The physiology of the circadian (daily) clock has been well studied in the unicellular eukaryote Chlamydomonas reinhardtii. Circadian rhythms of phototaxis, chemotaxis, cell division, UV sensitivity, and adherence to glass have been characterized in this green alga. Circadian phototaxis was even shown to operate in outer space! The related phenomenon of photoperiodic time measurement of germination has been demonstrated. The C. reinhardtii system now offers genetic and proteomic opportunities that make it an excellent unicellular eukaryotic model organism to study the circadian clock at all levels of organization. Several clock-controlled genes have been identified as well as a clock-controlled RNA-binding protein that acts on circadian output. A computer-based search in C. reinhardtii for components of the circadian system that are similar to those from other model species has shown that some phototransduction components and especially kinases and phosphatases are well conserved in this green alga, while their target proteins appear to be different. The first functional proteomic approaches have discovered novel components of the circadian system, including a protein disulfide isomerase and a tetratricopeptide repeat protein.
CIRCADIAN PROGRAMS IN C. REINHARDTII AND THEIR SIGNIFICANCE
Circadian (daily) rhythms are endogenous biological programs that control metabolic, physiological, and/or behavioral events to occur at optimal phases of the daily cycle. In addition to exhibiting a self-sustained oscillation in constant conditions that can be entrained to environmental cycles, circadian rhythms run at essentially the same rate at different ambient temperatures (i.e. they are “temperature compensated”). C. reinhardtii has long been a preferred model system for the analysis of circadian rhythms because of the genetic and molecular techniques that have been developed for C. reinhardtii and are described elsewhere in this issue (Harris, 1989, 2001; Johnson et al., 1992; Fuhrmann, 2002). The availability of its complete nuclear, chloroplast, and mitochondrial genome sequences and of more than 200,000 expressed sequence tags (ESTs) that have been assembled into approximately 10,000 unique cDNAs (Grossman et al., 2003) render it a highly attractive model system.
The circadian clock in C. reinhardtii is known to modulate a number of processes. As is the case for other flagellates, C. reinhardtii is able to orientate itself toward the light, a process known as photoaccumulation or phototaxis. This process is known to be rhythmically modulated in some species; Victor Bruce demonstrated circadian rhythms of photoaccumulation in C. reinhardtii more than 30 years ago (Bruce, 1970). The algae swim toward a supplied light source with maximum accumulation during the day phase. Computerized apparatuses can automatically monitor this rhythm (Mergenhagen, 1984; Kondo et al., 1991; Johnson et al., 1992). Mergenhagen and Mergenhagen (1987) used an automated apparatus aboard a spacecraft to confirm that the photoaccumulation rhythm persisted in outer space when the cells orbited the earth every 90 min! The rhythms continued for at least 6 d in microgravity without any terrestrial cue of the time of day. Therefore, these studies of C. reinhardtii showed conclusively that the circadian clock can run independently from daily cycles of gravity, magnetism, cosmic ray irradiation, and so on.
There are also circadian rhythms in C. reinhardtii that peak during the night phase, for example, chemotaxis to ammonium (Byrne et al., 1992). The cells swim maximally toward this nitrogen source during the middle of the night phase, even though the light-dependent uptake of ammonium does not occur until dawn. The uptake of nitrite also peaks at dawn, as does the activity of nitrite reductase (Pajuelo et al., 1995). Chemotaxis versus phototaxis (as well as the uptake and further metabolism of N sources) is an example of temporal separation of processes in algae. In other words, the daily biological clock organizes a temporal program that triggers biological events to occur at specific phases throughout the daily environmental cycle; phototaxis during daytime optimizes photosynthesis, and chemotaxis (to ammonium) in the nighttime allows C. reinhardtii to find and store nutrients when solar energy is not available.
Another circadian rhythm in C. reinhardtii that peaks during the night is the ability of the cells to adhere to a glass surface (Straley and Bruce, 1979). Yet another important rhythmic property is the temporal control of cell division, as first studied by Bruce (1970) and later by Goto and Johnson (1995). Several clock mutants that have an altered period were isolated by Victor Bruce using the phototaxis rhythm as a screen (Bruce, 1972; Bruce and Bruce, 1978). These mutants were called period (per), a name that was also given to a famous clock gene in Drosophila (but they are almost certainly unrelated since there are no putative homologs of the Drosophila per gene in the C. reinhardtii genome; see below). Victor Bruce demonstrated with one of the long-period mutants (per4) that three independent rhythms (phototaxis, adhesion to glass, and cell division) were affected by the mutation, implying that a component of the central oscillator may be defective in the per4 mutant (Straley and Bruce, 1979).
Evolution of Circadian Timers
The question of why organisms have endogenous temporal programs is closely linked with identifying the selective forces that encouraged the original evolution of these timers. Perhaps an initial driving force for the early evolution of circadian clocks could have been to phase cellular events that are inhibited by sunlight to occur in the night. This idea has been called the “escape from light” hypothesis (Pittendrigh, 1993). That speculation seems plausible when one considers the numerous examples of microorganisms with 24-h cell division cycles in which DNA replication and cell division occur during the night (Edmunds, 1988). Some of the events of the cell division cycle in these microorganisms might be sensitive to sunlight, e.g. replication during S phase.
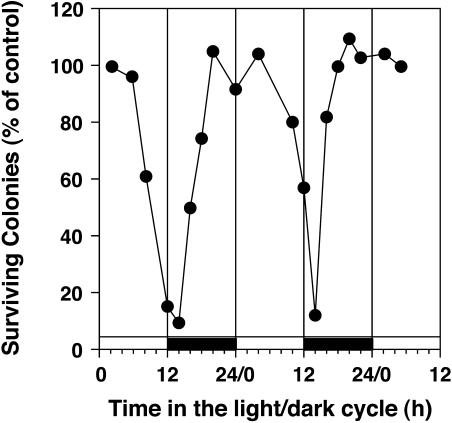
A prediction of this hypothesis would be that present-day organisms retain temporal regulation of light-sensitive processes to the night. Because the most generally deleterious wavelengths of sunlight are in the UV range, a daily rhythm of sensitivity to UV light in C. reinhardtii was tested (Nikaido and Johnson, 2000). As shown in Figure 1, these algae are more sensitive to UV light near sunset and into the early night. The rhythmic sensitivity persists in constant conditions. These data indicate that the circadian system in C. reinhardtii has programmed UV-sensitive processes to occur at phases of the daily cycle when UV levels will be low or absent in a manner that is consistent with the “escape from light” hypothesis (Nikaido and Johnson, 2000).
Figure 1.
Survival of Chlamydomonas cells after irradiation by UV light as a function of the time in a light/dark cycle. Chlamydomonas cultures were plated onto agar medium and treated with equal amounts of UV light at different phases of a 12-h-light/12-h-dark cycle. Survival was measured as the colony-forming ability of cells following treatment as compared to that of cells that were not irradiated with UV light (modified from Nikaido and Johnson, 2000).
Photoperiodism and Seasonal Responses
Plants and animals sense the season of the year by measuring the duration of the day and/or night in the natural environment and respond appropriately so as to adapt to seasonal changes in their environment (Thomas and Vince-Prue, 1997). This phenomenon is called photoperiodic time measurement (PTM). PTM is of fundamental importance to the biological adaptations of organisms to their environment, especially in the cases of reproduction and development. The model for PTM that has become generally accepted is that a circadian (daily) clock is the timer that somehow measures the length of the night and triggers the developmental events controlled by photoperiodism, as first proposed by Bünning (1936) and now supported by multiple lines of evidence (Thomas and Vince-Prue, 1997).
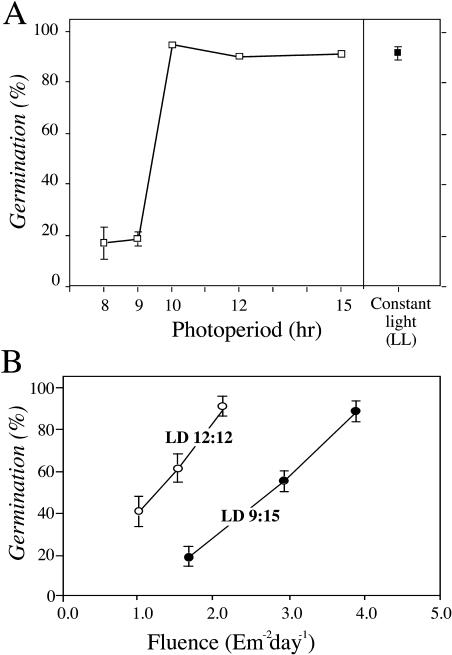
PTM is well documented in multicellular eukaryotes, but few examples exist for unicellular organisms. It seems logical that unicellular organisms might also benefit from being able to anticipate and respond to seasonal changes in their environment. One of the few reports of a photoperiodic response in unicells is a study of PTM in C. reinhardtii (Suzuki and Johnson, 2002). In the life cycle of Chlamydomonas, nonoptimal conditions (e.g. nitrogen deprivation) provoke the differentiation of haploid vegetative cells into gametes, which mate and form diploid zygospores. Zygospores are dormant cells that cannot divide until after they undergo meiosis and germinate into four haploid vegetative cells. Zygotes resist stressful environmental conditions, such as freezing, darkness, desiccation, and starvation, better than vegetative or gametic cells (Harris, 1989; Suzuki and Johnson, 2002). As shown in Figure 2A, the germination efficiency of Chlamydomonas zygospores is enhanced (approximately 90% germination) in long-day light/dark cycles, i.e. for photoperiods longer than 10 h, but is suppressed (<25% germination) in short-day light/dark cycles, i.e. for photoperiods of 8 to 9 h (Suzuki and Johnson, 2002). Comparisons of photoperiod versus fluence demonstrated that Chlamydomonas germination was a bona fide photoperiodic response and not merely a response to fluence (Fig. 2B). Thus, it is likely that the suppression of germination by short days is an adaptive response for the overwintering of Chlamydomonas.
Figure 2.
Germination in different photoperiods in Chlamydomonas. A, Photoperiodic response curve. The germination efficiencies 10 d after mating are shown as a function of the light duration in light/dark cycles of 24 h total duration (white squares). Constant light (LL, filled square) represents cells continuously exposed to light for 24 h a day. B, Germination efficiencies in two different photoperiods (LD 12:12 versus LD 9:15) at different light fluences. Data are plotted as the total light fluence in a day, calculated as irradiance (light intensity, μE m−2 s−1) × (duration of light treatment in seconds). A and B, Germinated cells were counted at 10 d after mating, all data are shown with SEM (modified from Suzuki and Johnson, 2002).
MOLECULAR “OUTPUTS” OF THE CIRCADIAN CLOCK
So far, most of the studies on the molecular basis of the circadian clock in C. reinhardtii have focused on clock-controlled genes. In particular, circadian control of transcriptional rate has been found for a number of genes, resulting in rhythms of RNA abundance over the circadian cycle. These data have been described in a recent review (Mittag and Wagner, 2003) and are summarized in Table I. Many of these clock-controlled genes encode proteins that are involved in photosynthesis and chloroplast metabolism, and most of these genes are expressed at a high rate during the subjective day. A few other genes, e.g. Arf1, GAS-3, HSP70B, and cytochrome c, are maximally expressed in the night. Most of these genes are encoded in the nuclear genome, while a few are encoded by chloroplast DNA (e.g. Tuf A, chl rRNA; Hwang et al., 1996). A possibly related observation is that the supercoiling status of chloroplast DNA itself oscillates over the daily cycle (Salvador et al., 1998). Because the expression of many promoters is sensitive to supercoiling status, gene expression within the chloroplast might be globally modulated by circadian changes of the topology of the chloroplast chromosome.
Table I.
Circadian changes in mRNA abundance and/or transcription rate in C. reinhardtii
References: Jacobshagen and Johnson (1994); Memon et al. (1995); Fujiwara et al. (1996); Hwang et al. (1996); Jacobshagen et al. (1996, 2001); Savard et al. (1996); and Lemaire et al. (1999).
| Time of Maximal Abundance (Day vs. Night) of mRNAs Encoding the Following Proteins | |
|---|---|
| Day Phase | |
| β-Subunit of ATPase | β-Tubulin |
| Carbonic anhydrase 1 | D1 of PSII (Psb A) |
| Elongation factor tu (Tuf A) | Ferredoxin |
| Ferredoxin-NADP reductase | Fru-biphosphate aldolase |
| LI818, new type of Lhcp | Light-harvesting complex protein (LhcpII) |
| Thioredoxin h | Thioredoxin m |
| Phosphoribulokinase | PSA A of PSI |
| Night Phase | |
| ADP-ribosylation factor (Arf 1) | Cytochrome c |
| GAS-3, gamete-specific protein | HSP70B, chloroplastic heat shock protein |
Other molecular studies concern the circadian binding activity of an RNA-binding protein called CHLAMY 1 (Mittag, 1996). This protein is an analog of CCTR (clock controlled translational regulator) from the dinoflagellate alga Lingulodinium polyedra (for review, see Mittag, 2003). Its binding activity increases at the end of the subjective day and stays at a high level until the middle of the subjective night. CHLAMY 1 interacts with mRNAs that have an UG repeat of at least seven units (Waltenberger et al., 2001), and we assume that it represses translation (Mittag, 2003). The UG-containing mRNAs encode mainly proteins of the nitrogen metabolic pathways (NRT2;3, a nitrite/nitrate transporter; nitrite reductase; Gln synthetase; arginino-succinate lyase) and carbon metabolic pathways (small subunit of Rubisco; LIP36-G1, a CO2 shuffling protein in the outer chloroplast membrane; CCM, a CO2 key regulator). CHLAMY 1 represents a novel type of heteromeric RNA-binding protein that consists of subunits comprising three RNA-recognition motifs and three Lys-homology motifs (Zhao et al., 2004). CHLAMY 1's circadian binding activity appears to be controlled at the posttranslational level by time-dependent formation of protein complexes. One of the subunits of CHLAMY 1, named C3, is conserved in mammals, where it appears to be involved in myotonic dystrophy (Zhao et al., 2004). Interestingly, patients suffering from myotonic dystrophy have disturbances in their circadian system (Okumura et al., 2002).
EXPLORING THE C. REINHARDTII GENOME WITH REGARD TO COMPONENTS OF THE CIRCADIAN SYSTEM
The entire nuclear genome (version 2) of C. reinhardtii has been sequenced by the U.S. Department of Energy, and the information is available at the Joint Genome Institute (JGI) Web site (http://genome.jgi-psf.org/chlre2/chlre2.home.html). We searched the C. reinhardtii nuclear genome for potential homologs to genes that are known to encode components of the circadian system in other organisms (Table II). These include the clock model systems of the prokaryotic cyanobacterium Synechococcus elongatus, the fungus Neurospora crassa, the angiosperm Arabidopsis (Arabidopsis thaliana), the fruit fly Drosophila melanogaster, and the mammals Mus musculus and human (Homo sapiens). We scanned for amino acid sequences (National Center for Biotechnology Information [NCBI] protein search) in C. reinhardtii that show any extended similarity to protein sequences that are involved in the circadian system of the aforementioned model organisms by using the JGI BLAST page (http://genome.jgi-psf.org/cgi-bin/runBlast?db=chlre2) in combination with tBLASTn (protein versus translated nucleotides). The results were presented in the form of scaffolds. The depicted sequences within these scaffolds were then used to search within ESTs of C. reinhardtii (http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html) in parallel with the genome browser site of the JGI BLAST page that shows predicted gene models. The proteins that were used for screening JGI were also analyzed within the NCBI conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). All domains are listed in Table II. If a domain of a protein appears to have been conserved in the C. reinhardtii sequence, it is in bold. Percent similarity was calculated by browsing the protein sequence from C. reinhardtii against an NCBI protein BLAST. Regions of similarity (amino acid range) as well as their e-values are indicated in Table II, and the percent similarity is given with respect to these regions. In some cases, only small regions of similarity are shown in Table II, so the significance of the findings must be tempered by the comparison of the extent of a potentially homologous region of the C. reinhardtii protein to the total number of amino acids in the candidate protein.
Table II.
Clock-related proteins from model organisms that are conserved in C. reinhardtii
Sequence analysis was carried out as explained in the first paragraph of the section “Exploring the C. reinhardtii Genome with Regard to Components of the Circadian System.” Briefly, the JGI BLAST page was used in combination with tBLASTn in addition to a NCBI conserved domain search and NCBI protein BLAST. Clock proteins that do not show any significant similarity (labeled as “no hits” in the output file of the JGI blast) to proteins of C. reinhardtii are: S. elongatus KAI A, KAI B, KAI C, LDPA, PEX, RPOD2, and SAS A; N. crassa FRQ; Arabidopsis ELF3, GI, PHYB, PHYE, PIF3, and TEJ; D. melanogaster CLK, CYC, LARK, PDP1, PER, TIM, and VRI; and mammal BMAL1, BMAL2, m-CLOCK, CREB, DBP, DEC1, DEC2, NPAS2, mPER1, mPER2, mPER3, REV-ERB, and mTIM. Other proteins that show only a very limited similarity to proteins of C. reinhardtii are: N. crassa VVD, WC1, and WC2; and Arabidopsis ADO1, ADO2 (LKP2), ADO3 (FKF1), APRR1, APRR3, APRR5, APRR7, APRR9, CCA1, CO1, and ZTL. “Very limited similarity” was defined as those cases when the percentage of identical amino acids (AA; including functional identical ones) was below 16% over the total length of the protein. %, Similarity in percent within the positives (AA range); AA Range, x/y = number of (functional) identical amino acids (x) from the amino acid area of similarity (y); e-value, expect value is a parameter that describes the number of hits one can expect to see just by chance when searching a database of a particular size (the lower the e-value, or the closer it is to 0, the more significant is the match); Total No. AA, total number of amino acids of the protein from the organism used for the similarity search.
| Organism/Protein | Domainsa and Characteristics | NCBI No. | Gene Model/ESTs | % | AA Range | Total No. AA | |
|---|---|---|---|---|---|---|---|
| e-value | |||||||
| Cyanobacteria (S. elongatus) | |||||||
| CIKA | Circadian input kinase | His kinase A, HATPase, REC and GAF domain; input pathway | AAF82192 | C_50007/5.114.1.5 | 57 | 127/220 (1e−35) | 754 |
| CPMA | Circadian phase modifier | Similar to NCAIR mutase (PurE)-related protein; output pathway | AAD29318 | C_250166/No EST | 48 | 112/230 (2e−21) | 260 |
| RPOD | RNA-polymerase sigma factor | Sigma70 regions 2, 3, and 4, FliA, RpoD and E domains | 2208419B | C_30171/3.55.1.51 | 62 | 194/308 (1e−54) | 391 |
| Fungi (N. crassa) | |||||||
| CKA | Casein kinase II catalytic subunit α | Ser/Thr kinase catalytic domain; component of central oscillator | AAM14624 | C_70148b/No EST | 58 | 144/245 (7e−58) | 336 |
| CKB1 | Casein kinase II regulatory subunit β 1 | Casein kinase II regulatory subunit; component of central oscillator | Q8TG12 | C_280152/28.27.1.0 | 59 | 121/20 (2e−41) | 333 |
| CKB2 | Casein kinase II regulatory subunit β 2 | Casein kinase II regulatory subunit; component of central oscillator | Q8TG11 | C_280152/28.27.1.0 | 65 | 132/203 (6e−54) | 285 |
| HHP1 | Casein kinase I homolog | Ser/Thr kinase catalytic domain; component of central oscillator | EAA36589 | C_70149/7.12.2.11 | 90 | 256/283 (1e−138) | 334 |
| PP1 | Protein phosphatase 1, catalytic subunit | PP2A catalytic, Metallphos and ApaH domains; component of central oscillator | Q9UW86 | C_2260011/226.1.2.11 | 92 | 278/300 (1e−155) | 308 |
| PP2A | Protein phosphatase 2A, regulatory subunit | B56 PP2A regulatory B subunit; component of central oscillator | CAC28812 | C_1270036/127.5.2.11 | 63 | 264/418 (1e−107) | 642 |
| Higher plants (Arabidopsis and Mesembryanthemum crystallinum in one case) | |||||||
| AtGRP7 (CCR2) | Arabidopsis Gly-rich protein (=cold circadian rhythm and RNA binding) | Gly rich, RNA-recognition motif; output pathway | AAM62447 | C_10096/1.211.1.5 | 67 | 53/79 (3e−11) | 175 |
| CCA1c | Circadian clock associated 1 | Myb-like DNA-binding domain; component of the central oscillator | NP_850460 | C_1060059/No EST | 84 | 63/75 (2e−22) | 608 |
| CKI | Casein kinase 1 | Ser/Thr kinase; component of the central oscillator | BAB10977 | C_70149/7.12.2.11 | 93 | 277/297 (1e−131) | 476 |
| CKIIA1 | Casein kinase 2, α-chain 1 | Ser/Thr kinase; component of the central oscillator | NP_201539 | C_970006b/97.66.3.52, 97.66.1.51 | 74 | 146/196 (3e−67) | 409 |
| CKIIA2 | Casein kinase 2, α-chain 2 | Ser/Thr kinase; component of the central oscillator | AAN41288 | C_970006b/97.66.3.52, 97.66.1.51 | 63 | 207/326 (2e−86) | 403 |
| CKIIB1 | Casein kinase 2, β-chain 1 | CKII regulatory subunit; component of the central oscillator | S47967 | C_280152/28.27.1.0 | 86 | 160/186 (4e−81) | 287 |
| CKIIB2 | Casein kinase 2, β-chain 2 | CKII regulatory subunit, DRP domain; component of the central oscillator | CAB78767 | C_280152/28.27.1.0 | 86 | 161/186 (3e−81) | 282 |
| COP1 | Constitutive photomorphogenic 1 | WD40 repeats, RING-finger, E3 ubiquitin ligase; input pathway | T01112 | C_1310019/131.28.1.5 | 64 | 196/305 (1e−70) | 675 |
| CRY1 (HY4) | Cryptochrome 1 apoprotein (=Flavin-type blue-light receptor) | FAD-binding domain 7, DNA-photolyase similar to PhrB-photolyases; input pathway | NP_567341 | C_190114b/19.29.1.51, 19.29.2.52 | 62 | 299/478 (1e−120) | 681 |
| CRY2 (PHH1) | Cryptochrome 2 apoprotein (=blue-light photoreceptor) | FAD-binding domain 7, DNA-photolyase similar to PhrB-photolyases; input pathway | AAL16377 | C_190114b/19.29.1.51, 19.29.2.52 | 61 | 288/469 (1e−120) | 612 |
| ELF4 | Early flowering 4 (M. crystallinum) | DUF1313 domain; function unclear | AAQ73526 | C_280056/28.99.2.51 | 72 | 39/54 (2e−7) | 139 |
| EPR1 | Early phytochrome responsive 1 | Myb-like DNA-binding domain; component of central oscillator | BAC98462 | C_1060059/No EST | 85 | 58/68 (8e−21) | 346 |
| LHYc | Late elongated hypocotyl | MYB-type DNA-binding domain; component of central oscillator | CAA07004 | C_1060059/No EST | 95 | 59/62 (2e−21) | 645 |
| NPH1 | Nonphototrophic hypocotyl protein 1 (Phototropin) | PAS/PAC, Ser/Thr kinase domains; input pathway (?) | NP_190164 | C_120056b/12.1.4.12, 12.1.2.51 | 63 | 337/532 (1e−130) | 996 |
| PHYAc | Phytochrome A | PAS, HisKA, HATPase_c, phytochrome, and GAF domains; input pathway | NP_172428 | C_210006b/No EST | 37 | 43/116 (0.056) | 1122 |
| PHYCc | Phytochrome C | PAS, HATPase_c, phytochrome, and GAF domains; input pathway | NP_198433 | C_210006b/No EST | 41 | 52/129 (1.2) | 1111 |
| PHYDc | Phytochrome D | PAS, HisKA, HATPase_c, phytochrome, and GAF domains; input pathway | NP_193360 | C_210006b/No EST | 39 | 44/111 (0.006) | 1164 |
| Fruit fly (D. melanogaster) | |||||||
| CKII α | Casein kinase 2, a-chain | CKII catalytic subunit; component of the central oscillator | AAA28429 | C_970006b/97.66.3.52, 97.66.1.51 | 70 | 140/199 (2e−58) | 336 |
| CKII β | Casein kinase 2, β-chain | CKII regulatory subunit; component of the central oscillator | AAA28430 | C_280152/28.27.1.0 | 75 | 143/190 (2e−64) | 215 |
| CRY | Cryptochrome | DNA photolyase, FAD-binding 7 and PhrB-photolyase domain; input pathway | AAC83828 | C_430042/43.52.2.51 | 59 | 148/249 (9e−51) | 542 |
| DBT | Doubletime casein kinase (protein zeste-white 3) | Ser/Thr kinase; component of the central oscillator | AAD27857 | C_70149/7.12.2.11 | 86 | 255/294 (1e−131) | 440 |
| PP2A_tws | Protein phosphatase 2A, subunit B (twin) | WD40 repeats and Ser/Thr-phosphase 2A regulatory subunit; component of central oscillator | NP_849681 | C_180129/18.24.2.11 | 70 | 346/488 (1e−158) | 500 |
| PP2A_wdb | Protein phosphatase 2A, subunit C (widerborst) | B56 protein phosphatase 2A regulatory B subunit; component of central oscillator | NP_651569 | C_1270036/127.5.2.11 | 67 | 289/431 (1e−119) | 524 |
| SGG | Protein kinase shaggy | Ser/Thr kinase; component of the central oscillator | CAA37419 | C_490046/49.49.6.11 | 79 | 270/340 (1e−128) | 514 |
| Mammals (Human, M. musculus) | |||||||
| CK1 epsilon | Casein kinase 1epsilon, ortholog to dDBT | Ser/Thr kinase; component of the central oscillator | NP_689407 | C_70149/7.12.2.11 | 91 | 268/294 (1e−144) | 416 |
| mCRY1 | Mus cryptochrome 1 | DNA-photolyase, FAD binding 7 and PhrB-photolyase domain; input pathway | NP_031797 | C_430042/43.52.2.51 | 63 | 311/489 (1e−134) | 606 |
| mCRY2 | Mus cryptochrome 2 | DNA-photolyase, FAD binding 7 and PhrB-photolyase domain; input pathway | NP_034093 | C_430042/43.52.2.51 | 64 | 315/489 (1e−133) | 592 |
| Hnat5 | Human N-acetyl-transferase 5 | GNAT family acetyltransferase 1 domain (1st step of melatonine biosynthesis); output pathway | NP_057184 | C_70196b/No EST | 81 | 45/55 (5e−18) | 178 |
| h_HIOMT (ASMT) | Human hydroxy-indole-O-methyltransferase (=Acetyl-serotonin-O-methyltransferase) | O-Methyltransferase 2 domain (2nd step of melatonine biosynthesis); output pathway | AAA75289 | C_1870014/187.14.2.31 | 61 | 71/116 (5e−22) | 298 |
Functional domains are written in (1) bold font if the domain is also present in the putative homolog in the C. reinhardtii genome and is situated within the depicted region of similarity, and (2) italic font if the domain is also present in the putative homolog in the C. reinhardtii genome but is situated outside of the depicted region of similarity.
Gene model still contains extended regions of “N” (nondetermined nucleotides).
Proteins that show only very limited similarity to proteins of C. reinhardtii but have been listed due to functional implications.
In addition, we have searched the C. reinhardtii chloroplast and mitochondrial genomes for potential homologs to the clock-related genes of S. elongatus mentioned in Table II. For this purpose, the chloroplast and mitochondrial sequences of C. reinhardtii were translated in all six open reading frames and compared by the BLAST 2 Sequence Tool (http:ww.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) with each single S. elongatus protein. However, no significant similarities were found to any of the clock-related proteins of S. elongatus.
The Significance of Potential Homologs with Regard to Their Circadian Function
In all circadian model systems studied so far, positive and negative feedback loops have been proposed to be key features (for review, see Dunlap, 1999; Harmer et al., 2001; Panda et al., 2002; Reppert and Weaver, 2002; Johnson, 2004). Within these loops, some molecular components are conserved among certain organisms with regard to their protein domain structure and their function. Thus, CLOCK and CYCLE act as transcription factors of clock components in flies and mammals. Casein kinases phosphorylate components of the endogenous oscillatory system in N. crassa, Arabidopsis, flies, and mammals, although the target proteins are often different. In some cases the protein domain architecture is conserved within a broad range of eukaryotes, but its function is different, as in the case of CRYPTOCHROME (CRY). CRY acts as blue-light photoreceptor in Arabidopsis and flies but is a component of the oscillatory loop in mammals. The similarity search can therefore suggest a clock-related C. reinhardtii component, but its function within the circadian system will have to be proven experimentally for each case.
Clock-Related Kinases and Phosphatases Are Well Conserved in C. reinhardtii
Progressive temporal phosphorylation of central oscillator proteins is thought to be crucial to the 24-h timing mechanism. In all eukaryotic examples (N. crassa, Arabidopsis, D. melanogaster, and mammals), casein kinases (CK1 and CK2) belonging to the Ser/Thr family of kinases are involved (Gorl et al., 2001; Panda et al., 2002; Yang et al., 2003; Daniel et al., 2004; Nawathean and Rosbash, 2004). While the phosphorylated target components are different depending on the organism (e.g. FREQUENCY [FRQ] in N. crassa, CCA1 in Arabidopsis, PERIOD [PER] in D. melanogaster and mammals, etc.), their temporal phosphorylation appears to be relevant for maintaining intact circadian rhythmicity. In C. reinhardtii, CK1 and CK2 (both subunits) are conserved (Table II). Furthermore, the Ser/Thr kinase SHAGGY, which has been shown to phosphorylate the interaction partners of PER and TIMELESS (TIM) in D. melanogaster, is also conserved in C. reinhardtii.
Recently, it has been demonstrated that the protein phosphatase 2A (PP2A) regulatory subunits “twin” and “widerborst” are also part of the oscillatory loop in D. melanogaster by virtue of the observation that they dephosphorylate PER in a circadian manner (Sathyanarayanan et al., 2004). Both subunits are well conserved in C. reinhardtii. Furthermore, distinct roles for the catalytic subunit of protein phosphatase 1 (PP1) and for a regulatory subunit of PP2A have been found in the Neurospora circadian clock (Yang et al., 2004). Both can act on the central clock protein FRQ. PP1 is also highly conserved in C. reinhardtii (Table II). PP1 and PP2A have been localized to specific regions within the Chlamydomonas flagellar axoneme: PP1 is part of the central pair mechanism that controls flagellar motility, and PP2A appears to be anchored to the doublet microtubules (Yang et al., 2000). Both PP1 and PP2A are promising candidates for clock regulation of circadian taxes in C. reinhardtii.
Therefore, kinases and phosphatases that could be involved in circadian systems are present within the C. reinhardtii genome. In fact, there are potential homologs in the C. reinhardtii nuclear genome to all known eukaryotic clock-related kinases and phosphatases. On the other hand, the situation is different with regard to the clock components that are targets of these kinases/phosphatases; there were no significant similarities found in the C. reinhardtii genome to the famous clock proteins FRQ, PER, or TIM.
Similarities of C. reinhardtii Genes to Clock Genes from Other Photosynthetic Organisms
It might be imagined that the likelihood of finding clock-homologous genes/proteins for C. reinhardtii would be greatest among its photosynthetic “sisters,” i.e. cyanobacteria and plants, represented by S. elongatus and Arabidopsis. However, if one excludes photoreceptor genes from the comparison, there are few central clock components that spring from the comparison. For example, there are no putative homologs in C. reinhardtii to the central cyanobacterial clock genes kaiA, kaiB, or kaiC of S. elongatus. The possible homologs that do appear are to cikA, cpmA, and rpoD. The cikA gene is a phytochrome-like gene that has a His-kinase domain that is similar to that of C_50007. The function of cpmA is unknown, and therefore its similarity to C_250166 is difficult to evaluate (also, no EST sequence exists). Finally, rpoD is a sigma factor for RNA polymerase and is therefore unlikely to be a clock-specific gene (and knockout of rpoD in cyanobacteria does not have a strong clock phenotype anyway).
Comparisons with plant clock genes (specifically those of Arabidopsis) likewise implicate similarities in phototransduction and kinase pathways, but no other specific clock insights. Potential kinase homologs were discussed above, and potential homologs to phototransduction genes will be discussed in the next section. The similarities to putative central clock components of Arabidopsis are limited. No putative homologs exist in the C. reinhardtii genome for the Arabidopsis clock components gi, elf3, or tej (Eriksson and Millar, 2003). Only limited similarity can be found to ztl, fkf1, lkp2, or the APRR quintet aprr1/toc1, aprr3, aprr5, aprr7, and aprr9. There are also very short sections of similarity to the central clock genes lhy, cca1, epr1, and elf4 (Table II), but these are probably just similarities to general-function domains found within these proteins (i.e. myb-like DNA binding domains in lhy, cca1, and epr1 or DUF1313 domains in elf4) rather than to the parts of these clock genes that confer the clock-specific functions. In addition, there is a short section of similarity between the RNA recognition motif of the output gene AtGRP7/ccr2 of Arabidopsis (Heintzen et al., 1997) and C_10096. Therefore, despite the close evolutionary relationship between green algae like Chlamydomonas and plants like Arabidopsis, the major correspondences between putative clock genes are in photoreceptor, kinase, and phosphatase genes but not in the genes that are considered to be central components of the clockwork.
Potential Photoreceptors within the Circadian Input Pathway of C. reinhardtii
Several years ago, action spectra from light-pulse-treated cells revealed that blue as well as red light could reset the phase of the circadian clock in C. reinhardtii (Johnson et al., 1991; Kondo et al., 1991). Relevant wavelengths were at 620 nm and 650 to 670 nm in the red and at 520 nm and 450 to 480 nm in the blue. Phytochrome was excluded as a red-light photoreceptor since red/far-red reversibility was not observed. The experimental data available at that time suggested that photosynthetic electron transport could be involved.
A potential blue-light photoreceptor(s) has not been identified. The C. reinhardtii genome suggests some candidates, however. Two classes of blue-light photoreceptors that have been characterized in other organisms were found in the C. reinhardtii genome. One class is the CRYs (Small et al., 1995), which can act as circadian photoreceptors (e.g. in Drosophila or Arabidopsis) or as a component of the circadian clockwork (e.g. in mammals; Harmer et al., 2001), as mentioned above. The similarity search identified two genes that encode potential CRYs in C. reinhardtii. When Arabidopsis CRYs (1 or 2) were used for the search, one gene model was obtained (C_190114; Table II). On the other hand, when either D. melanogaster or mammalian CRYs (1 or 2) were used for the search, a different gene model was the result (C_430042; Table II). For both gene models, proteins are encoded that bear all the characteristic domains of CRY. Thus, we conclude that C. reinhardtii has two CRYs, one of which is more closely related to Arabidopsis CRY and the other to animal CRY. Small and co-workers have sequenced a 7-kb region that includes the CRY protein according to their examinations (Small et al., 1995; Reisdorph and Small, 2004). If this sequence is taken and BLASTed against the C. reinhardtii genome, an extended region of similarity is found around gene model C_190114. But the predicted protein of this gene model is not fully identical with the protein sequence depicted by Small et al. (1995), resulting in some discrepancies between the sequences, presumably within the C_190114-bearing scaffold 19. Importantly, the second potential CRY deriving from gene model C_430042 has not been described and investigated up to now. C_430042 surely needs to be considered in experiments where CRY genes will be silenced since these CRY candidates could overlap functionally.
The other blue-light photoreceptor is phototropin (NPH1), an essential photoreceptor of the phototropic reaction of higher plants. NPH1 was discovered in Arabidopsis (Briggs and Christie, 2002; Kasahara et al., 2002) and was subsequently identified in C. reinhardtii (Huang et al., 2002). In C. reinhardtii, NPH1 controls multiple steps in its sexual life cycle, including changes in chemotaxis (Huang and Beck, 2003; Ermilova et al., 2004). Consistent with those effects, NPH1 can be found in the flagella of C. reinhardtii (Huang et al., 2004). The circadian clock in Arabidopsis controls the expression of NPH1 (Harmer et al., 2000). It remains open if CRY and NPH1 play a role in the circadian system of C. reinhardtii, but both remain prime candidates for an input component.
Over the years, there have been tantalizing suggestions for phytochrome-like transduction in C. reinhardtii, but until now neither physiological nor biochemical data provided persuasive support for a red/far-red photopigment like phytochrome (PHY) in C. reinhardtii. When the phy A, B, C, D, and E proteins from Arabidopsis were used for the homology search, only very limited similarities could be detected between C. reinhardtii proteins with phy A, C, and D, and essentially none to phy B or E. Also, we have used the phytochrome protein sequence from the green alga Mougeotia scalaris (NCBI no. P33529) and the PHY C sequence from Oryza sativa (NCBI no. Q9ZWI9) for the similarity search. But there were no positive hits when the Mougeotia and Oryza sequences were BLASTed against the translated C. reinhardtii nuclear genome sequences. These results suggest that there is indeed no PHY in C. reinhardtii or that these genes/proteins have diverged greatly between C. reinhardtii and other plant systems. Finally, another phototransduction/photomorphogenesis protein from Arabidopsis is COP1, which shows similarity in its WD40 repeats to C_1310019.
The Melatonin Pathway Seems To Occur in C. reinhardtii
Melatonin released by the pineal gland plays an important role within the circadian system regulating reproduction in vertebrates (Reiter, 1993; Goldman, 2001). However, its presence is not restricted to vertebrates, as it can be also found in nonvertebrates and in plants (Caniato et al., 2003; Hardeland and Poeggeler, 2003; Kolar et al., 2003). In animals, the last two steps in melatonin biosynthesis involve the enzymes N-acetyl transferase and hydroxy-indole-O-methyl-transferase (HIOMT). In C. reinhardtii, there are two sequences that show similarity to those animal enzymes (C_70196 and C_1870014; Table II). In the case of C_70196, the area of similarity could be broader than the narrow region reported in Table II because there is a large area of undetermined sequence within scaffold 7 immediately to the right of the C_70196 position. Thus, the possibility exists that C. reinhardtii includes enzymes that could contribute to a melatonin biosynthetic pathway.
ONE STEP BEYOND THE GENOME: APPLYING FUNCTIONAL PROTEOMICS TO FIND NOVEL COMPONENTS OF THE CIRCADIAN SYSTEM
C. reinhardtii offers excellent characteristics to facilitate the application of functional proteomics. Nuclear, chloroplast, and mitochondrial genome sequences as well as many EST sequences are available (Grossman et al., 2003). Although some proteins can now be identified at the picomole to femtomole range with modern mass spectrometry (MS), many regulatory proteins within a crude extract are not abundant enough to be unambiguously identified by MS. Therefore, enrichment of such proteins is a prerequisite for efficient proteome analysis, and C. reinhardtii can be easily and quickly grown in large quantities so as to efficiently implement biochemical purifications. Pioneering proteomic studies have already been carried out in C. reinhardtii with regard to the light-harvesting complex, the chloroplast 70S ribosome, and the circadian system (Stauber et al., 2003; Yamaguchi et al., 2003; Wagner et al., 2004).
In a first approach to apply functional proteomics to circadian-expressed proteins from C. reinhardtii, basic proteins were enriched by heparin affinity chromatography (Wagner et al., 2004). Cells that were oscillating in constant dim light were harvested throughout the subjective day and night. Comparative analyses were carried out using two-dimensional gel electrophoresis and a normalized spot volume procedure. Two proteins fulfilled a rigorous criterion of significance for a daily rhythm of abundance. These two proteins were digested with trypsin and identified by liquid chromatography-electrospray ionization-MS (Wagner et al., 2004). One of the proteins was a nuclear-encoded protein disulfide isomerase (PDI). Its amount was highest during the middle of the subjective night and lowest at the beginning of the subjective day. PDI is known to be an oxidoreductase that assists in the folding of newly synthesized proteins in the endoplasmic reticulum. It can function as a molecular chaperone (Freedman et al., 1994). PDI typically catalyzes the formation, reduction, and isomerization of disulfide bonds during protein folding. Interestingly, a chloroplast PDI had already been described in C. reinhardtii (named RB60) that is a major component of the psbA mRNA-binding protein complex encoding the D1 protein of PSII. Chloroplast PDI is implicated in the redox-responsive regulation of translation in C. reinhardtii chloroplasts in a light/dark-dependent manner (Danon and Mayfield, 1994; Kim and Mayfield, 1997; Trebitsh et al., 2001).
The other protein identified in the proteomic search was a TPR (tetratricopeptide repeat) protein (Wagner et al., 2004). Its highest amount also occurs in the middle of the subjective night, but its minimum is in the middle of the subjective day. TPR proteins have been proposed to interact preferably with WD40 domains [tandem repeats of about 40 residues, each containing a central Trp(W)-Asp(D) motif] (van der Voorn and Ploegh, 1992). Notably, the molecular mechanisms of the circadian clocks in D. melanogaster and N. crassa show a direct involvement of WD40 proteins in the regulation of the circadian oscillator loop (Ko et al., 2002; He et al., 2003). There are also TPR proteins described in C. reinhardtii that are suggested to regulate translation/initiation of chloroplast messages and that are part of multiprotein complexes including RNA (Boudreau et al., 2000; Vaistij et al., 2000).
CONCLUDING REMARKS
The conjunction of a wide spectrum of molecular, genetic, physiological, and biochemical techniques with the available genome and EST sequences renders C. reinhardtii to be an attractive eukaryotic model organism for in-depth studies of the circadian clock. The elucidation of the role of the circadian clock in regulating taxes (phototaxis and chemotaxis) is of particular interest. PP1 and PP2A have been suggested as potential regulatory factors based on the results of the similarity search and their presence within the flagella. Circadian mechanisms that control taxes might also be relevant to humans (e.g. sperm release/movement), especially when one considers that several proteins from the basal apparatus and the flagella of C. reinhardtii are well conserved in humans. Defects in the basal apparatus and the flagella of humans cause severe diseases (e.g. changes of left/right symmetry of organs, polycystic kidney disease, Bardet-Biedl syndrome, as well as a syndrome associated with obesity, hypertension, and diabetes [Olbrich et al., 2002; Li et al., 2004; Snell et al., 2004]).
We can make several suggestions based on the comparison of molecular components of the circadian oscillator from model organisms with potential candidates from C. reinhardtii. The clock kinases and phosphatases from fungi, plants, flies, and mammals are well conserved in this green alga, and it is possible that they may be also involved in its circadian system. Of course, this does not rule out the possibility that these kinases and phosphatases may also participate in other cellular processes. The same interpretation holds for the CRY sequences found in C. reinhardtii. In this context, it is interesting that there appears to be two CRY proteins, one of which is more closely related to plant CRYs while the other is more similar to animal CRYs. Also, COP1, a negative regulator of photomorphogenesis that interacts with CRY in Arabidopsis and mediates its signaling mechanism (Yang et al., 2001), appears to occur in C. reinhardtii. The central clock components that are targets of phosphorylation and are in most cases restricted to a subset of organisms (e.g. KaiC, FRQ, PER, TIM) are not obvious in the C. reinhardtii genome. For this reason, this green alga will probably have its “own” central clock components. The evolution of specific phospho-clock proteins that are key components of the central oscillator thus seems to have originated independently in different systems.
Acknowledgments
We thank Volker Wagner for suggestions on the manuscript. We are grateful for information supplied by the C. reinhardtii genome project of the U.S. Department of Energy.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Mi373/6–1, Mi373/7–1, and Mi373/8–1 to M.M.) and by the National Institute of Mental Health (grant nos. R01 MH43836 and K02 MH01179 to C.H.J.).
References
- Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix JD (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J 19: 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Bruce VG (1970) The biological clock in Chlamydomonas reinhardtii. J Protozool 17: 328–334 [Google Scholar]
- Bruce VG (1972) Mutants of the biological clock in Chlamydomonas reinhardtii. Genetics 70: 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG, Bruce NC (1978) Diploids of clock mutants of Chlamydomonas reinhardtii. Genetics 89: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünning E (1936) Die Endogene Tagesrhythmik als Grundlage der Photoperiodischen Reaktion. Ber Dtsch Bot Ges 54: 590–607 [Google Scholar]
- Byrne TE, Wells MR, Johnson CH (1992) Circadian rhythms of chemotaxis to ammonium and methylammonium uptake in Chlamydomonas. Plant Physiol 98: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniato R, Filippini R, Piovan A, Puricelli L, Borsarini A, Cappelletti EM (2003) Melatonin in plants. Adv Exp Med Biol 527: 593–597 [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP (1994) ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J 13: 2227–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Edmunds LN (1988) Cellular and Molecular Bases of Biological Clocks. Springer-Verlag, New York
- Eriksson ME, Millar AJ (2003) The circadian clock. A plant's best friend in a spinning world. Plant Physiol 132: 732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermilova EV, Zalutskaya ZM, Huang K, Beck CF (2004) Phototropin plays a crucial role in controlling changes in chemotaxis during the initial phase of the sexual life cycle in Chlamydomonas. Planta 19: 420–427 [DOI] [PubMed] [Google Scholar]
- Freedman RB, Hirst TR, Tuite MF (1994) Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci 19: 331–336 [DOI] [PubMed] [Google Scholar]
- Fuhrmann M (2002) Expanding the molecular toolkit for Chlamydomonas reinhardtii—from history to new frontiers. Protist 153: 357–364 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Ishida N, Tsuzuki M (1996) Circadian expression of the carbonic anhydrase gene, Cah 1, in Chlamydomonas reinhardtii. Plant Mol Biol 32: 745–749 [DOI] [PubMed] [Google Scholar]
- Goldman BD (2001) Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. J Biol Rhythms 16: 283–301 [DOI] [PubMed] [Google Scholar]
- Gorl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M (2001) A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J 20: 7074–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Johnson CH (1995) Is the cell division cycle gated by a circadian clock? The case of Chlamydomonas reinhardtii. J Cell Biol 129: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, et al (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2: 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland R, Poeggeler B (2003) Non-vertebrate melatonin. J Pineal Res 34: 233–241 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Satchidananda P, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17: 215–253 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego
- Harris EH (2001) Chlamydomonas as model organism. Annu Rev Plant Physiol Plant Mol Biol 52: 363–406 [DOI] [PubMed] [Google Scholar]
- He Q, Cheng R, Yang Y, He Q, Yu H, Liu Y (2003) FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J 22: 4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Beck CF (2003) Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Kunkel T, Beck CF (2004) Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol Biol Cell 15: 3605–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Merkle T, Beck C (2002) Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light responsive photoreceptor of the phototropin family. Physiol Plant 115: 613–622 [DOI] [PubMed] [Google Scholar]
- Hwang S, Kawazoe R, Herrin DL (1996) Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc Natl Acad Sci USA 93: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobshagen S, Johnson CH (1994) Circadian rhythms of gene expression in Chlamydomonas reinhardtii: circadian cycling of mRNA abundance of cab II, and possibly of β-tubulin and cytochrome c. Eur J Cell Biol 64: 142–152 [PubMed] [Google Scholar]
- Jacobshagen S, Kindle KL, Johnson CH (1996) Transcription of cab II is regulated by the biological clock in Chlamydomonas reinhardtii. Plant Mol Biol 31: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Jacobshagen S, Whetsine JR, Boling JM (2001) Many but not all genes in Chlamydomonas reinhardtii are regulated by the circadian clock. Plant Biol 3: 592–597 [Google Scholar]
- Johnson CH (2004) Precise circadian clocks in prokaryotic cyanobacteria. Curr Issues Mol Biol 6: 103–110 [PubMed] [Google Scholar]
- Johnson CH, Kondo T, Goto K (1992) Circadian rhythms in Chlamydomonas. In K Honma, S Honma, and T Hiroshige, eds, Circadian Clocks from Cell to Human: Proceedings of the Fourth Sapporo Symposium on Biological Rhythms. Hokkaido University Press, pp 139–155
- Johnson CH, Kondo T, Hastings JW (1991) Action spectrum for resetting the circadian phototaxis rhythm in the cw15 strain of Chlamydomonas. II. Illuminated cells. Plant Physiol 97: 1122–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, et al (2002) Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol 129: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mayfield SP (1997) Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278: 1954–1957 [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I (2002) Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678 [DOI] [PubMed] [Google Scholar]
- Kolar J, Johnson CH, Machackova I (2003) Exogenously applied melatonin (N-acetyl-5-methoxytryptamine) affects flowering of the short-day plant Chenopodium rubrum. Physiol Plant 118: 605–612 [Google Scholar]
- Kondo T, Johnson CH, Hastings JW (1991) Action spectrum for resetting the circadian phototaxis rhythm in the cw15 strain of Chlamydomonas. I. Cells in darkness. Plant Physiol 95: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Stein M, Isakidis-Bourguet E, Keryer E, Benoit VV, Pineau B, Gerard-Hirne C, Miginiac-Maslow M, Jacquot JP (1999) The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta 209: 221–229 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552 [DOI] [PubMed] [Google Scholar]
- Memon A, Hwang SB, Deshpande N, Thompson GA Jr, Herrin DL (1995) Novel aspects of the regulation of a cDNA (Arf1) from Chlamydomonas with high sequence identity to animal ADP-ribosylation factor 1. Plant Mol Biol 29: 567–577 [DOI] [PubMed] [Google Scholar]
- Mergenhagen D (1984) Circadian clock: genetic characterization of a short period mutant of Chlamydomonas reinhardtii. Eur J Cell Biol 33: 13–18 [PubMed] [Google Scholar]
- Mergenhagen D, Mergenhagen E (1987) The biological clock of Chlamydomonas reinhardtii in space. Eur J Cell Biol 43: 203–207 [PubMed] [Google Scholar]
- Mittag M (1996) Conserved circadian elements in phylogenetically diverse algae. Proc Natl Acad Sci USA 93: 14401–14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M (2003) The function of circadian RNA-binding proteins and their cis-acting elements in microalgae. Chronobiol Int 20: 529–541 [DOI] [PubMed] [Google Scholar]
- Mittag M, Wagner V (2003) The circadian clock of the unicellular eukaryotic model organism Chlamydomonas reinhardtii. Biol Chem 384: 689–695 [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M (2004) The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13: 213–223 [DOI] [PubMed] [Google Scholar]
- Nikaido SS, Johnson CH (2000) Daily and circadian variation in survival from ultraviolet radiation in Chlamydomonas reinhardtii. Photochem Photobiol 71: 758–765 [DOI] [PubMed] [Google Scholar]
- Okumura K, Aso Y, Tayama K, Yoshida N, Takiguchi Y, Takemura Y, Inukai T (2002) Myotonic dystrophy associated with variable circadian rhythms of serum cortisol and isolated thyrotropin. Am J Med Sci 324: 158–160 [DOI] [PubMed] [Google Scholar]
- Olbrich H, Häffner K, Kispert A, Völkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Omran H (2002) Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet 30: 143–144 [DOI] [PubMed] [Google Scholar]
- Pajuelo E, Pajuelo P, Clemente MT, Marquez AJ (1995) Regulation of the expression of ferredoxin-nitrite reductase in synchronous cultures of Chlamydomonas reinhardtii. Biochim Biophys Acta 1249: 72–78 [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA (2002) Circadian rhythms from flies to human. Nature 417: 329–335 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1993) Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 55: 17–54 [DOI] [PubMed] [Google Scholar]
- Reisdorph NA, Small GD (2004) The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol 134: 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RJ (1993) The melatonin rhythm: both a clock and a calendar. Experientia 49: 654–664 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L (1998) Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol Cell Biol 18: 7235–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A (2004) Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116: 603–615 [DOI] [PubMed] [Google Scholar]
- Savard F, Richard C, Guertin M (1996) The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the cab I/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol Biol 32: 461–473 [DOI] [PubMed] [Google Scholar]
- Small GD, Min B, Lefebvre PA (1995) Characterization of a Chlamydomonas reinhardtii gene encoding a protein of the DNA photolyase/blue photoreceptor family. Plant Mol Biol 28: 443–454 [DOI] [PubMed] [Google Scholar]
- Snell WJ, Pan J, Wang Q (2004) Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117: 693–697 [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, Hippler M (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot Cell 2: 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straley SC, Bruce VG (1979) Stickiness to glass: circadian changes in the cell surface of Chlamydomonas reinhardtii. Plant Physiol 63: 1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki L, Johnson CH (2002) Photoperiodic control of germination in the unicell Chlamydomonas. Naturwissenschaften 89: 214–220 [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D (1997) Photoperiodism In Plants, Ed 2. Academic Press, San Diego
- Trebitsh T, Meiri E, Ostersetzer O, Adam Z, Danon A (2001) The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J Biol Chem 276: 4564–4569 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD (2000) Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 97: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn L, Ploegh GH (1992) The WD-40 repeat. FEBS Lett 307: 131–134 [DOI] [PubMed] [Google Scholar]
- Wagner V, Fiedler M, Markert C, Hippler M, Mittag M (2004) Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett 559: 129–135 [DOI] [PubMed] [Google Scholar]
- Waltenberger H, Schneid C, Grosch JO, Bareiss A, Mittag M (2001) Identification of target mRNAs from C. reinhardtii for the clock-controlled RNA-binding protein Chlamy 1. Mol Genet Genomics 265: 180–188 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR III, Mayfield SP (2003) Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70S ribosome. J Biol Chem 278: 33774–33785 [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheng P, He Q, Wang L, Liu Y (2003) Phosphorylation of FREQUENCY protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol Cell Biol 23: 6221–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fox L, Colbran RJ, Sale WS (2000) Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J Cell Sci 113: 91–102 [DOI] [PubMed] [Google Scholar]
- Yang Y, He Q, Cheng P, Wrage P, Yarden O, Liu Y (2004) Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev 18: 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]