Abstract
Background and Aims:
Chronic HEV infections remain a serious problem in immunocompromised patients, as specifically approved antiviral drugs are unavailable. In 2020, a 24-week multicenter phase II pilot trial was carried out, evaluating the nucleotide analog sofosbuvir by treating nine chronically HEV-infected patients with sofosbuvir (Trial Number NCT03282474). During the study, antiviral therapy reduced virus RNA levels initially but did not lead to a sustained virologic response. Here, we characterize the changes in HEV intrahost populations during sofosbuvir treatment to identify the emergence of treatment-associated variants.
Approach and Results:
We performed high-throughput sequencing on RNA-dependent RNA polymerase sequences to characterize viral population dynamics in study participants. Subsequently, we used an HEV-based reporter replicon system to investigate sofosbuvir sensitivity in high-frequency variants. Most patients had heterogenous HEV populations, suggesting high adaptability to treatment-related selection pressures. We identified numerous amino acid alterations emerging during treatment and found that the EC50 of patient-derived replicon constructs was up to ~12-fold higher than the wild-type control, suggesting that variants associated with lower drug sensitivity were selected during sofosbuvir treatment. In particular, a single amino acid substitution (A1343V) in the finger domain of ORF1 could reduce susceptibility to sofosbuvir significantly in 8 of 9 patients.
Conclusions:
In conclusion, viral population dynamics played a critical role during antiviral treatment. High population diversity during sofosbuvir treatment led to the selection of variants (especially A1343V) with lower sensitivity to the drug, uncovering a novel mechanism of resistance-associated variants during sofosbuvir treatment.
INTRODUCTION
HEV is a single-stranded RNA virus that annually infects an estimated 20 million people worldwide and is one of the most common causes of viral hepatitis globally.1 HEV mostly leads to asymptomatic or acute self-limiting infections in healthy individuals2 but can also cause chronic hepatitis E (~3.3 million cases are symptomatic) in immunocompromised individuals.1 Especially, immunosuppressed patients, such as solid organ transplant recipients, stem cell transplant recipients, or HIV-infected individuals, are at high risk of developing persistent viral hepatitis, which can lead to aggravated disease outcomes, such as liver fibrosis, cirrhosis, and progression to end-stage liver disease.3–5 In addition, depending on the genotype, up to 30% of pregnant women infected with HEV-1 experience acute liver failure and fulminant hepatitis leading to fetal, neonatal, and maternal mortalities in the third trimester.6,7
Currently, there is no specific antiviral treatment for patients with chronic hepatitis E. The only available options are the off-label use of ribavirin or interferon-based regimens. However, treatment with ribavirin leads to sustained virologic response in only ~80% of patients and is often associated with severe adverse effects.8,9 In addition, patients with renal insufficiency or pregnant women are not eligible for ribavirin treatment,6 and organ transplant patients are at risk of graft rejection when treated with interferon.
A rapid, cost-effective approach that may prove promising for finding new antiviral agents against HEV infections is the repurposing of already approved drugs.10 Indeed, sofosbuvir, a direct-acting antiviral that was originally designed to inhibit the HCV polymerase, has been considered a potential antiviral drug against HEV infections in recent years. Sofosbuvir is a nucleotide analog administered as a prodrug, and once activated, it mimics uridine triphosphate in the target cell. The active compound can, thus, be incorporated into nascent RNA, which inhibits the replication of HCV by causing chain termination.11 Sofosbuvir is considered a pan-genotypic antiviral that efficiently inhibits multiple HCV genotypes12,13 with a high genetic barrier to resistance.14,15 Moreover, antiviral activity against other flaviviruses such as zika,16 dengue,17 yellow fever virus,18,19 viruses of the Togaviridae family20 and HEV21 has been demonstrated. So far, several studies have assessed sofosbuvir efficacy in patients chronically infected with HEV, albeit with conflicting study results.
In 8 case reports, the efficacy of sofosbuvir with or without ribavirin was limited because, although a transient reduction in HEV RNA levels was achieved, HEV was not eliminated.22–29 In contrast, patients treated with sofosbuvir and ribavirin or another HCV drug in a total of 4 studies achieved HEV clearance.30–33
Recently, Cornberg et al conducted a multicenter prospective pilot trial examining the efficacy of 400 mg sofosbuvir monotherapy for 24 weeks in 9 patients chronically infected with HEV.29 These patients had either previously failed or were not eligible for ribavirin treatment. The treatment outcome with sofosbuvir was comparable to those of previous studies, in which a sustained virologic response was not achieved. However, a significant decrease in viral RNA could be noted within the first weeks of treatment with a consecutive increase in viral load over time suggesting a possible escape mechanism of the virus.
Herein, we analyzed the longitudinal evolution of the viral population in these patients by high-throughput sequencing of the viral polymerase. Also, we investigated whether sofosbuvir could induce the emergence of resistance-associated variants, possibly explaining the recurrence of high HEV RNA levels. Identified viral variants were then assessed to sofosbuvir sensitivity by an HEV reporter replicon system.
METHODS
Cohort
Here, we analyzed the virus population of a previously published cohort of 9 patients infected with HEV who were treated daily with 400 mg of sofosbuvir for 24 weeks.29 Most of these patients (7/9) were infected with HEV-3c. The remaining 2 patients were infected with HEV-3i and HEV-3f, respectively (Supplemental Figure S1, http://links.lww.com/HEP/H882). Briefly, these patients were HEV-positive for at least 3 months and either failed previous ribavirin treatment or had a contraindication for treatment with ribavirin. Patients were monitored regularly until the end point of treatment (24 wk). At 12 weeks of follow-up, an additional sample was taken to determine the long-term efficacy of treatment.
Amplicon generation and sequencing
Total RNA from 200 μL serum was extracted using Cobas AmpliPrep total nucleic acid isolation kit (Roche, Basel, Switzerland). Complementary DNA (cDNA) synthesis and amplicon generation were performed according to Todt et al.34 Briefly, complementary DNA was synthesized from 2 to 8 μL purified total RNA using the SuperScript III first-strand synthesis system (Life Technologies, Carlsbad, CA, USA). Amplicons were generated in a 2-step touchdown PCR using the TaKaRa Ex Taq Hot Start Version (Dalian, China) polymerase.
Library preparation and sequencing
For NGS library generation, the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) was used according to the manufacturer’s recommendations. Library quality was assessed using the Agilent High Sensitivity DNA Kit (Agilent Technologies, Waldbronn, Germany) and a 2100 Bioanalyzer Instrument (Agilent Technologies). Any additional samples were normalized and quantified using the KAPA Library Quantification Kit for Illumina (Kapa Biosystems, Wilmington, MA, USA). The samples were sequenced on a MiSeq using a paired-end sequencing protocol with 2 × 250 bp.
Data analysis
Data analysis was based on the pipeline described by Gömer et al.35 Briefly, raw reads were trimmed using Trimmomatic 0.3936 and quality checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). In the first round, consensus sequences were called using Tanoti (https://github.com/vbsreenu/Tanoti) and Sam2Consesus v2.0 (https://github.com/vbsreenu/Sam2Consensus). The consensus sequence from each patient at baseline was used as a reference. Raw reads were then mapped to the respective reference using Tanoti. Subsequently, the mapped files were processed using SAMtools v1.937 and then analyzed using Diversitools (https://github.com/josephhughes/DiversiTools) and vNvS tools (https://github.com/rjorton/vnvs). The data were visualized using in-house R-scripts with the following packages: Tidyverse, ggpubr, and patchwork.38
The mean coverage for all samples was 231,137 (±SD 56,542, Supplemental Figure S2, http://links.lww.com/HEP/H882). For patients 7 and 8, there were residues with high variability at baseline (Supplemental Figure S3, http://links.lww.com/HEP/H882). At these positions, the nucleotide with the highest frequency was used as a reference. Structural models were generated using default settings in Colabfold v1.5.239 with HEV Kernow-C1 p6 clone as a reference (accession: JQ679013). The resulting PDB file is provided in the supplement. Coloring according to SNV data was performed using ChimeraX v1.5.40
Full genome HEV sequences were collected from the NCBI Nucleotide database using Entrez Direct v16.2. Sequences were sorted depending on genotype information in the GenBank files into fasta files for HEV-3. These were aligned using clustal omega v1.2.4 and analyzed with in-house R-scripts using the packages tidyverse, seqcombo, seqinr, Bios2cor, and Biostrings.
Eukaryotic cell culture
Human hepatoma cells (HepG2) were routinely cultured in 15-cm2 culture plates in Dulbecco’s Modified Eagle Medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 μg/mL of streptomycin, 100 IU/mL of penicillin (Invitrogen), 2 mM L-glutamine, and 1% nonessential amino acids (Invitrogen) at 37˚C in a humidified incubator at 5% CO2. Before plating cells, cell culture dishes were coated with rat collagen (SERVA Electrophoresis).
Compounds
Sofosbuvir was purchased from Medchemexpress (HY-15005), dissolved in DMSO, and stored according to the manufacturer’s recommendations. Ribavirin was purchased from Sigma Aldrich (R9644), dissolved in DMSO, and stored at −20°C.
Plasmid construction and in vitro transcription
When selecting patient-derived substitutions for generating replicon constructs resembling the most prominent amino acid substitutions per patient (patient-derived constructs), we made sure to cover as large a period as possible and to have as many different variants for each patient as possible. Determined mutation sequences for each patient were synthesized as gBlock Gene fragments (IDT, Coralville, Iowa, USA) or oligos and amplified by site-directed mutagenesis PCR. GBlock Gene fragments or PCR amplicons were then introduced into AflII, NsiI, and/or NruI-digested pBluescript backbone containing the coding sequence of subgenomic Kernow-C1 p6 coupled with a Gaussia luciferase reporter gene (p6-Gluc) by Gibson assembly (NEB, New England, USA).
For in vitro transcription, generated plasmids were first linearized by digestion with MluI and purified using the Qiaquick Spin Mini Kit (Qiagen). Linearized plasmids were then in vitro transcribed according to Todt et al and Meister et al.41,42 Briefly, in vitro transcription mix containing 2 µg linearized DNA template, 80 U T7 RNA polymerase, and 5 mM Ribo m7G Cap Analog (m7G Cap analog, Promega) was incubated for 2 hours at 37°C. Subsequently, 2 µL T7 RNA polymerase was added and incubated for another 2 hours at 37°C, and the DNA template was digested by the addition of DNase and incubation for 30 minutes at 37°C. In vitro transcripts were purified with the NucleoSpin RNA Clean-Up Kit (Macherey & Nagel), and RNA integrity was ensured by agarose gel electrophoresis.
Gaussia luciferase reporter assay
To screen patient-derived amino acid variants for altered sofosbuvir sensitivity, in vitro transcribed RNA was delivered to human hepatoma cells by electroporation according to Todt et al.43 In brief, 5 × 106 human hepatoma cells were resuspended in 400 μL Cytomix and electroporated with 5 μg of respective in vitro transcribed HEV RNA. Cells were electroporated with the Gene Pulser Xcell apparatus (Bio-Rad) and seeded at a density of 20,000 cells/well in a 96-well format. For dose-response assays, cells were treated with triplicate 3-fold serial dilutions of sofosbuvir and with triplicate 2-fold serial dilutions of ribavirin at concentrations ranging from 0.015 to 100 µM and 0.39 to 100 µM, respectively. Supernatants were sampled 72 hours posttreatment, and Gaussia luciferase activity was determined by the detection of luminescence using a Centro XS3 LB 960 luminometer (Berthold Technologies). The luciferase reaction was carried out on a 96-well LUMITRAC 600 plate and by dispensing 50 µL of coelenterazine substrate onto 20 µL harvested cell culture supernatant/well, shaking for 1 second and reading for 1 second.
Cell viability assay
Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were incubated with 5 mg/mL MTT substrate in Dulbecco’s Modified Eagle Medium complete media for 1 hour at 37 °C and 5% CO2. For solubilization, the MTT substrate was removed, and 50 µL DMSO was added. Absorbance was measured at 570 nm on a microplate absorbance reader (Tecan). Cells treated with 70% ethanol for 10 minutes served as background control.
Statistical analysis
Statistical tests on titration data were performed using the nonlinear 4 parametric log-logistic models based on the Hill equation implemented in GraphPad Prism 9.4.1 (GraphPad Software, La Jolla, CA, USA). All biological replicates were used to fit dose-response curves, and results are presented as EC50 values and the respective 95% CIs. Differences in EC50 values were tested using the sum-of-squares F-test for significance. To test the significance of differences in the mean of replication data, one-way ANOVA followed by Dunnett’s multiple comparison tests was used. The Pearson correlation analysis was conducted in R.
RESULTS
Effects of sofosbuvir on viral sequence diversity of the HEV RNA-dependent RNA polymerase
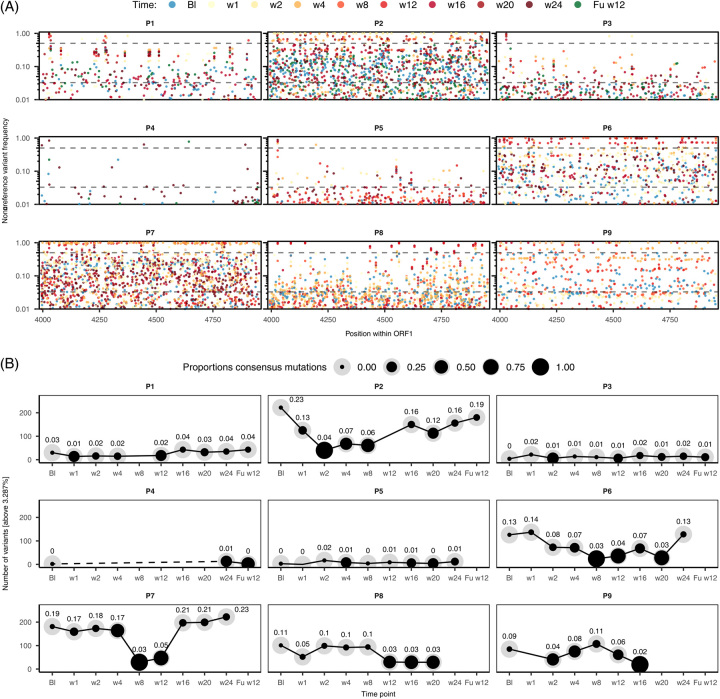
To assess the influence of sofosbuvir on HEV population diversity in chronically-infected patients, we used an amplicon-based sequencing approach and determined the longitudinal nucleotide diversity within the HEV RNA-dependent RNA polymerase (RdRp) (position 3996–4953 of ORF1) (Figure 1). The cutoff value for variants with biological relevance was set to 3.287% (error cutoff; bottom dashed line), while variants above 50% indicate consensus sequence changes (top dashed line).
FIGURE 1.
Viral diversity in patients chronically infected with HEV under sofosbuvir treatment. (A) Frequency of nonreference variants within the RdRp of HEV ORF1 compared to the time of treatment initiation (baseline). The cutoff for biological relevance (lower dashed line) was set at 3.287% according to Illumina error rates.44 The top dashed line (50%) indicates consensus sequence mutations. Each dot represents a variant at a given time point, which is color coded; (B) quantification of variants for each patient and time point that occur above the error cutoff. The size ratio of the dot indicates the percentage of consensus sequence changes compared to the total number of mutations. Labels indicate size normalized variation. Abbreviations: RdRp, RNA-dependent RNA polymerase.
Overall, nucleotide heterogeneity was high, as shown by the number of variants above the error cutoff at different time points, particularly in patients treated with ribavirin prior to the sofosbuvir pilot study (P1–P3, P6–P9, Figure 1A). Notably, low nucleotide heterogeneity observed for P4 should be interpreted with caution, as this may be due to the lack of sample sequences or input RNA rather than low heterogeneity. At all-time points under treatment, multiple variants were above the 50% frequency threshold, indicating that high-frequency mutations resulted in consensus sequence changes.
The number of mutations above the error cutoff was quantified for each patient and time point to highlight differences in viral heterogeneity (black line; Figure 1B). Patients that have not been treated previously with ribavirin (P4, P5) and patients P1 and P3 had the lowest number of variants (0–43 variants, 0%–4% diversity, defined as RdRp amplicon positions that carried mutations above cutoff). Patients P6, P8, and P9 had intermediately variable virus populations with 19–137 variants (3%–14% diversity). The most diverse populations were in P2 and P7 (29–222 variants) where high viral heterogeneity was already present at baseline (23%, 19% diversity). For both viral populations, the total number of variants decreased during treatment, (down to 3%–4%), which coincided with an increase in the number of consensus sequence changes: 40 and 83 consensus variants, respectively. From this point of decreased heterogeneity, HEV diversified again in both patients (19%–23%).
In summary, sofosbuvir did not change the total number of sites exhibiting nucleotide variants, but was associated with changes in the consensus sequence in multiple patients.
Identification of sofosbuvir resistance-associated substitutions
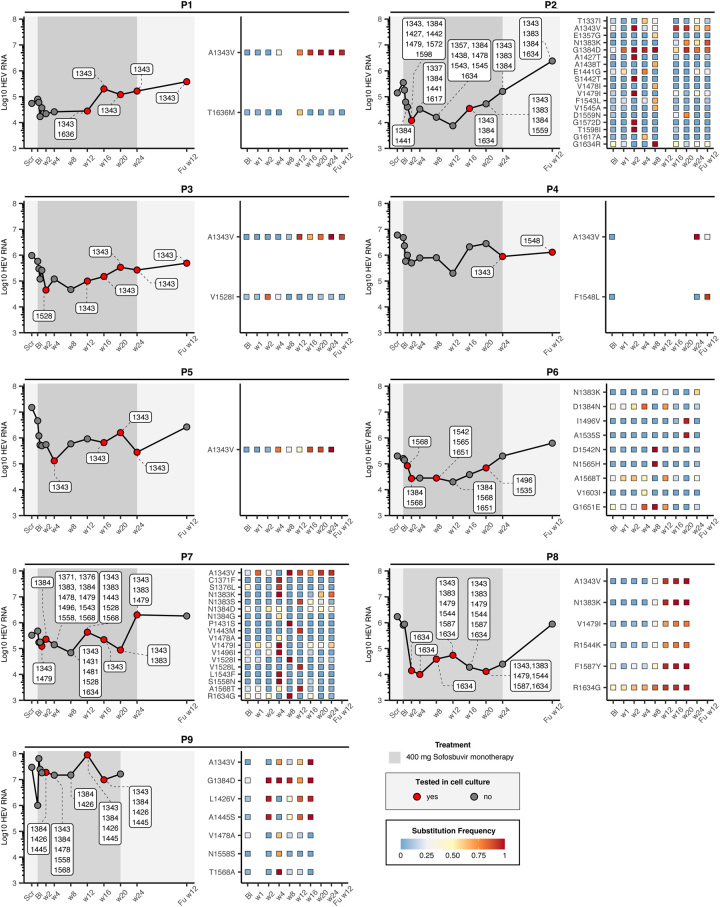
Because nucleotide diversity is not necessarily related to biological processes at the protein level, we examined nonsynonymous substitutions that occurred during sofosbuvir treatment (Figure 2). HEV RNA levels in study participants decreased by at least 1 log from baseline to week 2 in all patients after initiation of sofosbuvir monotherapy. In patient P1, only a minor reduction in HEV RNA from 80,000 IU/ml to 22,000 IU/ml (0.6 log10) was achieved by sofosbuvir, whereas, in P8, RNA decreased from 805,000 IU/ml to 14,000 IU/ml (1.76 log10), which was the maximum decline observed in this study. However, HEV RNA levels increased again between the fourth week and the end of treatment. At the 12-week follow-up, HEV RNA returned to baseline, suggesting that sofosbuvir had only a transient effect. The pattern of heterogeneity was similar to that at the nucleotide level, with P2 and P7 having the most diverse HEV populations with up to 18 amino acid positions with consensus substitutions, whereas patients P4 and P5 had the lowest number with 2 and 1, respectively (Figure 2, Figure S4, http://links.lww.com/HEP/H882). For any of the identified substitutions, the frequency over time was illustrated for each patient. Some of the variants accumulated in a time-dependent manner, for example, a substitution from alanine to valine at position 1343, which was selected for in multiple patients, increased on average from baseline (5.5%) more than 70% at later time points during treatment. An increase in frequency was also observed for other substitutions, for example, at position 1383 in patients P2, P6, P7, and P8. For some substitutions, only a transient increase in frequency was observed, for example, at position 1528 in patient P7, where the initial frequency of 0.22 (Bl) increased to 0.99 at week 8 and then declined again to 0.008 in week 16, reflecting the dynamics of the viral population under sofosbuvir treatment in these patients.
FIGURE 2.
Identification of patient-derived consensus substitution. The left panel shows HEV RNA quantity over the treatment period. The dark gray highlighted area refers to the time during which patients were treated with sofosbuvir. Red dots indicate time points at which variants were engineered into the replicon system (Figure 3). The heatmap (right side) shows all changes in the amino acid consensus sequence and their respective frequency in percent per time point.
Impact of patient-derived variants on sofosbuvir sensitivity and viral replication in vitro
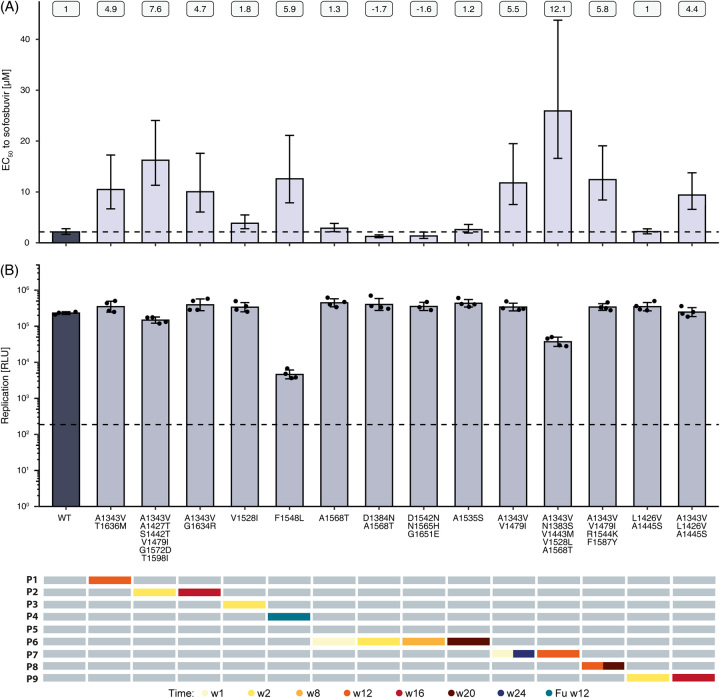
To test whether substitutions could alter sofosbuvir sensitivity in vitro, we introduced patient-derived substitutions at defined time points (Figure 2, red dots) into an HEV-3 Gaussia luciferase reporter system. A total of 14 patient-derived variant constructs were generated and used in dose-response assays to determine the EC50 and replicative capacity under sofosbuvir treatment (Figure 3, Supplemental Table S1, http://links.lww.com/HEP/H882). In most constructs (8/14), EC50 values increased at least 4.4-fold compared to p6 wild-type control (p6-WT) (P1-w12, P2-w2, P2-w16, P4-Fuw12, P7-w1,24, P7-w12, P8-w12,20, and P9-w16), suggesting reduced sofosbuvir sensitivity of these variants (Figure 3A). The highest-fold increase (12.1-fold) in EC50 was observed for a variant found in P7-w12 (A1343V/N1383S/V1443M/V1528L/A1568T).
FIGURE 3.
Impact of patient-derived consensus variants on sofosbuvir sensitivity and viral fitness. Amino acid substitutions found in P1–P4 and P6–P9 at specific time points during sofosbuvir treatment (bottom panel) were introduced into a p6 Kernow-C1 replicon harboring a Gaussia luciferase reporter gene and delivered into HepG2 cells by electroporation. For subsequent dose-response experiments, cells were treated with sofosbuvir concentrations ranging from 100 µM to 0.015 µM for 72 hours. (A) EC50 values were determined by nonlinear regression. Numbers at the top indicate fold changes compared to wild-type. Error bars = 95% CI. (B) Replication capacity of variants was determined by plotting the measurement of RLU of vehicle-treated (DMSO) cells in GraphPad Prism (middle panel). Error bars = ± SD. Dashed lines represent wild-type control (A) and replication-deficient control (GAA) (B). n ≥ 3. Abbreviation: HepG2, human hepatoma cells; RLU, relative light units.
Interestingly, 7 out of 8 variants with at least a 4.4-fold increase in EC50 carried the A1343V substitution, and 3 of them additionally harbored the V1479I substitution as consensus (P2-w2 [7.6-fold], P7-w1,24 [5.5-fold], and P8-w12,20 [5.8-fold]). Other variants showed ~1.7-fold decrease in EC50 values (P6-w2 [1.7-fold] and P6-w8 [1.6-fold]) or did not display altered sofosbuvir sensitivity (P6-w1 [1.3-fold], P6-w20 [1.2-fold], and P9-w2 [1.0-fold]).
Viral replication capacity was not affected in most of the constructs (12/14) compared to wild-type p6-Gluc replicon (Figure 3B). However, reduced viral replication fitness was observed in both the P4-Fuw12 and P7-w12 variants where luminescence was decreased by 1.7 and 0.8 logs, respectively. Overall, cell viability was not affected during sofosbuvir treatment [half-maximum cytotoxic concentration (CC50) values between 91.41 µM – > 100 µM] (Supplemental Figure S5B, http://links.lww.com/HEP/H882, top panel; Supplemental Table S1, http://links.lww.com/HEP/H882).
To investigate whether patient-derived substitutions altered ribavirin sensitivity (Figure S5A, http://links.lww.com/HEP/H882, B), dose-response assays were additionally performed under ribavirin treatment. The majority of the constructs did not alter ribavirin sensitivity or replication capacity. However, substitutions found in P4-Fuw12 and P7-w12 increased sensitivity to ribavirin (P4-Fuw12 [-2.2-fold] and P7-w12 [-4.9-fold]) and reduced replication capacity by 1.9 and 1 logs, respectively. Similar to sofosbuvir treatment, ribavirin did not result in a strong reduction in cell viability in any of the tested variants (CC50 97.54 µM- >100 µM) (Supplemental Figure S5B, http://links.lww.com/HEP/H882, bottom panel, Supplemental Table S2, http://links.lww.com/HEP/H882).
In summary, many patient-derived variants showed a decrease in sofosbuvir sensitivity while maintaining replication capacity. Of these, A1343V-containing variants showed the strongest decrease in sofosbuvir sensitivity and no loss in replication fitness.
The emergence of A1343V as a sofosbuvir resistance-associated substitution
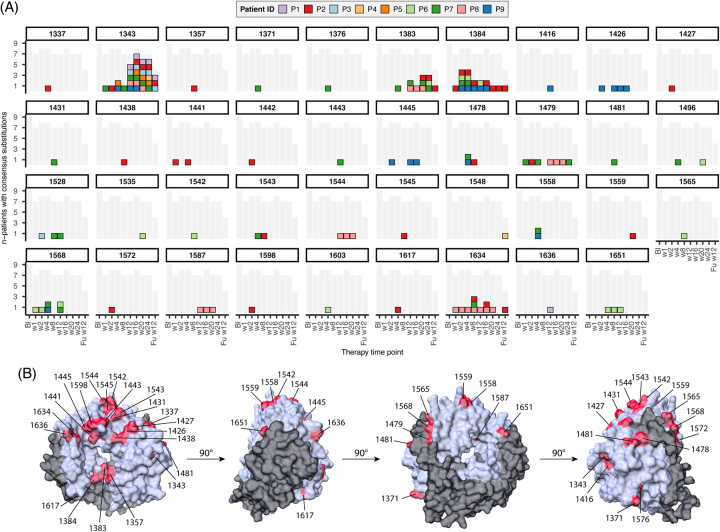
Following the observation that A1343V-containing variants showed the strongest decrease in sofosbuvir sensitivity, next, we evaluated the impact of single amino acid substitution on both replication capacity and sofosbuvir sensitivity. For most patients, alanine to valine substitution at position 1343 was almost absent at baseline (mean frequency ~5%) but occurred during treatment in at least 8 of 9 patients (Figure 4A). Only patients P2 and P7 had a subpopulation encoding for valine (GTT) at baseline (on average 23%, Supplemental Figure S6, http://links.lww.com/HEP/H882). In all other patients at baseline, the GCG, GCT, or GCC codons, all translating into alanine, were present at 99% on average. Notably, substituting alanine (GCN) into valine (GTN) requires only 1 mutation: the transition of the cytosine at the second position of the codon, resulting in thymidine. In most patients, this transition was made by weeks 12–16. Although A1343V became the consensus sequence in most patients (Figure 4), other substitutions that occurred at high frequency in these patients had a similar, but less pronounced pattern, namely at position 1383, 1384, or 1634 (Figure 4A).
FIGURE 4.
Emergence of A1343V as a resistance-associated substitution. (A) All positions with at least 1 substitution above 50%. Each box represents a patient with a substitution at that position; see the color code in the legend. The grey shaded area refers to all samples sequenced at that time point. (B) Alpha-fold model of the HEV RdRp (based on Kernow-C1 p6 nucleotide sequence). Residues with consensus sequence changes are highlighted in red. Abbreviation: RdRp, RNA-dependent RNA polymerase.
The location of all substitutions that became dominant during treatment in either one of the patients was plotted onto an alpha-fold predicted protein structure of the HEV RdRp. Interestingly, some of these substitutions (1383, 1384, and 1634) were located near the tunnel in which the RdRp may interact with the RNA (Figure 4B, Supplemental Figure S7, http://links.lww.com/HEP/H882). In contrast, the A1343V substitution was located at the outside surface of the RdRp.
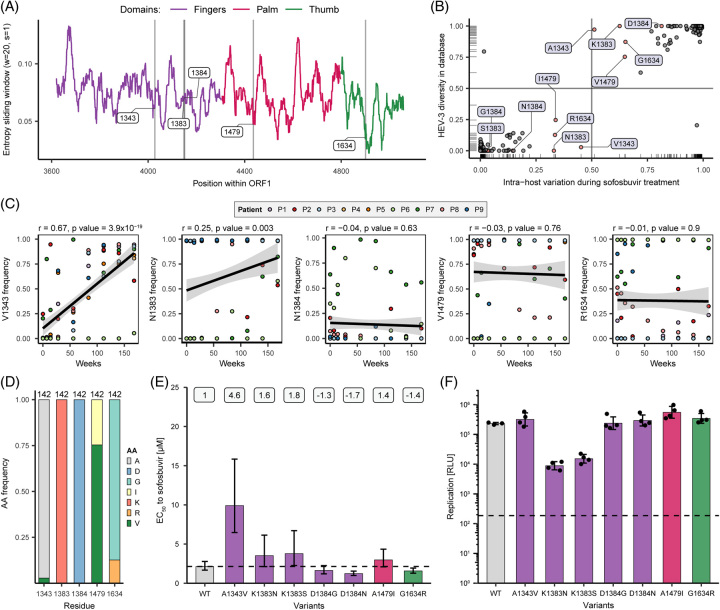
Subsequently, the frequency of these and other variants were assessed from public databases. In total, 142 HEV-3 sequences covering the RdRp amplicon were assessed. The Shannon entropy (sliding window, window = 20, and step = 1) indicated that heterogeneity was not evenly distributed (Figure 5A). Next, variant frequencies from publicly available sequencing data were compared to the intrahost data of variants identified in this study, to determine whether variants detected here were sofosbuvir treatment-specific (Figure 5B). The frequency distribution of HEV-3 amino acid variants of interest (at positions 1343, 1383, 1384, and 1634) from public databases were plotted against their intrahost frequency during sofosbuvir treatment (Figure 5B). At position 1343, either alanine was encoded, which occurred in both datasets at a frequency above 50% or valine. V1343 was, however, almost explicitly present in the patient cohorts but not in the databases (4/142 sequences, Figure 5A, B). Moreover, V1343 emergence correlated significantly with treatment time (r = 0.67 and p value < 0.05), while the correlation was nonsignificant for other variants (Figure 5C). Overall, most amino acids clustered at either corner of the plots, indicating similar sequence distribution between the cohort and public database (Figure 5B). In addition, A1343, K1383, and D1384 were highly conserved in the database, while they showed considerable heterogeneity in the patient cohort (Figure 5B). Heterogeneities at position 1479, either isoleucine (I) or valine (V), and at position 1634, either glycine (G) or arginine (R), were similar in the database and the cohort (Figure 5D).
FIGURE 5.
Impact of single amino acid variants on sofosbuvir sensitivity and viral fitness. (A) Shannon entropy of HEV-3 RdRp sequences (sliding window = 20 and step = 1) from publicly available sequences was downloaded from NCBI. Colored areas correspond to polymerase domains: purple, finger; pink, palm; green, thumb. (B) Frequency of amino acids at positions of interest (1343, 1383, 1384, 1479, and 1634; highlighted in red and with label) in the publicly available datasets and the intrahost diversity data. (C) Pearson correlation (r) of SNV data at baseline and during treatment. p values below 0.05 were considered significant. (D) HEV-3 amino acid sequence variability in the proportion of encoded amino acids of selected residues from the public database. (E) In vitro analysis of single amino acid variants on viral sensitivity to sofosbuvir treatment (error bars = 95% CI), and numbers at the top indicate fold changes compared to wild-type, and (F) replication capacity (error bars = ± SD). HepG2 cells were transfected with single amino acid variants harboring a Gaussia luciferase gene and treated with various doses (100 µM–0.015 µM) of sofosbuvir in vitro. Supernatants were sampled after 72 hours, and variant replication was measured through reporter luciferase read-out and normalized to the respective vehicle-treated control (DMSO). EC50 values and replication capacity were determined by the 4-parameter logistic model and measurement of relative light units of vehicle-treated (DMSO) cells, respectively. Dashed lines represent wild-type control (E) and replication-deficient control (GAA) (F). n ≥ 3. Abbreviations: HepG2, human hepatoma cells; RdRp, RNA-dependent RNA polymerase; SNV, single nucleotide variant.
To test whether single amino acid variants that were selected in multiple patients could confer decreased sensitivity to sofosbuvir in vitro, 7 single substitutions were introduced into the HEV replicon system (Figure 5E, F). The resistance profile to sofosbuvir showed that substitutions at amino acid positions 1383, 1384, 1479, and 1634 marginally altered sofosbuvir sensitivity compared to wild-type p6-Gluc replicon (EC50 2.14 µM, dashed line) (Figure 5E, Table S1, http://links.lww.com/HEP/H882). The strongest effect was observed for the A1343V variant, where the EC50 increased to 9.9 µM, representing a 4.6-fold shift and reduced sofosbuvir sensitivity in vitro (Figure 5E, Figure S8A, http://links.lww.com/HEP/H882). Remarkedly, replication capacity increased from 19.4% (p6-WT) to 35.1% (A1343V) at maximum sofosbuvir concentration (100 µM) (Figure S8A, http://links.lww.com/HEP/H882, middle panel). Furthermore, dose-response assays under ribavirin treatment were performed to test whether single substitutions additionally altered ribavirin sensitivity (Figure S8, http://links.lww.com/HEP/H882). The substitutions K1383N (EC50 0.86 µM), K1383S (EC50 3.83 µM), D1384N (EC50 6.59 µM), and A1343V (EC50 7.46 µM) displayed increased ribavirin sensitivity compared to wild-type (EC50 10.34 µM) (Figure S8A, B, http://links.lww.com/HEP/H882, Table S2, http://links.lww.com/HEP/H882). Overall, most single amino acid variants showed a comparable replication fitness to wild-type p6-Gluc replicon (Figure 5F, Figure S8B, http://links.lww.com/HEP/H882). However, replication capacity was reduced only for substitutions at position 1383, where viral replication decreased by ~1.5 logs. Cell viability was not affected by sofosbuvir or ribavirin treatment (Figure S8C, http://links.lww.com/HEP/H882).
In summary, subsets of variants from different time points were capable of conferring higher sofosbuvir resistance. Of these, a single substitution, A1343V, which occurred in the majority of patients, resulted in a strong decrease in sofosbuvir sensitivity while maintaining replication capacity.
DISCUSSION
In this study, we analyzed the intrahost evolution of HEV in 9 chronically-infected patients who received 400 mg of sofosbuvir daily for 24 weeks but failed to achieve HEV RNA elimination after an initial decline in viral load.29 As high viral heterogeneity has been shown to increase the likelihood of resistance-associated variants emerging during treatment for several viruses,45,46 we first assessed virus population diversity dynamics. Overall, we observed higher nucleotide and amino acid heterogeneity in patients treated with ribavirin before sofosbuvir treatment (P1–P3 and P6–P9) compared to patients not pretreated with ribavirin and detected the emergence of several resistance-associated variants. Ribavirin is known to increase viral diversity34,47,48 and might have already promoted viral heterogeneity and an accumulation of resistance-associated variants prior to sofosbuvir treatment (eg, the alanine to valine substitution at position 1343 of the RdRp). The 2 patients (P1 and P7) with the highest diversity among the patients pretreated with ribavirin were the only ones that had a subpopulation of valine at position 1343 encoding variants at baseline and were among the fastest to establish this substitution as a consensus. In contrast, in the 2 patients, not previously treated with ribavirin, the A1343V substitution still emerged de novo, but on average later. Collectively, these observations show that the A1343V, and other substitutions, may arise de novo in patients treated with sofosbuvir, and ribavirin may be preselecting and accelerating resistance-associated variants.
Next, we introduced patient-derived and single consensus substitutions into a Gaussia luciferase reporter replicon and found decreased sofosbuvir sensitivity for the majority of these constructs. In particular, the A1343V substitution was able to decrease sofosbuvir sensitivity without reduced viral replication fitness. Similarly, substitutions in the NS5B polymerase of HCV have been found to confer decreased sensitivity to sofosbuvir.12,49 The most prominent effect was mediated by the S282T substitution, which is located close to the polymerase active site and has been found to increase the IC50 about 10-fold12 by preventing sofosbuvir from entering the polymerase active site. This substitution has been described to decrease the affinity for nucleotide analogs but at the cost of replication fitness.50,51 Similar mechanisms have also been discussed for substitutions that confer ribavirin resistance for multiple viruses.52–54 For HEV, substitutions at positions 1383 and 1384 are thought to lead to increased polymerase fidelity and, thus, decrease affinity for nucleoside analogs.34,55,56 In accordance with the finding from this study, these substitutions were found to lead to decreased replication fitness in vitro. The identification of A1343V in treatment resistance sheds new light on the mode of action of sofosbuvir in HEV. Although the alpha-fold model predicted that A1343V is most likely not located within the RNA tunnel, a sterical clash may also be caused by V1343 altering the overall conformation of the polymerase.
Independent of this report, A1343V has been identified after sofosbuvir treatment in one patient with chronic hepatitis E who was treated with a combination of ribavirin and sofosbuvir.27 This patient also had variants including A1343V, K1383N, D1384N, V1479I, and G1634R, which were similar to those detected in this study. Therefore, it is likely that these substitutions are associated with sofosbuvir resistance and, alone or in combination, may have a significant impact on HEV RNA levels in vivo. In support of this, the low prevalence of V1343 in public databases identified in this study may suggest that it is not frequently selected for and may be specific to sofosbuvir treatment. Of the total 142 entries, 4 (2.8%) accounted for the variant. It should be noted that sequencing databases for hepatitis E may underrepresent the true diversity of HEV and most likely contain sampling biases. Therefore, it remains to be seen how frequently A1343V circulates in HEV-infected individuals and what impact it has on current and future treatment approaches.
Sofosbuvir is considered a pan-genotype anti-HCV drug that has been shown to inhibit other flaviviruses and togaviruses. These viruses, similar to HEV, can replicate in different cell types. Therefore, it is important to note that sofosbuvir activation may not be limited to the liver: Indeed, activation has been demonstrated in nonhepatic cells, including neuronal cells.20,57 This may be a crucial point for the treatment of patients infected with HEV because extrahepatic reservoirs may have an important role in viral recurrence, even after the virus is undetectable in the blood.58
For HCV, sofosbuvir treatment success was associated with the dose of treatment: 400 mg was highly effective, and seldomly resistant-associated mutations occurred. However, lower doses (200 mg) increased the risk of selecting resistant-associated variants.59 This may explain why the emergence of A1343V in HEV occurred at such a high frequency. The effective dose in inhibiting HEV replication was about 10-fold lower than what is known for HCV.21 Thus, the standardized dose for HCV, 400 mg daily for 24 weeks, may not be sufficient to repress resistant-associated variants, and higher doses may be required. Sofosbuvir has, in general, a good safety and toxicity profile, and also supratherapeutic doses of up to 1200 mg were well tolerated in patient trials.60 Hence, it may be worth re-evaluating the dosing of sofosbuvir for the treatment of patients infected with HEV. In addition, future trials should investigate the efficacy of combination therapy with sofosbuvir and ribavirin. Most importantly, close monitoring of circulating variants during or even prior to treatment can be a useful tool to assess response to treatment and guide treatment decisions.
In summary, viral variants were selected during sofosbuvir treatment that altered sensitivity to the drug. In particular, A1343V, which was found to be specific in patients treated with sofosbuvir, showed a profound reduction of sensitivity.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Klaus Sure for kindly providing technical support and the HepNet Study-House/German Liver Foundation and Gilead for kindly providing patient samples. They also thank all the patients, family members, and staff from all the units that participated in the study. Moreover, they thank all members of the Department for Molecular and Medical Virology, Ruhr University Bochum, for helpful support, suggestions, and discussions.
Footnotes
Abbreviations: CC50, half-maximum cytotoxic concentration; HepG2, human hepatoma cells; Gluc, Gaussia luciferase; RBV, ribavirin; RdRp, RNA-dependent RNA polymerase; SNV, single nucleotide variant.
André Gömer, Mara Klöhn and Michelle Jagst contributed equally.
Eike Steinmann and Daniel Todt share senior authorship.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.hepjournal.com.
Contributor Information
André Gömer, Email: andre.goemer@rub.de.
Mara Klöhn, Email: mara.kloehn@rub.de.
Michelle Jagst, Email: michelle.jagst@rub.de.
Maximilian K. Nocke, Email: maximilian.nocke@rub.de.
Sven Pischke, Email: s.pischke@uke.de.
Thomas Horvatits, Email: t.horvatits@uke.de.
Julian Schulze zur Wiesch, Email: julianszw@googlemail.de.
Svenja Hardtke, Email: s.hardtke@uke.de.
Markus Cornberg, Email: cornberg.markus@mh-hannover.de.
Heiner Wedemeyer, Email: wedemeyer.heiner@mh-hannover.de.
Patrick Behrendt, Email: Patrick.behrendt@twincore.de.
Eike Steinmann, Email: eike.steinmann@ruhr-uni-bochum.de.
Daniel Todt, Email: daniel.todt@rub.de.
AUTHORS CONTRIBUTIONS
The study was designed and the protocol was written by André Gömer, Mara Klöhn, Michelle Jagst, Eike Steinmann, and Daniel Todt; the study was coordinated by André Gömer, Mara Klöhn, Michelle Jagst, Heiner Wedemeyer, Patrick Behrendt, Eike Steinmann, and Daniel Todt; patients were recruited and treated by Thomas Horvatits, Julian Schulze zur Wiesch, Tobias Müller, Sven Pischke, Markus Cornberg, Heiner Wedemeyer, and Patrick Behrendt; data analysis and statistics were performed by André Gömer, Mara Klöhn, Michelle Jagst, Maximilian K. Nocke, and Daniel Todt; and HEV sequencing data were provided by André Gömer and Daniel Todt. Drafting of the manuscript was done by André Gömer, Mara Klöhn, Michelle Jagst, Eike Steinmann, and Daniel Todt; critical revision of the manuscript was performed by all authors; and administrative support: Svenja Hardtke, Heiner Wedemeyer, Eike Steinmann, and Daniel Todt.
FUNDING INFORMATION
Daniel Todt was supported by grants from the German Ministry of Education and Research (BMBF, project VirBio; 01KI2106). Eike Steinmann was supported by grants from the German Federal Ministry of Health (ZMVI1-2518FSB705), the German Research Foundation (DFG, grant number: 398066876-GRK 2485/1), and the German Ministry of Education and Research (BMBF, project SILVIR: 16GW0202). Markus Cornberg, Heiner Wedemeyer, Patrick Behrendt, and Eike Steinmann were funded by a grant from the German Centre for Infection Research (DZIF). Heiner Wedemeyer and Eike Steinmann were funded by the German Ministry of Education and Research (HepEDiaSeq. 01EK2106A/B). All authors had access to the study data, and reviewed and approved the final manuscript.
CONFLICTS OF INTEREST
Thomas Horvatits is on the speakers’ bureau for Microgen diagnostics. Julian Schulze zur Wiesch is on the speakers‘ bureau for Gilead. Markus Cornberg advises and is on the speakers’ bureau for AbbVie, Gilead, and MSD. He advises AiCuris, GlaxoSmithKline, Jansen-Cilag, Novartis, Roche, and Swedish Orphan Biovitrum. He is on the speakers’ bureau for Falk. Heiner Wedemeyer consults, is on the speakers’ bureau, received grants, and has other interests with Gilead. He consults, received grants, and has other interests with Falk. He consults and has other interests with MSD. He consults and advises Roche. He consults and is on the speakers’ bureau for Pfizer. He is on the speakers’ bureau for the Falk Foundation. The remaining authors have no conflicts to report.
REFERENCES
- 1.World Health Organization. Hepatitis E Fact Sheets [Internet]. 2022 [Accessed cited 2023 Jan 3]; https://www.who.int/news-room/fact-sheets/detail/hepatitis-e
- 2.Velavan TP, Pallerla SR, Johne R, Todt D, Steinmann E, Schemmerer M, et al. Hepatitis E: An update on One Health and clinical medicine. Liver Int. 2021;41:1462–1473. [DOI] [PubMed] [Google Scholar]
- 3.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of Hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361:1025–1027. [DOI] [PubMed] [Google Scholar]
- 4.Singh GKJ, Ijaz S, Rockwood N, Farnworth SP, Devitt E, Atkins M, et al. Chronic Hepatitis E as a cause for cryptogenic cirrhosis in HIV. J Infect. 2013;66:103–106. [DOI] [PubMed] [Google Scholar]
- 5.Gérolami R, Moal V, Colson P. Chronic Hepatitis E with cirrhosis in a kidney-transplant recipient. N Engl J Med. 2008;358:859–860. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Gracia MT, Suay-García B, Mateos-Lindemann ML. Hepatitis E and pregnancy: current state. Rev Med Virol. 2017;27:e1929. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A, Beniwal M, Kar P, Sharma JB, Murthy NS. Hepatitis E in pregnancy. Int J Gynecol Obstet. 2004;85:240–244. [DOI] [PubMed] [Google Scholar]
- 8.Dalton HR, Kamar N, Baylis SA, Moradpour D, Wedemeyer H, Negro F. EASL Clinical Practice Guidelines on hepatitis E virus infection. J Hepatol. 2018;68:1256–1271. [DOI] [PubMed] [Google Scholar]
- 9.Kamar N, Abravanel F, Behrendt P, Hofmann J, Pageaux GP, Barbet C, et al. Ribavirin for Hepatitis E virus infection after organ transplantation: A Large European Retrospective Multicenter Study. Clin Infect Dis. 2020;71:1204–1211. [DOI] [PubMed] [Google Scholar]
- 10.Kinast V, Burkard TL, Todt D, Steinmann E. Hepatitis E Virus Drug Development. Viruses. 2019;11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, et al. Discovery of a β-d-2′-Deoxy-2′-α-fluoro-2′-β-C-methyluridine Nucleotide Prodrug (PSI-7977) for the treatment of Hepatitis C Virus. J Med Chem. 2010;53:7202–7218. [DOI] [PubMed] [Google Scholar]
- 12.Lam AM, Espiritu C, Bansal S, Micolochick Steuer HM, Niu C, Zennou V, et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of Hepatitis C Virus. Antimicrob Agents Chemother. 2012;56:3359–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Torres M. Sofosbuvir (GS-7977), a pan-genotype, direct-acting antiviral for hepatitis C virus infection. Expert Rev Anti Infect Ther. 2013;11:1269–1279. [DOI] [PubMed] [Google Scholar]
- 14.McQuaid T, Savini C, Seyedkazemi S. Sofosbuvir, a significant paradigm change in HCV treatment. J Clin Transl Hepatol. 2015;3:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keating GM. Sofosbuvir: A review of its use in patients with Chronic Hepatitis C. Drugs. 2014;74:1127–1146. [DOI] [PubMed] [Google Scholar]
- 16.Bullard-Feibelman KM, Govero J, Zhu Z, Salazar V, Veselinovic M, Diamond MS, et al. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017;137:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H-T, Colby-Germinario SP, Hassounah SA, Fogarty C, Osman N, Palanisamy N, et al. Evaluation of Sofosbuvir (β-D-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine) as an inhibitor of Dengue virus replication #. Sci Rep. 2017;7:6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendes ÉA, Pilger DRB, Santos Nastri ACS, Malta FM, Pascoalino BDS, Carneiro D’Albuquerque LA, et al. Sofosbuvir inhibits yellow fever virus in vitro and in patients with acute liver failure. Ann Hepatol. 2019;18:816–824. [DOI] [PubMed] [Google Scholar]
- 19.Freitas CS de, Higa LM, Sacramento CQ, Ferreira AC, Reis PA, Delvecchio R, et al. Yellow fever virus is susceptible to sofosbuvir both in vitro and in vivo. PLoS Negl Trop Dis. 2019;13:e0007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira AC, Reis PA, de Freitas CS, Sacramento CQ, Villas Bôas Hoelz L, Bastos MM, et al. Beyond members of the flaviviridae family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother. 2019;63:e01389–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dao Thi VL, Debing Y, Wu X, Rice CM, Neyts J, Moradpour D, et al. Sofosbuvir inhibits Hepatitis E Virus replication in vitro and results in an additive effect when combined with ribavirin. Gastroenterology. 2016;150:82–85.e4. [DOI] [PubMed] [Google Scholar]
- 22.Donnelly MC, Imlach SN, Abravanel F, Ramalingam S, Johannessen I, Petrik J, et al. Sofosbuvir and daclatasvir Anti–Viral Therapy Fails to Clear HEV Viremia and Restore Reactive T Cells in a HEV/HCV Co-Infected Liver Transplant Recipient. Gastroenterology. 2017;152:300–301. [DOI] [PubMed] [Google Scholar]
- 23.Sastre L, García-López M, Pérez-del-Pulgar S, Lens S, Costa J, Navasa M, et al. The challenge of chronic hepatitis E in liver transplant recipients: Failure of sofosbuvir plus ribavirin therapy. Gastroenterol Hepatol. 2020;43:136–137. [DOI] [PubMed] [Google Scholar]
- 24.van Wezel EM, de Bruijne J, Damman K, Bijmolen M, van den Berg AP, Verschuuren EAM, et al. Sofosbuvir add-on to ribavirin treatment for Chronic Hepatitis E Virus infection in solid organ transplant recipients does not result in sustained virological response. Open Forum Infect Dis. 2019;6:ofz346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraga M, Gouttenoire J, Sahli R, Chtioui H, Marcu C, Pascual M, et al. Sofosbuvir add-on to ribavirin for chronic hepatitis E in a cirrhotic liver transplant recipient: a case report. BMC Gastroenterol. 2019;19:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todesco E, Mazzola A, Akhavan S, Abravanel F, Poynard T, Roque-Afonso A-M, et al. Chronic hepatitis E in a heart transplant patient: sofosbuvir and ribavirin regimen not fully effective. Antivir Ther. 2018;23:463–465. [DOI] [PubMed] [Google Scholar]
- 27.Schulz M, Papp CP, Bock C-T, Hofmann J, Gerlach UA, Maurer MM, et al. Combination therapy of sofosbuvir and ribavirin fails to clear chronic hepatitis E infection in a multivisceral transplanted patient. J Hepatol. 2019;71:225–227. [DOI] [PubMed] [Google Scholar]
- 28.Valk M, van der, Zaaijer HL, Kater AP, Schinkel J. Sofosbuvir shows antiviral activity in a patient with chronic hepatitis E virus infection. J Hepatol. 2017;66:242–243. [DOI] [PubMed] [Google Scholar]
- 29.Cornberg M, Pischke S, Müller T, Behrendt P, Piecha F, Benckert J, et al. Sofosbuvir monotherapy fails to achieve HEV RNA elimination in patients with chronic hepatitis E – The HepNet SofE pilot study. J Hepatol. 2020;73:696–699. [DOI] [PubMed] [Google Scholar]
- 30.Wahid B. Successful treatment of HBV, HCV, & HEV, with 12-week long use of tenofovir, sofosbuvir, daclatasvir, and ribavirin: A case report. J Infect Public Health. 2020;13:149–150. [DOI] [PubMed] [Google Scholar]
- 31.Drinane M, Jing Wang X, Watt K. Sofosbuvir and ribavirin eradication of refractory Hepatitis E in an immunosuppressed kidney transplant recipient. Hepatology. 2019;69:2297–2299. [DOI] [PubMed] [Google Scholar]
- 32.De Martin E, Antonini TM, Coilly A, Pittau G, Vibert E, Duclos-Vallée J-C, et al. HCV and HEV recurrence after liver transplantation: One antiviral therapy for two viruses. Transpl Int. 2017;30:318–319. [DOI] [PubMed] [Google Scholar]
- 33.Biliotti E, Franchi C, Spaziante M, Garbuglia AR, Volpicelli L, Palazzo D, et al. Autochthonous acute hepatitis E: treatment with sofosbuvir and ribavirin. Infection. 2018;46:725–727. [DOI] [PubMed] [Google Scholar]
- 34.Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, et al. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut. 2016;65:1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gömer A, Brown RJP, Pfaender S, Deterding K, Reuter G, Orton R, et al. Intra-host analysis of hepaciviral glycoprotein evolution reveals signatures associated with viral persistence and clearance. Virus Evol. 2022;8:veac007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, et al. Welcome to the tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 39.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: Making protein folding accessible to all. Nat Methods. 2022;19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci Publ Protein Soc. 2021;30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meister TL, Klöhn M, Steinmann E, Todt D. A cell culture Model for producing high titer Hepatitis E Virus stocks. J Vis Exp. 2020;160:e61373. [DOI] [PubMed] [Google Scholar]
- 42.Todt D, Friesland M, Moeller N, Praditya D, Kinast V, Brüggemann Y, et al. Robust hepatitis E virus infection and transcriptional response in human hepatocytes. Proc Natl Acad Sci. 2020;117:1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todt D, François C, Anggakusuma, Behrendt P, Engelmann M, Knegendorf L, et al. Antiviral activities of different interferon types and subtypes against Hepatitis E virus replication. Antimicrob Agents Chemother. 2016;60:2132–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoler N, Nekrutenko A. Sequencing error profiles of Illumina sequencing instruments. NAR Genom Bioinform. 2021;3:lqab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howe AYM, Rodrigo C, Cunningham EB, Douglas MW, Dietz J, Grebely J, et al. Characteristics of hepatitis C virus resistance in an international cohort after a decade of direct-acting antivirals. JHEP Rep. 2022;4:100462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487–503. [DOI] [PubMed] [Google Scholar]
- 47.Todt D, Meister TL, Steinmann E. Hepatitis E virus treatment and ribavirin therapy: viral mechanisms of nonresponse. Curr Opin Virol. 2018;32:80–87. [DOI] [PubMed] [Google Scholar]
- 48.Todt D, Walter S, Brown RJP, Steinmann E. Mutagenic effects of ribavirin on Hepatitis E Virus—viral extinction versus selection of fitness-enhancing mutations. Viruses. 2016;8:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu S, Doehle B, Rajyaguru S, Han B, Barauskas O, Feng J, et al. In vitro selection of resistance to sofosbuvir in HCV replicons of genotype-1 to -6. Antivir Ther. 2017;22:587–597. [DOI] [PubMed] [Google Scholar]
- 50.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–49170. [DOI] [PubMed] [Google Scholar]
- 51.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. [DOI] [PubMed] [Google Scholar]
- 52.Zeng J, Wang H, Xie X, Yang D, Zhou G, Yu L. An increased replication fidelity mutant of foot-and-mouth disease virus retains fitness in vitro and virulence in vivo. Antiviral Res. 2013;100:1–7. [DOI] [PubMed] [Google Scholar]
- 53.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci U S A. 2003;100:7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Curti E, Jaeger J. Residues Arg283, Arg285, and Ile287 in the nucleotide binding pocket of bovine viral diarrhea virus NS5B RNA polymerase affect catalysis and fidelity. J Virol. 2013;87:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Debing Y, Ramière C, Dallmeier K, Piorkowski G, Trabaud M-A, Lebossé F, et al. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol. 2016;65:499–508. [DOI] [PubMed] [Google Scholar]
- 56.Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, et al. A Mutation in the Hepatitis E Virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology. 2014;147:1008–1011.e7. [DOI] [PubMed] [Google Scholar]
- 57.Sacramento CQ, de Melo GR, de Freitas CS, Rocha N, Hoelz LVB, Miranda M, et al. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci Rep. 2017;7:40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalton HR, Kamar N, van Eijk JJJ, Mclean BN, Cintas P, Bendall RP, et al. Hepatitis E virus and neurological injury. Nat Rev Neurol. 2016;12:77–85. [DOI] [PubMed] [Google Scholar]
- 59.Rose L, Bias TE, Mathias CB, Trooskin SB, Fong JJ. Sofosbuvir: A Nucleotide NS5B inhibitor for the treatment of Chronic Hepatitis C Infection. Ann Pharmacother. 2014;48:1019–1029. [DOI] [PubMed] [Google Scholar]
- 60.https://www.gilead.ca/-/media/gilead-canada/pdfs/medicines/sovaldi_english_pm_e162880-gs-009.pdf Gilead Sciences. Product monograph, Pr sovaldi (sofosbuvir) tablets 400 mg sofosbuvir [Internet]. 2021 [Accessed cited 2023 Feb 6];