Abstract
Simulations have long played an important role in neurobiology education. This paper describes the design-research process that led to development of two popular simulation-based neurobiology modules used in undergraduate biology classes. Action Potentials Explored, and the more in-depth and quantitative Action Potentials Extended, are the third generation of neurobiology teaching simulations the author has helped develop. The paper focuses on how we used the idea of constraining simulations as a way of tuning the modules to different student populations. Other designers of interactive educational materials may also find constraint a useful lens through which to view designs.
Keywords: educational simulation, virtual experiments, neuroscience education, inquiry-based learning constraint
With calls to teach science in more student-centered ways (AAAS 2011; NGSS 2013) and the high availability of computers, activities using simulations have become common in biology classrooms. Simulations have repeatedly been shown as effective teaching tools (Hillmayr et al., 2020). A major benefit of simulation-based activities is they often allow students to focus on conceptual aspects of a topic by abstracting away some of the technical aspects (Putambekar et al., 2020). In this way they can complement wet labs, or better prepare students for lecture or other activities in a class. The literature has now gone beyond the simple question of “do simulations help students learn” to explore how particular choices in the design and features of simulation-based learning materials influence student learning. Rutten et al. (2012), for instance, summarize research on how quality of visualizations and types of instructional support impact the educational effectiveness of simulation-based teaching tools. In our own research, we’ve explored how constraints interact with scaffolding and feedback to promote efficient learning (Meir, 2022).
Simulations have a long history in neurobiology. The Hodgkin-Huxley model of action potentials is a founding moment for the modern field and much of neuroscience research since has been driven by modeling of neurons and neuronal systems. Perhaps because of this, neurobiology also has a long history of simulations used as teaching tools. Among the most prominent in the past two decades are programs that try to cover large portions of the field such as Neurons in Action (Moore and Stuart, 2007), NerveWorks (Meir et al., 2005), and Neurosim (Heitler, 2022, this JUNE issue). Each of these programs, in different ways, let students explore the roles of ion concentrations, channels and channel properties, neurotransmitters and synapses, and other aspects of neurons on neuronal function. Many instructors have also built their own smaller scale teaching simulations (e.g., Schettino, 2014; Ali, 2020).
I have participated in developing many simulation-based teaching modules across the undergraduate biology curriculum. This paper will focus on the process for developing two popular simulation-based neurobiology modules targeted at introductory biology and introductory neuroscience classes. Action Potentials Explored and Action Potentials Extended introduce students to neuron function, resting and action potentials, and problems that can affect signal transmission in neurons such as demyelination and channel blockers. My hope is that this example of a design process will aid others in thinking about how to build successful educational simulations.
Constraining Simulations to Aid Learning
From an educational research and pedagogy perspective, among the most interesting stories of our design process was a general reduction in the open-ended nature of the simulations. We would now call this the addition of constraints to the simulations. Constraint in this context describes limiting the choices students have when doing some activity. Scalise and Gifford (2006), for instance, categorized question formats in terms of degree of constraint. Multiple choice questions would be highly constrained, while essay questions would be low constraint. They point to other question formats as being intermediate in degree of constraint, with more available choices or freedom for students to express themselves than a typical multiple choice, but more constrained than a completely free-form answer like an essay. More recently, I was part of a team who showed that asking questions in an intermediate constraint format can have learning benefits for students compared to less constrained question formats (Meir et al., 2019). Similar ideas have gained traction at larger scales too, such as making whole classes highly structured to help students study better (Penner, 2018; Freeman et al., 2011).
Other activities, such as simulation-based exercises, can also be considered in terms of constraint. In a simulation, for instance, one can change the degree of constraint by exposing or hiding underlying parameters, giving students more or less flexibility to pick parameter values, and modifying the ways in which students can manipulate other aspects of the simulation. In a study involving another of our modules, Understanding Experimental Design, we explicitly tested how constraining a simulation might impact student learning. There, we found hints that intermediate degrees of constraint may aid learning in simulations just as it does in question formats (Meir, 2022; Pope et al., in review). As well, constraint in both question formats and simulations can help introduce specific, immediate feedback (Kim et al., 2017; Meir, 2022), which itself is a powerful boost to learning efficiency (Hattie and Timperley, 2007; Van der Kleij et al., 2015). As I describe our iterations of neurophysiology labs, you’ll see a theme of considering and adjusting the degree of constraint.
RESULTS
Our Earlier Open-Ended Simulation Framework for Learning Neuroscience: Nerveworks
Prior to the Action Potentials modules that are the focus here, I helped develop another neurobiology teaching simulation called NerveWorks (Meir et al., 2005). That experience influenced how we wrote the Action Potentials modules, so I’ll start by briefly describing NerveWorks.
The driving idea of NerveWorks (Figure 1A) was to give students a playground within which they could build their own neurons (and in later versions, networks of neurons) as a way of learning how neurons operate. The program presented students with a cell into whose membrane they could place channels through a drag and drop interface (Figure 1B). They could define the properties of each channel, including what ions could pass through, any voltage gating properties, overall conductance through that channel in the cell, and other properties. Students could also define concentrations of each ion inside and outside of the cell, and they could build multiple cells and draw connections (synapses) between them.
Figure 1.
Screenshots from NerveWorks, showing where students conduct experiments (A), the neuron construction window where users could drag and drop channels and change concentrations to build a neuron (B), and an interface where users could put together a recording rig, including wiring together all the components (C).
Once a cell was designed, the student had access to another area of the program where they could build a typical (for the time) neurophysiology rig to conduct cellular neurophysiology experiments. In an interface shaped like a rack of instruments, students could add an oscilloscope, stimulators, amplifiers, spritzers, and so on (Figure 1A). Next to that was a microscope dish containing the students constructed cell, where they could place electrodes. Pushing a button showed the backside of all of the instruments, allowing the student to run wires between the electrodes and the instruments (Figure 1C). Students could also construct drugs by specifying the properties of each drug, for instance specifying a channel that the drug blocks.
This setup created a vast playground within which students could conduct a multitude of simulated experiments or just explore. Pre-written labs guided students through skills like wiring up a simple recording setup or manipulating ion concentrations to learn how membrane potential is set. Part of each activity was somewhat technical as the program realistically represented what happens if you wire the equipment incorrectly, or specify a channel with different properties than intended. So, just as in real life, there was an aspect of debugging in using NerveWorks. At least one study showed the program was effective at helping students learn neurobiology as part of a broader curriculum (Bisch and Schleidt, 2008).
In some ways, NerveWorks represents much of what animated me to develop simulation-based biology teaching tools. It allowed a very constructivist type of learning where a student could experiment with simple up to quite sophisticated models that they themselves built. There was no preconceived notion of what would be interesting to explore, rather NerveWorks was built to have basic components where, like wooden blocks for younger kids, the construction was limited only by the user’s imagination. With a basic understanding of neurophysiology and curiosity, using NerveWorks was really fun, like playing an intellectual game.
That said, the approach was not fully successful. Because of how open-ended the environment was, most students needed a lot of guidance. We pre-wrote a number of labs targeted at both introductory biology and upper-level classes that appeared in an instruction window within the program to help scaffold students, but the flexibility of the program made it more challenging to write polished, concise tutorials. Much of each tutorial was devoted to guiding students through the mechanics of working within the program – how to wire together instruments, how to build a channel, and so on. Though doing these processes in a simulation might help students who were then going to use the real equipment in lab, they also potentially took attention away from the concepts being taught, like the mechanisms behind resting or action potentials. The open environment could be a hindrance as much as a learning aid, and students had a tendency to get lost and find the software difficult to use (Bish and Schleidt, 2008). While used in dozens of classes per year at both introductory and more advanced undergraduate levels, unfortunately, NerveWorks did not have enough users to support updates so it is no longer available. The experience of designing this program, however, informed our next learning modules.
Action Potentials Module
With the NerveWorks experience in mind, at SimBio we set out to design a lab specifically on membrane and action potentials, which we titled Action Potentials. In contrast to NerveWorks, whose flexibility we imagined would allow it to appeal to a broad range of undergraduate and graduate classes, with Action Potentials we targeted primarily majors introductory biology and 2nd year introduction to neuroscience classes. Before discussing some of the ways we used our previous experience with NerveWorks in building the new module, here is a brief description of the original Action Potentials module.
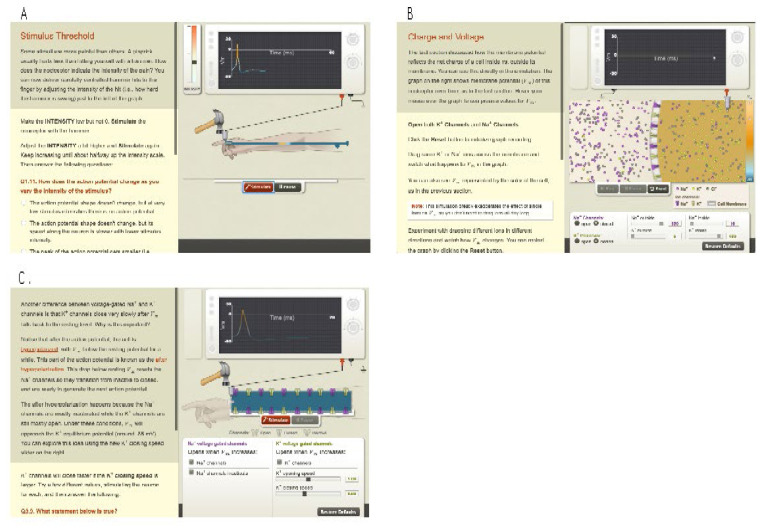
Action Potentials was divided into three main sections, all written around a central storyline about pain receptors. The first section introduced students to neurons and axons at a larger scale, including neural anatomy, what a membrane potential is and its movement through axons, stimulus and thresholds, and the way that neurons communicate using action potential firing frequency. Several simulation activities use macro-scale visualizations of a person’s finger getting hit by a hammer, generating action potentials, to let students play with those ideas (Figure 2A).
Figure 2.
Examples of simulations in the Action Potentials module. The first part (A) introduces neurons and neuronal action at a larger scale, the second (B) explores electrochemical gradients, and the third part (C) examines how ionic concentration differences and voltage-gated channels form action potentials.
The second section focused on electrochemical gradients. The section revolved around a simulation of molecular movement through an axonal membrane (Figure 2B). Using this simulation, students were guided to play with concentrations and conductances of Na+ and K+ inside and outside the axon to discover how both diffusion and electrical gradients combine to generate a membrane potential. While the Nernst equation for calculating membrane potential is itself only mentioned parenthetically, by the end of the section students have a solid introduction to the physics behind the Nernst equation and a feel for how changing ionic concentrations and conductances determine the membrane potential.
The third section brings together the first two to explain the generation of action potentials. A new simulation shows a cross section of an axon, to which students can add channels and control the properties of those channels. By adding the right mix of channels with the right properties, students build an axon that transmits an action potential (Figure 2C). Along the way they are introduced to voltage-gated channels and channel properties such as activation and inactivation parameters. The section ends with some applications of what they’ve learned, including exploring the effects of myelination and multiple sclerosis and the actions of several drugs.
Adding Constraint to Target Simulations to an Introductory Student Population
We did not have as sophisticated an understanding of the role of constraint when designing the Action Potentials module as we do now, but intuitively we increased the degree of constraint in the simulations presented to the students as a way of making the module more effective for less prepared students. For instance, there were displays of membrane potential over time (Figure 2), but those were not labeled as oscilloscopes, nor were there controls for gain or speed, wiring, electrodes, or other research equipment as there were in NerveWorks. Rather than an open-ended toolbox from which students could build just about any neurophysiological model they wished, the simulations in Action Potentials had limited numbers of parameters available to the student. In the second section of Action Potentials, as an example, students could set Na+ and K+ concentrations inside and outside the cell, and manipulate conductance of each. This is in contrast to NerveWorks where students could design almost any channel they wished involving any of the common ions (and even making up new ions).
By placing constraints on the simulations, we reduced the cognitive load associated with technical aspects of using the simulation, reserving brainpower for learning about electrochemical gradients. This is a trade-off. For students who would be going on to do wet lab experiments with real recording equipment, simulating the equipment they would encounter likely would help them understand the real lab equipment better. For students with a more advanced understanding of the theoretical concepts, removing the freedom to play to the extent available in NerveWorks potentially limited the depth to which they could learn the material and made the program less engaging. But for more introductory students who were in classes with learning outcomes focused on understanding electrochemical gradients, the more constrained simulations in Action Potentials were likely to lead to more efficient and effective learning of that material.
We did not completely abandon more open-ended simulations. In a final “playground” section we provided students with the simulations from the Action Potentials module but exposed more parameters. Though not as flexible as in NerveWorks, students that wished (or instructors that wanted to challenge their students) could use the additional parameters to explore further.
Feedback Indicated Two Separate Student Populations that Could Not Be Served by the Same Learning Module
When we released Action Potentials, it was one of our most popular module releases to that point. There were lots of things we got right. In a post-use survey, instructors wrote comments such as “Action potentials is a difficult concept to grasp. This tutorial was great at visually representing what occurs. “ and “Students were eager to try a hands-on activity and take a break from traditional lectures.” Students had similar positive comments in an end-of-module survey. While we did not have data on learning gains, students were able to successfully answer most questions on a conceptually-focused quiz placed at the end of the module.
But despite the positives, the feedback was much more mixed than is usual for our beta releases. We had a number of sources of data to explore why this might be. Twenty six instructors responded to a feedback survey after using the module in their class. When asked how challenging the tutorial was for their students the instructors were almost evenly split between a group who selected “it was just about right” (10 instructors) and a group responding “it was a little too hard, but they still learned from it” (11 instructors). Very few said it was too easy (4) and even less that it was way too hard (1). Many instructors in the too hard group elaborated with comments such as, “It is too detailed and complicated for a freshman level class”.
We corroborated this with data from 606 students. The quiz at the end of Action Potentials had 11 multiple choice questions designed to test student understanding of the material in the module, and though it was not a research-grade assessment, we thought the scores might show whether instructor intuition had merit. Indeed, when we compared students in classes where the instructor thought the module was too hard to those whose instructors thought it was just right, the “just right” population answered on average one more question correct than the “too hard” population (p << 0.01 on a t-test; Cohen’s D effect size = 0.43). Students in the “too hard” classes also self-reported enjoying the module less, learning less, and taking longer to complete it.
Based on this data, we realized there were two separate audiences interested in Action Potentials. One was instructors teaching classes that spent a small amount of time on neurophysiology and wanted their students to have a very basic understanding of how a nerve works (“shorter-time population”). The second were instructors teaching classes that spend more time on the material and want students to have a more in-depth understanding of how membrane and action potentials are formed. Since it didn’t seem possible to meet the needs of both audiences with the same material, we forked the module into two versions. Action Potentials Extended was similar to the original version with some smaller modifications based on the beta-testing. Action Potentials Explored became a shorter, less quantitative version targeted at classes that spent less time on the material.
In re-designing the module for the shorter-time population, we did several things. First, we took out much of the material introducing reversal potential, and made what remained less quantitative. Where Action Potentials Extended leads students to an understanding of equilibrium potential and prepares them for the Nernst equation, Action Potentials Explored is limited to exploring how changing conductances to Na+ and K+ affects membrane potential. This reduced the length of the module by over a third, also helpful for classes spending less time on neurophysiology. From a biology education perspective, this amounts to reducing the number and scope of learning objectives for the Explored version compared to the Extended version.
Adding Constraint To Target Shorter-Time Student Population
As with the initial Action Potentials compared to NerveWorks, the most interesting thrust of our revisions from a pedagogical perspective was increasing constraint on the simulations to aid the short time population. We did this primarily by hiding more parameters from the simulations, and by limiting the range of values students could choose of those parameters left exposed. This is shown for two examples in Figure 3. The top two panels (Figure 3 A&B) show a molecular-level simulation where students can see how conductance affects membrane potential. Both versions allow students to manipulate Na+ and K+ conductance in the membrane, but the Explored version only allows an “open” and a “closed” state for each channel type, whereas in Extended the student is allowed to vary conductance continuously. In addition, in the Extended version students also have access to change the concentrations of both ions, where this is fixed and hidden in Explored.
Figure 3.
Two examples of how constraint was added to simulations from Action Potentials Extended (A and C) to make the shorter and more introductory Action Potentials Explored (B and D).
The bottom two panels in Figure 3 (C&D) show a similar situation for a simulation of how voltage-gated channels can produce action potentials. Here the Explored version provides checkboxes for students to add or remove three aspects of the neuron’s voltage gated channels. The same simulation in the Extended version has six continuously manipulable parameters related to Na+ and K+ voltage-gated channels.
As I argue elsewhere, imposing higher levels of constraint acts as an unstated scaffolding for students, guiding them through to successful experiments with the simulations while still allowing them to discover what to do for themselves (Meir, 2022). The cost is that students with a more advanced understanding may not be challenged to experiment and learn as much as they could with lower constraints. This tradeoff mirrors the needs of the two populations we were trying to help with the simulation.
Interestingly, use of the two Action Potentials versions has been roughly equivalent, with about equal numbers of students in any given year using each version. The split is not by class level, as many introductory biology classes use the longer, less constrained Extended version. The difference really appears to be how much time and depth the instructor plans to spend on the topic. While we do not have formal research to support this, a reasonable hypothesis is that most college biology students are capable of the higher-level Extended version given enough time and assistance from the instructors. Alternatively, with less time and assistance devoted to the topic, the Explored version, with the differences discussed above that make it faster and easier, is more appropriate.
(For those wishing to see the two versions of the lab we describe as they read this paper, Action Potentials Explored and Action Potentials Extended are available for review to instructors and researchers in free evaluation software from SimBio which can be requested at http://simbio.com.)
Formal Evaluation Of Learning Suggests Large Learning Gains
In 2016 we partnered with a research group at the University of Washington who were developing an assessment of student understanding of electrophysiology (Cerchiara et al., 2019). They were interested in validating their assessment, called the Electrochemical Gradient Assessment Device (EGAD), by trying it with a curriculum that taught principles of electrophysiology. The EGAD is the most comprehensive assessment we are aware of on the topic and as Cerchiara and colleagues report, the EGAD has good validity by a number of metrics for diverse students, especially for lower and medium-performing students. For those interested in their students’ understanding of electrochemical gradients and how those are used to generate action potentials, the EGAD is well worth considering.
Our research used the EGAD as a pre/post test in a large introductory biology class that spent a week covering topics in electrophysiology. Part of the coverage was having students use Action Potentials Extended in their lab period, as well as answering questions in groups at a white board related to the simulations. The instructors also used active learning teaching methods in the lectures (Freeman et al., 2014). We found an effect size of ~1.5 between the pre- and post-tests, showing the combination of activities, including Action Potentials Explored as a central component, likely led to substantial learning gains on the material (Cerchiara et al., 2019). One interesting aspect of the EGAD is most of the questions are structured in pairs. The first question in those pairs asks for factual information, such as “What causes membrane potential to be more positive than −70mV at the point labeled X?” in a diagram of an action potential, with multiple-choice answers. The second question in each pair asks for an explanation, such as, “Those ions are moving into the neuron because,” again with multiple choice answers but focused on conceptual explanations. As one might expect, the factual questions were all easier than their paired conceptual explanation questions, but students improved both on the lower Blooms level (Crowe et al., 2008) factual questions and the higher Blooms level conceptual questions.
DISCUSSION
Here we’ve shown the process we took to developing popular simulation-based modules on electrophysiology. We are hopeful that our description of how the module evolved may aid others who are building learning materials. In particular, there is currently a large and deserved emphasis on backward design where developing educational materials starts with learning outcomes and works backwards from those to the design of the activity itself. The process of developing the Action Potentials modules shows that learning outcomes are not enough. One must also consider how an activity targets those learning outcomes with a high degree of efficiency for a particular population of students. Our experience with these electrophysiology modules points to constraint as a strong lever for doing such targeting.
Footnotes
NerveWorks was conceived by E. Meir and written with a team at the University of Washington including Anna Davis, Edwin Munro, William Moody, Bill Sunderland, and Scott Votaw. Kerry Kim was the lead author at SimBio of the Action Potentials modules, working with a team including Susan Maruca, Steve Allison-Bunnell, Erik Harris, Josh Quick, Eleanor Steinberg, Jennifer Wallner, and others. Thanks to Kathleen Barry and Amy Gallagher for help preparing this manuscript and to Kerry Kim for reviewing an earlier draft.
REFERENCES
- American Association for the Advancement of Science. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, DC: AAAS; 2011. [DOI] [Google Scholar]
- Ali D. Neuromembrane: A Simulator for Teaching Neuroscience. Presentation at FUN Summer Virtual Meeting: Teaching, Learning, and Mentoring Across Distances, July 30 – August. 2020;1:2020. Abstract available at https://www.funfaculty.org/files/FUN2020SVMProgram.pdf. [Google Scholar]
- Bish JP, Schleidt S. Effective Use of Computer Simulations in an Introductory Neuroscience Laboratory. J Undergrad Neurosci Ed. 2008;6(2):A64–A67. Available at https://pubmed.ncbi.nlm.nih.gov/23493239/ [PMC free article] [PubMed] [Google Scholar]
- Cerchiara JA, Kim KJ, Meir E, Wenderoth MP, Doherty JH. A new assessment to monitor student performance in introductory neurophysiology: Electrochemical Gradients Assessment Device. Adv Physiol Educ. 2019;43:211–220. doi: 10.1152/advan.00209.2018. [DOI] [PubMed] [Google Scholar]
- Crowe A, Dirks C, Wenderoth MP. Biology in Bloom: Implementing Bloom’s Taxonomy to Enhance Student Learning in Biology. CBE Life Sci Ed. 2008;7(4):368–381. doi: 10.1187/cbe.08-05-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S, Haak D, Wenderoth MP. Increased Course Structure Improves Performance in Introductory Biology. CBE Life Sciences Ed. 2011;10(2):175–186. doi: 10.1187/cbe.10-08-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. Active learning increases student performance in science, engineering, and mathematics. PNAS. 2014;111(23):8410–15. doi: 10.1073/pnas.1319030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattie J, Timerperley H. The power of feedback. Rev Ed Research. 2007;77(1):81–112. doi: 10.3102/003465430298487. [DOI] [Google Scholar]
- Heitler WJ. Neurosim: some thoughts on using computer simulation in teaching electrophysiology. J Undergrad Neurosci Educ. 2022;20(2):A282–A289. doi: 10.59390/JGIP5297. Available at https://www.funjournal.org/wp-content/uploads/2022/08/june-20-282.pdf?x36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmayr D, Ziernwald L, Reinhold F, Hofer SI, Reiss KM. The potential of digital tools to enhance mathematics and science learning in secondary schools: A context-specific meta-analysis. Computers & Education. 2020;153:103897. doi: 10.1016/j.compedu.2020.103897. [DOI] [Google Scholar]
- Kim KJ, Pope DS, Wendel D, Meir E. WordBytes: Exploring an intermediate constraint format for rapid classification of student answers on constructed response assessments. Journal of Educational Data Mining. 2017;9(2):45–71. Available at https://jedm.educationaldatamining.org/index.php/JEDM/article/view/209. [Google Scholar]
- Meir E. Strategies for targeting the learning of complex skills like experimentation to different student levels: The intermediate constraint hypothesis. In: Pelaez NJ, Gardner SM, Anderson TR, editors. Trends in Teaching Experimentation in Life Sciences. Switzerland AG, Cham, Switzerland: Springer Nature; 2022. [DOI] [Google Scholar]
- Meir E, Davis A, Munro EM, Sunderland W, Votaw S, Moody WJ. NerveWorks. Missoula, MT: SimBiotic Software, Inc; 2005. [Google Scholar]
- Meir E, Wendel D, Pope DS, Hsiao L, Chen D, Kim KJ. Are intermediate constraint question formats useful for evaluating student thinking and promoting learning in formative assessments? Computers & Education. 2019;142:103606. doi: 10.1016/j.compedu.2019.103606. [DOI] [Google Scholar]
- Moore JW, Stuart AE. Neurons in Action. Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- NGSS Lead States. Next Generation Science Standards: for States, By States. Washington, DC: The National Academies Press; 2013. [Google Scholar]
- Penner MR. Building an Inclusive Classroom. J Undergraduate Neurosci Ed. 2018;16(3):A268–A272. Available at https://pubmed.ncbi.nlm.nih.gov/30254542/ [PMC free article] [PubMed] [Google Scholar]
- Puntambekar S, Gnesdilow D, Dornfeld Tissenbaum C, Narayanan NH, Rebello NS. Supporting middle school students’ science talk: A comparison of physical and virtual labs. J Res Sci Teaching. 2020;58:392–419. doi: 10.1002/tea.21664. [DOI] [Google Scholar]
- Rutten N, van Joolingen WR, van der Veen JT. The learning effects of computer simulations in science education. Computers & Education. 2012;58(2012):136–153. doi: 10.1016/j.compedu.2011.07.017. [DOI] [Google Scholar]
- Scalise K, Gifford B. Computer-based assessment in e-learning: A framework for constructing “intermediate constraint” questions and tasks for technology platforms. The Journal of Technology, Learning, and Assessment. 2006;4(6):1–45. Available at http://files.eric.ed.gov/fulltext/EJ843857.pdf. [Google Scholar]
- Schettino LF. NeuroLab: A set of graphical computer simulations to support neuroscience instruction at the high school and undergraduate level. J Undergrad Neurosci Educ. 2014;12(2):A123–A129. Available at https://pubmed.ncbi.nlm.nih.gov/24693259/ [PMC free article] [PubMed] [Google Scholar]
- Van der Kleij FM, Feskens RCW, Eggen TJHM. Effects of feedback in a computer-based learning environment on students’ learning outcomes: A meta-analysis. Rev Ed Research. 2015;85(4):1–37. doi: 10.3102/0034654314564881. [DOI] [Google Scholar]