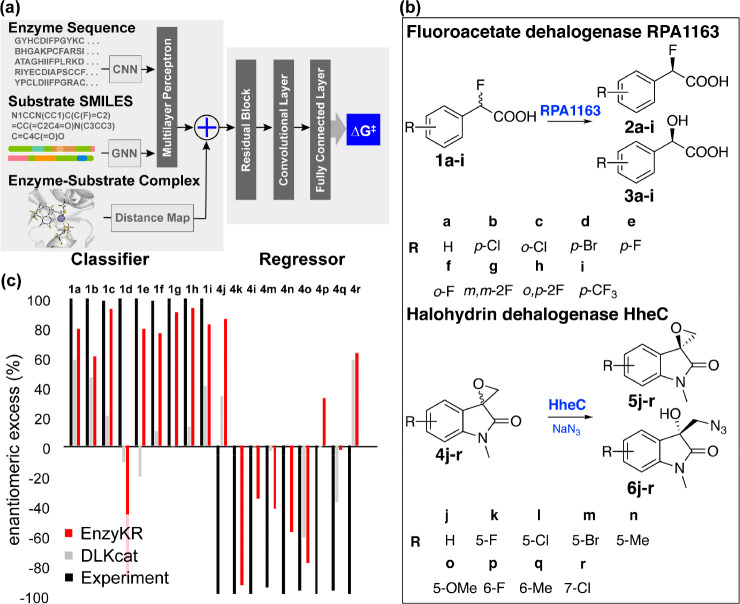
Design and application of EnzyKR, a deep learning model for predicting the enantiomeric outcome of hydrolase-catalyzed kinetic resolution. (a) EnzyKR consists of a classifier and a regressor. Three types of input data for the classifier involve the enzyme–substrate complex structure, enzyme sequence, and simplified molecular-input line-entry system (SMILES) string. The distance map derived from the complex structure is encoded using a 2D convolutional neural network (CNN). The multiple sequence alignments (MSA) of the enzyme sequences are also encoded by a 2D CNN model. The substrate SMILES strings are encoded by a graph neural network (GNN) model. The embeddings from the classifier and the interaction maps are used as input for the regressor. The regressor involves one module of cross-attention, followed by residual blocks consisting of three 2D dilated convolution layers, one 2D batch norm layer, and one ReLU layer. Two layers of a fully connected neural network (i.e., multiple-layer perceptron) are employed to conduct regression between the extracted feature and the activation free energy. (b) The test reactions used to assess the ability of EnzyKR to predict the outcomes of kinetic resolution. The test set involves 18 enantioselective hydrolytic reactions catalyzed by two hydrolases. RPA1163 is a fluoroacetate dehalogenase that catalyzes the C–F bond hydrolysis in 9 fluoroacetic acid derivatives labeled from a to i. HheC is a halohydrin dehalogenase that catalyzes the stereoselective epoxide ring-opening in 9 spiro-epoxyoxindoles derivatives labeled from j to r. (c) The predicted enantiomeric excess (ee%) values of EnzyKR (red) and the baseline model DLKcat (gray) for 18 enantiomer pairs in hydrolase-catalyzed reactions. The experimental ee% value is shown in black. A positive ee% value indicates that the S-configuration is favored.
To evaluate the significance of different features in EnzyKR, we conducted a baseline evaluation by removing specific features and observing their impact on the predictive accuracy of the regressor. Eliminating the atomic distance map leads to reductions in both Pearson and Spearman R values to 0.53 and 0.51, respectively, accompanied by an MAE increase to 2.2 kcal/mol. Similarly, removal of SMILES strings of substrates from the classifier input results in lowered Pearson and Spearman R values of 0.6, and an MAE increase to 2.0 kcal/mol. These findings indicate that both the distance map and substrate SMILES strings are indispensable features positively contributing to the predictive accuracy of the EnzyKR regressor.
We further tested EnzyKR for its ability to differentiate enantiomeric reactions using 18 hydrolytic reactions catalyzed by FAcD RPA1163154 and halohydrin HheC155 (Figure 6b). These reactions were separately curated from literature sources. The performance of EnzyKR was compared against DLKcat, a deep learning kcat predictor.152Figure 6c shows that compared to the experimental results (black), EnzyKR (red) correctly predicts the favored enantiomer and outperforms DLKcat (gray) in 13 out of 18 reactions (i.e., 1a-c, 1e-i, 4k-o). In more than half of the test cases, DLKcat predicts an enantiomeric excess (ee%) value lower than 50%. Due to the lack of chirality encoding in the model, the overall predictive performance of DLKcat appears to be similar to a random guess. Despite a decent performance, we should note that the limitation of EnzyKR lies in the small size of data set and potentially inadequate representation of chirality. We are addressing these issues in our ongoing works.
The second deep learning model is DeepLasso. In recent decades, lasso peptides, such as microcin J25,156 ubonodin,157 cloacaenodin,158 and so on,18 have emerged as promising candidates for stemming the tide of the antimicrobial crisis. However, the development of computational models to facilitate the engineering of lasso peptide mutants with enhanced antimicrobial activities lag far behind the pace of lasso peptide discovery. To fill in the void, we collaborated with the Link lab and developed DeepLasso to predict the antibiotic activity (i.e., enrichment value) for ubonodin variants (Figure 7). The training and test data involve ∼90,000 mutants of lasso peptide ubonodin that were collected from experimental high-throughput screening and next-generation sequencing of single and double mutant library constructed by site-saturation mutagenesis. The antimicrobial activity of a ubonodin mutant is represented by an enrichment value, which is the base-2 logarithm of the ratio of the mutant’s frequency at a specific step of the screen relative to the mutant’s frequency in the cloning transformation library.60 Negative enrichment values indicate that the variant likely inhibits RNAP. Dropout mutants are those with very high RNAP inhibitory activity and their the enrichment values are annotated as “not available”.
Figure 7.
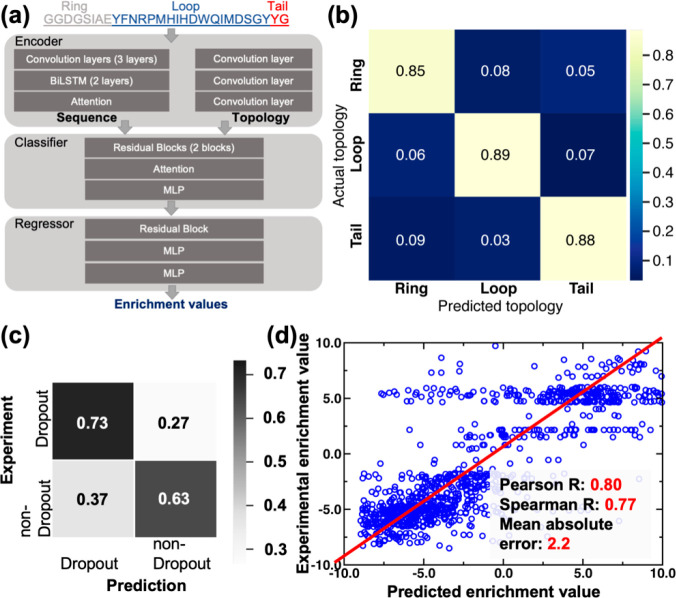
Design architecture of DeepLasso, a deep learning model for predicting the antibiotic activity of lasso peptide ubonodin mutants. (a) The architecture of DeepLasso consists of an encoder, classifier, and regressor. The sequence encoder is constructed by three layers of a convolutional neural network (CNN), two layers of a bidirectional long–short-term memory network, and one attention layer. The topology encoder is constructed by three layers of CNN with each layer used to learn a specific topological region of lasso peptide sequence (i.e., ring, loop, or tail). The classifier involves a sequential layout of two residual blocks, one attention layer, and one layer of multilayer perceptron (MLP). The regressor involves a sequential layout of one residual block and two layers of MLP. The tensors derived from the encoder are concatenated and fed into the classifier for prediction; the resulting tensor from the classifier is then used in the regressor for prediction. (b) Confusion matrix analysis for the classifier of DeepLasso. The matrix shows classification of sequence regions of ubonodin variants (ring, loop, and tail). The color scale is used to represent the magnitude of hit rate (i.e., high: yellow; low: dark blue). (c) Confusion matrix analysis for the classifier of DeepLasso. The matrix shows binary classification of dropout versus nondropout variants. Grayscale is used to represent the magnitude of hit rate (i.e., high: black; low: white). (d) Regression analysis for the nondropout variants with enrichment values. The linear correlation between experimental vs predicted enrichment values is shown along with Pearson correlation coefficient, Spearman correlation coefficient, and mean absolute error. Adapted with permission from ref (60). Copyright 2023 American Chemical Society.
Similar to EnzyKR, DeepLasso also adopts a classifier-regressor architecture (Figure 7a). Using a ubonodin variant sequence as input, the classifier first predicts whether the variant is likely to be a dropout variant. If the sequence is deemed likely to be a nondropout variant, the regressor predicts an enrichment value for the variant. DeepLasso employs a sequence encoder to learn the pattern of the ubonodin amino acid sequence as well as a topology encoder to represent the sequence regions for the ring, loop, and tail of the lasso peptide (Figure 7b). Unlike existing deep learning models for antimicrobial peptide prediction,159 the topology encoder we implemented in DeepLasso can potentially improve the learning efficiency because the topology of lasso peptides is known to be essential in the inhibition of RNAP.160−162 To evaluate the accuracy of DeepLasso, we performed confusion matrix analysis for the classifier (Figure 7c) and linear regression analysis for the regressor (Figure 7d). The results show that DeepLasso achieves a 73% hit rate for the dropout variants and 63% for nondropout variants (Figure 7c). The higher accuracy for predicting dropout variants is desired because these variants are the most likely to exhibit strong antibiotic activity. For nondropout variants, the predicted enrichment values are correlated to the experimental value with a Pearson correlation R of 0.80, a Spearman rank correlation R of 0.77, and a MAE of 2.2. The regressor allows us to score the nondropout variants for their RNAP inhibitory activity.
DeepLasso provides a computational tool to map out the fitness landscape of ubonodin variants as potential antibiotics. Though trained with mostly single and double mutants, DeepLasso is capable of identifying higher order ubonodin mutants with enhanced antimicrobial activity. One critical aspect that has yet to be considered here is the ability to predict permeability of ubonodin variants through the membrane of target bacteria. The permeability through cell membrane is independent from RNAP inhibition but should weigh in as an important factor for development of the next version of DeepLasso. Besides, the magnitude to which we can generalize DeepLasso for the antimicrobial prediction of other types of lasso peptides remains a valuable question for investigation.
5. Applications
The preceding sections present the core technical components underlying Mutexa, including an integrated structure–function database (Section 2), software packages for high-throughput modeling of protein mutants (Section 3), and scoring functions for predicting the sequence-structure–function relationships (Section 4). In this section, we will demonstrate three applications where these new computational tools are leveraged to determine the conditions for computational convergence in enzyme modeling,61 to gain a statistical view across members of the methyltransferase family,62 and to deepen the understanding of dynamic effects in enzyme catalysis.63
The first case applies the high-throughput enzyme modeling workflow of Mutexa (i.e., EnzyHTP56) to investigate the boundary conditions that should be used in enzyme modeling for a reliable description of mutation effects. In computational protein engineering, functional descriptors have been calculated from molecular simulations to aid the search for beneficial enzyme variants.163−166 However, the optimal size of the active-site region for computing these descriptors across multiple enzyme variants has not yet been investigated. Using EnzyHTP, we conducted convergence tests on 18 Kemp eliminase variants,167,168 evaluating functional descriptors in six active-site regions with varying distances from the substrate. The assessed descriptors include the dynamic fluctuation of the active-site (represented by root-mean-square deviation, or RMSD), the substrate positioning index (represented by the SASA ratio between the substrate and the active site), and the electric field index (represented by the projection of the electric field on the reacting C–H bond). Both molecular mechanics and multiscale quantum mechanics/molecular mechanics methods have been used to compute the descriptors. The descriptor values were determined for each of the 18 Kemp eliminase variants. Spearman correlation matrices were employed to identify the condition for the region size beyond which further expansion of the boundary does not significantly alter the ranking of descriptor values. Our results show that dynamics-derived descriptors, specifically the dynamic fluctuation and substrate positioning index, reached convergence at a distance cutoff of 5 Å from the substrate. The electric field descriptor exhibits convergence at 6 Å when employing molecular mechanics methods with truncated enzyme models, and at 4 Å when utilizing quantum mechanics/molecular mechanics methods with the entire enzyme model. This study serves as a reference for selecting descriptors in predictive modeling of enzyme engineering.
The second case uses EnzyHTP to study the convergent catalytic behaviors of S-adenosyl methionine (SAM)-dependent methyl transferases (MTases). MTases are a ubiquitous class of enzymes catalyzing dozens of reactions in the life processes.169−172 Despite targeting a large variety of substrates with diverse intrinsic reactivity, MTases demonstrate similar catalytic efficiency.53,54,173 To elucidate the evolutionary adaptations that allow SAM MTases to accommodate the diverse chemical features of their respective substrates, we curated 91 SAM MTases from the protein databank (PDB) and conducted a comprehensive computational analysis using EnzyHTP to gain insights into how specific properties, such as electric field strength and active site volumes, contribute to achieving similar catalytic efficiency across substrates with different reactivity levels. When looking at O-, N- and even C-targeting MTases, we found that there was not a significant difference in cavity volumes but the electric field strengths have largely adjusted to enhance the target atom’s ability to accept a methyl group. For MTases targeting RNA/DNA and histone proteins, the electric field strength accommodates the formal hybridization state. Our study also shows that metal ions in MTases contribute negatively to electric field strength for methyl donation and enzyme scaffolds likely offset these contributions.
The last case integrates the workflow software EnzyHTP with a scoring function of Mutexa, substrate positioning index (SPI, discussed in the Section 4.1), to investigate the behavior of nonelectrostatic dynamics in enzyme catalysis. The dynamic positioning of substrates within the active site, known as substrate positioning dynamics (SPD), plays a crucial role in facilitating enzyme catalysis by aligning the substrate in a reactive conformation.125,164,174−186 However, as conformational changes often coincide with alterations in the electrostatic environment inside the enzyme, it remains unclear whether SPD involves a nonelectrostatic component that independently influences catalysis or primarily arises from perturbations in the enzyme’s internal electrostatics.184,187,188 To answer this question, we integrated computational and experimental approaches to investigate the nonelectrostatic component of SPD using Kemp eliminase as a model enzyme. We employed substrate positioning index to quantify the impact of protein dynamics on substrate positioning. Using EnzyHTP, we selected seven variants for kinetic evaluation, which exhibited significantly different SPD while maintaining similar enzyme interior electrostatics. Our analysis revealed a valley-shaped, two-segment piecewise linear correlation between experimentally determined activation free energies and substrate positioning index values. This trend was further validated using previously reported kinetic data from the Head-Gordon group.189 Notably, an optimal SPI value, corresponding to the lowest activation free energy, was observed for the R154W variant, a surface mutation located distantly from the active site. Compared to the wild type, the R154W variant displayed favorable SPD, resulting in an increased proportion of reactive conformations for substrate deprotonation. These findings indicate the existence of a nonelectrostatic component in SPD, serving as a factor that mediates catalysis by modulating the population of reactive conformations.
6. Next Steps
In this Review, we have discussed the construction and applications of Mutexa as a computational ecosystem to facilitate protein engineering. Below we will discuss specific aspects to further develop Mutexa.
Selector of Beneficial Mutations
The immediate next step is to build a locator of beneficial mutants to enhance catalytic efficiency, mediate selectivity, and expand substrate scope in enzyme engineering. We expect the locator to contain three computational modules that separately evaluate the impact of mutations on 1) enzyme biophysics (i.e., thermal stability, solubility, etc.), 2) enzyme–substrate binding affinity, and 3) enzyme specificity and selectivity. For each of the modules, the proper computational readouts, either derived from data-driven modeling or physics-based simulations, remain a question of investigation. Besides being predictive about the functions, these readouts must be computed with a balanced accuracy and efficiency for better compatibility with a high-throughput computational workflow. Designing a computational protocol within these constrains presents the first challenge.
Complex Mutations
Another challenge faced in the community is how to achieve the modeling or prediction of complex mutants that go beyond single amino acid substitution. Complex mutants that contain multiple mutations, insertions, or deletions are commonly seen in protein engineering. However, the data-driven and molecular modeling approaches for describing and predicting these complex mutants are significantly underdeveloped. With increasing joint efforts in computational and experimental protein engineering, we are hopeful that Mutexa will provide more solutions for predicting the functional effects of complex mutations.
Throughput Capability
In contrast to experimental ultrathroughput screening tools capable of assessing 108 mutants per week, the EnzyHTP-based high-throughput enzyme modeling allows the computation for only 102 to 104 variants within the same time frame. Enhancing the throughput capacity stands as an urgent need to amplify its utility within the scientific community. The primary bottleneck of computational efficiency resides in the resource-intensive nature of conformational sampling in MD simulations and of electronic structure computation in QM modeling. With the ongoing development of enhanced sampling algorithms,190−194 deep learning-based energy functions,195−197 and deep generative model-enabled conformational samplers,198,199 we expect the substituent molecular modeling engines to be further accelerated, thus increasing the throughput capacity of enzyme modeling.
Prediction Accuracy and Benchmarks
Accuracy serves as a metric that gauges the extent to which computational predictions conform to the standard. To comprehensively assess the accuracy of a computational protein engineering tool, it is critical to establish the standard for computational predictions by specifically defining the scope of prediction tasks and constructing corresponding benchmark sets. The breadth of prediction tasks encompasses properties of functional proteins such as thermostability, catalytic activity, substrate promiscuity, selectivity, binding affinity, solubility, and antimicrobial activity, among others. Furthermore, protein engineering frequently involves a multiobjective optimization task, striving to simultaneously improve multiple properties for academic or industrial applications.
For a specific prediction task, a benchmark should be employed to develop and evaluate physics-based or data-driven scoring functions that predict protein properties from their structures or sequences. The accuracy of these scoring functions can be evaluated through well-established statistical metrics, such as Pearson or Spearman correlation coefficient, mean average error, root-mean-square error, and so on. To enhance functional prediction accuracy, it is vital to create a benchmark set with large amounts of high-quality experimental data encompassing sequence, structure, and function. Equally important as data set size is the balance of mutation types in the benchmark set, which has been shown to improve the classification accuracy for protein thermostability prediction,200 and it is likely critical for prediction of other protein properties.
The creation of a protein engineering benchmark set aimed at evaluating the accuracy of methods designed to identify beneficial mutations remains elusive. Different from scoring functions that are used for functional assessment of mutation effects, these methods are designed to improve the outcome of identifying function-enhancing mutations. The accuracy of these methods is generally represented by the “hit rate” – the proportion of beneficial mutants identified from mutation library screening. Presumably, the benchmark for beneficial mutant identification should encompass numerous instances of protein engineering tasks under the same category of prediction task. These testing cases should cover proteins with different sequence identity, structural scaffold, stages of directed evolution, types of binding substrates (if applicable), numbers of mutations (single, double, etc.), and spatial distribution of mutations (active site, surface, etc.).
Constructing such protein engineering benchmark sets will allow us to systematically evaluate the accuracy of Mutexa in identifying mutation hotspots, categorizing mutations based on their functional impacts, and prioritizing mutations by the degree of functional enhancement or reduction. Notably, using Kemp eliminase KE07 as the exclusive test scenario, the hit rate of the EnzyHTP-based directed evolution protocol was computed to be 12.5%, surpassing the frequency of naturally occurring beneficial mutations (∼1%).55 As discussed in our prior research,55 we expect to further optimize several components of the computational protein engineering protocol, including mutation engine, algorithms for smart library construction, MD sampling engine, substrate-binding configurations, and the scoring functions, and the reaction mechanism. These advancements will pave the way for Mutexa’s transformation into a tool capable of discovering, generating, designing, predicting function-enhancing variants, thereby empowering biocatalysts to accelerate new-to-nature chemical synthesis, industrial enzymes to degrade environmental pollutants, binder proteins for early stage tumor detection, peptides with antimicrobial activities, and therapeutic proteins to address food allergies.
Acknowledgments
This research was supported by the startup grant from Vanderbilt University and the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM146982. Z.J.Y. thanks the sponsorship from the Dean’s Faculty Fellowship in the College of Arts and Science at Vanderbilt. Z.J.Y. thanks the support from Rosetta Commons Seed grant. S.L.S. acknowledges financial support from the Vanderbilt Undergraduate Summer Research Program and the Department of Computer Science. R.J.J. thanks the financial support from the National Institutes of Health Molecular Biophysics Training Grant (MBTP T32 GM008320). C.J. thanks the financial support from the National Institutes of Health Vanderbilt Chemistry-Biology Interface Training Grant (T32GM065086).
The authors declare no competing financial interest.
References
- Arnold F. H.; Volkov A. A. Directed evolution of biocatalysts. Curr. Opin Chem. Biol. 1999, 3 (1), 54–9. 10.1016/S1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- Packer M. S.; Liu D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet 2015, 16 (7), 379–94. 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- Arnold F. H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem., Int. Ed. Engl. 2018, 57 (16), 4143–4148. 10.1002/anie.201708408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Xue P.; Cao M.; Yu T.; Lane S. T.; Zhao H. Directed Evolution: Methodologies and Applications. Chem. Rev. 2021, 121 (20), 12384–12444. 10.1021/acs.chemrev.1c00260. [DOI] [PubMed] [Google Scholar]
- Kolkman J. A.; Stemmer W. P. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 2001, 19 (5), 423–8. 10.1038/88084. [DOI] [PubMed] [Google Scholar]
- Akbulut N.; Tuzlakoglu Ozturk M.; Pijning T.; Issever Ozturk S.; Gumusel F. Improved activity and thermostability of Bacillus pumilus lipase by directed evolution. J. Biotechnol. 2013, 164 (1), 123–9. 10.1016/j.jbiotec.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Reetz M. T.; Bocola M.; Carballeira J. D.; Zha D.; Vogel A. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem., Int. Ed. Engl. 2005, 44 (27), 4192–6. 10.1002/anie.200500767. [DOI] [PubMed] [Google Scholar]
- Reetz M. T.; Carballeira J. D.; Peyralans J.; Hobenreich H.; Maichele A.; Vogel A. Expanding the substrate scope of enzymes: combining mutations obtained by CASTing. Chemistry 2006, 12 (23), 6031–8. 10.1002/chem.200600459. [DOI] [PubMed] [Google Scholar]
- Yi D.; Bayer T.; Badenhorst C. P. S.; Wu S.; Doerr M.; Hohne M.; Bornscheuer U. T. Recent trends in biocatalysis. Chem. Soc. Rev. 2021, 50 (14), 8003–8049. 10.1039/D0CS01575J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.; Ishqi H. M.; Husain Q. Enzyme engineering: Reshaping the biocatalytic functions. Biotechnol. Bioeng. 2020, 117 (6), 1877–1894. 10.1002/bit.27329. [DOI] [PubMed] [Google Scholar]
- Min K.; Kim H.; Park H. J.; Lee S.; Jung Y. J.; Yoon J. H.; Lee J. S.; Park K.; Yoo Y. J.; Joo J. C. Improving the catalytic performance of xylanase from Bacillus circulans through structure-based rational design. Bioresour. Technol. 2021, 340, 125737. 10.1016/j.biortech.2021.125737. [DOI] [PubMed] [Google Scholar]
- Cecchini D. A.; Pepe O.; Pennacchio A.; Fagnano M.; Faraco V. Directed evolution of the bacterial endo-beta-1,4-glucanase from Streptomyces sp. G12 towards improved catalysts for lignocellulose conversion. AMB Express 2018, 8 (1), 74. 10.1186/s13568-018-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelRe C.; Jiang Y.; Kang P.; Kwon J.; Hall A.; Jayapurna I.; Ruan Z.; Ma L.; Zolkin K.; Li T.; Scown C. D.; Ritchie R. O.; Russell T. P.; Xu T. Near-complete depolymerization of polyesters with nano-dispersed enzymes. Nature 2021, 592 (7855), 558–563. 10.1038/s41586-021-03408-3. [DOI] [PubMed] [Google Scholar]
- Tournier V.; Topham C. M.; Gilles A.; David B.; Folgoas C.; Moya-Leclair E.; Kamionka E.; Desrousseaux M. L.; Texier H.; Gavalda S.; Cot M.; Guemard E.; Dalibey M.; Nomme J.; Cioci G.; Barbe S.; Chateau M.; Andre I.; Duquesne S.; Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580 (7802), 216–219. 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Li Z.; Jiang Y.; Guengerich F. P.; Ma L.; Li S.; Zhang W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295 (3), 833–849. 10.1016/S0021-9258(17)49939-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.; Grathwol C. W.; Aslan-Uzel A. S.; Wu S.; Link A.; Pavlidis I. V.; Badenhorst C. P. S.; Bornscheuer U. T. Directed Evolution of a Halide Methyltransferase Enables Biocatalytic Synthesis of Diverse SAM Analogs. Angew. Chem., Int. Ed. Engl. 2021, 60 (3), 1524–1527. 10.1002/anie.202013871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J. L.; Dunn M. K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26 (10), 2700–2707. 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Cheung-Lee W. L.; Link A. J. Genome mining for lasso peptides: past, present, and future. Journal of Industrial Microbiology and Biotechnology 2019, 46 (9–10), 1371–1379. 10.1007/s10295-019-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Li F.; Qiu W.; Xu B.; Li Y.; Lian X.; Yu H.; Zhang Z.; Wang J.; Li Z.; Xue W.; Zhu F. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022, 50 (D1), D560–D570. 10.1093/nar/gkab926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Diolintzi A.; Storch J. Fatty acid-binding proteins: functional understanding and diagnostic implications. Current Opinion in Clinical Nutrition & Metabolic Care 2019, 22 (6), 407–417. 10.1097/MCO.0000000000000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B. A.; Stubbs H. E.; Agarwal R.; Yamakawa I.; Luong K.; Solakyildirim K.; Yu H.; Hadadianpour A.; Castro M. A.; Fialkowski K. P.; Morrison K. M.; Wawrzak Z.; Chen X.; Lebrilla C. B.; Baudry J.; Smith J. C.; Sullam P. M.; Iverson T. M. Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation. Nat. Commun. 2022, 13 (1), 2753. 10.1038/s41467-022-30509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan H.; Dingfelder F.; Butté A.; Lorenzen N.; Sokolov M.; Arosio P. Machine Learning for Biologics: Opportunities for Protein Engineering, Developability, and Formulation. Trends Pharmacol. Sci. 2021, 42 (3), 151–165. 10.1016/j.tips.2020.12.004. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Ran X.; Yang Z. J. Data-driven enzyme engineering to identify function-enhancing enzymes. Protein Engineering, Design and Selection 2023, 36, gzac009. 10.1093/protein/gzac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka A.; Chovancova E.; Damborsky J. HotSpot Wizard: a web server for identification of hot spots in protein engineering. Nucleic Acids Res. 2009, 37 (suppl_2), W376–W383. 10.1093/nar/gkp410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborsky J.; Brezovsky J. Computational tools for designing and engineering enzymes. Curr. Opin. Chem. Biol. 2014, 19, 8–16. 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Romero P. A.; Arnold F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10 (12), 866–876. 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A.; Rogov P.; Wang L.; Gnirke A.; Mikkelsen T. S.. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. bioRxiv, June 10, 2014. ver. 1. 10.1101/004317. [DOI] [PMC free article] [PubMed]
- Fowler D. M.; Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods 2014, 11 (8), 801–807. 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C. L.; Fowler D. M. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011, 29 (9), 435–442. 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kries H.; Blomberg R.; Hilvert D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 2013, 17 (2), 221–228. 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Hilvert D. Design of Protein Catalysts. Annu. Rev. Biochem. 2013, 82 (1), 447–470. 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- Bunzel H. A.; Garrabou X.; Pott M.; Hilvert D. Speeding up enzyme discovery and engineering with ultrahigh-throughput methods. Curr. Opin. Struct. Biol. 2018, 48, 149–156. 10.1016/j.sbi.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Zeymer C.; Hilvert D. Directed Evolution of Protein Catalysts. Annu. Rev. Biochem. 2018, 87 (1), 131–157. 10.1146/annurev-biochem-062917-012034. [DOI] [PubMed] [Google Scholar]
- Yeh A. H.-W.; Norn C.; Kipnis Y.; Tischer D.; Pellock S. J.; Evans D.; Ma P.; Lee G. R.; Zhang J. Z.; Anishchenko I.; Coventry B.; Cao L.; Dauparas J.; Halabiya S.; DeWitt M.; Carter L.; Houk K. N.; Baker D. De novo design of luciferases using deep learning. Nature 2023, 614 (7949), 774–780. 10.1038/s41586-023-05696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.; Kortemme T. Recent advances in de novo protein design: Principles, methods, and applications. J. Biol. Chem. 2021, 296, 100558. 10.1016/j.jbc.2021.100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korendovych I. V.; DeGrado W. F. De novo protein design, a retrospective. Q. Rev. Biophys. 2020, 53, e3. 10.1017/S0033583519000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S.; Boyken S. E.; Baker D. The coming of age of de novo protein design. Nature 2016, 537 (7620), 320–327. 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Xun G.; Feng Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnology Advances 2019, 37 (4), 530–537. 10.1016/j.biotechadv.2018.10.011. [DOI] [PubMed] [Google Scholar]
- Rosenfeld L.; Heyne M.; Shifman J. M.; Papo N. Protein Engineering by Combined Computational and In Vitro Evolution Approaches. Trends Biochem. Sci. 2016, 41 (5), 421–433. 10.1016/j.tibs.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Vaissier Welborn V.; Head-Gordon T. Computational Design of Synthetic Enzymes. Chem. Rev. 2019, 119 (11), 6613–6630. 10.1021/acs.chemrev.8b00399. [DOI] [PubMed] [Google Scholar]
- Khersonsky O.; Lipsh R.; Avizemer Z.; Ashani Y.; Goldsmith M.; Leader H.; Dym O.; Rogotner S.; Trudeau D. L.; Prilusky J.; Amengual-Rigo P.; Guallar V.; Tawfik D. S.; Fleishman S. J. Automated Design of Efficient and Functionally Diverse Enzyme Repertoires. Mol. Cell 2018, 72 (1), 178–186.e5. 10.1016/j.molcel.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso V. A.; Romero-Rivera A.; Gutierrez-Rus L. I.; Ortega-Muñoz M.; Santoyo-Gonzalez F.; Gavira J. A.; Sanchez-Ruiz J. M.; Kamerlin S. C. L. Enhancing a de novo enzyme activity by computationally-focused ultra-low-throughput screening. Chemical Science 2020, 11 (24), 6134–6148. 10.1039/D0SC01935F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petřek M.; Otyepka M.; Banáš P.; Košinová P.; Koča J.; Damborský J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics 2006, 7 (1), 316. 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Mehmood R.; Wang M.; Qi H. W.; Steeves A. H.; Kulik H. J. Revealing quantum mechanical effects in enzyme catalysis with large-scale electronic structure simulation. Reaction Chemistry & Engineering 2019, 4 (2), 298–315. 10.1039/C8RE00213D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X.; Kazemi M.; Planas F.; Himo F. Modeling Enzymatic Enantioselectivity using Quantum Chemical Methodology. ACS Catal. 2020, 10 (11), 6430–6449. 10.1021/acscatal.0c00983. [DOI] [Google Scholar]
- Kiss G.; Çelebi-Ölçüm N.; Moretti R.; Baker D.; Houk K. N. Computational Enzyme Design. Angew. Chem., Int. Ed. 2013, 52 (22), 5700–5725. 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- Sheng X.; Himo F. The Quantum Chemical Cluster Approach in Biocatalysis. Acc. Chem. Res. 2023, 56 (8), 938–947. 10.1021/acs.accounts.2c00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeiros-Borja C. E.; Surpeta B.; Brezovsky J. Recent advances in user-friendly computational tools to engineer protein function. Briefings in Bioinformatics 2021, 22 (3), bbaa150. 10.1093/bib/bbaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh J.; Saraogee A.; Romero P. A.. Data-driven Protein Engineering. In Protein Engineering: Tools and Applications; Zhao H., Ed.; Wiley-VCH, 2021; pp 133–151. [Google Scholar]
- Yang K. K.; Wu Z.; Arnold F. H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16 (8), 687–694. 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Verma D.; Sheridan R. P.; Liaw A.; Ma J.; Marshall N. M.; McIntosh J.; Sherer E. C.; Svetnik V.; Johnston J. M. Deep Dive into Machine Learning Models for Protein Engineering. J. Chem. Inf. Model. 2020, 60 (6), 2773–2790. 10.1021/acs.jcim.0c00073. [DOI] [PubMed] [Google Scholar]
- Alley E. C.; Khimulya G.; Biswas S.; AlQuraishi M.; Church G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16 (12), 1315–1322. 10.1038/s41592-019-0598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B.; Ran X.; Gollu A.; Cheng Z.; Zhou X.; Chen Y.; Yang Z. J. IntEnzyDB: an Integrated Structure–Kinetics Enzymology Database. J. Chem. Inf. Model. 2022, 62 (22), 5841–5848. 10.1021/acs.jcim.2c01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B.; Ran X.; Jiang Y.; Torrence S. K.; Yuan L.; Shao Q.; Yang Z. J. Rate-Perturbing Single Amino Acid Mutation for Hydrolases: A Statistical Profiling. J. Phys. Chem. B 2021, 125 (38), 10682–10691. 10.1021/acs.jpcb.1c05901. [DOI] [PubMed] [Google Scholar]
- Shao Q.; Jiang Y.; Yang Z. J. EnzyHTP Computational Directed Evolution with Adaptive Resource Allocation. J. Chem. Inf. Model. 2023, 63, 5650. 10.1021/acs.jcim.3c00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q.; Jiang Y.; Yang Z. J. EnzyHTP: A High-Throughput Computational Platform for Enzyme Modeling. J. Chem. Inf. Model. 2022, 62 (3), 647–655. 10.1021/acs.jcim.1c01424. [DOI] [PubMed] [Google Scholar]
- Juarez R. J.; Jiang Y.; Tremblay M.; Shao Q.; Link A. J.; Yang Z. J. LassoHTP: A High-Throughput Computational Tool for Lasso Peptide Structure Construction and Modeling. J. Chem. Inf. Model. 2023, 63 (2), 522–530. 10.1021/acs.jcim.2c00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Yan B.; Chen Y.; Juarez R. J.; Yang Z. J. Molecular Dynamics-Derived Descriptor Informs the Impact of Mutation on the Catalytic Turnover Number in Lactonase Across Substrates. J. Phys. Chem. B 2022, 126 (13), 2486–2495. 10.1021/acs.jpcb.2c00142. [DOI] [PubMed] [Google Scholar]
- Ran X.; Jiang Y.; Shao Q.; Yang Z. J., EnzyKR: A Chirality-Aware Deep Learning Model for Predicting the Outcomes of the Hydrolase-Catalyzed Kinetic Resolution. ChemRxiv, May 2023, 2023, ver. 2. [DOI] [PMC free article] [PubMed]
- Thokkadam A.; Do T.; Ran X.; Brynildsen M. P.; Yang Z. J.; Link A. J. High-Throughput Screen Reveals the Structure–Activity Relationship of the Antimicrobial Lasso Peptide Ubonodin. ACS Central Science 2023, 9 (3), 540–550. 10.1021/acscentsci.2c01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Stull S. L.; Shao Q.; Yang Z. J. Convergence in determining enzyme functional descriptors across Kemp eliminase variants. Electronic Structure 2022, 4 (4), 044007 10.1088/2516-1075/acad51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurich C.; Yang Z. J. High-throughput computational investigation of protein electrostatics and cavity for SAM-dependent methyltransferases. Protein Sci. 2023, 32 (7), e4690. 10.1002/pro.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Ding N.; Shao Q.; Stull S.; Cheng Z.; Yang Z.. Investigating the Non-Electrostatic Component of Substrate Positioning Dynamics. ChemRxiv, June 23, 2023, ver. 2. [DOI] [PMC free article] [PubMed]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28 (1), 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A.; Darsow M.; Degtyarenko K.; Fleischmann W.; Boyce S.; Axelsen K. B.; Bairoch A.; Schomburg D.; Tipton K. F.; Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004, 32, D434–D437. 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.; Jeske L.; Ulbrich S.; Hofmann J.; Koblitz J.; Schomburg I.; Neumann-Schaal M.; Jahn D.; Schomburg D. BRENDA, the ELIXIR core data resource in 2021: new developments and updates. Nucleic Acids Res. 2021, 49 (D1), D498–D508. 10.1093/nar/gkaa1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig U.; Kania R.; Golebiewski M.; Rey M.; Shi L.; Jong L.; Algaa E.; Weidemann A.; Sauer-Danzwith H.; Mir S.; Krebs O.; Bittkowski M.; Wetsch E.; Rojas I.; Müller W. SABIO-RK—database for biochemical reaction kinetics. Nucleic Acids Res. 2012, 40 (D1), D790–D796. 10.1093/nar/gkr1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y.; Chang P. M.; Ary M. L.; Allen B. D.; Chica R. A.; Mayo S. L.; Olafson B. D. ProtaBank: A repository for protein design and engineering data. Protein Sci. 2018, 27 (6), 1113–1124. 10.1002/pro.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Kim D.; Huang P.; Vater A.; Siegel J. B.. Design to Data for mutants of β-glucosidase B from Paenibacillus polymyxa: Q22T, W123R, F155G, Y169M, W438D, V401A. bioRxiv, Dec. 29, 2019, ver. 1. 10.1101/2019.12.23.887380 [DOI]
- Li F.; Zhang X.; Renata H. Enzymatic CH functionalizations for natural product synthesis. Curr. Opin. Chem. Biol. 2019, 49, 25–32. 10.1016/j.cbpa.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott B. C.; Erickson E.; Allen M. D.; Gado J. E.; Graham R.; Kearns F. L.; Pardo I.; Topuzlu E.; Anderson J. J.; Austin H. P.; Dominick G.; Johnson C. W.; Rorrer N. A.; Szostkiewicz C. J.; Copié V.; Payne C. M.; Woodcock H. L.; Donohoe B. S.; Beckham G. T.; McGeehan J. E. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (41), 25476–25485. 10.1073/pnas.2006753117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorrer N. A.; Nicholson S.; Carpenter A.; Biddy M. J.; Grundl N. J.; Beckham G. T. Combining Reclaimed PET with Bio-based Monomers Enables Plastics Upcycling. Joule 2019, 3 (4), 1006–1027. 10.1016/j.joule.2019.01.018. [DOI] [Google Scholar]
- Wang J.-B.; Ilie A.; Yuan S.; Reetz M. T. Investigating Substrate Scope and Enantioselectivity of a Defluorinase by a Stereochemical Probe. J. Am. Chem. Soc. 2017, 139 (32), 11241–11247. 10.1021/jacs.7b06019. [DOI] [PubMed] [Google Scholar]
- Goldman P. The Carbon-Fluorine Bond in Compounds of Biological Interest. Science 1969, 164 (3884), 1123–1130. 10.1126/science.164.3884.1123. [DOI] [PubMed] [Google Scholar]
- Gan J.; Siegel J. B.; German J. B. Molecular annotation of food – Towards personalized diet and precision health. Trends in Food Science & Technology 2019, 91, 675–680. 10.1016/j.tifs.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr S.; Harvey M. J.; Noé F.; De Fabritiis G. HTMD: High-Throughput Molecular Dynamics for Molecular Discovery. J. Chem. Theory Comput. 2016, 12 (4), 1845–1852. 10.1021/acs.jctc.6b00049. [DOI] [PubMed] [Google Scholar]
- Godwin R. C.; Melvin R.; Salsbury F. R.. Molecular Dynamics Simulations and Computer-Aided Drug Discovery. Springer: New York, 2015; pp 1–30. [Google Scholar]
- Parton D. L.; Grinaway P. B.; Hanson S. M.; Beauchamp K. A.; Chodera J. D. Ensembler: Enabling High-Throughput Molecular Simulations at the Superfamily Scale. PLoS Comput. Biol. 2016, 12 (6), e1004728 10.1371/journal.pcbi.1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein B. A.; Steffen-Munsberg F.; Szeler I.; Purg M.; Kulkarni Y.; Kamerlin S. C. L. CADEE: Computer-Aided Directed Evolution of Enzymes. IUCrJ. 2017, 4 (1), 50–64. 10.1107/S2052252516018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H.; Mehrabi P.; Ren Z.; Sljoka A.; Ing C.; Bezginov A.; Ye L.; Pomès R.; Prosser R. S.; Pai E. F. The role of dimer asymmetry and protomer dynamics in enzyme catalysis. Science 2017, 355 (6322), eaag2355. 10.1126/science.aag2355. [DOI] [PubMed] [Google Scholar]
- Makinen M. W.; Fink A. L. Reactivity and Cryoenzymology of Enzymes in the Crystalline State. Annu. Rev. Biophys. Bioeng. 1977, 6 (1), 301–343. 10.1146/annurev.bb.06.060177.001505. [DOI] [PubMed] [Google Scholar]
- Mehrabi P.; Di Pietrantonio C.; Kim T. H.; Sljoka A.; Taverner K.; Ing C.; Kruglyak N.; Pomès R.; Pai E. F.; Prosser R. S. Substrate-Based Allosteric Regulation of a Homodimeric Enzyme. J. Am. Chem. Soc. 2019, 141 (29), 11540–11556. 10.1021/jacs.9b03703. [DOI] [PubMed] [Google Scholar]
- Mehrabi P.; Schulz E. C.; Dsouza R.; Müller-Werkmeister H. M.; Tellkamp F.; Miller R. J. D.; Pai E. F. Time-resolved crystallography reveals allosteric communication aligned with molecular breathing. Science 2019, 365 (6458), 1167–1170. 10.1126/science.aaw9904. [DOI] [PubMed] [Google Scholar]
- Schulz E. C.; Mehrabi P.; Müller-Werkmeister H. M.; Tellkamp F.; Jha A.; Stuart W.; Persch E.; De Gasparo R.; Diederich F.; Pai E. F.; Miller R. J. D. The hit-and-return system enables efficient time-resolved serial synchrotron crystallography. Nat. Methods 2018, 15 (11), 901–904. 10.1038/s41592-018-0180-2. [DOI] [PubMed] [Google Scholar]
- Yue Y.; Fan J.; Xin G.; Huang Q.; Wang J.-B.; Li Y.; Zhang Q.; Wang W. Comprehensive Understanding of Fluoroacetate Dehalogenase-Catalyzed Degradation of Fluorocarboxylic Acids: A QM/MM Approach. Environ. Sci. Technol. 2021, 55 (14), 9817–9825. 10.1021/acs.est.0c08811. [DOI] [PubMed] [Google Scholar]
- Weber W.; Fischli W.; Hochuli E.; Kupfer E.; Weibel E. K. Anantin-a peptide antagonist of the atrial natriuretic factor (ANF). Journal of antibiotics 1991, 44 (2), 164–171. 10.7164/antibiotics.44.164. [DOI] [PubMed] [Google Scholar]
- Salomon R. A.; Farías R. N. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. Journal of bacteriology 1992, 174 (22), 7428–7435. 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine K. L.; Friedrichs M. S.; Detlefsen D.; Nishio M.; Tsunakawa M.; Furumai T.; Ohkuma H.; Oki T.; Hill S.; Bruccoleri R. E.; et al. High-resolution solution structure of siamycin II: novel amphipathic character of a 21-residue peptide that inhibits HIV fusion. Journal of biomolecular NMR 1995, 5 (3), 271–286. 10.1007/BF00211754. [DOI] [PubMed] [Google Scholar]
- Tsunakawa M.; Hu S.-L.; Hoshino Y.; Detlefson D. J.; Hill S. E.; Furumai T.; White R. J.; Nishio M.; Kawano K.; Yamamoto S.; et al. Siamycins I and II, new anti-HIV peptides: I. Fermentation, isolation, biological activity and initial characterization. Journal of antibiotics 1995, 48 (5), 433–434. 10.7164/antibiotics.48.433. [DOI] [PubMed] [Google Scholar]
- Pan S. J.; Cheung W. L.; Fung H. K.; Floudas C. A.; Link A. J. Computational design of the lasso peptide antibiotic microcin J25. Protein Engineering, Design and Selection 2011, 24 (3), 275–282. 10.1093/protein/gzq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braffman N. R.; Piscotta F. J.; Hauver J.; Campbell E. A.; Link A. J.; Darst S. A. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (4), 1273. 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbiati J. O.; Ciaccio M.; Farías R. N.; González-Pastor J. E.; Moreno F.; Salomón R. A. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. Journal of bacteriology 1999, 181 (8), 2659–2662. 10.1128/JB.181.8.2659-2662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Lee W. L.; Link A. J. Genome mining for lasso peptides: past, present, and future. J. Ind. Microbiol. Biotechnol. 2019, 1–9. 10.1007/s10295-019-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T.; Link A. J. Protein Engineering in Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs). Biochemistry 2023, 62, 201–209. 10.1021/acs.biochem.1c00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen A. R.; Katelyn V. C.; Asim K. B.; Alex K.; Simon K.; Joshmyn De La C.; Victor A.; Guangfeng Z.; Frank D.; Sergey O.; Gaurav B.. Cyclic peptide structure prediction and design using AlphaFold. bioRxiv, Feb. 26, 2023, ver. 1. 10.1101/2023.02.25.529956 [DOI]
- Case D.A.; Cerutti D. S.; Cheatham T.E. III; Darden T. A.; Duke R.E.; Giese T.J.; Gohlke H.; Goetz A.W.; Greene D.; Homeyer N.; Izadi S.; Kovalenko A.; Lee T.S.; LeGrand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Mermelstein D.; Merz K. M.; Monard G.; Nguyen H.; Omelyan I.; Onufriev A.; Pan F.; Qi R.; Roe D.R.; Roitberg A.; Sagui C.; Simmerling C. L.; Botello-Smith W. M.; Swails J.; Walker R.C.; Wang J.; Wolf R.M.; Wu X.; Xiao L.; York D.M.; Kollman P.A.. AMBER 2017; University of California, San Francisco, 2017. [Google Scholar]
- Hegemann J. D.; Fage C. D.; Zhu S.; Harms K.; Di Leva F. S.; Novellino E.; Marinelli L.; Marahiel M. A. The ring residue proline 8 is crucial for the thermal stability of the lasso peptide caulosegnin II. Molecular BioSystems 2016, 12 (4), 1106–1109. 10.1039/C6MB00081A. [DOI] [PubMed] [Google Scholar]
- Zong C.; Wu M. J.; Qin J. Z.; Link A. J. Lasso Peptide Benenodin-1 Is a Thermally Actuated [1]Rotaxane Switch. J. Am. Chem. Soc. 2017, 139 (30), 10403–10409. 10.1021/jacs.7b04830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Lee W. L.; Parry M. E.; Jaramillo Cartagena A.; Darst S. A.; Link A. J. Discovery and structure of the antimicrobial lasso peptide citrocin. J. Biol. Chem. 2019, 294 (17), 6822–6830. 10.1074/jbc.RA118.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe T. A.; Manzenrieder F.; Mas-Moruno C.; Linne U.; Sasse F.; Kessler H.; Xie X.; Marahiel M. A. Introducing lasso peptides as molecular scaffolds for drug design: engineering of an integrin antagonist. Angew. Chem., Int. Ed. 2011, 50 (37), 8714–8717. 10.1002/anie.201102190. [DOI] [PubMed] [Google Scholar]
- Metelev M.; Tietz J. I.; Melby J. O.; Blair P. M.; Zhu L.; Livnat I.; Severinov K.; Mitchell D. A. Structure, Bioactivity, and Resistance Mechanism of Streptomonomicin, an Unusual Lasso Peptide from an Understudied Halophilic Actinomycete. Chemistry & Biology 2015, 22 (2), 241–250. 10.1016/j.chembiol.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T.; Thokkadam A.; Leach R.; Link A. J. Phenotype-Guided Comparative Genomics Identifies the Complete Transport Pathway of the Antimicrobial Lasso Peptide Ubonodin in Burkholderia. ACS Chem. Biol. 2022, 17, 2332. 10.1021/acschembio.2c00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Lee W. L.; Parry M. E.; Zong C.; Cartagena A. J.; Darst S. A.; Connell N. D.; Russo R.; Link A. J. Discovery of ubonodin, an antimicrobial lasso peptide active against members of the Burkholderia cepacia complex. ChemBioChem. 2020, 21 (9), 1335–1340. 10.1002/cbic.201900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann J. D.; Zimmermann M.; Zhu S.; Steuber H.; Harms K.; Xie X.; Marahiel M. A. Xanthomonins I–III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring. Angew. Chem., Int. Ed. 2014, 53 (8), 2230–2234. 10.1002/anie.201309267. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Hajlasz N.; Kulik H. J. Computational Modeling of Conformer Stability in Benenodin-1, a Thermally Actuated Lasso Peptide Switch. J. Phys. Chem. B 2022, 126 (18), 3398–3406. 10.1021/acs.jpcb.2c00762. [DOI] [PubMed] [Google Scholar]
- Young T. A.; Silcock J. J.; Sterling A. J.; Duarte F. autodE: Automated Calculation of Reaction Energy Profiles— Application to Organic and Organometallic Reactions. Angew. Chem. 2021, 133 (8), 4312–4320. 10.1002/ange.202011941. [DOI] [PubMed] [Google Scholar]
- An Q.; Shen Y.; Fortunelli A.; Goddard W. A. QM-Mechanism-Based Hierarchical High-Throughput in Silico Screening Catalyst Design for Ammonia Synthesis. J. Am. Chem. Soc. 2018, 140 (50), 17702–17710. 10.1021/jacs.8b10499. [DOI] [PubMed] [Google Scholar]
- Colón Y. J.; Snurr R. Q. High-throughput computational screening of metal–organic frameworks. Chem. Soc. Rev. 2014, 43 (16), 5735–5749. 10.1039/C4CS00070F. [DOI] [PubMed] [Google Scholar]
- Gan Y.; Miao N.; Lan P.; Zhou J.; Elliott S. R.; Sun Z. Robust Design of High-Performance Optoelectronic Chalcogenide Crystals from High-Throughput Computation. J. Am. Chem. Soc. 2022, 144 (13), 5878–5886. 10.1021/jacs.1c12620. [DOI] [PubMed] [Google Scholar]
- McInnes C. Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 2007, 11 (5), 494–502. 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Li Z.; Li X.; Huang Y.-Y.; Wu Y.; Liu R.; Zhou L.; Lin Y.; Wu D.; Zhang L.; Liu H.; Xu X.; Yu K.; Zhang Y.; Cui J.; Zhan C.-G.; Wang X.; Luo H.-B. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (44), 27381–27387. 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welborn V. V.; Head-Gordon T. Fluctuations of Electric Fields in the Active Site of the Enzyme Ketosteroid Isomerase. J. Am. Chem. Soc. 2019, 141 (32), 12487–12492. 10.1021/jacs.9b05323. [DOI] [PubMed] [Google Scholar]
- Bhowmick A.; Sharma S. C.; Head-Gordon T. The Importance of the Scaffold for de Novo Enzymes: A Case Study with Kemp Eliminase. J. Am. Chem. Soc. 2017, 139 (16), 5793–5800. 10.1021/jacs.6b12265. [DOI] [PubMed] [Google Scholar]
- Vaissier V.; Sharma S. C.; Schaettle K.; Zhang T.; Head-Gordon T. Computational Optimization of Electric Fields for Improving Catalysis of a Designed Kemp Eliminase. ACS Catal. 2018, 8 (1), 219–227. 10.1021/acscatal.7b03151. [DOI] [Google Scholar]
- Yang Z.; Liu F.; Steeves A. H.; Kulik H. J. Quantum Mechanical Description of Electrostatics Provides a Unified Picture of Catalytic Action Across Methyltransferases. J. Phys. Chem. Lett. 2019, 10 (13), 3779–3787. 10.1021/acs.jpclett.9b01555. [DOI] [PubMed] [Google Scholar]
- Bím D.; Alexandrova A. N. Local Electric Fields As a Natural Switch of Heme-Iron Protein Reactivity. ACS Catal. 2021, 11 (11), 6534–6546. 10.1021/acscatal.1c00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari J.; Schaller K.; Molina G. A.; Borch K.; Westh P. The Sabatier principle as a tool for discovery and engineering of industrial enzymes. Curr. Opin. Biotechnol. 2022, 78, 102843. 10.1016/j.copbio.2022.102843. [DOI] [PubMed] [Google Scholar]
- Arnling Bååth J.; Jensen K.; Borch K.; Westh P.; Kari J. Sabatier Principle for Rationalizing Enzymatic Hydrolysis of a Synthetic Polyester. JACS Au 2022, 2 (5), 1223–1231. 10.1021/jacsau.2c00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller K. S.; Molina G. A.; Kari J.; Schiano-di-Cola C.; Sørensen T. H.; Borch K.; Peters G. H.J.; Westh P. Virtual Bioprospecting of Interfacial Enzymes: Relating Sequence and Kinetics. ACS Catal. 2022, 12 (12), 7427–7435. 10.1021/acscatal.2c02305. [DOI] [Google Scholar]
- Vaissier Welborn V.; Head-Gordon T. Computational Design of Synthetic Enzymes. Chem. Rev. 2019, 119 (11), 6613–6630. 10.1021/acs.chemrev.8b00399. [DOI] [PubMed] [Google Scholar]
- Mehmood R.; Vennelakanti V.; Kulik H. J. Spectroscopically Guided Simulations Reveal Distinct Strategies for Positioning Substrates to Achieve Selectivity in Nonheme Fe(II)/α-Ketoglutarate-Dependent Halogenases. ACS Catal. 2021, 11 (19), 12394–12408. 10.1021/acscatal.1c03169. [DOI] [Google Scholar]
- Broom A.; Rakotoharisoa R. V.; Thompson M. C.; Zarifi N.; Nguyen E.; Mukhametzhanov N.; Liu L.; Fraser J. S.; Chica R. A. Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico. Nat. Commun. 2020, 11 (1), 4808. 10.1038/s41467-020-18619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. B.; Zanghellini A.; Lovick H. M.; Kiss G.; Lambert A. R.; St.Clair J. L.; Gallaher J. L.; Hilvert D.; Gelb M. H.; Stoddard B. L.; Houk K. N.; Michael F. E.; Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 2010, 329 (5989), 309–13. 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O.; Kiss G.; Rothlisberger D.; Dym O.; Albeck S.; Houk K. N.; Baker D.; Tawfik D. S. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (26), 10358–63. 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S.; Bruice T. C. The near attack conformation approach to the study of the chorismate to prephenate reaction. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (21), 12015–12020. 10.1073/pnas.1534873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkcuoglu Z.; Bakan A.; Kocaman D.; Bahar I.; Doruker P. Coupling between Catalytic Loop Motions and Enzyme Global Dynamics. PLOS Computational Biology 2012, 8 (9), e1002705 10.1371/journal.pcbi.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q.; Kulkarni Y.; Sengupta U.; Petrović D.; Mulholland A. J.; van der Kamp M. W.; Strodel B.; Kamerlin S. C. L. Loop Motion in Triosephosphate Isomerase Is Not a Simple Open and Shut Case. J. Am. Chem. Soc. 2018, 140 (46), 15889–15903. 10.1021/jacs.8b09378. [DOI] [PubMed] [Google Scholar]
- Masterson J. E.; Schwartz S. D. Evolution alters the enzymatic reaction coordinate of dihydrofolate reductase. J. Phys. Chem. B 2015, 119 (3), 989–96. 10.1021/jp506373q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Antoniou D.; Schwartz S. D.; Schramm V. L. Hydride Transfer in DHFR by Transition Path Sampling, Kinetic Isotope Effects, and Heavy Enzyme Studies. Biochemistry 2016, 55 (1), 157–66. 10.1021/acs.biochem.5b01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. T.; Layfield J. P.; Stewart R. J. 3rd; French J. B.; Hanoian P.; Asbury J. B.; Hammes-Schiffer S.; Benkovic S. J. Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase. J. Am. Chem. Soc. 2014, 136 (29), 10349–60. 10.1021/ja5038947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. T.; Francis K.; Layfield J. P.; Huang X.; Hammes-Schiffer S.; Kohen A.; Benkovic S. J. Escherichia coli dihydrofolate reductase catalyzed proton and hydride transfers: temporal order and the roles of Asp27 and Tyr100. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (51), 18231–6. 10.1073/pnas.1415940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitakrishnan R. P.; Zaborowski E.; McElheny D.; Benkovic S. J.; Dyson H. J.; Wright P. E. Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle. Biochemistry 2004, 43 (51), 16046–55. 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- Bhabha G.; Ekiert D. C.; Jennewein M.; Zmasek C. M.; Tuttle L. M.; Kroon G.; Dyson H. J.; Godzik A.; Wilson I. A.; Wright P. E. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat. Struct Mol. Biol. 2013, 20 (11), 1243–9. 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.; Thompson E. J.; Barrow S. L.; Zhang W.; Iavarone A. T.; Klinman J. P. Hydrogen-Deuterium Exchange within Adenosine Deaminase, a TIM Barrel Hydrolase, Identifies Networks for Thermal Activation of Catalysis. J. Am. Chem. Soc. 2020, 142 (47), 19936–19949. 10.1021/jacs.0c07866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunzel H. A.; Kries H.; Marchetti L.; Zeymer C.; Mittl P. R. E.; Mulholland A. J.; Hilvert D. Emergence of a Negative Activation Heat Capacity during Evolution of a Designed Enzyme. J. Am. Chem. Soc. 2019, 141 (30), 11745–11748. 10.1021/jacs.9b02731. [DOI] [PubMed] [Google Scholar]
- Afriat L.; Roodveldt C.; Manco G.; Tawfik D. S. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 2006, 45 (46), 13677–86. 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- Hiblot J.; Gotthard G.; Elias M.; Chabriere E. Differential active site loop conformations mediate promiscuous activities in the lactonase SsoPox. PLoS One 2013, 8 (9), e75272 10.1371/journal.pone.0075272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng F. S.; Wright D. M.; Seah S. Y. Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 2011, 77 (4), 1181–6. 10.1128/AEM.01642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdrenga J.; Daude D.; Remy B.; Jacquet P.; Plener L.; Elias M.; Chabriere E. Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact 2017, 267, 104–115. 10.1016/j.cbi.2016.05.028. [DOI] [PubMed] [Google Scholar]
- Billot R.; Plener L.; Jacquet P.; Elias M.; Chabriere E.; Daude D. Engineering acyl-homoserine lactone-interfering enzymes toward bacterial control. J. Biol. Chem. 2020, 295 (37), 12993–13007. 10.1074/jbc.REV120.013531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar R.; Elias M. Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: a review of recent advances. Expert Rev. Anti-Infe 2020, 18 (12), 1221–1233. 10.1080/14787210.2020.1794815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonar K.; Mancera R. L. Characterization of the Conformations of Amyloid Beta 42 in Solution That May Mediate Its Initial Hydrophobic Aggregation. J. Phys. Chem. B 2022, 126 (40), 7916–7933. 10.1021/acs.jpcb.2c04743. [DOI] [PubMed] [Google Scholar]
- Pinheiro M. P.; Rios N. S.; Fonseca T. d. S.; Bezerra F. d. A.; Rodríguez-Castellón E.; Fernandez-Lafuente R.; Carlos de Mattos M.; Dos Santos J. C.; Gonçalves L. R. Kinetic resolution of drug intermediates catalyzed by lipase B from Candida antarctica immobilized on immobead-350. Biotechnol. Prog. 2018, 34 (4), 878–889. 10.1002/btpr.2630. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U. T.; Kazlauskas R. J.. Hydrolases in organic synthesis: regio-and stereoselective biotransformations; John Wiley & Sons: 2006. [Google Scholar]
- Lee J.; Oh Y.; Choi Y. K.; Choi E.; Kim K.; Park J.; Kim M.-J. Dynamic kinetic resolution of diarylmethanols with an activated lipoprotein lipase. ACS Catal. 2015, 5 (2), 683–689. 10.1021/cs501629m. [DOI] [Google Scholar]
- Bassegoda A.; Nguyen G. S.; Schmidt M.; Kourist R.; Diaz P.; Bornscheuer U. T. Rational protein design of Paenibacillus barcinonensis esterase EstA for kinetic resolution of tertiary alcohols. ChemCatChem. 2010, 2 (8), 962–967. 10.1002/cctc.201000053. [DOI] [Google Scholar]
- Kazlauskas R. J.; Weissfloch A. N.; Rappaport A. T.; Cuccia L. A. A rule to predict which enantiomer of a secondary alcohol reacts faster in reactions catalyzed by cholesterol esterase, lipase from Pseudomonas cepacia, and lipase from Candida rugosa. Journal of Organic Chemistry 1991, 56 (8), 2656–2665. 10.1021/jo00008a016. [DOI] [Google Scholar]
- Tomić S.; Kojić-Prodić B. A quantitative model for predicting enzyme enantioselectivity: application to Burkholderia cepacia lipase and 3-(aryloxy)-1, 2-propanediol derivatives. Journal of Molecular Graphics and Modelling 2002, 21 (3), 241–252. 10.1016/S1093-3263(02)00148-1. [DOI] [PubMed] [Google Scholar]
- Cadet F.; Fontaine N.; Li G.; Sanchis J.; Ng Fuk Chong M.; Pandjaitan R.; Vetrivel I.; Offmann B.; Reetz M. T. A machine learning approach for reliable prediction of amino acid interactions and its application in the directed evolution of enantioselective enzymes. Sci. Rep. 2018, 8 (1), 16757. 10.1038/s41598-018-35033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann D.; Lloyd C. J.; Mih N.; Ha Y.; Zielinski D. C.; Haiman Z. B.; Desouki A. A.; Lercher M. J.; Palsson B. O. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models. Nat. Commun. 2018, 9 (1), 5252. 10.1038/s41467-018-07652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Yuan L.; Lu H.; Li G.; Chen Y.; Engqvist M. K.; Kerkhoven E. J.; Nielsen J. Deep learning-based k cat prediction enables improved enzyme-constrained model reconstruction. Nature Catalysis 2022, 5 (8), 662–672. 10.1038/s41929-022-00798-z. [DOI] [Google Scholar]
- Jiang Y.; Ran X.; Yang Z. J. Data-driven enzyme engineering to identify function-enhancing enzymes. Protein Engineering, Design and Selection 2022, gzac009. 10.1093/protein/gzac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Tian S.; Yue Y.; Li M.; Tong W.; Xu G.; Chen B.; Ma M.; Li Y.; Wang J.-b. Semirational design of fluoroacetate dehalogenase RPA1163 for kinetic resolution of α-fluorocarboxylic acids on a gram scale. ACS Catal. 2020, 10 (5), 3143–3151. 10.1021/acscatal.9b04804. [DOI] [Google Scholar]
- Zhang F.-R.; Wan N.-W.; Ma J.-M.; Cui B.-D.; Han W.-Y.; Chen Y.-Z. Enzymatic Kinetic Resolution of Bulky Spiro-Epoxyoxindoles via Halohydrin Dehalogenase-Catalyzed Enantio-and Regioselective Azidolysis. ACS Catal. 2021, 11 (15), 9066–9072. 10.1021/acscatal.1c02345. [DOI] [Google Scholar]
- Braffman N. R.; Piscotta F. J.; Hauver J.; Campbell E. A.; Link A. J.; Darst S. A. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (4), 1273–1278. 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Lee W. L.; Parry M. E.; Zong C.; Cartagena A. J.; Darst S. A.; Connell N. D.; Russo R.; Link A. J. Discovery of Ubonodin, an Antimicrobial Lasso Peptide Active against Members of the Burkholderia cepacia Complex. ChemBioChem. 2020, 21 (9), 1335–1340. 10.1002/cbic.201900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson D. V.; Patiño M.; Elashal H. E.; Cartagena A. J.; Zhang Y.; Whitley M. E.; So L.; Kayser-Browne A. K.; Earl A. M.; Bhattacharyya R. P.; Link A. J. Cloacaenodin, an Antimicrobial Lasso Peptide with Activity against Enterobacter. ACS Infectious Diseases 2023, 9 (1), 111–121. 10.1021/acsinfecdis.2c00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S. N.; Alvarez J. A. E.; Zabetakis D.; Walper S. A.; Malanoski A. P. PepVAE: variational autoencoder framework for antimicrobial peptide generation and activity prediction. Frontiers in microbiology 2021, 12, 725727. 10.3389/fmicb.2021.725727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan I.; Zirah S.; Mehmood S.; Choudhury H. G.; Goulard C.; Li Y.; Robinson C. V.; Rebuffat S.; Beis K. Structural basis for hijacking siderophore receptors by antimicrobial lasso peptides. Nat. Chem. Biol. 2014, 10 (5), 340–342. 10.1038/nchembio.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braffman N.; Piscotta F. J.; Hauver J.; Campbell E. A.; Link A. J.; Darst S. A. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 1273–1278. 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. A.; Kalkum M.; Ottesen J.; Yuzenkova J.; Chait B. T.; Landick R.; Muir T.; Severinov K.; Darst S. A. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 2003, 125 (41), 12475–83. 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- Fried S. D.; Boxer S. G. Electric Fields and Enzyme Catalysis. Annu. Rev. Biochem. 2017, 86, 387–415. 10.1146/annurev-biochem-061516-044432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Yan B.; Chen Y.; Juarez R. J.; Yang Z. J. Molecular Dynamics-Derived Descriptor Informs the Impact of Mutation on the Catalytic Turnover Number in Lactonase Across Substrates. J. Phys. Chem. B 2022, 126 (13), 2486–2495. 10.1021/acs.jpcb.2c00142. [DOI] [PubMed] [Google Scholar]
- Bartlett G. J.; Porter C. T.; Borkakoti N.; Thornton J. M. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 2002, 324 (1), 105–21. 10.1016/S0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- Lodola A.; Sirirak J.; Fey N.; Rivara S.; Mor M.; Mulholland A. J. Structural Fluctuations in Enzyme-Catalyzed Reactions: Determinants of Reactivity in Fatty Acid Amide Hydrolase from Multivariate Statistical Analysis of Quantum Mechanics/Molecular Mechanics Paths. J. Chem. Theory Comput 2010, 6 (9), 2948–60. 10.1021/ct100264j. [DOI] [PubMed] [Google Scholar]
- Rothlisberger D.; Khersonsky O.; Wollacott A. M.; Jiang L.; DeChancie J.; Betker J.; Gallaher J. L.; Althoff E. A.; Zanghellini A.; Dym O.; Albeck S.; Houk K. N.; Tawfik D. S.; Baker D. Kemp elimination catalysts by computational enzyme design. Nature 2008, 453 (7192), 190–5. 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Hong N. S.; Petrovic D.; Lee R.; Gryn’ova G.; Purg M.; Saunders J.; Bauer P.; Carr P. D.; Lin C. Y.; Mabbitt P. D.; Zhang W.; Altamore T.; Easton C.; Coote M. L.; Kamerlin S. C. L.; Jackson C. J. The evolution of multiple active site configurations in a designed enzyme. Nat. Commun. 2018, 9 (1), 3900. 10.1038/s41467-018-06305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bügl H.; Fauman E. B.; Staker B. L.; Zheng F.; Kushner S. R.; Saper M. A.; Bardwell J. C.; Jakob U. RNA methylation under heat shock control. Mol. Cell 2000, 6 (2), 349–60. 10.1016/S1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Nai Y.-S.; Huang Y.-C.; Yen M.-R.; Chen P.-Y. Diversity of Fungal DNA Methyltransferases and Their Association With DNA Methylation Patterns. Frontiers in Microbiology 2021, 11, 616922. 10.3389/fmicb.2020.616922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S.; Syeda F.; Park L. DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int. J. Biochem Mol. Biol. 2011, 2 (1), 58–66. [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Lang Z.; Zhu J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19 (8), 489–506. 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- Sousa S. F.; Calixto A. R.; Ferreira P.; Ramos M. J.; Lim C.; Fernandes P. A. Activation Free Energy, Substrate Binding Free Energy, and Enzyme Efficiency Fall in a Very Narrow Range of Values for Most Enzymes. ACS Catal. 2020, 10 (15), 8444–8453. 10.1021/acscatal.0c01947. [DOI] [Google Scholar]
- Norberg A. L.; Dybvik A. I.; Zakariassen H.; Mormann M.; Peter-Katalinić J.; Eijsink V. G. H.; So̷rlie M. Substrate positioning in Chitinase A, a processive chito-biohydrolase from Serratia marcescens. FEBS Lett. 2011, 585 (14), 2339–2344. 10.1016/j.febslet.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Hamre A. G.; Jana S.; Reppert N. K.; Payne C. M.; Sorlie M. Processivity, Substrate Positioning, and Binding: The Role of Polar Residues in a Family 18 Glycoside Hydrolase. Biochemistry 2015, 54 (49), 7292–7306. 10.1021/acs.biochem.5b00830. [DOI] [PubMed] [Google Scholar]
- Patra N.; Ioannidis E. I.; Kulik H. J. Computational Investigation of the Interplay of Substrate Positioning and Reactivity in Catechol O-Methyltransferase. PLoS One 2016, 11 (8), e0161868. 10.1371/journal.pone.0161868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S. S.; Offenbacher A. R.; Thompson E. M.; Gee C. L.; Wilcoxen J.; Carr C. A. M.; Prigozhin D. M.; Yang V.; Alber T.; Britt R. D.; Fraser J. S.; Klinman J. P. Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The ″Undoing″ of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network. J. Am. Chem. Soc. 2019, 141 (4), 1555–1567. 10.1021/jacs.8b10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood R.; Qi H. W.; Steeves A. H.; Kulik H. J. The Protein’s Role in Substrate Positioning and Reactivity for Biosynthetic Enzyme Complexes: The Case of SyrB2/SyrB1. ACS Catal. 2019, 9 (6), 4930–4943. 10.1021/acscatal.9b00865. [DOI] [Google Scholar]
- Yabukarski F.; Biel J. T.; Pinney M. M.; Doukov T.; Powers A. S.; Fraser J. S.; Herschlag D. Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (52), 33204–33215. 10.1073/pnas.2011350117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood R.; Vennelakanti V.; Kulik H. J. Spectroscopically Guided Simulations Reveal Distinct Strategies for Positioning Substrates to Achieve Selectivity in Nonheme Fe(II)/alpha-Ketoglutarate-Dependent Halogenases. ACS Catal. 2021, 11 (19), 12394–12408. 10.1021/acscatal.1c03169. [DOI] [Google Scholar]
- Ruscio J. Z.; Kohn J. E.; Ball K. A.; Head-Gordon T. The Influence of Protein Dynamics on the Success of Computational Enzyme Design. J. Am. Chem. Soc. 2009, 131 (39), 14111–14115. 10.1021/ja905396s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. B.; Zanghellini A.; Lovick H. M.; Kiss G.; Lambert A. R.; St.Clair J. L.; Gallaher J. L.; Hilvert D.; Gelb M. H.; Stoddard B. L.; Houk K. N.; Michael F. E.; Baker D. Computational Design of an Enzyme Catalyst for a Stereoselective Bimolecular Diels-Alder Reaction. Science 2010, 329 (5989), 309–313. 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg R.; Kries H.; Pinkas D. M.; Mittl P. R. E.; Grutter M. G.; Privett H. K.; Mayo S. L.; Hilvert D. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 2013, 503 (7476), 418–421. 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- Broom A.; Rakotoharisoa R. V.; Thompson M. C.; Zarifi N.; Nguyen E.; Mukhametzhanov N.; Liu L.; Fraser J. S.; Chica R. A. Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico. Nat. Commun. 2020, 11 (1), 4808. 10.1038/s41467-020-18619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haataja T.; Gado J. E.; Nutt A.; Anderson N. T.; Nilsson M.; Momeni M. H.; Isaksson R.; Valjamae P.; Johansson G.; Payne C. M.; Stahlberg J. Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation. Febs J. 2023, 290 (2), 379–399. 10.1111/febs.16602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenbacher A. R.; Sharma A.; Doan P. E.; Klinman J. P.; Hoffman B. M. The Soybean Lipoxygenase-Substrate Complex: Correlation between the Properties of Tunneling-Ready States and ENDOR-Detected Structures of Ground States. Biochemistry 2020, 59 (7), 901–910. 10.1021/acs.biochem.9b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. F.; Fried S. D.; Boxer S. G. A Preorganized Electric Field Leads to Minimal Geometrical Reorientation in the Catalytic Reaction of Ketosteroid Isomerase. J. Am. Chem. Soc. 2020, 142 (22), 9993–9998. 10.1021/jacs.0c00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten R.; Padua R. A. P.; Bunzel H. A.; Nguyen V.; Pitsawong W.; Patterson M.; Sui S.; Perry S. L.; Cohen A. E.; Hilvert D.; Kern D. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 2020, 370 (6523), 1442–1446. 10.1126/science.abd3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick A.; Sharma S. C.; Honma H.; Head-Gordon T. The role of side chain entropy and mutual information for improving the de Novo design of Kemp eliminases KE07 and KE70. Phys. Chem. Chem. Phys. 2016, 18 (28), 19386–96. 10.1039/C6CP03622H. [DOI] [PubMed] [Google Scholar]
- Zuckerman D. M.; Chong L. T. Weighted Ensemble Simulation: Review of Methodology, Applications, and Software. Annual Review of Biophysics 2017, 46 (1), 43–57. 10.1146/annurev-biophys-070816-033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwary P.; Parrinello M. From Metadynamics to Dynamics. Phys. Rev. Lett. 2013, 111 (23), 230602. 10.1103/PhysRevLett.111.230602. [DOI] [PubMed] [Google Scholar]
- Miao Y.; Feher V. A.; McCammon J. A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11 (8), 3584–3595. 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.; Zhu Q.; Song R.; Ma J.; Dong H. A Two-Ended Data-Driven Accelerated Sampling Method for Exploring the Transition Pathways between Two Known States of Protein. J. Chem. Theory Comput. 2020, 16 (7), 4631–4640. 10.1021/acs.jctc.9b01184. [DOI] [PubMed] [Google Scholar]
- Yang Y. I.; Shao Q.; Zhang J.; Yang L.; Gao Y. Q. Enhanced sampling in molecular dynamics. J. Chem. Phys. 2019, 151 (7), 070902 10.1063/1.5109531. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Han J.; Wang H.; Car R.; E W. Deep Potential Molecular Dynamics: A Scalable Model with the Accuracy of Quantum Mechanics. Phys. Rev. Lett. 2018, 120 (14), 143001. 10.1103/PhysRevLett.120.143001. [DOI] [PubMed] [Google Scholar]
- Gao X.; Ramezanghorbani F.; Isayev O.; Smith J. S.; Roitberg A. E. TorchANI: A Free and Open Source PyTorch-Based Deep Learning Implementation of the ANI Neural Network Potentials. J. Chem. Inf. Model. 2020, 60 (7), 3408–3415. 10.1021/acs.jcim.0c00451. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Lei Y.-K.; Zhang Z.; Chang J.; Li M.; Han X.; Yang L.; Yang Y. I.; Gao Y. Q. A Perspective on Deep Learning for Molecular Modeling and Simulations. J. Phys. Chem. A 2020, 124 (34), 6745–6763. 10.1021/acs.jpca.0c04473. [DOI] [PubMed] [Google Scholar]
- Noé F.; Olsson S.; Köhler J.; Wu H. Boltzmann generators: Sampling equilibrium states of many-body systems with deep learning. Science 2019, 365 (6457), eaaw1147. 10.1126/science.aaw1147. [DOI] [PubMed] [Google Scholar]
- Strokach A.; Kim P. M. Deep generative modeling for protein design. Curr. Opin. Struct. Biol. 2022, 72, 226–236. 10.1016/j.sbi.2021.11.008. [DOI] [PubMed] [Google Scholar]
- Frenz B.; Lewis S. M.; King I.; DiMaio F.; Park H.; Song Y. Prediction of Protein Mutational Free Energy: Benchmark and Sampling Improvements Increase Classification Accuracy. Frontiers in Bioengineering and Biotechnology 2020, 8, 558247. 10.3389/fbioe.2020.558247. [DOI] [PMC free article] [PubMed] [Google Scholar]