Abstract
Eukaryotic cell cycles are driven by a set of regulators that have undergone lineage-specific gene loss, duplication, or divergence in different taxa. It is not known to what extent these genomic processes contribute to differences in cell cycle regulatory programs and cell division mechanisms among different taxonomic groups. We have undertaken a genome-wide characterization of the cell cycle genes encoded by Chlamydomonas reinhardtii, a unicellular eukaryote that is part of the green algal/land plant clade. Although Chlamydomonas cells divide by a noncanonical mechanism termed multiple fission, the cell cycle regulatory proteins from Chlamydomonas are remarkably similar to those found in higher plants and metazoans, including the proteins of the RB-E2F pathway that are absent in the fungal kingdom. Unlike in higher plants and vertebrates where cell cycle regulatory genes have undergone extensive duplication, most of the cell cycle regulators in Chlamydomonas have not. The relatively small number of cell cycle genes and growing molecular genetic toolkit position Chlamydomonas to become an important model for higher plant and metazoan cell cycles.
It is well established that eukaryotic cell cycles are controlled by a conserved set of proteins. Central among these are cyclin-dependent kinases (CDKs; Nasmyth, 1996; Sherr, 1996; Johnson and Walker, 1999; Mironov et al., 1999; Pavletich, 1999). CDKs are subject to multiple regulatory mechanisms that cause their activity and substrate specificity to oscillate and thereby drive cell cycle phase transitions. These regulatory mechanisms include binding of cyclins that are obligatory activating subunits of CDKs, binding of inhibitory proteins, and activating or inhibitory phosphorylation (Morgan, 1995). Research over the past two decades has highlighted the remarkable conservation of these mechanisms, indicating that eukaryotes share a fundamentally conserved cell cycle (Morgan, 1997; Obaya and Sedivy, 2002; De Veylder et al., 2003; Dewitte and Murray, 2003).
Despite the unity in underlying principles of cell cycle regulation, there are some important differences that remain to be elucidated. First, cell cycles that deviate in some way from the best characterized G1-S-G2-M progression occur in some unicellular eukaryotes and are essential for the proper development of most multicellular eukaryotes, but the mechanisms required to generate altered cell cycles are not fully understood (Joubes and Chevalier, 2000; Moser and Russell, 2000; Schwab et al., 2000; Edgar and Orr-Weaver, 2001; Coffman, 2004). Second, the mechanics of cell division in different taxa vary considerably, particularly the mechanisms used for cytokinesis (Guertin et al., 2002). It is not known to what extent CDKs became adapted to their new substrates, or vice versa, as separate division mechanisms evolved. Third, it is not clear why multicellular eukaryotes, such as plants and animals, contain multiple CDKs, whereas in budding yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe), a single CDK is sufficient to drive the cell cycle. Until a broader spectrum of unicellular eukaryotes is examined, this question cannot be addressed.
The cell cycle regulators from the sequenced genomes of animals, plants, and fungi show distinct patterns of divergence, duplication, and gene loss. Higher plants are interesting because the CDK and cyclin family proteins have duplicated and diverged in them, thus giving rise to a novel CDK, CDKB (Mironov et al., 1999; Joubes et al., 2000; Vandepoele et al., 2002), and several new cyclin families, including a plant-specific D-type cyclin family that appears to be functionally related to the animal D-cyclins (Meijer and Murray, 2000; Oakenfull et al., 2002). Unlike the fungi that lack the retinoblastoma (RB) pathway, homologs of RB and its binding partners E2F/DP are found in higher plants and probably function in a similar manner to those of animals (Xie et al., 1996; Huntley et al., 1998; Ramirez-Parra et al., 1999; Albani et al., 2000; Boniotti and Gutierrez, 2001; Mariconti et al., 2002). The extensive duplications within higher plant cell cycle gene families have complicated the task of reverse genetics. It is therefore of great interest to determine the extent to which cell cycle regulators in higher plants are conserved in simpler representatives of the plant kingdom.
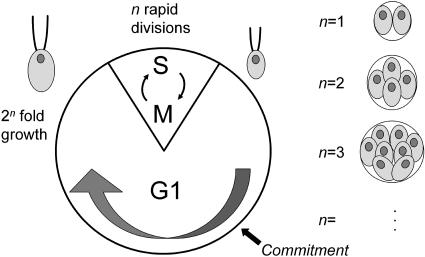
Chlamydomonas reinhardtii is a chlorophyte alga that has served as a model for plant cell biology and physiology (Goodenough, 1992; Rochaix, 1995; Gutman and Niyogi, 2004), and whose genome has recently been sequenced (http://genome.jgi-psf.org/chlre2/chlre2.home.html). Cell division in Chlamydomonas occurs by a noncanonical mechanism, termed multiple fission (Fig. 1; Setlik and Zachleder, 1984; Donnan et al., 1985; John, 1987). The multiple fission cell cycle is characterized by a prolonged G1 period during which cells may grow to many times their original size. This growth phase is followed by a rapid series of (n) alternating S (DNA synthesis) and M (mitotic) phases (Coleman, 1982), producing 2n daughter cells of uniform size. Size homeostasis is maintained by two related mechanisms. A mitotic sizer governs mother cell division number (n) so that daughter cell size is similar regardless of mother cell size. A second sizer operates during early G1 to control passage through Commitment (Donnan and John, 1983, 1984), a cell cycle control point that is conceptually similar to Start in yeasts or the Restriction Point in animal cells (John, 1984, 1987). Cells that have passed Commitment will divide at least one time during the S/M phase. A major difference between Commitment and Start/Restriction is that, after passage through Commitment, Chlamydomonas cells remain in G1 for an additional 5 to 8 h, whether or not they continue to grow, and only begin the S/M phase after this delay. Under physiological conditions of alternating light and dark periods, Chlamydomonas cells become highly synchronized so that growth occurs during the light period and S/M occurs during a brief interval in the dark period (Fig. 2).
Figure 1.
Diagram of the multiple fission cell cycle. A clock-type diagram depicts the phases of the Chlamydomonas cell cycle. Most of the cycle is spent in G1, which is divided into two periods demarcated by the Commitment point (see text for details). At the end of G1, a rapid series of alternating S and M cycles generates 2n daughter cells of uniform size. The value of n is variable and is related to mother cell size. Postmitotic mother cells with different values of n are depicted to the right with 2, 4, 8, etc., daughters.
Figure 2.
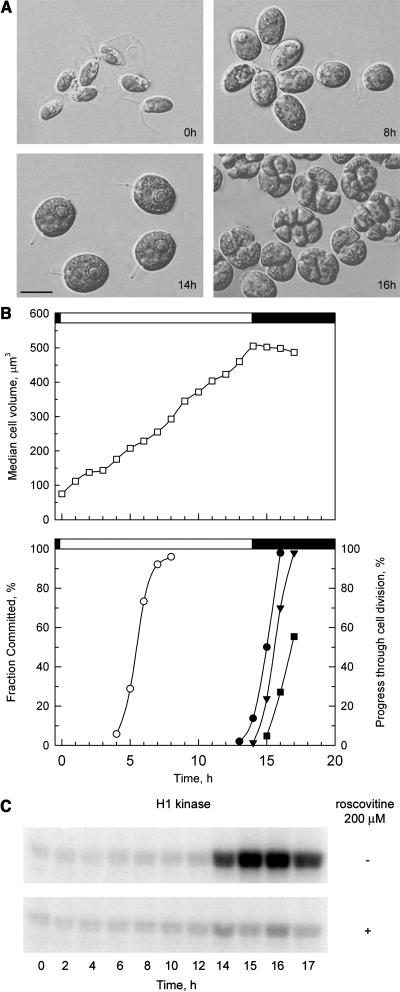
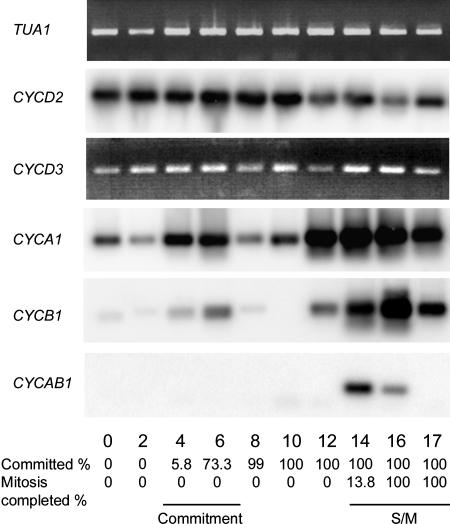
Synchronization of the cell cycle with alternating light and dark periods. A, Photomicrographs of cells at different time points with a scale bar in the bottom left corresponding to 10 μm. B, Graph of synchronized cultures. The top image depicts cell size (white squares). The bottom image depicts the fraction of cells that have passed Commitment (white circles), and progression through cell division with mother cells that have undergone one (black circles), two (black triangles), and three (black squares) rounds of division. Division was complete by 17 h with approximately 60% of the cells dividing three times into eight daughters and approximately 40% dividing twice into four daughters. The light-dark phasing is indicated by the white or black bars above the graphs. C, CKS1-purified histone H1 kinase activity assayed from extracts prepared at different time points. Each image shows an autoradiograph indicating the extent of histone H1 phosphorylation in the presence and absence of the CKI roscovitine.
Given its specialized cell cycle, its unicellular lifestyle, and its estimated divergence time from higher plants (Chlamydomonas and Arabidopsis [Arabidopsis thaliana] may have shared a common ancestor approximately 1.1 billion years ago [Hedges, 2002]), it might be expected that the cell cycle genes in Chlamydomonas would have diverged considerably from those in higher plants and other eukaryotes. In this article, we have identified and profiled the expression of the core cell cycle regulators from Chlamydomonas. Contrary to our initial expectation, we have found that Chlamydomonas encodes orthologs of the major plant CDK and cyclin families. Chlamydomonas also encodes two CDK subtypes and two cyclin subtypes that are not found in higher plants, fungi, or animals. Besides cyclins and CDKs, we have found orthologs of wee1 kinase, a negative regulator of CDKs, CKS1/suc1, a CDK subunit, and potential CDC25 homologs. As expected from the presence of an RB-related gene, MAT3 (Umen and Goodenough, 2001), we have also identified Chlamydomonas E2F and DP orthologs. Unlike higher plants and animals, most of the core cell cycle regulatory genes in Chlamydomonas are present in single copy. We discuss these results in light of their evolutionary implications and with respect to Chlamydomonas as a model for higher plant and metazoan cell cycles.
RESULTS
Annotation Strategy
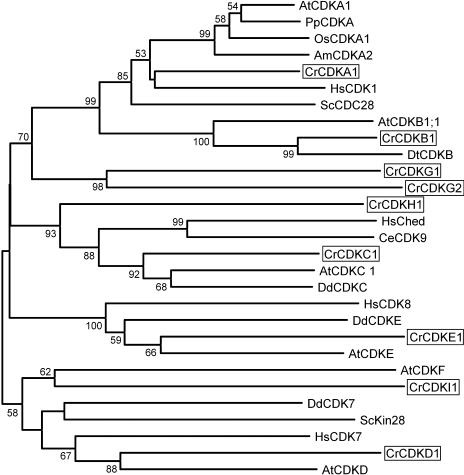
The gene families for which we carried out comprehensive searches were CDK, cyclin, RB-related, E2F/DP, CKS1/suc1, wee1, CDC25, and CDK inhibitors (CKIs). We first made use of automated annotations based on genome-wide BLAST searches carried out as part of the Chlamydomonas genome project and available on the genome Web site (http://genome.jgi-psf.org/chlre2/chlre2.home.html). This initial search yielded a few high-scoring positives but mostly low-scoring or misannotated genes. A more productive strategy involved the use of low-stringency reciprocal BLAST searches with representatives from each gene class to query the conceptually translated Chlamydomonas genome sequence: Typically, one or more BLAST hits from Chlamydomonas could be found, and then the BLAST search was repeated with the Chlamydomonas candidate genes against the National Center for Biotechnology Information (NCBI) nonredundant protein database and against the Chlamydomonas genome to identify potential duplications or additional family members. When possible, gene model and expressed sequence tag (EST) evidence were used to assemble predicted coding regions in order to improve the search. Reverse transcription (RT)-PCR was used in some cases to confirm gene models and to test for expression. Using these strategies, we found that a reciprocal orthology relationship could be established for most of the cell cycle genes. For example, the CDKB genes from Arabidopsis show the highest similarity to a single gene model in Chlamydomonas, and that gene model in Chlamydomonas shows the highest degree of similarity to the CDKB genes from higher plants (Fig. 3; Table I). To complement this approach, we also carried out BLAST searches against the conceptually translated Chlamydomonas EST database that includes more than 100,000 sequences. No cell cycle genes were identified in our EST search that had not already been identified in the sequenced genome, suggesting that our search is essentially complete. Low-stringency Southern blots of several genes were also used to confirm that the key cell cycle regulators are single copy (Supplemental Fig. 1). Phylogenetic analyses were carried out to more objectively place each family member within its proper clade. The results from individual gene families are discussed below, and the results of our annotation are summarized in Table I. For a more complete description of gene models and annotation evidence, see Supplemental Table I.
Figure 3.
Neighbor-joining tree of CDKs. Bootstrap values of 50% or higher are shown for each clade. Am, Antirrhinum majus; At, Arabidopsis; Cr, C. reinhardtii; Ce, Caenorhabditis elegans; Dd, Dictyostelium discoideum; Dt, Dunaliella tertiolecta; Hs, Homo sapiens; Os, Oryza sativa; Sc, budding yeast; Sp, fission yeast; Pp, Physcomitrella patens. See Supplemental Table II for GenBank accession numbers.
Table I.
Summary of annotated cell cycle genes in Chlamydomonas
Gene model names are according to Chlamydomonas genome version 2 (http://genome.jgi-psf.org/chlre2/chlre2.home.html). na, Not applicable; Yes, successful amplification and subcloning of RT-PCR fragment; No, amplification was tried but was not successful; nd, RT-PCR was not done.
| Chlamydomonas Gene Model | Gene Name | Protein Family | Protein Subfamily | Signature Motif | Putative Function(s) | EST | RT-PCR2 |
|---|---|---|---|---|---|---|---|
| C_1630009 | CDKA1 | CDK | CDKA | PSTAIRE | G1/S, G2/M | Yes | Yes |
| C_1340024 | CDKB1 | CDK | CDKB | PSTTLRE | G2/M | Yes | Yes |
| C_1730005 | CDKC1 | CDK | CDKC/CDK9 | PITAIRE | Unknown | Yes | Yes |
| C_700049 | CDKD1 | CDK | CDKD/CDK7 | DPTLARE | CAK | Yes | Yes |
| C_1700010/11 | CDKE1 | CDK | CDKE/CDK8 | SPTAIRE | Unknown | No | Yes |
| C_270112 | CDKG1 | CDK | Novel | SDSTIRE | Unknown | Yes | Yes |
| C_1020028 | CDKG2 | CDK | Novel | AASTLRE | Unknown | Yes | No |
| C_980026 | CDKH1 | CDK | Novel | PVTSIRE | Unknown | No | Yes |
| C_740039 | CDKI1 | CDK | Novel | PDVVVRE | Unknown | Yes | nd |
| C_120118 | CYCA1 | Cyclin | A | LVEVSEEY | S/G2/M | Yes | Yes |
| C_1420018 | CYCB1 | Cyclin | B | HLKF | G2/M | Yes | Yes |
| C_460084 | CYCC1 | Cyclin | C | Transcription | No | No | |
| C_140186 | CYCD1 | Cyclin | D | G1/S | No | No | |
| C_290120 | CYCD2 | Cyclin | D | LQCDE | G1/S | Yes | Yes |
| C_1460039 | CYCD3 | Cyclin | D | LFCGE | G1/S | Yes | Yes |
| C_570097 | CYCL1 | Cyclin | L | Unknown | Yes | nd | |
| C_320095 | CYCM1 | Cyclin | Novel | Unknown | Yes | nd | |
| C_660038 | CYCU1 | Cyclin | U | Unknown | Yes | nd | |
| C_1740016 | CYCT1 | Cyclin | T | Unknown | No | nd | |
| C_1630021 | CYCAB1 | Cyclin | Novel A/B related | Unknown | No | Yes | |
| C_1840020 | MAT3 | RBR | na | G1/S, S/M | Yes | Yes | |
| C_180138 | E2F1 | E2F | E2F | G1/S, S/M | No | Yes | |
| C_570078 | DP1 | E2F | DP | G1/S, S/M | Yes | Yes | |
| C_210071 | E2FR1 | E2F | E2F? | Unknown | No | No | |
| C_980002 | WEE1 | WEE1 | na | G2/M | Yes | Yes | |
| Nonea | CKS1 | CKS1 | na | CDK subunit | Yes | Yes | |
| C_980016 | RDP1 | cdc25-like | na | HCHGSKVRGP | G2/M | No | Yes |
| C_410005 | RDP2 | cdc25-like | na | HCMFSQQRGP | G2/M | No | Yes |
| C_100077 | RDP3 | cdc25-like | na | HCHFSKVRGP | G2/M | Yes | nd |
Gene is located between gene models C_860084 and C_860085.
Synchronization and Expression Profiling
A major advantage of using Chlamydomonas to investigate cell cycle regulation is the ease with which cultures can be synchronized under physiological conditions. By growing cells phototrophically in alternating periods of light and dark, we were able to synchronize our cultures so that they passed Commitment during the middle of the light period, entered the S/M phase of the cell cycle at the end of the light period, and completed division during the dark period (Fig. 2).
We used semiquantitative RT-PCR to examine the expression pattern of a subset of the genes predicted to be cell cycle regulators. Expression levels for each gene were determined in RNA samples prepared from synchronous cultures. The cultures from which RNA was prepared were simultaneously monitored for cell size (Fig. 2, A and B), passage through Commitment (Fig. 2B), passage through mitosis (Fig. 2, A and B), and histone H1 kinase activity (Fig. 2C). Besides the annotated cell cycle regulatory genes, we also examined expression of some genes encoding proteins required for S phase that were expected to be cell cycle regulated.
CDK Family
CDKs are Ser-Thr kinases that function in cell cycle regulation and in other processes such as transcription. The most widely conserved CDKs possess a canonical PSTAIRE motif in the C-helix (De Bondt et al., 1993). In both fission and budding yeast, a single PSTAIRE CDK is sufficient to regulate all the cell cycle phases (Mendenhall and Hodge, 1998; Moser and Russell, 2000). By contrast, metazoans and plants encode several variant CDKs with functions in cell cycle regulation. In humans, there are three PSTAIRE CDKs (CDK1/cdc2, CDK2, and CDK3) and a variant CDK4/6 subfamily with a P(I/L)ST(V/I)RE motif, all of which function in cell cycle regulation (Meyerson et al., 1992; Pines, 1995; Reed, 1997; Lee and Yang, 2003). Higher plants encode only one PSTAIRE CDK, designated CDKA, that can functionally substitute for its yeast orthologs, cdc2/CDC28 (Ferreira et al., 1991; Hirt et al., 1991). Plant CDKs C, D, and E are variants that have metazoan counterparts CDK9, CDK7, and CDK8, respectively (Mironov et al., 1999; Joubes et al., 2000; Vandepoele et al., 2002). Plant-specific kinase CDKB has been found in all plants and CDKF only in Arabidopsis. B-type CDKs are expressed during the G2/M transition when they are thought to function (Mironov et al., 1999; Porceddu et al., 2001; Lee et al., 2003; Boudolf et al., 2004). D- and F-type CDKs are CDK-activating kinases (CAK) that serve to activate A-type CDKs (Umeda et al., 1998; Yamaguchi et al., 1998, 2000; Shimotohno et al., 2003). CDKD binds cyclin H to form a CAK complex (Yamaguchi et al., 2000), while CDKF, a distant relative of CDKD, is able to function without any binding partner. Animal CDK8 and CDK9, as well as plant C-type CDKs, have been implicated in transcriptional control (Oelgeschlager, 2002; Barroco et al., 2003), and no function has yet been determined for E-type CDKs in plants (Magyar et al., 1997).
Chlamydomonas encodes a single ortholog for each of the plant-type CDKs A, B, C, D, and E (genes were designated CDKA1, B1, C1, D1, and E1, respectively) but does not encode an F-type CDK (Fig. 3; Table I; Supplemental Fig. 1). In addition, Chlamydomonas encodes four novel members of the CDK family that are not orthologous to any known CDKs in plants or other CDKs in databases. Two of these novel CDKs encoded by genes designated CDKG1 and CDKG2 are related to each other, but are significantly diverged in their predicted C-helices (SDSTIRE and AASTLRE, respectively). The third locus that we have designated CDKH1 encodes a protein with a PVSTIRE motif and forms a sister group with the CDKC family (Fig. 3). However, CDKH1 is more distantly related to Chlamydomonas CDKC1 than are the plant, metazoan, and slime mold CDKC orthologs, indicating that its duplication preceded the divergence of these taxa. The fourth CDK, encoded by the CDKI1 gene, is the most diverged CDK that we identified and is very distantly related to Arabidopsis CDKF, but it does not appear to be a CDKF ortholog. It does not contain the N-terminal insertion that is characteristic of CDKF, and it does not show a reciprocal best-hit relationship with CDKF in BLAST searches.
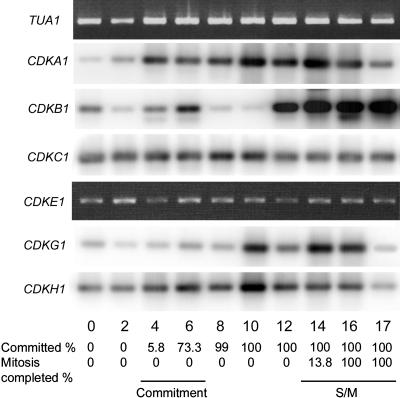
The expression profiles of Chlamydomonas CDKs are similar to those of their plant counterparts. mRNA for CDKA1 was present constitutively during the cell cycle with expression increasing as cells entered the growth phase at the beginning of the light period and increasing further around the time of S/M phase (Fig. 4). Abundance of presumed Chlamydomonas CDKA protein (reacting with anti-PSTAIR antibody) was relatively constant during the cell cycle with phosphorylation-induced isoforms appearing during S/M phase (John et al., 1989). Higher plant CDKA message and protein levels are also relatively constant during the cell cycle (Martinez et al., 1992; Hemerly et al., 1993; Magyar et al., 1997; Richard et al., 2001; Sorrell et al., 2001; Menges et al., 2003). mRNA for CDKB1 shows two peaks of expression, one corresponding to passage through commitment and a second, very strong peak during S/M phase (Fig. 4). Elevated expression at the time of mitosis was described for other members of the CDKB subfamily in plants (Magyar et al., 1997; Richard et al., 2001; Sorrell et al., 2001; Menges et al., 2002; Menges and Murray, 2002) and is consistent with a role for CDKB in the regulation of the G2/M transition (Porceddu et al., 2001; Lee et al., 2003; Boudolf et al., 2004).
Figure 4.
Expression of Chlamydomonas CDK homologs. RT-PCR products were detected by Southern blotting (CDKA1, B1, C1, G1, H1) or ethidium bromide staining (CDKE1). Fraction of cells that had passed Commitment and completed cell division when the sample was prepared is indicated at the bottom. α-Tubulin transcript (TUA1) detected by ethidium bromide staining serves as an internal control.
CDKC1 and E1 are expressed constitutively as are their orthologs in higher plants and in animals (De Luca et al., 1997; Magyar et al., 1997; Garriga et al., 1998). The Chlamydomonas-specific CDKs, G1 and H1, are also expressed constitutively at a low level, with CDKG1 message levels rising somewhat after Commitment (Fig. 4). Although we were able to amplify CDKD1, its abundance appears to be extremely low, and the variability of amplification prevents us from drawing any conclusions about its expression pattern during the cell cycle. We have not detected expression of CDKG2 by RT-PCR, but there is a single EST that corresponds to the predicted CDKG2 mRNA (Supplemental Table I). We have not examined expression of CDKI1, but there is EST evidence supporting its expression (Supplemental Table I).
Cyclin Family
Cyclins bind CDKs, and formation of cyclin-CDK heterodimers is essential for kinase activity. In plants and animals, there are three main classes of cyclins that regulate the cell cycle. D-type cyclins function in G1, A-type cyclins function in S phase, and B-type cyclins function in mitosis. The plant A and B cyclins have orthologs in animals, while the D-type cyclins from plants and animals are divergent (for review, see Renaudin et al., 1996; Murray, 2004). However, the D-cyclins of plants and animals share some structural similarities in that they contain a conserved N-terminal LXCXE motif. The LXCXE peptide binds to the pocket region of RB-related proteins (RBRs) and targets RBRs for CDK phosphorylation. In animals, D-cyclins associate with CDK4/6, whereas in plants they associate with CDKA (Xiong et al., 1992; Bates et al., 1994; Meyerson and Harlow, 1994; Nakagami et al., 1999; Boniotti and Gutierrez, 2001; Healy et al., 2001).
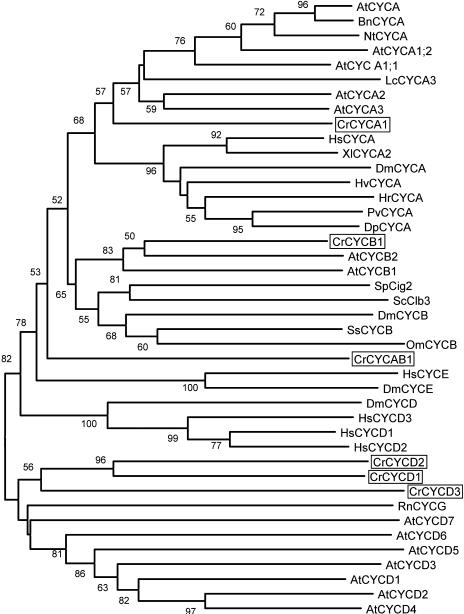
We identified a single Chlamydomonas ortholog for each of the A- and B-type cyclins, designated CYCA1 and CYCB1, respectively, and three D-type cyclins, CYCD1, CYCD2, and CYCD3 (Fig. 5; Table I; Supplemental Fig. 1). Both CYCA1 and CYCB1 have a destruction box (D-box) in their N-terminal domains (RAMLGDLTN and RRALGDLSN, respectively; Supplemental Table I). The D-box is a characteristic feature of A- and B-type cyclins that targets their degradation by ubiquitin-dependent proteolysis (Glotzer et al., 1991). The three Chlamydomonas D-type cyclin homologs cluster with the plant D-type cyclin group but are not closely related to any of the higher plant D-type cyclin subtypes, indicating that the Chlamydomonas D-type cyclin family probably arose as a result of lineage-specific gene duplication. Both CYCD2 and CYCD3 have a LXCXE motif in their N-terminal domains that supports their classification as D-type cyclins. CYCD1 does not have an LXCXE motif in its N-terminal domain, but it is possible that this is due to misannotation. There is an LXCXE coding sequence approximately 500 bp upstream from the annotated start for CYCD1 that is not part of a predicted exon; however, we have not yet been able to determine whether CYCD1 is expressed and therefore have no information on its putative mRNA structure.
Figure 5.
Neighbor-joining tree of A-, B-, and D-cyclin groups. Bootstrap values of 50% or higher are shown for each clade. At, Arabidopsis; Cr, C. reinhardtii; Bn, Brassica napus; Dp, Dreissena polymorpha; Dm, Drosophila melanogaster; Hv, Hydra viridis; Hr, Helobdella robusta; Hs, Homo sapiens; Lc, Lycopersicon esculentum; Nt, Nicotiana tabacum; Om, Oncorhynchus mykiss; Os, Oryza sativa; Pv, Patella vulgata; Rn, Rattus norvegicus; Sc, budding yeast; Sp, fission yeast; Ss, Spisula solidissima; Xl, Xenopus laevis. See Supplemental Table II for GenBank accession numbers.
There is a novel cyclin in Chlamydomonas that we have designated CYCAB1 (Fig. 5; Table I; Supplemental Fig. 1). This novel cyclin does not have signature motifs that place it definitively within the A or B cyclin families, but its closest homologs are in the cyclin A and cyclin B clades. Like A and B cyclins, CYCAB1 has a D-box motif (RATLVDWLSE) in its N-terminal domain, indicating that it may undergo destruction via the ubiquitination pathway.
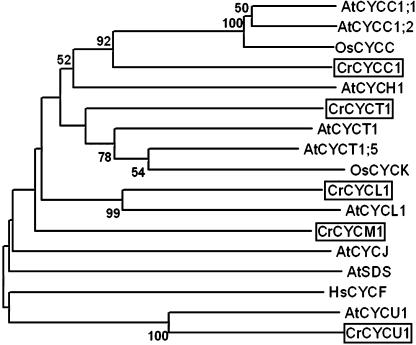
Poorly characterized groups of cyclins include L, T, C, and H families from plants and animals, and some divergent groups of plant cyclins designated SOLO DANCERS (Azumi et al., 2002), J (Abrahams et al., 2001), and either U (Wang et al., 2004) or P (Torres Acosta et al., 2004). Some of the U/P family members can bind CDKA in a yeast two-hybrid assay (Torres Acosta et al., 2004), but there is no direct evidence for their cell cycle involvement. Cyclin C can bind CDK3 and direct phosphorylation of pRb in humans (Ren and Rollins, 2004), and cyclin H forms a CAK complex with CDK7/CDKD (animals/plants; Fisher and Morgan, 1994; Makela et al., 1994; Yamaguchi et al., 2000). Chlamydomonas encodes one representative of each of the L, T, C, and U/P classes that we have designated CYCL1, CYCT1, CYCC1, and CYCU1, respectively (Fig. 6). We have not identified an H-type cyclin, the significance of which is discussed below. Chlamydomonas encodes one highly divergent cyclin that does not appear to be related to any previously characterized group and has been designated CYCM1 (Fig. 6).
Figure 6.
Neighbor-joining tree of divergent cyclin groups (C, H, L, T, J, F, U, SDS). Bootstrap values of 50% or higher are shown for each clade. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Hs, Homo sapiens; Os, Oryza sativa. See Supplemental Table II for GenBank accession numbers.
The expression profiles of both CYCA1 and CYCB1 resemble that of CDKB1 with elevated levels of mRNA appearing at Commitment and during S/M phase (Fig. 7). Whereas the expression during S/M phase is expected based on the predicted function of these genes, the early G1 increase and then suppression of expression after Commitment was not anticipated. This G1 expression pattern may be related to the cell cycle regulatory mechanisms used by Chlamydomonas (see “Discussion”). We analyzed expression of CYCD2 and CYCD3 but were unable to amplify CYCD1. CYCD2 and CYCD3 are expressed constitutively during the entire cell cycle (Fig. 7), as has been observed for some plant and animal D-cyclins (Fuerst et al., 1996; Sorrell et al., 1999; Riou-Khamlichi et al., 2000; Menges and Murray, 2002; Sa et al., 2002; Sherr, 2002). CYCAB1 expression was difficult to detect, suggesting that the mRNA may be very low in abundance. CYCAB1 cDNA could only be amplified from S/M phase samples (Fig. 7).
Figure 7.
Expression of Chlamydomonas cyclins. RT-PCR products were detected by Southern blotting (CYCD2, A1, B1, AB1) or ethidium bromide staining (CYCD3). Fraction of cells that had passed Commitment and completed cell division when the sample was prepared is indicated at the bottom. α-Tubulin transcript (TUA1) detected by ethidium bromide staining serves as an internal control.
We have not looked at expression of any of divergent Chlamydomonas cyclins, but EST evidence indicates that CYCL1, CYCM1, and CYCU1 are expressed (Supplemental Table I).
CDK/Cyclin-Binding and Regulatory Proteins
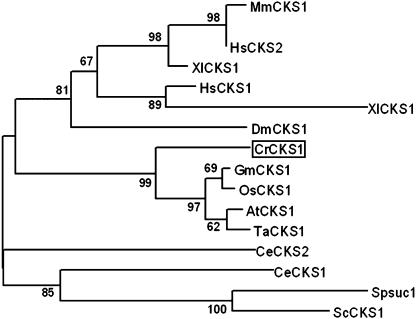
CKS (cyclin-dependent kinase subunit) proteins are orthologs of the fission yeast protein suc1p13 that mediate interactions of CDK with their substrates and with other regulatory proteins (Endicott and Nurse, 1995). Budding yeast CKS was recently shown to be involved in promoting mitosis by activating the APC ubiquitin ligase (Morris et al., 2003). CKS proteins are also widely used as a means to purify CDKs. Chlamydomonas has a single CKS ortholog, encoded by the CKS1 gene (Fig. 8), whose expression is constitutive during the cell cycle (Fig. 13). The presence of this protein was previously inferred from the existence of a Chlamydomonas protein that cross-reacted with antibodies specific for fission yeast suc1p13 (John et al., 1991). We used recombinant Chlamydomonas CKS1 to purify complexes of CDKs from crude protein extracts. The CKS1-purified CDK activity peaked during S/M phase and displayed sensitivity to the CKI roscovitine (Fig. 2C).
Figure 8.
Neighbor-joining tree of CKS1 homologs. Bootstrap values of 50% or higher are shown for each clade. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Gm, Glycine max; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Sc, budding yeast; Sp, fission yeast; Ta, Triticum aestivum; Xl, Xenopus laevis. See Supplemental Table II for GenBank accession numbers.
Figure 13.
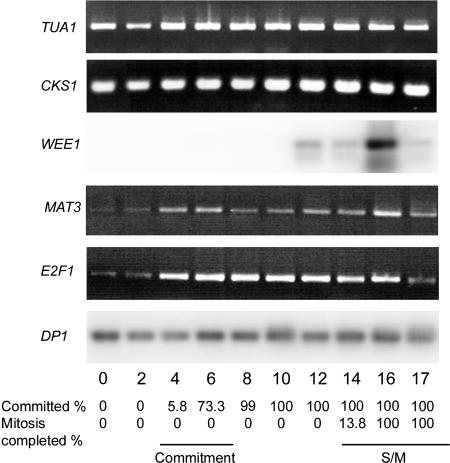
Expression of Chlamydomonas CKS1, WEE1, MAT3, E2F1, and DP1. RT-PCR products were detected by Southern blotting (WEE1, DP1) or ethidium bromide staining (CKS1, MAT3, E2F1). Fraction of cells that had passed Commitment and completed cell division when the sample was prepared is indicated at the bottom. α-Tubulin transcript (TUA1) detected by ethidium bromide staining serves as an internal control.
CDK/cyclin complexes are negatively regulated by wee1 kinases that phosphorylate a conserved Tyr residue of the CDK subunit. This negative regulation is necessary for the proper timing of mitosis (Gould and Nurse, 1989; Jin et al., 1996). Chlamydomonas has a single WEE1 ortholog encoded by the WEE1 gene (Fig. 9), whose expression is up-regulated during S/M phase but not detectable at earlier stages (Fig. 13).
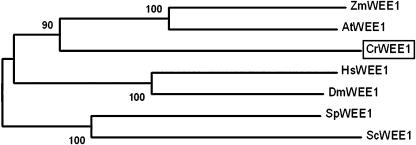
Figure 9.
Neighbor-joining tree of wee1 kinases. Bootstrap values of 50% or higher are shown for each clade. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Sc, budding yeast; Sp, fission yeast; Zm, Zea mays. See Supplemental Table II for GenBank accession numbers.
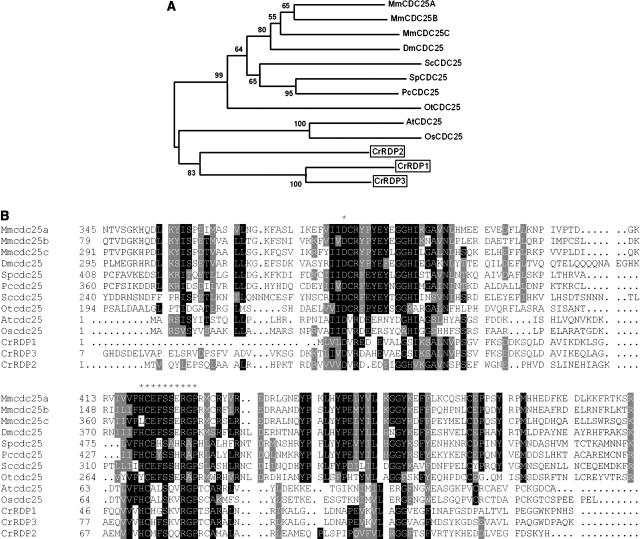
The CDC25 genes encode dual-specificity phosphatases that activate CDKs by opposing the activity of wee1 family kinases (Russell and Nurse, 1986; Kumagai and Dunphy, 1991). Ostreococcus tauri was found to encode a functional CDC25 that shows a close match to the CDC25 consensus indicating that the gene is present in the green algal/land plant lineage (Khadaroo et al., 2004), but no obvious CDC25-encoding genes are present in the Chlamydomonas or Arabidopsis genome sequences. Recently, however, a small CDC25-like dual-specificity phosphatase was found in Arabidopsis. The Arabidopsis CDC25-like protein contains a functional phosphatase domain with a rhodanese fold, but not an N-terminal regulatory domain that is typical for fungal and metazoan CDC25s (Landrieu et al., 2004). Three predicted proteins in the Chlamydomonas genome sequence, designated RDP1, RDP2, and RDP3 (rhodanese domain phosphatase), show homology to the conserved rhodanese domains from the CDC25 family and contain the core catalytic motifs (HCXXSXXRGP and aspartic acid residue) necessary for phosphatase activity (Denu and Dixon, 1995; Fauman and Saper, 1996; Fig. 10B). A closer analysis of RDP1 and RDP2 using the KOG database (Koonin et al., 2004) revealed that these two Chlamydomonas proteins are homologous to small phosphatases in fission yeast, Arabidopsis, and other plants (data not shown). A phylogenetic analysis indicates that RDP1 to RDP3 are closely related to CDC25 proteins and form a clade with them (Fig. 10A). We were able to amplify cDNAs for both RDP1 and RDP2 and found that transcript levels were constitutive during the cell cycle (data not shown). EST evidence indicates that RDP3 is also expressed (Supplemental Table I).
Figure 10.
A, Neighbor-joining tree of cdc25 phosphatase. Bootstrap values of 50% or higher are shown for each clade. B, ClustalW alignment of phosphatase domains of Chlamydomonas RDP1, 2, 3 with CDC25 from different organisms. Conserved residues are shaded; asterisks indicate position of catalytic site (HCXXSSSRGP) and conserved Asp residue. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster; Mm, Mus musculus; Os, Oryza sativa; Ot, Ostreococcus tauri, Pc, Pneumocystis carinii; Sc, budding yeast; Sp, fission yeast. See Supplemental Table II for GenBank accession numbers.
We searched for homologs of the conserved CKI family represented by Cip/Kip proteins in mammals and ICK/KRP proteins in plants (Sherr and Roberts, 1995, 1999; Wang et al., 1997; Stals et al., 2000; De Veylder et al., 2001), but we have yet to identify any likely candidates in Chlamydomonas.
RBR, E2F/DP, and S-Phase Genes
RBRs and the E2F/DP family of transcription factors are a central part of the pathway regulating DNA replication in animals and likely play a similar role in plants (Weinberg, 1995; Gutierrez, 1998). E2F and DP form a heterodimeric transcription factor that binds to promoters of many S-phase-regulated genes. These target genes include those necessary for initiation of and progression through S phase. Hypophosphorylated RB binds to E2F/DP complexes at promoters and inhibits transcription. At the G1/S-phase transition, RB is hyperphosphorylated by CDKs and released from promoter complexes, thereby allowing the activation of E2F/DP target genes. Animals and plants encode one or more RBRs and multiple E2F/DP family members (Gutierrez, 1998, 2002; Harbour and Dean, 2000; Classon and Harlow, 2002; De Veylder et al., 2002, 2003; Dewitte and Murray, 2003). E2Fs can serve as transcriptional activators or repressors depending on the presence or absence of an activation domain (Muller and Helin, 2000; Bracken et al., 2004). Another class of E2F/DP-related proteins is represented by DEL (DP-E2F-like) in plants (Kosugi and Ohashi, 2002; Vandepoele et al., 2002) and E2F7 in animals (de Bruin et al., 2003; Di Stefano et al., 2003; Logan et al., 2004). DEL/E2F7 proteins contain a tandem DNA-binding domain and no RBR interaction or transcriptional activation domains. DEL/E2F7 proteins are thus thought to function as transcriptional repressors (Di Stefano et al., 2003; Ramirez-Parra et al., 2004).
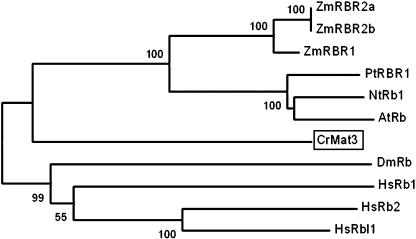
Chlamydomonas has a single RBR encoded by the MAT3 gene (Fig. 11). Loss of MAT3 leads to deregulated cell size and cell cycle control (Umen and Goodenough, 2001). The MAT3 message is expressed constitutively during the cell cycle (Fig. 13), and the protein level is also constant (K. Bisova, unpublished data).
Figure 11.
Neighbor-joining tree of RB and RBR proteins. Bootstrap values of 50% or higher are shown for each clade. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Nt, Nicotiana tabacum; Pt, Populus tremula × Populus tremuloides; Zm, Zea mays. See Supplemental Table II for GenBank accession numbers.
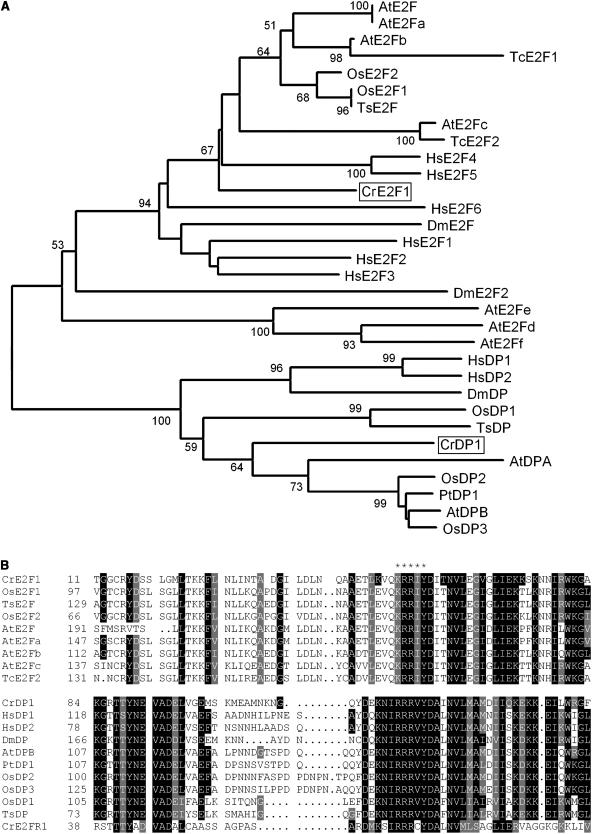
In searching the Chlamydomonas genome, we found three loci that can encode proteins with DNA-binding domains related to the E2F/DP family. One of these, designated E2F1, encodes an E2F ortholog, and a second one, designated DP1, encodes a DP ortholog (Fig. 12A; Supplemental Fig. 1). The third locus, designated E2FR1, potentially encodes a protein with partial similarity to E2F/DP-like DNA-binding domains, but with no other domains that are found in the E2F and DP families (Fig. 12B). Interestingly, neither E2F1 nor DP1 displays recognizable sequence homology to the conserved C-terminal RBR-binding and transcriptional activation domains present in plant and animal E2Fs. Nonetheless, genetic evidence places both E2F1 and DP1 downstream of MAT3 as activators of cell cycle progression (S.-C. Fang and J.G. Umen, unpublished data). mRNAs for both E2F1 and DP1 are present constitutively at low abundance and do not appear to be cell cycle regulated (Fig. 13). The mRNA of E2FR1 was very low abundance and also present constitutively during the cell cycle (data not shown). We have found no DEL/E2F7 homologs in Chlamydomonas.
Figure 12.
A, Neighbor-joining tree of E2F and DP families. Bootstrap values of 50% or higher are shown for each clade. B, ClustalW alignment of DNA-binding domains of Chlamydomonas E2F/DP with E2F/DP from different organisms. Conserved residues are shaded; asterisks indicate position of DNA-binding motif. At, Arabidopsis; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster; Hs, Homo sapiens; Os, Oryza sativa; Pt, Populus tremula × Populus tremuloides; Tc, Thlaspi caerulescens; Ts, Triticum sp. See Supplemental Table II for GenBank accession numbers.
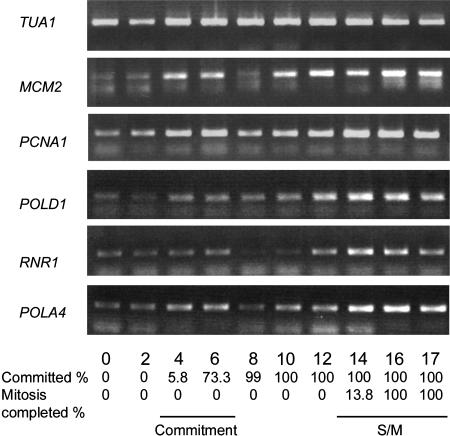
We examined the expression of several Chlamydomonas genes whose plant or animal orthologs are under the control of the RB-E2F pathway and whose expression is up-regulated during S phase (Ramirez-Parra et al., 2003; Bracken et al., 2004). These include ribonucleotide reductase large subunit (RNR1), DNA polymerase delta subunit (POLD1), DNA primase (POLA4), proliferating cell nuclear antigen (PCNA1), and minichromosome maintenance protein 2 (MCM2). Most of the genes showed an increase in message levels at the time of Commitment. Up-regulation was seen again during the S/M phase, although none of these genes was as strongly regulated as CYCA1, CYCB1, and CDKB1 (Fig. 14).
Figure 14.
Expression of S-phase genes MCM2, PCNA1, POLD1, RNR1, and POLA4. RT-PCR products were detected by ethidium bromide staining. Fraction of cells that had passed Commitment and completed cell division when the sample was prepared is indicated at the bottom. α-Tubulin transcript (TUA1) detected by ethidium bromide staining serves as an internal control.
DISCUSSION
This work represents a comprehensive description and expression profiling of cell cycle regulators in the green algae. The only comparable expression and annotation information available for cell cycle regulators from the plant kingdom was from Arabidopsis (Richard et al., 2001; Menges et al., 2002, 2003; Menges and Murray, 2002; Vandepoele et al., 2002; Hennig et al., 2003). A parallel annotation project on the green microalga O. tauri has also been carried out independently (H. Moreau, personal communication). The genome of a more primitive acidophilic red alga, Cyanidioschyzon merolae, has recently been sequenced (Matsuzaki et al., 2004), but to our knowledge the C. merolae cell cycle genes have not yet been annotated. Below we compare the cell cycle regulators from Chlamydomonas with those in higher plants and metazoans and discuss the implications of our findings for understanding eukaryotic cell cycles.
A Single-Celled Plant?
Although there are some differences between Chlamydomonas and higher plants, the most striking features are the similarities. These include (1) similarity in both sequence and expression pattern of the major cell cycle CDKs, CDKA and CDKB, the former being constitutively expressed and the latter up-regulated during G2/M; (2) the presence of and similarity between the other CDK family members (C–E); (3) the presence of all three major classes of cyclin genes (A, B, and D) that are also found in metazoans but are much more diverged in the fungi; and (4) the retention of the RB-E2F pathway, also present in metazoans but lost from the fungi.
There are several classes of regulators that we expected to find but that were either missing or ambiguous. These include cyclin H, CDC25, and CKI proteins. H-type cyclins bind CDKD to form a CAK complex (Fisher and Morgan, 1994; Makela et al., 1994; Yamaguchi et al., 2000). The presence of a CDKD ortholog in Chlamydomonas and the conservation of a CAK phosphorylation site in CDKA suggest that there is a cyclin H-like activity that interacts with CDKD1. The gene encoding this cyclin H-like activity could be missing from this genome sequence, diverged beyond recognition, or one of the other cyclin genes. One possibility for a cyclin H substitute is the C-type cyclin CYCC1, which belongs to a closely related sister clade. A second possibility is that a novel cyclin, such as CYCM1, has taken over the role of cyclin H. Direct testing will ultimately be required to identify the composition of the predicted CAK activity in Chlamydomonas.
There are three genes in Chlamydomonas—RDP1, RDP2, and RDP3—that encode RDPs related to CDC25 but lack the N-terminal extension typical for metazoan CDC25s. It has been argued for higher plants that the presence of a WEE1-encoded activity means that an opposing CDC25-like activity must exist (Vandepoele et al., 2002), and the same can be argued for Chlamydomonas. The CDC25-like protein described in Arabidopsis is orthologous to the RDPs and has been shown to have phosphatase activity that can activate CDKs purified from plant extracts (Landrieu et al., 2004). The RDPs, therefore, are the best candidates for CDC25 in Chlamydomonas. A more definitive conclusion about CDC25 function in Chlamydomonas and the role of the RDP proteins can be drawn after biochemical and genetic data become available.
The only conserved activity that appears to be missing from Chlamydomonas is the CKIs. This is not surprising given the low similarity between different members of this family (Wang et al., 1997). Since CKIs are a widespread group, they are probably also present in Chlamydomonas, but their identification will require either more sensitive sequence search methods or direct biochemical assays.
There is one major difference between Chlamydomonas and higher plants that is especially noteworthy. Whereas many of the key cell cycle regulators have undergone multiple duplications in higher plants (Mironov et al., 1999; Joubes et al., 2000; Vandepoele et al., 2002), most of the Chlamydomonas cell cycle regulatory genes are single copy. The genome of C. reinhardtii (approximately 100 Mb, approximately 20,000 predicted genes) is comparable in size to that of Arabidopsis (approximately 120 Mb, approximately 26,000–29,000 predicted genes) and is about 10-fold larger than the genome of O. tauri (approximately 10–12 Mb). Therefore, differences in genome size or gene number cannot explain the differing degrees of cell cycle gene duplication in green algae versus land plants. It is tempting to speculate that the cell cycle gene duplications in higher plants are coupled to increased developmental complexity, but it remains to be determined whether the correlation of copy number with developmental complexity holds for plants other than angiosperms. In any case, its lack of apparent genetic redundancy and simple unicellular life cycle are advantageous properties that position Chlamydomonas as a good model for the eukaryotic cell cycle.
The most notable exception to the single-copy rule is the D-cyclin family that has three members in Chlamydomonas. The three D-type cyclins in Chlamydomonas are not closely related to any of the seven Arabidopsis D-cyclin subfamilies, suggesting that CYCD1, 2, and 3 are the result of independent duplication in Chlamydomonas (Fig. 5). It is not clear whether this duplication of the D-cyclin family is unique to Chlamydomonas since there is only a single cyclin D in O. tauri (H. Moreau, personal communication). It has been suggested that the proliferation of D-cyclins in plants is related to an elaboration of G1 control mechanisms (Meijer and Murray, 2000), and the same may be true in Chlamydomonas. It remains to be determined whether the Chlamydomonas D-cyclins have separate, overlapping, or redundant roles in cell cycle regulation.
Chlamydomonas-Specific Genes
It is interesting that, besides the higher plant repertoire of CDKs and cyclins, Chlamydomonas encodes four novel CDKs (G1, G2, H1, and I1), two novel cyclins (CYCAB1 and CYCM1), and a novel E2F-related protein (E2FR1). These novel classes are missing from O. tauri (H. Moreau, personal communication), meaning either that they were present early in the green algal/land plant ancestor and lost from some lineages, or that they evolved later in a subgroup of algae that includes Chlamydomonas. The fact that O. tauri is missing these genes may suggest that they carry out a specialized function in Chlamydomonas, but it is equally possible that Ostreococcus, which has a very small genome, might be the more derived species due to gene loss. More information on the cell division mechanism used by Ostreococcus and a broader sampling of genomes from green eukaryotes might shed light on these differences.
Although CDKH1 belongs to the plant/animal CDKC/CDK9 family, it appears to be an outlier in the CDKC/CDK9 clade. The fact that Chlamydomonas encodes another protein that appears to be a bona fide CDKC ortholog suggests that CDKH1 may have a novel, specialized function as compared to the other CDKC/CDK9 proteins.
CYCAB1 is intriguing. Even though it lacks the canonical signature motifs found in either A- or B-type cyclins (Table I), it is clearly related to the A- and B-type cyclin families that play a conserved role in eukaryotic cell cycles (Fig. 5). Importantly, CYCAB1 has a putative destruction box, suggesting that its proteolysis may be linked to cell cycle progression. Moreover, the cell cycle-regulated message abundance of CYCAB1 suggests a role for CYCAB1 in regulating S phase and/or mitosis.
E2FR1 is clearly related to the E2F/DP family of transcription factors, but has diverged considerably and does not cluster with any known E2F or DP family members in phylogenetic trees (data not shown). Most other E2F-containing organisms have at least one repressor E2F-related gene, so an interesting possibility is that E2FR1 acts as a novel type of repressor E2F in Chlamydomonas. However, it remains to be determined whether E2FR1 interacts with the RB-E2F pathway in Chlamydomonas.
Implications for the Chlamydomonas Cell Cycle
The finding of potential Chlamydomonas-specific cell cycle regulators raises the question of whether these novel proteins contribute to aspects of cell division that are unique to Chlamydomonas. Many chlorophyte algae utilize a multiple fission cell cycle similar to that of C. reinhardtii. Although multiple fission appears unusual, it incorporates cell cycle modifications that are quite common in animals and plants. Egg cells grow in the absence of cell division to achieve their characteristic large size, and the early embryos of some species undergo a series of rapid S/M cycles in the absence of growth (Saucedo and Edgar, 2002). Therefore, it is possible that Chlamydomonas and other algae evolved multiple fission by altering the regulation of core conserved cell cycle genes and not by utilization of novel genes. In any case, it will require more direct tests of gene function to resolve this question.
There are at least two major modifications required to generate a Chlamydomonas-like multiple fission cell cycle. The first modification involves suppression of S-phase entry after passage through Commitment. This suppression generates an extended G1 period during which cells can grow to many times their original size before initiating the S/M part of the cell cycle. The second modification allows for mother cells to undergo rapid successive rounds of S phase and mitosis with either greatly reduced or absent G1 and G2 intervals. Exit from these rapid S/M cycles is connected to cell size through the RB-E2F pathway (Umen and Goodenough, 2001), but may also involve other types of regulation. The Chlamydomonas-specific CYCAB1 gene is expressed during S/M phase and is conceivably involved in regulating entry into or exit from S phase or mitosis.
It is intriguing that expression of a subset of Chlamydomonas genes (e.g. CYCA1, CYCB1, CDKB1, RNR1, POLA4), most of which are likely to be RB-E2F pathway targets, is up-regulated prior to Commitment and down-regulated just after passage through Commitment. These same genes are up-regulated a second time during S/M phase, which is when they are likely to function. Although we cannot rule out a function for these genes during Commitment, to our knowledge there is no evidence for DNA synthesis or increased DNA repair in G1 cells as they pass Commitment. Instead, we favor an alternative explanation for the transient up-regulation of these genes at Commitment. In a conventional cell cycle, passage through the equivalent of Commitment entails the immediate preparation for S phase and subsequent mitosis, and therefore necessitates the expression of S/M-phase genes. It is possible that the transient increase in S/M-phase genes that we see in Chlamydomonas cells as they pass Commitment is an atavistic signature of the conventional cell cycle from which multiple fission evolved. When a G1 delay period arose in algae as a modification required for multiple fission, the expression of S/M-phase genes at Commitment became either unnecessary or detrimental, and their expression was suppressed. However, the suppression was incomplete, so we still observe a temporary derepression of S/M-phase genes during Commitment.
Passage through Commitment in Chlamydomonas is under the control of the RB-E2F pathway (Umen and Goodenough, 2001), and loss of MAT3 (RBR) presumably leads to deregulated E2F activity. However, a normal post-Commitment G1 interval was observed in mat3 deletion strains, indicating that the post-Commitment S/M-phase transcriptional suppression mechanism still operates in these cells. These genetic data, taken together with the expression profiles of prospective RB-E2F transcriptional targets, suggest the existence of a G1 transcriptional control mechanism that is activated after Commitment and that suppresses expression of some RB-E2F pathway genes independently of MAT3. The mechanism by which this RB-independent suppression is achieved will be an interesting area for future investigation.
CONCLUSIONS AND PERSPECTIVES
A simplistic analogy based on the divergence between yeasts and animals would have predicted a large divergence between single-celled algae and higher plants. Contrary to this prediction, the majority of cell cycle regulatory genes in higher plants have orthologs in Chlamydomonas, and the overall conservation between Chlamydomonas and metazoan cell cycle regulators is substantial. The sequence similarities that we have observed suggest that, outside the fungal kingdom, there are greater underlying similarities between the cell cycle regulators of unicellular and multicellular eukaryotes than has previously been appreciated. Importantly, cell cycle regulatory genes in Chlamydomonas have not undergone the extensive duplications seen in higher plants and vertebrates. Its relatively small number of cell cycle genes, combined with its extensive molecular genetic toolkit (for review, see Grossman et al., 2003), make Chlamydomonas an attractive model for eukaryotic cell cycles.
MATERIALS AND METHODS
Tree Construction and Phylogenetic Analysis
Protein sequences were aligned by ClustalW using default parameters (Thompson et al., 1994) and then optimized manually. The resulting alignments were used to construct phylogenetic trees in Mega 2.1 (Kumar et al., 2001). Unrooted trees were produced by the neighbor-joining method using p-distance for distance matrix and 1,000 replicas for bootstrapping. Bootstrap values are presented as a percentage; values lower than 50% are not shown on the trees.
Culture Conditions and Cell Cycle Analysis
Chlamydomonas reinhardtii cultures of wild-type strain 21gr (CC-1690; Chlamydomonas Genetics Center, Duke University, Durham, NC) were synchronized by growth in Erlenmeyer flasks in inorganic high-salt medium (HSM) aerated with 0.5% CO2 in air, alternating 14/10-h light/dark cycles, at 250 μmol m−2 s−1, 24°C. Cell density was maintained between 105 cells/mL and 106 cells/mL by dilution into fresh media at the beginning of each light phase. Passage through Commitment was evaluated at hourly intervals by spreading a 1-mL aliquot of culture on an HSM plate, decanting excess liquid, and then incubating the plate in the dark to prevent further growth. After 12- to 16-h dark incubation, the fraction of committed cells (those that produced 2, 4, 8, or more daughters) was determined by microscopic examination. Cell volumes were measured with a Coulter counter (MULTISIZER 3; Beckman Coulter, Miami) in samples fixed with glutaraldehyde (0.2% final concentration). The percentage of cells that completed cell division was determined in fixed samples that were observed microscopically.
RNA and DNA Preparation and Southern Analysis
Cell pellets containing 2 × 107 cells were harvested at each time point, fast frozen in liquid nitrogen, and stored at −70°C. RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions for one set of samples or according to a previously described protocol for a second set of samples (Verwoerd et al., 1989).
For isolation of genomic DNA, 1 × 107 cells were harvested and resuspended in 10 mm Tris, pH 8.0, 10 mm EDTA, 150 mm NaCl, lysed by adding SDS and sarcosyl to final concentrations of 2% (w/v) and pronase to final concentration 2.5 mg/mL, and incubated 40 min at 37°C. DNA was extracted with phenol/chloroform and precipitated with 2 volumes of ethanol 2 h at −20°C. The DNA pellet was washed with 70% ethanol and resuspended in 10 mm Tris, pH 7.4, 1 mm EDTA. Approximately 3 μg of DNA was restriction digested overnight in the recommended buffer supplemented with 2 mm spermidine and 0.1 mg/mL RNase A. Southern blotting and hybridization were carried out as described previously (Church and Gilbert, 1984) using Hybond-XL membranes (Amersham Pharmacia Biotech, Uppsala) and a UV cross-linker (UV STRATALINKER 1800; Stratagene, La Jolla, CA) to attach DNA to membranes. The stringency was reduced by lowering the wash temperature from 65°C to room temperature.
cDNA Synthesis and RT-PCR
Five micrograms of total RNA were treated with RQ1 RNase-free DNase (Promega, Madison, WI), according to the manufacturer's instructions. DNA-free RNA was reverse transcribed using the ThermoScript RT-PCR system (Invitrogen), according to the manufacturer's instructions, in a total volume of 40 μL using oligo(dT) primers. One microliter of cDNA was used for each RT-PCR reaction. PCR fragments were amplified by ExTaq DNA polymerase (TAKARA, Shiga, Japan), according to the manufacturer's instructions, in a final volume 20 μL in the presence of 1 μm primers and 2% DMSO. Primers and conditions used for RT-PCR are listed in Supplemental Table III. Each PCR product was subcloned into pGEM-T easy vector (Promega). For each primer set, different cycle numbers were tested to ensure that the products were in the linear range for amplification. For the products that were detected by Southern blot (all except CDKE, CYCD3, CKS1, MAT3, E2F1, S-phase genes, and internal controls), the lowest number of cycles yielding a signal on an ethidium bromide-stained gel was determined, and five fewer cycles than this minimum were used for the Southern-blot gels. PCR products were separated in 1% agarose gels containing ethidium bromide and photographed. PCR fragments were subjected to Southern transfer under neutral conditions (Sambrook and Russell, 2001) onto Hybond XL membranes (Amersham Pharmacia Biotech), and transferred DNA was fixed by UV cross-linking (UV STRATALINKER 1800; Stratagene). Membranes were probed and washed as described (Church and Gilbert, 1984), and radioactive signals were visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Preparation of CrCKS1 Beads and Affinity Purification of CDKs
A cDNA coding for the C. reinhardtii CKS1/suc1p13 homolog, CKS1, was amplified from oligo(dT)-primed cDNA by PCR with primers (5′-GGAATTCATGGCACAGAACAACATTCA-3′, 5′-GCGCTCGAGTTACTGAACCTGCAGAGCCT-3′) and cloned into pGEM-T easy vector (Promega). The sequence was verified and the fragment was subcloned into the EcoRI-XhoI sites of pGEX-4T-1 (Amersham Pharmacia Biotech), yielding pGEX-CrCKS1. pGEX-CrCKS1 was transformed into BL21-CodonPlus(DE3)-RP cells (Stratagene). Cells were grown at 37°C until OD600 0.4, then transferred to 20°C for 30 min and induced with 0.5 mm isopropyl β-d-thiogalactoside for 2 h. The recombinant protein was purified using glutathione-Sepharose (Amersham Pharmacia Biotech), according to the manufacturer's instructions.
At intervals during synchronous growth, 2 × 107 Chlamydomonas cells were harvested, fast frozen in liquid nitrogen, and stored at −70°C. Cell pellets were mixed with 300 μL of RIPA buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 0.1% SDS, 1% NP-40) containing 1× protease inhibitor cocktail (SIGMA P9599; Sigma, St. Louis), 1 mm Na3VO4, 1 mm benzamidine, 10 mm NaF, and 150 μL zirconium beads (diameter 1.0 mm; Biospec, Bartlesville, OK) and vortexed 7 min at 4°C. Protein lysates were cleared by centrifugation 15 min at 14,000 rpm (EPPENDORF 5417 R). Twenty microliters of Chlamydomonas protein lysate were diluted 10-fold with RIPA buffer and incubated 1 h at 4°C with 20 μL of 50% CrCKS1 bead slurry. Unbound proteins were washed out by four consecutive washes with RIPA buffer and two washes with kinase buffer (20 mm HEPES, pH 7.5, 15 mm MgCl2, 5 mm EGTA, 1 mm DTT; Brizuela et al., 1987; Draetta et al., 1987).
H1 Kinase Assay
Histone H1 kinase activity was assayed by a previously described method (Langan et al., 1989) with the following modifications. The kinase activity was assayed in a final volume of 10 μL in assay buffer (20 mm HEPES, pH 7.5; 15 mm MgCl2, 5 mm EGTA, 1 mm DTT, 0.1% histone [SIGMA H4524], 0.1 mm ATP, and 0.185 MBq of [γ-32P] ATP). Roscovitine was added where indicated to a final concentration 200 μm just prior to addition of radioactive ATP. The reaction was incubated at room temperature for 30 min and terminated by adding 5 μL of 5× SDS sample buffer (250 mm Tris-HCl, pH 6.8, 50% [w/v] glycerol, 10% SDS, 100 mm dithiothreitol, 0.5% [w/v] bromphenol blue), boiled for 2 min, and immediately cooled. Proteins were separated by SDS-PAGE in 12% gels (Laemmli, 1970). Phosphorylated histone bands were detected with a PhosphorImager (Molecular Dynamics).
Note Added in Proof
The data referred to here as H. Moreau, personal communication, are due to be published in Molecular Biology and Evolution as S. Robbens, B. Khadaroo, A. Camasses, E. Derelle, C. Ferraz, D. Inzé, Y. Van de Peer, and H. Moreau, Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri.
Acknowledgments
We thank Su-Chiung Fang, Tony Hunter, Gerard Manning, and Eugene Koonin for their helpful advice and comments on this manuscript. We thank Hiep Le and Diana Goyena for their excellent technical assistance.
This work was supported by generous contributions to the laboratory of J.G.U. from the H.N. and Frances C. Berger Foundation, the Joe W. and Dorothy Dorsett Brown Foundation, the Fritz B. Burns Foundation, The Arthur Vining Davis Foundations, The Lebensfeld Foundation, the John Stacy Lyons Memorial Foundation, The Gertrude E. Skelly Charitable Foundation, and the Irving A. Hansen Memorial Foundation.
The online version of this article contains Web-only data.
References
- Abrahams S, Cavet G, Oakenfull EA, Carmichael JP, Shah ZH, Soni R, Murray JAH (2001) A novel and highly divergent Arabidopsis cyclin isolated by complementation in budding yeast. Biochim Biophys Acta 1539: 1–6 [DOI] [PubMed] [Google Scholar]
- Albani D, Mariconti L, Ricagno S, Pitto L, Moroni C, Helin K, Cella R (2000) DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J Biol Chem 275: 19258–19267 [DOI] [PubMed] [Google Scholar]
- Azumi Y, Liu D, Zhao D, Li W, Wang G, Hu Y, Ma H (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J 21: 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroco RM, De Veylder L, Magyar Z, Engler G, Inze D, Mironov V (2003) Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell Mol Life Sci 60: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G (1994) CDK6(PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9: 71–79 [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, de Almeida Engler J, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L (2004) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K (2004) E2F target genes: unraveling the biology. Trends Biochem Sci 29: 409–417 [DOI] [PubMed] [Google Scholar]
- Brizuela L, Draetta G, Beach D (1987) p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. EMBO J 6: 3507–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Harlow E (2002) The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2: 910–917 [DOI] [PubMed] [Google Scholar]
- Coffman JA (2004) Cell cycle development. Dev Cell 6: 321–327 [DOI] [PubMed] [Google Scholar]
- Coleman AW (1982) The nuclear cell cycle in Chlamydomonas (Chlorophyceae). J Phycol 18: 192–195 [Google Scholar]
- De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH (1993) Crystal structure of cyclin-dependent kinase 2. Nature 363: 595–602 [DOI] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G (2003) Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 278: 42041–42049 [DOI] [PubMed] [Google Scholar]
- De Luca A, Esposito V, Baldi A, Claudio PP, Fu Y, Caputi M, Pisano MM, Baldi F, Giordano A (1997) CDC2-related kinase PITALRE phosphorylates pRb exclusively on serine and is widely expressed in human tissues. J Cell Physiol 172: 265–273 [DOI] [PubMed] [Google Scholar]
- Denu JM, Dixon JE (1995) A catalytic mechanism for the dual-specific phosphatases. Proc Natl Acad Sci USA 92: 5910–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van der Schueren E, Jacqumard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras P, Landrieu I, Van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Joubes J, Inze D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6: 536–543 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K (2003) E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22: 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnan L, Carvill EP, Gilliland TJ, John PCL (1985) The cell cycles of Chlamydomonas and Chlorella. New Phytol 99: 1–40 [Google Scholar]
- Donnan L, John PCL (1983) Cell cycle control by timer and sizer in Chlamydomonas. Nature 304: 630–633 [DOI] [PubMed] [Google Scholar]
- Donnan L, John PCL (1984) Timer and sizer controls in the cell cycles of Chlamydomonas and Chlorella. In P Nurse, E Streiblova, eds, The Microbial Cell Cycle. CRC Press, Boca Raton, FL, pp 231–251
- Draetta G, Brizuela L, Potashkin J, Beach D (1987) Identification of p34 and p13, human homologs of the cell cycle regulators of fission yeast encoded by cdc2+ and suc1+. Cell 50: 319–325 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL (2001) Endoreplication cell cycles: more for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Endicott JA, Nurse P (1995) The cell cycle and suc1: from structure to function? Structure 3: 321–325 [DOI] [PubMed] [Google Scholar]
- Fauman EB, Saper MA (1996) Structure and function of the protein tyrosine phosphatases. Trends Biochem Sci 21: 413–417 [DOI] [PubMed] [Google Scholar]
- Ferreira P, Hemerly AS, Villarroel R, Van Montagu M, Inze D (1991) The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Morgan DO (1994) A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78: 713–724 [DOI] [PubMed] [Google Scholar]
- Fuerst RA, Soni R, Murray JA, Lindsey K (1996) Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol 112: 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Peng J, Parreno M, Price DH, Henderson EE, Grana X (1998) Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 17: 3093–3102 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Goodenough UW (1992) Green yeast. Cell 70: 533–538 [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P (1989) Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342: 39–45 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, et al (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryotic Cell 2: 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D (2002) Cytokinesis in eukaryotes. Microbiol Mol Biol Rev 66: 155–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C (1998) The retinoblastoma pathway in plant cell cycle and development. Curr Opin Plant Biol 1: 492–497 [DOI] [PubMed] [Google Scholar]
- Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC (2002) G1 to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5: 480–486 [DOI] [PubMed] [Google Scholar]
- Gutman BL, Niyogi KK (2004) Chlamydomonas and Arabidopsis. A dynamic duo Plant Physiol 135: 607–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14: 2393–2409 [DOI] [PubMed] [Google Scholar]
- Healy JMS, Menges M, Doonan JH, Murray JAH (2001) The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J Biol Chem 276: 7041–7047 [DOI] [PubMed] [Google Scholar]
- Hedges SB (2002) The origin and evolution of model organisms. Nat Rev Genet 3: 838–849 [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inze D (1993) cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5: 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Menges M, Murray JA, Gruissem W (2003) Arabidopsis transcript profiling on Affymetrix GeneChip arrays. Plant Mol Biol 53: 457–465 [DOI] [PubMed] [Google Scholar]
- Hirt H, Pay A, Gyorgyey J, Bako L, Nemeth K, Borge L, Schweyen R, Heberle-Bors E, Dudits D (1991) Complementation of a yeast cell cycle mutant by an alfalfa cDNA encoding a protein kinase homologous to p34cdc2. Proc Natl Acad Sci USA 88: 1636–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, et al (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37: 155–169 [DOI] [PubMed] [Google Scholar]
- Jin P, Gu Y, Morgan DO (1996) Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol 134: 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John PCL (1984) Control of the cell division cycle in Chlamydomonas. Microbiol Sci 1: 96–101 [PubMed] [Google Scholar]
- John PCL (1987) Control points in the Chlamydomonas cell cycle. In W Wiesnar, DG Robinson, RC Starr, eds, Algal Development. Molecular and Cellular Aspects. Springer-Verlag, Berlin, pp 9–16
- John PCL, Sek FJ, Hayles J (1991) Association of the plant p34cdc2 like protein with p13suc1—implications for control of cell division cycles in plants. Protoplasma 161: 70–74 [Google Scholar]
- John PC, Sek FJ, Lee MG (1989) A homolog of the cell cycle control protein p34cdc2 participates in the division cycle of Chlamydomonas, and a similar protein is detectable in higher plants and remote taxa. Plant Cell 1: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Walker CL (1999) Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol 39: 295–312 [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C (2000) Endoreduplication in higher plants. Plant Mol Biol 43: 735–745 [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudin J-P (2000) CDK-related protein kinases in plants. Plant Mol Biol 43: 607–620 [DOI] [PubMed] [Google Scholar]
- Khadaroo B, Robbens S, Ferraz C, Derelle E, Eychenie S, Cooke R, Peaucellier G, Delseny M, Demaille J, Van de Peer Y, et al (2004) The first green lineage cdc25 dual-specificity phosphatase. Cell Cycle 3: 513–518 [PubMed] [Google Scholar]
- Koonin E, Fedorova N, Jackson J, Jacobs A, Krylov D, Makarova K, Mazumder R, Mekhedov S, Nikolskaya A, Rao B, et al (2004) A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J Biol Chem 277: 16553–16558 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (1991) The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell 64: 903–914 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Landrieu I, da Costa M, De Veylder L, Dewitte F, Vandepoele K, Hassan S, Wieruszeski J-M, Faure J-D, Van Montagu M, Inze D, et al (2004) A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan TA, Gautier J, Lohka M, Hollingsworth R, Moreno S, Nurse P, Maller J, Sclafani RA (1989) Mammalian growth-associated H1 histone kinase: a homologue of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol Cell Biol 9: 3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimiya H, Umeda M (2003) Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J 34: 417–425 [DOI] [PubMed] [Google Scholar]
- Lee MH, Yang HY (2003) Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev 22: 435–449 [DOI] [PubMed] [Google Scholar]
- Logan N, Delavaine L, Graham A, Reilly C, Wilson J, Brummelkamp TR, Hijmans EM, Bernards R, La Thangue NB (2004) E2F-7: a distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene 23: 5138–5150 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Meszaros T, Miskolczi P, Deak M, Feher A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, et al (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela TP, Tassan JP, Nigg EA, Frutiger S, Hughes GJ, Weinberg RA (1994) A cyclin associated with the CDK-activating kinase MO15. Nature 371: 254–257 [DOI] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana—novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Martinez MC, Jorgensen JE, Lawton MA, Lamb CJ, Doerner PW (1992) Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc Natl Acad Sci USA 89: 7360–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima S-y, Mori T, Nishida K, Yagisawa F, Nishida K, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Meijer M, Murray JAH (2000) The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol Biol 43: 621–633 [DOI] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, Nelson C, Harlow E, Tsai LH (1992) A family of human cdc2-related protein kinases. EMBO J 11: 2909–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Harlow E (1994) Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol 14: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inze D (1999) Cyclin-dependent kinases and cell division in plants—the nexus. Plant Cell 11: 509–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (1995) Principles of CDK regulation. Nature 374: 131–134 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI (2003) Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Moser BA, Russell P (2000) Cell cycle regulation in Schizosaccharomyces pombe. Curr Opin Microbiol 3: 631–636 [DOI] [PubMed] [Google Scholar]
- Muller H, Helin K (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta 1470: M1–M12 [DOI] [PubMed] [Google Scholar]
- Murray AW (2004) Recycling the cell cycle. Cyclins revisited Cell 116: 221–234 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (1996) Viewpoint: putting the cell cycle in order. Science 274: 1643–1645 [DOI] [PubMed] [Google Scholar]
- Oakenfull EA, Riou-Khamlichi C, Murray JAH (2002) Plant D-type cyclins and the control of G1 progression. Philos Trans R Soc Lond B Biol Sci 357: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaya AJ, Sedivy JM (2002) Regulation of cyclin-Cdk activity in mammalian cells. Cell Mol Life Sci 59: 126–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager T (2002) Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J Cell Physiol 190: 160–169 [DOI] [PubMed] [Google Scholar]
- Pavletich N (1999) Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 287: 821–828 [DOI] [PubMed] [Google Scholar]
- Pines J (1995) Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J 308: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheldt JP, Segers G, De Veylder L, Barroco RD, Casteels P, Van Montagu M, Inze D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G(2)/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Frundt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Xie Q, Boniotti M, Gutierrez C (1999) The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res 27: 3527–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI (1997) Control of the G1/S transition. Cancer Surv 29: 7–23 [PubMed] [Google Scholar]
- Ren S, Rollins BJ (2004) Cyclin C/Cdk3 promotes Rb-dependent G0 exit. Cell 117: 239–251 [DOI] [PubMed] [Google Scholar]
- Renaudin J-P, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inze D, Jacobs T, Kouchi H, Rouze P, Sauter M, et al (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32: 1003–1018 [DOI] [PubMed] [Google Scholar]
- Richard C, Granier C, Inze D, De Veylder L (2001) Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J Exp Bot 52: 1625–1633 [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JMS, Murray JAH (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cel Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J (1995) Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet 29: 209–230 [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P (1986) Cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45: 145–153 [DOI] [PubMed] [Google Scholar]
- Sa G, Hitomi M, Harwalkar J, Stacey AW, Gc GC, Stacey DW (2002) Ras is active throughout the cell cycle, but is able to induce cyclin D1 only during G2 phase. Cell Cycle 1: 50–58 [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA (2002) Why size matters: altering cell size. Curr Opin Genet Dev 12: 565–571 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Southern blotting: capillary transfer of DNA to membranes. In J Sambrook, DW Russel, eds, Molecular Cloning: A Laboratory Manual, Ed 3, Vol 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schwab B, Folkers U, Ilgenfritz H, Hulskamp M (2000) Trichome morphogenesis in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 355: 879–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlik I, Zachleder V (1984) The multiple fission cell reproductive patterns in algae. In P Nurse, E Streiblova, eds, The Microbial Cell Cycle. CRC Press, Boca Raton, FL, pp 253–279
- Sherr C (2002) D1 in G2. Cell Cycle 1: 36–38 [PubMed] [Google Scholar]
- Sherr CJ (1996) Cancer cell cycles. Science 274: 1672–1677 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Shimotohno A, Matsubayashi S, Yamaguchi M, Uchimiya H, Umeda M (2003) Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett 534: 69–74 [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Combettes B, Chaubet-Gigot N, Gigot C, Murray JA (1999) Distinct cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco bright yellow-2 cells. Plant Physiol 119: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell DA, Menges M, Healy JMS, Deveaux Y, Amano C, Su Y, Nakagami H, Shinmyo A, Doonan JH, Sekine M, et al (2001) Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar bright yellow-2 cells. Plant Physiol 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stals H, Casteels P, van Montagu M, Inze D (2000) Regulation of cyclin-dependent kinases in Arabidopsis thaliana. Plant Mol Biol 43: 583–593 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Acosta JA, de Almeida Engler J, Raes J, Magyar Z, De Groot R, Inze D, De Veylder L (2004) Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol Life Sci 61: 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Bhalerao RP, Schell J, Uchimiya H, Koncz C (1998) A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 5021–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J, Goodenough U (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T, Dekker B, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, dePamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135: 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fowke LC, Crosby WL (1997) A plant cyclin-dependent kinase inhibitor gene. Nature 386: 451–452 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutierrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Zhang H, Beach D (1992) D-type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell 71: 505–514 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Fabian T, Sauter M, Bhalerao RP, Schrader J, Sandberg G, Umeda M, Uchimiya H (2000) Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J 24: 11–20 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Umeda M, Uchimiya H (1998) A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J 16: 613–619 [DOI] [PubMed] [Google Scholar]