Abstract
Diatoms are one of the most important constituents of phytoplankton communities in aquatic environments, but in spite of this, only recently have large-scale diatom-sequencing projects been undertaken. With the genome of the centric species Thalassiosira pseudonana available since mid-2004, accumulating sequence information for a pennate model species appears a natural subsequent aim. We have generated over 12,000 expressed sequence tags (ESTs) from the pennate diatom Phaeodactylum tricornutum, and upon assembly into a nonredundant set, 5,108 sequences were obtained. Significant similarity (E < 1E-04) to entries in the GenBank nonredundant protein database, the COG profile database, and the Pfam protein domains database were detected, respectively, in 45.0%, 21.5%, and 37.1% of the nonredundant collection of sequences. This information was employed to functionally annotate the P. tricornutum nonredundant set and to create an internet-accessible queryable diatom EST database. The nonredundant collection was then compared to the putative complete proteomes of the green alga Chlamydomonas reinhardtii, the red alga Cyanidioschyzon merolae, and the centric diatom T. pseudonana. A number of intriguing differences were identified between the pennate and the centric diatoms concerning activities of relevance for general cell metabolism, e.g. genes involved in carbon-concentrating mechanisms, cytosolic acetyl-Coenzyme A production, and fructose-1,6-bisphosphate metabolism. Finally, codon usage and utilization of C and G relative to gene expression (as measured by EST redundance) were studied, and preferences for utilization of C and CpG doublets were noted among the P. tricornutum EST coding sequences.
The unicellular brown algal class Bacillariophyceae (diatoms) is among the most successful and diversified groups of photosynthetic eukaryotes, with probably over 100,000 extant species (Round et al., 1990) widespread in all kinds of aquatic and humid environments from the poles to the equator. The contribution of diatom photosynthesis to marine primary productivity has been estimated to be between 30% and 40% (Nelson et al., 1995; Raven and Waite, 2004). As a consequence of their ecological success, diatoms sustain several large fisheries and contribute a large proportion of the oxygen we breathe.
One of the most distinctive features of diatoms is their ability to precipitate soluble silicic acid into a finely patterned silica cell wall. Diatom silicon metabolism is an important regulator of biogeochemical cycling of silicon in the oceans and is also crucial to diatom survival and life histories. Nonetheless, these particular metabolic pathways remain largely unexplored and constitute a clear potential source for discovery of novel protein functions. Indeed, the first identified components of diatom cell wall synthesis, such as silaffins and silicon transporters (Hildebrand et al., 1998; Kroger et al., 1999; Poulsen et al., 2003), have no similarity to previously known peptides and so far have only been found in diatoms.
More generally, diatoms have a peculiar genetic makeup in that, unlike the green and red lineages of photosynthetic organisms, they are likely to have emerged from a secondary endosymbiosis between a photosynthetic eukaryote, most probably red algal-like, and a heterotrophic eukaryote (Falkowski et al., 2004). Diatoms therefore carry genetic information derived from a host that contained a nucleus and some mitochondria, and from an endosymbiont that carried nuclear, mitochondrial, and plastid genomes. Eukaryotes with a secondary endosymbiotic origin also comprise other important algal groups, such as dinoflagellates and coccolithophores, as well as several human pathogens, e.g. the apicomplexan genera Plasmodium and Toxoplasma, and represent for the most part the least-explored groups among the eight major eukaryotic crown groups (Baldauf, 2003), especially concerning gene sequence and gene expression.
The complete nuclear, mitochondrial, and plastid genome sequences of the centric diatom Thalassiosira pseudonana have recently become available (Armbrust et al., 2004). In addition, sequencing of the genome of the pennate diatom Phaeodactylum tricornutum is under way and will become available in the course of 2005. The availability of complete genome sequences for centric and pennate model diatom species should boost the use of molecular biological approaches to understand the ecophysiological success of these organisms, particularly because, until recently, such studies have only been performed on a limited scale (e.g. Scala et al., 2002).
T. pseudonana has a small genome (34 Mb) and the genus Thalassiosira is of significant ecological importance, with some species being major components of the total phytoplankton biomass in their ecosystems. P. tricornutum is instead thought to be of little ecological relevance and, unlike other diatoms, it does not have an obligate requirement for silicon and can undergo morphological transitions between three possible morphotypes (Borowitzka and Volcani, 1978). However, this species has been extensively studied physiologically, and phylogenetic analyses position it firmly in the middle of the raphid clade (Medlin et al., 1996; Kooistra et al., 2003). In addition, P. tricornutum appears to have a small genome (less than 30 Mb; Veldhuis et al., 1997; Scala et al., 2002), and it is currently the only diatom that can be routinely transformed (Apt et al., 1996; Falciatore et al., 1999). For these reasons, P. tricornutum is emerging as a model species for dissecting diatom biology (Falciatore and Bowler, 2002).
Besides T. pseudonana, the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschyzon merolae are currently the only eukaryotic microalgae whose genomes are publicly available (Matsuzaki et al., 2004; http://www.biology.duke.edu/chlamy_genome). C. merolae grows in acidic water at temperatures above 45°C in hot springs and has a small, highly evolving nuclear genome of about 16 Mb (Matsuzaki et al., 2004). The order Cyanidiales is only distantly related to other rhodophytes (Yoon et al., 2002), so the relevance of C. merolae for marine red algae may therefore be limited. On the other hand, C. reinhardtii is a freshwater unicellular alga that has long been used as a model organism for photosynthesis research, although its genome has an unusually high GC content (62.1%). Notwithstanding these peculiarities, both C. merolae and C. reinhardtii are unicellular, photosynthetic, and eukaryotic, and together with T. pseudonana, they constitute a valuable starting point for comparative genome analyses to investigate differences, commonalities, and phylogenetic relationships between single-celled photoautotrophs whose ability to photosynthesize was derived from lateral transfer (endocytobiosis) events.
We report here the generation and analysis of 12,136 expressed sequence tags (ESTs) from the pennate diatom P. tricornutum and their subsequent comparison with the whole-genome predicted proteomes of T. pseudonana, C. reinhardtii, and C. merolae. This analysis greatly extends an initial study of 997 P. tricornutum ESTs (Scala et al., 2002) and constitutes an attempt to perform comparative genomics of eukaryotic algae using large numbers of protein coding sequences. Our EST analysis pipeline was automated and applied also to a donated set of T. pseudonana ESTs, and the ensemble was organized into a queryable database with a Web interface, the Diatom EST database (http://avesthagen.sznbowler.com), which represents the first such publicly available database for these organisms. In addition, codon usage was derived from a subset of P. tricornutum and T. pseudonana EST sequences and a number of differences in nucleotide utilization were detected between the two diatoms.
RESULTS AND DISCUSSION
Assembly and Functional Characterization of 12,136 Phaeodactylum tricornutum ESTs
Over 12,000 P. tricornutum cDNAs from a previously described cDNA library (Scala et al., 2002) were 5′-end sequenced, and after filtering for vector sequence, polyA tails, and poor-quality runs (see “Materials and Methods”), an EST collection of 12,136 sequences was obtained. We proceeded to reduce the initial EST collection to a nonredundant set (NRS) by means of the contig assembly program CAP3 (Liang et al., 2000). A total of 8,271 sequences could be assembled into 1,243 contigs, whereas the remaining 3,865 ESTs were found to be uniquely represented within the collection and are referred to as singletons (Table I). Nearly one-half of the contigs (590, 47.5%) consist of only two ESTs, although the most redundant were represented by several dozen and up to a few hundred ESTs (see later). Because the majority of the contigs have only two or three sequences, the consensus sequences derived from them are tentative sequences rather than confirmed transcript sequences. Individual ESTs within each contig, instead, are more likely to represent real transcript sequences, albeit with some errors. The longest EST within each of the contigs was therefore picked as representative of its cluster, and these, together with the singletons, constitute an NRS of 5,108 sequences, hereafter termed NRS sequences or NRS ESTs. In some cases, a single mRNA may give rise to different NRS sequences because nonoverlapping ESTs were obtained from it. This effect, however, is minimized in directionally cloned, single-end sequenced EST collections. Therefore, although our nonredundant collection does not correspond to a unigene collection or collection of tags for different genes, the real number of different genes represented in the redundant raw collection is probably not far below 5,108, which is the maximum possible value. Taking the 11,242 predicted genes of T. pseudonana as a reference (Armbrust et al., 2004), we may have detected about one-third of the P. tricornutum genes.
Table I.
Number of P. tricornutum ESTs and NRS sequences
| Category | No. of Sequences | Average Length |
|---|---|---|
| bp | ||
| Total ESTs | 12,136 | 653 |
| Singletons | 3,865 | 691 |
| Contigs | 1,243 | 931 |
| NRS | 5,108 | 709 |
We subsequently proceeded to functionally annotate the P. tricornutum NRS using two local alignment search programs. First, individual ESTs and contig consensi were searched for similarity to proteins in GenBank by means of the BLASTX algorithm (Altschul et al., 1997). Only 2,298 NRS sequences (45%) retrieved known proteins with E < 1E-04. Second, the reverse position-specific (RPS)-BLAST algorithm (Marchler-Bauer et al., 2003) was used to detect (E < 1E-04) protein family (Pfam) domains and profiles of clusters of orthologous groups (COGs; Tatusov et al., 2000) among the P. tricornutum NRS sequences. A total of 1,896 NRS ESTs (37.1%) contained Pfam protein domains. Domains characteristic of protein kinases, light-harvesting complex components, and proteasome subunits were the most abundant in the nonredundant collection (data not shown). On the other hand, only 1,098 NRS ESTs (21.5%) were found to contain COG profiles and could be sorted into COG functional categories. Functions related to energy production are highly represented in the nonredundant collection, as are protein metabolism categories (Maheswari et al., 2005).
An identical analysis was performed with 15,171 ESTs from T. pseudonana, kindly provided by Mark Hildebrand (Scripps Institution of Oceanography, San Diego) and Diego Martinez (Department of Energy Joint Genome Institute, Walnut Creek, CA). We estimated that this collection contains around 5,500 nonredundant sequences (data not shown).
A queryable relational database, The Diatom EST Database (http://avesthagen.sznbowler.com), was constructed with all the sequence information and functional analyses described above, organized into PtDB (for P. tricornutum) and TpDB (for T. pseudonana; Maheswari et al., 2005). At this URL, the NRS sequences are cataloged by different criteria (redundancy, COG and gene ontology functional classifications, sequence identity [ID]), and a variety of keyword and BLAST searches can be performed.
Comparison of the P. tricornutum Nonredundant Set with Other Microalgal Genomes
The genomes of the green alga C. reinhardtii, the red alga C. merolae, and the centric diatom T. pseudonana, estimated to be 90%, 99.98%, and 98% complete, respectively, have recently become available (Armbrust et al., 2004; Matsuzaki et al., 2004; http://www.biology.duke.edu/chlamy_genome). We considered these sequences as a useful set to initiate the task of pinpointing and analyzing differences between centric and pennate diatom species and to assess how they compare with photosynthetic eukaryotes derived from a primary cyanobacterial endosymbiosis. We therefore compared the P. tricornutum nonredundant EST collection with the predicted protein sequences of these organisms. It should be noted that a caveat of this approach is that the current genome browsers are not complete genome sequences and the gene-finding software that generates protein predictions can miss existing genes or include false gene models. The C. reinhardtii genome browser is the least complete of the genomes, but it has an impressive cDNA coverage (over 200,000 ESTs; Shrager et al., 2003), which minimizes gene-finding errors.
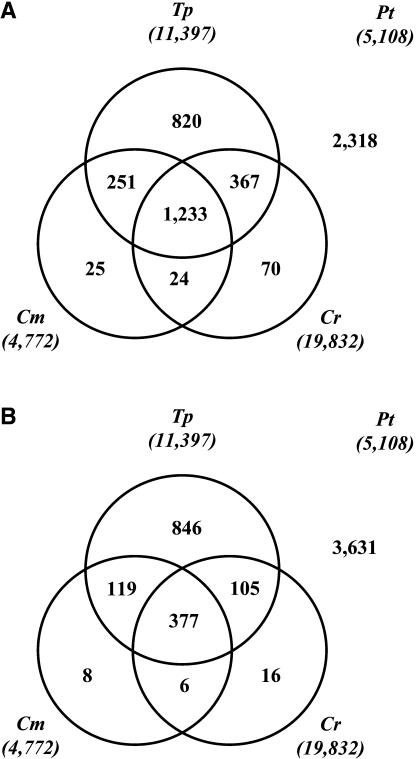
A P. tricornutum NRS sequence was considered to have a similar representative in a genome if it showed similarity to at least one of its putative encoded proteins with E < 1E-04. A nonstringent E-value threshold was used to ensure that proteins sorted as being absent in a certain genome were really absent in it (no hits with E < 1E-04), because it was our ultimate goal to identify P. tricornutum nonredundant sequences that are absent in T. pseudonana (see below). From these analyses, it was apparent that the category with the most sequences was “no similarities in any of the three genomes” (2,318 sequences, 45% of the NRS), followed by “similarities in all three” (1,233 sequences, 24% of the NRS; Fig. 1A). This was perhaps not surprising given the probable abundance of untranslated transcript regions among our ESTs and that conserved genes are always abundant when comparing virtually any two photosynthetic eukaryotes. A total of 820 P. tricornutum NRS sequences (16%) were found to have similarities in T. pseudonana, but not in the other two genomes, while for C. merolae and C. reinhardtii, these values were just 25 and 70, respectively. Similar results were obtained by Armbrust et al. (2004) in analogous comparisons of T. pseudonana with the C. merolae and Arabidopsis (Arabidopsis thaliana) genomes. For comparison, we repeated this sorting with an E-value threshold of E-30 which, contrary to a threshold of E-04, ensures a low error rate in cataloging homologs as being present rather than absent. A similar scheme was obtained (Fig. 1B), although some differences expected from an increase in stringency were noticeable (e.g. more P. tricornutum nonredundant sequences are sorted as “no similar peptides in any of the three genomes” and less are found to be common to all three).
Figure 1.
Sorting of the P. tricornutum (Pt) nonredundant EST set into categories according to the presence of putative orthologs in either of three microalgal genomes or combinations of them. Two E-value thresholds were applied, one ensuring a low error rate when classifying sequences as absent (E < 1E-04; Fig. 1A) and one minimizing the error when defining sequences as present (E < 1E-30; Fig. 1B). See Tables II to IV for recognizable proteins within the subsets of sequences encoding peptides similar to C. merolae (Cm) and/or C. reinhardtii (Cr) predicted proteins but with no similarity to T. pseudonana (Tp) predicted proteins, and Supplemental Table II for sequences with similarities in T. pseudonana but not in C. merolae or C. reinhardtii. The predicted proteomes of Cm, Cr, and Tp were downloaded from their respective genome browsers. Total numbers of predicted proteins (NRS sequences for Pt) are indicated in parentheses.
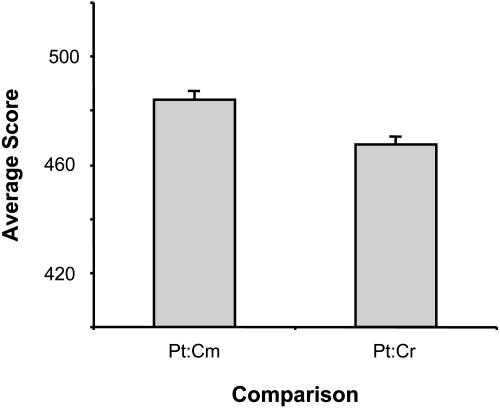
The fact that the two diatoms have more genes in common with C. reinhardtii than with C. merolae (367 versus 251; Fig. 1A) might prompt one to conclude that diatoms are more closely related to the green lineage than they are to the red branch. This would contradict the commonly accepted theory that diatoms arose by endocytobiosis of a red algal ancestor by a heterotrophic eukaryote (Falkowski et al., 2004). The sorting with a threshold of E-30 gives a more understandable comparison in this regard (105 versus 119; Fig. 1B). This indicates that many of the putative similar protein predictions in C. reinhardtii may be spurious hits with low E-values, which are more likely to occur in the green alga than in the red because of its larger proteome. We therefore selected the P. tricornutum NRS sequences that were most highly conserved in all three genomes (i.e. those whose encoded proteins showed over 40% ID for at least 100 amino acids in alignments with their most similar proteins in all 3 genomes) and calculated the average score of the diatom-red and diatom-green alignments (Fig. 2). According to this subset of 108 sequences (Supplemental Table I), such proteins are on average more highly conserved between P. tricornutum and the red alga than between P. tricornutum and the green alga, although this difference did not quite pass a test of statistical significance (P = 0.07; paired t test).
Figure 2.
Similarity of P. tricornutum (Pt) NRS sequences to related sequences in C. merolae (Cm) and C. reinhardtii (Cr). Average score of 108 pairwise alignments between P. tricornutum translated ESTs and green or red algal proteins that are most highly conserved (>40% ID over at least 100 amino acids) in the 4 species represented in Figure 1. Error bars indicate se.
Pennate diatoms are believed to have arisen from the polar centric diatoms (Medlin et al., 2000; Kooistra et al., 2003) and they are first found in the fossil record about 70 million years ago (Raven and Waite, 2004). The most ancient fossil records for silicified centric diatoms are about 120 million years old (Raven and Waite, 2004). Given that molecular data places the origin of diatoms at around 200 million years ago (Medlin et al., 2000; Kooistra et al., 2003), it is likely that a significant fraction of the evolutionary history of diatoms passed between the initial endosymbiotic event and the acquisition of silicified walls (Raven and Waite, 2004). Although cell symmetry is the most prominent difference between centric and pennate diatoms, other traits distinguish the two types, such as mode of sexual reproduction and mechanisms for cell motility (Round et al., 1990). Such fundamental differences between the two types would be expected to be reflected in their gene content. We therefore examined the pool of P. tricornutum sequences showing similarity to proteins in C. merolae and/or C. reinhardtii but not to T. pseudonana proteins.
P. tricornutum Sequences Present in C. reinhardtii But Not in T. pseudonana
We detected 70 P. tricornutum NRS sequences with similarities in the green algal genome but not in T. pseudonana or C. merolae (Fig. 1A). Among these, 13 have similarities in the SwissProt database (E < 1E-10) and only 1 is represented more than 3 times in our EST collection (Table II). This redundant transcript, PTMM01656, encodes a β-type carbonic anhydrase (CA) that has been characterized previously (Satoh et al., 2001). Additionally, singleton PTMM08526 shows over 80% nucleotide identity (89% at the amino acid level) to the contig represented by PTMM01656. The trace file associated with this EST indicates that the sequence is reliable (data not shown). P. tricornutum may therefore possess two expressed copies of a β-type CA originating from a recent duplication event.
Table II.
P. tricornutum NRS sequences encoding peptides that are absent in the centric diatom but present in C. reinhardtii and that can be assigned a tentative SwissProt annotation (E < 1E-10)
| ESTa | Redundancy | BLASTX E-Value SwissProt | BLASTX E-Value Cr Proteome | SwissProt Definition | tBLASTX E-Value Tp Genome | tBLASTX E-Value Cm Genome |
|---|---|---|---|---|---|---|
| PTAM00956 | 1 | 9E-31 | 7E-34 | Calcium/calmodulin-dependent protein kinase type II δ-chain (EC 2.7.1.123) | >E-04 | >E-04 |
| PTMM04630 | 1 | 2E-29 | 6E-05 | Extracellular Ser proteinase precursor (EC 3.4.21.) | >E-04 | >E-04 |
| PTMM01656 | 9 | 3E-24 | 6E-13 | Carbonic anhydrase 2 (EC 4.2.1.1) | >E-04 | >E-04 |
| PTMM03675 | 1 | 6E-22 | 6E-07 | Aqualysin I precursor (EC 3.4.21.) | >E-04 | >E-04 |
| PTMM08526 | 1 | 6E-21 | 6E-11 | Carbonic anhydrase 2 (EC 4.2.1.1) | >E-04 | >E-04 |
| PTMM07339 | 1 | 1E-18 | 5E-12 | Cuticle-degrading protease precursor (EC 3.4.21.-; PR1; chymoelastase) | >E-04 | >E-04 |
| PTMM09029 | 1 | 1E-17 | 1E-09 | Lysosomal protective protein precursor (EC 3.4.16.5; cathepsin A) | >E-04 | >E-04 |
| PTMM04888 | 1 | 7E-15 | 3E-16 | Cell cycle control protein cwf16 | >E-04 | >E-04 |
| PTMM00034 | 1 | 3E-14 | 6E-09 | Guanine deaminase (EC 3.5.4.3; guanase; guanine aminase) | >E-04 | >E-04 |
| PTMM02006 | 1 | 9E-14 | 4E-05 | Glycerol uptake facilitator protein | >E-04 | >E-04 |
| PTMM05103 | 3 | 4E-11 | 2E-07 | Probable sulfate transporter 4.2. | >E-04 | >E-04 |
| PTAM00539 | 1 | 1E-10 | 4E-05 | Sensor protein luxQ (EC 2.7.3.) | >E-04 | >E-04 |
| PTMM09702 | 1 | 2E-10 | 1E-05 | Eukaryotic translation initiation factor 2C 1 (eIF2C 1; RNA-binding protein Q99) | 1E-16 | >E-04 |
Sequences commented on in the text are highlighted in bold.
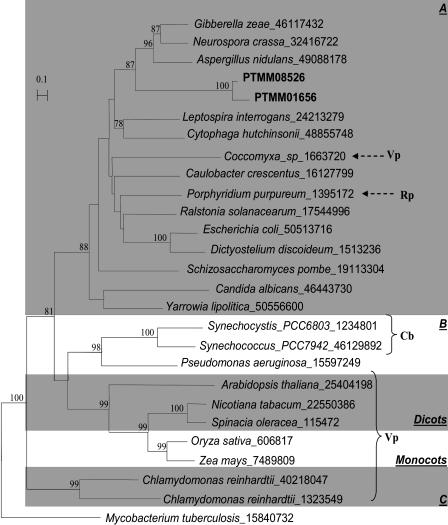
To understand whether these putative CAs may have been derived from the secondary endosymbiont or were present in the nuclear or mitochondrial genomes of the ancestral heterotrophic host, we aligned the two P. tricornutum ESTs with the most similar CA-like genes in photosynthetic and heterotrophic eukaryotes and prokaryotes, and derived a neighbor-joining tree (Fig. 3). The two diatom sequences clustered with β-type CAs of fungi and then with heterotrophic bacteria rather than with plant or cyanobacterial orthologs, suggesting that this gene may have been present in the genome of the host when the endosymbiotic event that gave rise to diatoms occurred. Nonetheless, this gene does not appear to be present in the T. pseudonana genome (see below).
Figure 3.
Phylogenetic analysis of putative β-type carbonic anhydrases (CAs) from P. tricornutum. A neighbor-joining tree is shown of putative or described β-type CAs from several lineages within the eukarya and eubacteria, including the two P. tricornutum β-type CA-like NRS sequences (in bold). The sequences were selected to represent 5 out of the 6 major clades of β-type CAs that contain photosynthetic or eukaryotic orthologs (A, B, C, monocots, and dicots; Smith et al., 1999) and trimmed to a conserved core of 191 amino acids. The Mycobacterium tuberculosis sequence, a distant ortholog of the sixth clade D, was chosen to root the tree. Bootstrap values above 70% (of 1,000 replicates) are shown. The GenBank GI sequence identifiers of the proteins used are shown following the species name. Members of the Rhodophyta (Rp), Cyanobacteria (Cb), and Viridiplantae (Vp) are indicated. Scale bar = 0.1 substitutions/site.
CAs catalyze the reversible dehydration of bicarbonate into CO2 and in photosynthetic organisms are fundamental players in carbon-concentrating mechanisms (CCM), which ensure a high concentration of CO2 in the vicinity of the Rubisco enzyme (Kaplan and Reinhold, 1999). Three widespread types of CA are known, termed α, β, and γ, all of which require a zinc atom as ligand for catalytic activity. Studies of carbon utilization in the centric diatom Thalassiosira weissflogii revealed the existence of two novel CAs, a zinc-dependent enzyme encoded by TWCA1 (Morel et al., 1994) and a cadmium-dependent enzyme (Lane and Morel, 2000) whose sequence has not yet been reported. The T. pseudonana genome does not contain any genes encoding β-type CAs but does contain putative α- and γ-type CA genes, along with a probable TWCA1 ortholog (Armbrust et al., 2004).
None of the putative CAs encoded in the T. pseudonana genome appears to contain a plastid-targeting sequence, which could support the provocative hypothesis of Morel and colleagues (Reinfelder et al., 2000) that diatoms utilize a CCM based on C4 photosynthesis, proposed upon physiological studies under CO2 and zinc limitation with T. weissflogii. Interestingly, Satoh et al. (2001) previously noted that the N-terminal region of the β-type CA encoded by PTMM01656 is similar to that of diatom fucoxanthin, chlorophyll a/c-binding proteins (FCPs), known to be targeted to the plastid. It also shows similarity to the bipartite plastid-targeting sequence of P. tricornutum AtpC, published later (Apt et al., 2002). Moreover, some of the general features concluded upon analysis of 67 plastid-targeted peptides encoded in the T. pseudonana genome (Armbrust et al., 2004), such as an enrichment in Ser and the presence of a Pro residue downstream of the cleavage site between the signal (endoplasmic reticulum translocation) and the transit (plastid-targeting) peptides, are also present. If the β-type CAs identified in Table II are indeed plastid localized, a CCM for P. tricornutum could be explained more conventionally than the C4-based mechanism proposed by Reinfelder et al. (2000). Therefore, it will clearly be of interest to assess the intracellular localization of these enzymes in P. tricornutum and to determine how widespread they are among centric and pennate diatoms.
On the other hand, PTMM00034 in Table II is highly similar (41% ID) to the N-terminal portion of a mammalian guanine deaminase (GDA) that has been characterized biochemically (Yuan et al., 1999). BLAST searches in GenBank showed that similar proteins are present in heterotrophic bacteria, fungi, insects, and vertebrates. In mammals, this enzyme hydrolyzes guanine to xanthine, releasing an ammonium group, and xanthine is then used for synthesis of uric acid. However, no GDA-like coding sequence was detected in the T. pseudonana genome by tBLASTN searches with the P. tricornutum GDA-like sequence (Table II) or with similar proteins from mouse, insects, or C. reinhardtii (data not shown). It is also surprising that such a gene is present in C. reinhardtii, as GenBank protein database BLASTP searches with the P. tricornutum GDA-like ESTs or with similar sequences from mouse, insects, and notably, C. reinhardtii did not give any significant matches in the Viridiplantae (data not shown).
It is worth noting that our starting datasets and sorting procedures may have caused one sequence, PTMM09702, to be improperly classified in Table II. A highly similar region of the T. pseudonana genome was detected by tBLASTX (1E-16; Table II), but the gene-finding software failed to predict any coding sequence in this region. Furthermore, the sequence presents poor similarity to a C. reinhardtii model (1E-05; Table II), just below the threshold established, for which this gene may be present in T. pseudonana and absent in C. reinhardtii rather than vice versa. As discussed above, such errors occur as an intrinsic limitation of bioinformatic approaches. In curated annotations or individual case studies, additional manual inspections should therefore be conducted.
P. tricornutum Sequences Present in C. merolae But Not in T. pseudonana
Among the 25 translated P. tricornutum NRS sequences showing similarity to red algal proteins but not to T. pseudonana or C. reinhardtii proteins (Fig. 1A), only 3 had significant similarity to SwissProt accesions (E < 1E-10), and none of these are highly redundant in the P. tricornutum EST collection (Table III). The only sequence with a high degree of similarity to a SwissProt entry (PTSS00077, over 60% ID to a C-terminal region of animal and fungal UDP-protein glucosyltransferases) covers only about 100 amino acids.
Table III.
P. tricornutum NRS sequences encoding peptides that are absent in the centric diatom but present in C. merolae and that can be assigned a tentative SwissProt annotation (E < 1E-10)
| ESTa | Redundancy | BLASTX E-Value SwissProt | BLASTX E-Value Cm proteome | SwissProt Definition | tBLASTX E-Value Tp Genome | tBLASTX E-Value Cr Genome |
|---|---|---|---|---|---|---|
| PTSS00077 | 1 | 9E-40 | 4E-35 | UDP-Glc:glycoprotein glucosyltransferase precursor (EC 2.4.1.-) | >E-04 | >E-04 |
| PTMM03212 | 1 | 3E-13 | 7E-05 | Glycerol uptake facilitator protein (Aquaglyceroporin) | >E-04 | >E-04 |
| PTMM08334 | 1 | 1E-10 | 3E-13 | Sericin-1 (Silk gum protein 1; fragment) | >E-04 | 1E-05 |
Sequences commented on in the text are highlighted in bold.
P. tricornutum Sequences Present in Both C. reinhardtii and C. merolae But Not in T. pseudonana
Twenty-four NRS sequences were sorted as being present in both red and green algal genomes but not in T. pseudonana (Fig. 1A). Nine of these could be annotated by BLASTX analysis against SwissProt (E < 1E-10; Table IV), only three of which have one or two redundant sequences in the raw collection. Like PTMM00034 in Table II, PTMM09692 was similar to mammalian guanine deaminases but to a lesser extent (24%–30% ID) and to a more C-terminal region. It appears in Table IV rather than Table II because this more downstream region was weakly similar to a C. merolae predicted protein (E = 8E-09; Table IV) that is most similar to bacterial chlorohydrolases, which are close relatives of guanine deaminases (most are classified as “cytosine deaminases and related hydrolases” in the COG database). The sequence PTMM09692 is more similar to the C. reinhardtii putative GDA gene than to the C. merolae chlorohydrolase-like sequence, and it is possible that PTMM00034 and PTMM09692 are nonoverlapping ESTs from a unique cDNA.
Table IV.
P. tricornutum NRS sequences encoding peptides that are absent in the centric diatom but present in both C. reinhardtii and C. merolae and that can be assigned a tentative SwissProt annotation (E < 1E-10)
| ESTa | Redundancy | BLASTX E-Value SwissProt | BLASTX E-Value Cr Proteome | BLASTX E-Value Cm Proteome | SwissProt Definition | tBLASTX E-Value Tp Genome |
|---|---|---|---|---|---|---|
| PTMM05217 | 1 | 9E-71 | 9E-56 | 2E-51 | ATP-citrate synthase (EC 2.3.3.8; ATP-citrate [pro-S-]-lyase) | >E-04 |
| PTMM07433 | 1 | 7E-64 | 5E-54 | 6E-67 | Fru-bisphosphate aldolase 2 (EC 4.1.2.13; ALDO-2) | >E-04 |
| PTMM08369 | 2 | 1E-48 | 4E-23 | 2E-48 | Phenylethylamine oxidase precursor (EC 1.4.3.6; amine oxidase) | >E-04 |
| PTMM08609 | 1 | 3E-46 | 2E-05 | 6E-25 | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 ζ; eIF3d; p66) | >E-04 |
| PTMM09692 | 1 | 2E-27 | 3E-23 | 8E-09 | Guanine deaminase (EC 3.5.4.3; guanase; guanine aminase) | >E-04 |
| PTMM00972 | 3 | 3E-24 | 1E-35 | 6E-28 | Leucoanthocyanidin reductase (EC 1.-.-.-; LAR) | >E-04 |
| PTMM07394 | 1 | 3E-22 | 3E-06 | 5E-09 | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20; diglyceride acyltransferase) | >E-04 |
| PTAM00559 | 1 | 7E-19 | 2E-07 | 1E-04 | Probable small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1; Sm-D1) | 1E-14 |
| PTMM08844 | 2 | 5E-16 | 1E-10 | 6E-05 | Vacuolar ATP synthase subunit G 2 (EC 3.6.3.14; V-ATPase G-subunit 2) | 1E-35 |
Sequences commented on in the text are highlighted in bold.
On the other hand, Table IV includes a sequence, PTMM05217, which shows over 60% ID to the C terminus of mammalian and insect cytosolic ATP-citrate lyases (ACLs). We identified another NRS sequence, PTMM07214, which was similar to a more upstream region of animal ACLs by tBLASTN searching the P. tricornutum EST collection with a complete human ACL. This additional EST does not appear in Tables II to IV because the central region of ACLs shows similarity to succinyl-CoA ligases present in T. pseudonana. It may nonetheless represent a nonoverlapping region of the same mRNA as PTMM05217. However, no obvious ACLs are present in T. pseudonana, as manually tested by searching the T. pseudonana genome with plant and animal sequences using tBLASTN.
ACL (EC 2.3.3.8) is a multimeric enzyme that catalyzes the generation of acetyl-CoA and oxaloacetate from citrate and CoA and constitutes a major source of cytoplasmic acetyl-CoA in mammals, plants, and oleaginous yeasts (Rangasamy and Ratledge, 2000; Fatland et al., 2002). Cytosolic acetyl-CoA is used for synthesis of a wide array of compounds (e.g. waxes, isoprenoids, and flavonoids in plants, sterols, and fatty acids in vertebrates), but the exact biosynthetic pathways and their regulation are not yet fully understood (Fatland et al., 2000). Green plants, a glaucophyte, fungi, and a photosynthetic prokaryote have been found to have heteromeric ACL complexes consisting of α- and β-subunits of approximately 400 and 600 amino acids in length, respectively (Nowrousian et al., 2000; Fatland et al., 2002), whereas animals have a single subunit of up to 1,100 amino acids forming homomeric complexes (Kim et al., 1994). It was therefore concluded that, in the ancestral eukaryote, the ACL complex likely consisted of two different subunits that fused into a single gene early in the animal lineage (Fatland et al., 2002). Nonoleaginous eukaryotic microorganisms do not usually have any version of this enzyme (e.g. Saccharomyces cerevisiae) and their reduced requirements for cytosolic acetyl-CoA are thought to be fulfilled by other means (Ratledge, 2002). The fact that this enzyme was found in P. tricornutum but not in T. pseudonana is in agreement with the high capability of the former to accumulate lipids (Domergue et al., 2003).
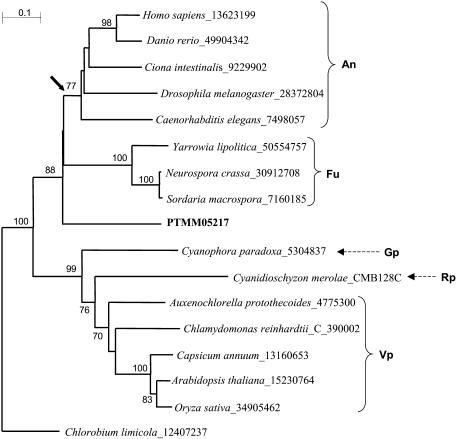
No α-ACL-like sequences were found among the P. tricornutum ESTs by tBLASTN searches with various plant and fungal sequences. To gain some insight as to whether diatom ACLs are animal- or fungal/plant-like, we aligned the most downstream ACL-like P. tricornutum EST, PTMM05217, with 8 ACLs from fungi and animals and 7 β-ACLs from a range of photosynthetic eukaryotes and derived a neighbor-joining tree (Fig. 4). The C-terminal region of animal ACL contains the catalytic center of this enzyme and is therefore highly conserved. The diatom ACL clustered with good bootstrap support with animal and fungal ACLs rather than with ACLs of photosynthetic organisms, but the bootstrap values did not resolve which of the two Opisthokont clades is most closely related to the diatom sequences. The putative P. tricornutum ACL-encoding nonredundant sequences are heterokont ortholog(s) and the topology of the tree obtained by us did not contradict the gene-fusion hypothesis put forward by Fatland et al. (2002). We therefore expect that, as genomes of other members of major eukaryotic crown groups (Baldauf, 2003) are sequenced, this hypothesis will be confirmed.
Figure 4.
Phylogenetic analysis of putative ATP:citrate lyases (ACLs) from P. tricornutum. The figure shows a neighbor-joining tree of putative or described ACLs representing the major lineages in which ACL-like genes are known, including the most highly conserved ACL-like sequence in P. tricornutum (in bold). The sequences were trimmed to a conserved core of 185 amino acids. The GenBank GI sequence identifiers of the proteins used are shown following the species name. The C. reinhardtii and C. merolae sequences were obtained from their respective genome browsers rather than from GenBank and their predicted gene identification numbers are given. Bootstrap values above 70% (of 1,000 replicates) are shown. Members of the Animalia (An), Fungi (Fu), Glaucocystophyta (Gp), Rhodophyta (Rp), and Viridiplantae (Vp) are indicated. The Chlorobium limicola ACL is the sole prokaryotic ortholog known to date and was used to root the tree. The arrow indicates the time of fusion of α- and β-ACLs into a single gene as proposed by Fatland et al. (2002). Scale bar = 0.1 substitutions/site.
Table IV also includes singleton PTMM07433, which likely encodes a eukaryotic type I Fru-1,6-bisphosphate aldolase (FBA) and implies a further complication in the already complex phylogeny of FBAs. FBAs are amphibolic enzymes involved in glycolysis, gluconeogenesis, and Calvin cycle reactions. Two types of evolutionarily unrelated FBAs are known, and both can catalyze the cleavage of FBA to glyceraldehyde-3-P plus dihydroxyacetone-P in catabolic functions and the reverse condensation in anabolic pathways. FBA-encoding genes have undergone numerous lateral transfers, gene duplications, and gene product recompartmentalizations throughout their phylogenetic history (Rogers and Keeling, 2004) and provide evidence for a common secondary endosymbiont ancestor for all chromalveolate present-day plastids (Patron et al., 2004). An initial gene replacement is thought to have occurred early after the primary endosymbiotic event, because cyanobacteria typically contain type II FBAs, whereas the green lineage and red algae generally contain type I plastidial FBAs considered to have arisen from their cytoplasmic paralogs. A further gene replacement apparently occurred early in the lineage that appeared upon endocytobiosis of a red alga by a heterotrophic eukaryote, given that heterokonts, haptophytes, dinoflagellates, and cryptomonads all seem to contain type II enzymes in their plastids (Patron et al., 2004).
In their recent summary, Patron et al. (2004) identified a bacterial type I plus two plastidial type II FBAs in P. tricornutum. In our EST collection, we identified these 3 proteins plus the above-mentioned conventional eukaryotic type I sequence, PTMM07433 (Table IV), and a third type II-like sequence, PTMM07123 (Table V). Neighbor-joining trees paralleling those in Patron et al. (2004) suggest that the type I enzyme may be plastid localized because the P. tricornutum sequence clusters with plastid-localized counterparts from two red algae and a cryptomonad (Table V; Supplemental Fig. 1A). If confirmed, the presence of a plastid-localized eukaryotic type I FBA in the pennate, but not in the centric diatom, as well as in a cryptophyte complicates the picture of protein recompartmentalizations and gene deletions that FBAs must have undergone to result in their present-day distribution across taxa. It should be noted, however, that the plastidial type I is the least redundant of the P. tricornutum FBAs. Given that for many chromalveolate lineages only EST collections of moderate size are available, it may be that a plastid type I FBA is more widespread among the chromists but has not been noticed so far due to low expression levels.
Table V.
Fructose-1,6-bisphosphate aldolases (FBAs) of P. tricornutum identified in the nonredundant EST collection
FBA-like sequences within the nonredundant EST set were identified by BLAST and keyword searches. The presence of a putative leader sequence for translocation into the endoplasmic reticulum as detected by SignalP (http://www.cbs.dtu.dk/services/SignalP) is indicated.
| EST | Redundancy | FBA Type | Present in (Fig. 1) | Organisms in Same Clade (Suppl. Fig. 1) | Localization of Proteins in Same Clade (Suppl. Fig. 1) | SignalPb |
|---|---|---|---|---|---|---|
| PTMM00206a | 13 | I | Tp | Prokaryotes only | ? | No |
| PTMM07433 | 1 | I | Cm, Cr | Red algae Viridiplantae | Plastid | Truncated N terminus |
| PTMM05813a | 13 | II | Tp | Diatom-specific | Plastid | Yes |
| PTMM07123 | 6 | II | Tp | Fungi, Chromalveolates, Euglenoids | Cytoplasm | (?)c |
| PTMM08077a | 6 | II | Tp | Chromista | Plastid | No |
EST encoding a peptide sequence included in the study of Patron et al. (2004).
The protein sequence was inferred as far upstream as possible using other ESTs in the redundant cluster.
Signal peptide residues are detected, but not putative cleavage sites.
On the other hand, the typically prokaryotic type I sequence PTMM00206, a probable case of lateral transfer (Patron et al., 2004), was found to contain a homolog in the centric T. pseudonana (53% ID), for which reason this lateral transfer may date back far enough to affect most diatoms. Diatoms seem to have acquired a second diatom-specific feature concerning FBA genes, a variant (PTMM05813) of the common chromalveolate plastid-localized type II FBA (PTMM08077; Supplemental Fig. 1B), as pointed out by Patron et al. (2004). Understanding the biological significance of such differences and commonalities between the pennate and the centric diatoms, experimentally or by further in silico surveys for related enzymes, will be of interest given the importance of the metabolic pathways involved.
Further Differences between Pennate and Centric Diatoms and Identification of Diatom-Specific Genes
We next attempted to identify P. tricornutum NRS sequences with no related sequences in T. pseudonana or the other 2 algal genomes but that can be recognized based on their similarity to proteins in the SwissProt database. Following initial BLASTX analyses against the predicted T. pseudonana proteome, 2,437 NRS ESTs (47.7%) were found to have no matches (E > 1E-04), and 550 of these were represented by more than 1 EST (data not shown). We filtered this set further by a tBLASTX search against the T. pseudonana genome sequence to double check for possible T. pseudonana genes that may have been missed by the gene-finding software used for T. pseudonana genome annotation (Armbrust et al., 2004). Up to 847 translated NRS sequences showed similarity to some region of the translated T. pseudonana genome, and 220 of these were represented more than once in our P. tricornutum EST collection. The remaining 1,588 nonredundant sequences (31%), with no obvious counterparts in T. pseudonana, were compared to sequences in SwissProt using BLASTX (E < 1E-10). After removal of sequences already shown in Tables II to IV, five additional sequences were found (Table VI). Only two of these were found to have meaningful SwissProt annotations upon inspection of their alignments; PTMM06865 probably encodes a tryptophanase with orthologs in two archaeal species and six eubacteria (as represented in COG3033 in the COG database), and PTMM04006 appears to encode a protein with similarity to palmitoyl thioesterases from mammals.
Table VI.
P. tricornutum NRS sequences encoding peptides that are absent in T. pseudonana, C.merolae, and C. reinhardtii, which can be assigned a tentative SwissProt annotation (E < 1E-10)
| ESTa | Redundancy | BLASTX E-Value SwissProt | SwissProt Definition | tBLASTX E-Value Tp Genome | tBLASTX E-Value Cr Genome | tBLASTX E-Value Cm Genome |
|---|---|---|---|---|---|---|
| PTMM06865 | 3 | 3E-48 | Tryptophanase (EC 4.1.99.1) | >E-04 | >E-04 | >E-04 |
| PTMM04006 | 2 | 8E-15 | Palmitoyl-protein thioesterase 1 precursor (EC 3.1.2.22) | >E-04 | >E-04 | >E-04 |
| PTMM09746 | 1 | 1E-15 | ATP-binding cassette, subfamily A, member 1 | >E-01 | >E-04 | >E-04 |
| PTMM01729 | 5 | 1E-10 | Hypothetical 70.5-kD protein in AGP3-DAK3 intergenic region | >E-04 | >E-04 | >E-04 |
| PTMM01736 | 1 | 1E-10 | Hypothetical 70.5-kD protein in AGP3-DAK3 intergenic region | >E-04 | 8E-08 | >E-04 |
Sequences commented on in the text are highlighted in bold.
Among the 820 sequences common in the two diatoms but absent in the green and red algae, genes that are usually absent in primary endosymbiosis-derived photosynthetic organisms but are known in eukaryotic heterotrophs or prokaryotes can be identified, along with known brown lineage-specific genes. As many as 119 of the sequences in this subset (14.5%) could be assigned a tentative SwissProt annotation (E < 1E-10; Supplemental Table II). Among the best 60 SwissProt annotations in this table (supported by E < 1E-20), the abundance of the phaeophyte-specific FCP proteins (five sequences) and of sequences related to nitrogen metabolism (11 sequences) or to utilization of citrate (four sequences) is particularly striking. The detection of several components of a urea cycle, typically a metazoan feature, was one of the major surprises from the recent T. pseudonana genome analysis (Armbrust et al., 2004), and traces of it can be found in this table (e.g. the arginase II-like PTMM07144 or the typically prokaryotic peptide of similar function, agmatinase, probably encoded by PTMM03490). Among the 119 tentatively annotated sequences of this subset, the most redundant are FCP proteins, as expected (see below), followed by 2 probable transporters, PTMM09485 (101 redundant sequences in the redundant EST set) and PTMM02857 (19 redundant sequences), which, according to their BlASTX GenBank search output, contain putative orthologs in prokaryotes and metazoans, respectively.
In an independent analysis, we identified putative novel diatom-specific sequences by selecting those that did not present similarity to any protein in SwissProt (E < 1E-04) and then searching the T. pseudonana genome using BLASTX (E < 1E-20). A more stringent E-value threshold was set for the T. pseudonana BLASTX analysis in this case to decrease the number of false positives (regions of weak similarity that are not likely to represent related genes). Up to 3,949 translated NRS sequences did not resemble any sequence in SwissProt, and 1,028 of these had potential orthologs encoded in the T. pseudonana genome (Supplemental Table III). This suggests that multiple novel gene families are bound to be identified among diatoms, a possibility that will probably be confirmed with the forthcoming P. tricornutum genome and other diatom cDNA sequencing projects. The function of such sequences being unknown, we can only note that 36 of them are rather highly expressed (more than 10 ESTs) and constitute an interesting, manageable set for benchwork aimed at discovering novel gene functions.
Expression and Base Composition
The cDNA library employed to generate the ESTs discussed here was created from a culture grown in a single condition (standard exponential growth conditions), so many genes that are expressed only under particular environmental conditions will not be represented in our sequence dataset. In addition, the cDNA library was not normalized or subtracted for any sequences. These manipulations are known to improve the rate of gene discovery, as they reduce the amount of nontarget cDNAs (e.g. highly redundant sequences; Bonaldo et al., 1996). On the other hand, in nonnormalized libraries, EST redudancy is likely to more accurately reflect gene expression than in more manipulated libraries. The most highly represented transcripts in the P. tricornutum EST collection are therefore likely to be genes that are highly expressed in exponentially growing cultures.
The 20 most redundant P. tricornutum NRS sequences are represented by between 52 and 315 ESTs (Table VII). Among these highly represented transcripts, we found known sequences encoding P. tricornutum FCPs (Bhaya and Grossman, 1993), a glyceraldehyde 3-phosphate dehydrogenase (Liaud et al., 2000), and several other recognizable proteins that were also present in the T. pseudonana EST collection, albeit with lower redundancy (Table VII). The frustulin encoded by contig C0013, in contrast, is highly expressed in P. tricornutum and is present in multiple copies in the T. pseudonana genome, but no frustulin-like transcripts could be found among the T. pseudonana ESTs. Very interestingly, out of the 20 most redundant P. tricornutum NRS sequences, 8 retrieved no significant matches (E > 1E-04) in the GenBank protein database by BLASTX searches. Four of them could be aligned with different regions of the T. pseudonana genome by tBLASTX (E < 1E-10), and two of these are also present in the T. pseudonana EST set (Table VII).
Table VII.
The most redundant ESTs in the P. tricornutum EST collection
| Pt Contig | NRS EST | Redundancy | BLASTX E-Value GenBank | GenBank Description | Putative Copies in Tp Genome (tBLASTX, E<1E-10) | Redundancy in Tp EST Set (cDNAs) |
|---|---|---|---|---|---|---|
| C0001 | PTMM09600 | 315 | 4E-94 | FCP | >5 | >5 |
| C0002 | PTMM04145 | 206 | 3E-08 | Unknown protein | 0 | 0 |
| C0003 | PTMM06831 | 184 | >E-04 | No clear hits 1 | 1a | 0 |
| C0004 | PTMM03311 | 181 | 1E-88 | FCP | >5 | >5 |
| C0005 | PTMM06833 | 127 | >E-04 | No clear hits 2 | 0 | 0 |
| C0006 | PTMM09837 | 125 | 8E-64 | FCP | >5 | >5 |
| C0007 | PTMM09485 | 101 | 4E-57 | Probable transporter | 1 | 4 |
| C0008 | PTMM05394 | 91 | 1E-179 | Glyceraldehyde-3-phosphate dehydrogenase | 5 | 4b |
| C0009 | PTMM05069 | 71 | >E-04 | No clear hits 3 | 0 | 0 |
| C0010 | PTMM07864 | 70 | >E-04 | No clear hits 4 | 1 | 1 |
| C0011 | PTMM06363 | 69 | 3E-16 | Secreted metalloprotease | >5c | 3 |
| C0012 | PTMM04398 | 68 | 1E-19 | Light-harvesting protein | 4 | 1 |
| C0013 | PTMM01606 | 63 | 2E-56 | Frustulin | 4 | 0 |
| C0014 | PTMM02781 | 60 | 2E-05 | Weakly similar to ferredoxin components | 1 | 1 |
| C0015 | PTMM05909 | 58 | >E-04 | No clear hits 5 | 0 | 0 |
| C0016 | PTMM02742 | 58 | >E-04 | No clear hits 6 | 4d | 3 |
| C0017 | PTMM08324 | 55 | >E-04 | No clear hits 7 | 0 | 0 |
| C0018 | PTMM03334 | 55 | 1E-30 | Light-harvesting protein | 4 | 3 |
| C0019 | PTMM07490 | 53 | 1E-95 | Vacuolar H+ translocating phosphatase | 2 | 2 |
| C0020 | PTMM03689 | 52 | >E-04 | No clear hits 8 | 1 | 0 |
Not modeled.
From two copies.
Weak similarity.
One not modeled.
We next aimed to derive codon usage tables for both species, which required the selection of a subset of sequences with a known protein coding frame. We chose to assign a sound frame when a translated EST sequence showed similarity to known proteins (GenBank protein database BLASTX) with more than 50% coverage and more than 35% identity. We thus obtained subsets of 859 P. tricornutum and 465 T. pseudonana NRS sequences, with which codon usage was tabulated (Supplemental Table IV; also available at http://avesthagen.sznbowler.com). Inspection of P. tricornutum codon usage tables revealed that C is the preferred third-codon-position nucleotide in 13 out of 16 triplets. This is not the case for T. pseudonana, in which C is used in only 9 out of 16 cases. Interestingly, Arg is preferentially encoded by CGN codons in P. tricornutum, whereas T. pseudonana avoids this quartet in favor of the duet AG(A/G). Such differential use of synonymous codons in the two diatoms may be related to tRNA abundance, as previously suggested for Escherichia coli and yeast (Akashi, 2001).
Because the latter observations could not be explained by a marked difference in GC content, we analyzed the content of CpG dinucleotides in the two diatoms. The C of such doublets is a widespread target for DNA methylation, and because mC is converted to T upon deamination, these sites are particularly susceptible to mutation (Jabbari et al., 1997; Jabbari and Bernardi, 2004). Deamination of unmethylated C results in U, which is usually detected efficiently by the DNA repair machinery.
Upon analysis of the subsets of ESTs with an assigned frame, the occurrence of CpG doublets was found to be significantly higher (P = 0.0001; unpaired t test) in P. tricornutum than in T. pseudonana (Table VIII). The two species contain these doublets more often between codons in which a mutation of the C is more likely to lead to a synonymous change than in positions 1,2 or 2,3. It is also in position 3,1 that the difference between CpG usage in the two diatoms is most pronounced. These data may therefore suggest that T. pseudonana is more susceptible to the CpG to TpG transition than P. tricornutum. This could be due to differences in genome methylation levels or in robustness of DNA repair mechanisms between the two diatom species. Other factors related to coding constraints and, consequently, to base compositional correlations along coding sequences could also contribute to these differences in dinucleotide composition.
Table VIII.
Utilization of individual bases and of dinucleotides involving C and G as derived from highly conserved NRS sequences
| P. tricornutum | T. pseudonana | |
|---|---|---|
| % A | 24.5 | 26.0 |
| % C | 25.6 | 23.3 |
| % T | 23.5 | 24.1 |
| % G | 26.3 | 26.5 |
| % GpC total | 6.5 | 5.3 |
| % CpG total | 7.6 | 5.3 |
| % CpG (1,2) | 5.7 | 3.9 |
| % CpG (2,3) | 6.2 | 4.6 |
| % CpG (3,1) | 10.9 | 7.4 |
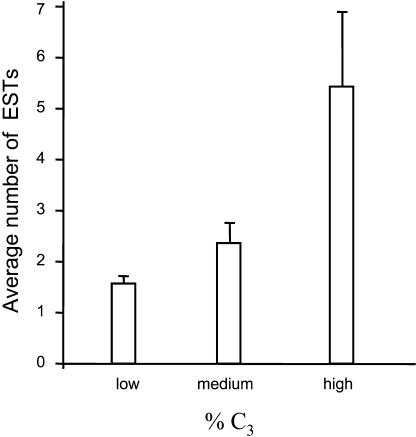
Because the majority of preferred codons in P. tricornutum are C-ending (13 out of 16), we plotted C3% NRS sequences against the number of ESTs in its cluster. The 859 NRS sequences used to derive the P. tricornutum codon usage table were sorted into three C3% categories (containing equal numbers of sequences) and their average redundancy was calculated (Fig. 5). NRS sequences with a low C3% were represented on average by less than two redundant sequences, while those with a high C3% were typically represented by more than five redundant ESTs. The differences observed between the three C3% categories are statistically significant (P = 0.035; Kruskal-Wallis nonparametric test). This is in agreement with the preference for C at the third codon position noted above, because abundant codons have been shown to be recognized and translated more quickly, and with fewer errors, than their less frequent counterparts (Andersson and Kurland, 1990; Dong et al., 1996; Kanaya et al., 1999).
Figure 5.
Correlation of expression and %C3 content of P. tricornutum transcripts. P. tricornutum NRS sequences with soundly assigned frames were classified by increasing %C3, divided into 3 groups with equal numbers of sequences, and their average redundancy within the redundant EST collection was calculated. Bars indicate the se.
CONCLUDING REMARKS
The EST project reported here may have identified an important number of the total P. tricornutum genes. Comparative analyses of these sequences with other eukaryotic algae, and in particular with T. pseudonana, has brought to light a number of interesting features and has allowed a preliminary comparison of gene repertoires between pennate and centric diatoms.
The P. tricornutum genome is currently being sequenced by the Joint Genome Institute (Walnut Creek, CA). Once sequencing and assembly have been completed, the genes will be modeled with the help of an additional 60,000 ESTs generated from a range of cDNA libraries by Genoscope (Evry, France). The availability of such large amounts of sequence information will open new horizons in our studies of diatom phylogeny, ecology, physiology, and molecular biology. This study should serve as a useful platform for these new initiatives and as a basis for hypothesis-driven research aimed at dissecting the novel features of diatom biology and the molecular differences between the two major classes of diatoms. The putative genes encoding CAs, ACLs, and FBAs identified here are interesting cases in point.
MATERIALS AND METHODS
Generation of 12,136 ESTs
Over 12,000 bacterial clones were picked at random from a previously described cDNA library (Scala et al., 2002) from the pennate diatom Phaeodactylum tricornutum Bohlin clone CCMP632 (Provasoli-Guillard National Center for Culture of Marine Phytoplankton, Bigelow, West Boothbay Harbor, ME) and sequenced from the 5′ end with the T3 primer. PolyA tails, vector sequences, and poor-quality reads were trimmed by means of the Trimest, Trimseq, and Vectorstrip programs of EMBOSS (European Molecular Biology Open Software Suite), and short (<50 bp) sequences were discarded. A total of 12,136 partial cDNA sequences were obtained for subsequent analyses. The sequencing work was performed in 3 batches between 1999 and 2002. An initial set of 997 ESTs has been reported previously (Scala et al., 2002) and was labeled PTSS0001 to PTSS0997, another 1,131 ESTs were sequenced by MWG Biotech (Ebersberg, Germany) and were denoted PTAM00001 to PTAM01131, and, most recently, 10,008 ESTs were generated by Avestha Gengraine Technologies (Bangalore, India), denoted PTMM00001 to PTMM10008. All P. tricornutum EST sequences generated in this study were deposited in National Center for Biotechnology Information (NCBI) dbEST, PTSS sequences in December 2001, PTAM in June 2002, and PTMM in May 2003.
Contig Assembly and Functional Annotation
The initial EST collection was assembled into an NRS by means of the CAP3 algorithm (Huang and Madan, 1999). Two ESTs were considered to be contiguous if they showed at least 95% ID over an overlapping region longer than 30 bp. All EST and contig sequences were compared to the GenBank nonredundant protein database by means of the BLASTX algorithm (Altschul et al., 1997). COG and Pfam domains were identified among NRS sequences by RPS-BLAST comparisons with the Conserved Domain database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd; Marchler-Bauer et al., 2003). A tentative annotation was assigned to an EST or contig if it showed similarity to some entry in the corresponding database with E < 1E-04.
Database Construction
All EST sequences and contigs, together with alignments of redundant clusters and functional annotations, were organized into the twin databases PtDB and TpDB for P. tricornutum and Thalassiosira pseudonana, respectively, together forming The Diatom EST Database (http://avesthagen.sznbowler.com). The T. pseudonana clone CCMP1335 ESTs were a kind gift from Mark Hildebrand (Scripps Institution of Oceanography, San Diego) and Diego Martinez (Department of Energy Joint Genome Institute, Walnut Creek, CA). The programming is the work of Uma Maheswari and the database is maintained at the Avestha Gengraine facilities under contract. Technical details on the construction of this database have been published elsewhere (Maheswari et al., 2005).
Comparison of the P. tricornutum Nonredundant EST Collection with Microalgal Genomes
The P. tricornutum NRS was compared to the predicted proteins of Chlamydomonas reinhardtii (http://www.biology.duke.edu/chlamy_genome), Cyanidioschyzon merolae (http://merolae.biol.s.u-tokyo.ac.jp), and T. pseudonana (http://spider.jgi-psf.org/JGI_microbial/html) using BLASTX. A pennate EST was considered to be related to a protein in a different genome if it displayed similarity to it in any of its translated frames with E < 1E-04. After sorting the P. tricornutum sequences as being present in one, two, or all three genomes, tentative annotations were performed by using the BLASTX algorithm against SwissProt, as described in “Results and Discussion.”
Generation of Neighbor-Joining Trees
Neighbor-joining trees were derived in selected cases to evaluate the degree of relatedness of P. tricornutum sequences to sequences from other organisms. In each case, all sequences to be included in a tree were aligned by means of the ClustalX program (Thompson et al., 1997) and trimmed to their conserved core. The alignment output was used to derive a distance matrix (Tajima-Nei correction) and subsequently generate a neighbor-joining tree with 1,000 bootstrap replicates by means of the TreeCon package (Van de Peer and Dewachter, 1994).
Codon and GC Usage
Translated NRS sequences that displayed similarity to a GenBank protein entry with an ID level above 35% and a coverage of the subject protein of at least 50% were assigned a sound frame (i.e. the protein sequence and function can be predicted with high degrees of confidence). Codon usage tables for P. tricornutum and T. pseudonana were derived from these subsets, amounting to 859 P. tricornutum sequences and 465 T. pseudonana sequences. Base usage at the third codon position and its correlation with redundancy of the sequence in the P. tricornutum EST collection were studied, along with the abundance of the methylation target dinucleotide CpG, as more specifically described in “Results and Discussion.”
Supplementary Material
Acknowledgments
We are grateful to the sequencing and bioinformatics teams of Avestha Gengraine Technologies, to Nicola Patron and Patrick Keeling for their comments and kind contributions to the analysis of the novel P. tricornutum FBAs, and to Margherita Groeben, Andrew Allen, Angela Falciatore, and Assaf Vardi for their help and suggestions. The T. pseudonana ESTs were a kind gift from Mark Hildebrand and Diego Martinez.
This work was supported in part by the European Union-funded Margenes (grant no. QLRT–2001–01226 to C.B.) and Marine Genomics (grant no. 505403 to C.B.) projects.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052829.
References
- Akashi H (2001) Gene expression and molecular evolution. Curr Opin Genet Dev 11: 660–666 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Kurland CG (1990) Codon preferences in free-living microorganisms. Microbiol Rev 54: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt KE, Kroth-Pancic PG, Grossman AR (1996) Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet 252: 572–579 [DOI] [PubMed] [Google Scholar]
- Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG (2002) In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci 115: 4061–4069 [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JB, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86 [DOI] [PubMed] [Google Scholar]
- Baldauf SL (2003) The deep roots of eukaryotes. Science 300: 1703–1706 [DOI] [PubMed] [Google Scholar]
- Bhaya D, Grossman AR (1993) Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Res 21: 4458–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res 6: 791–806 [DOI] [PubMed] [Google Scholar]
- Borowitzka MA, Volcani BE (1978) The polymorphic diatom Phaeodactylum tricornutum: ultrastructure of its morphotypes. J Phycol 14: 10–21 [Google Scholar]
- Domergue F, Spiekermann P, Lerchl J, Beckmann C, Kilian O, Kroth PG, Boland W, Zähringer U, Heinz E (2003) New insight into Phaeodactylum tricornutum fatty acid metabolism. Cloning and functional characterization of plastidial and microsomal delta12-fatty acid desaturases. Plant Physiol 131: 1648–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HJ, Nilsson L, Kurland CG (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Falciatore A, Bowler C (2002) Revealing the molecular secrets of marine diatoms. Annu Rev Plant Biol 53: 109–130 [DOI] [PubMed] [Google Scholar]
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C (1999) Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY) 1: 239–251 [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR (2004) The evolution of modern eukaryotic phytoplankton. Science 305: 354–360 [DOI] [PubMed] [Google Scholar]
- Fatland B, Anderson M, Nikolau BJ, Wurtele ES (2000) Molecular biology of cytosolic acetyl-CoA generation. Biochem Soc Trans 28: 593–595 [PubMed] [Google Scholar]
- Fatland BL, Ke JS, Anderson MD, Mentzen WI, Cui LW, Allred CC, Johnston JL, Nikolau BJ, Wurtele ES (2002) Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol 130: 740–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M, Dahlin K, Volcani BE (1998) Characterization of a silicon transporter gene family in Cylindrotheca fusiformis: sequences, expression analysis, and identification of homologs in other diatoms. Mol Gen Genet 260: 480–486 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G (2004) Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 333: 143–149 [DOI] [PubMed] [Google Scholar]
- Jabbari K, Caccio S, de Barros JPP, Desgres J, Bernardi G (1997) Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene 205: 109–118 [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, Ikemura T (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143–155 [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L (1999) CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50: 539–570 [DOI] [PubMed] [Google Scholar]
- Kim KS, Park SW, Moon YA, Kim YS (1994) Organization of the 5′ region of the rat ATP citrate lyase gene. Biochem J 302: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra WHCF, DeStefano M, Mann DG, Medlin LK (2003) The phylogeny of the diatoms. In WEG Müller, ed, Progress in Molecular and Subcellular Biology, Vol 33. Springer-Verlag, Berlin, pp 63–97 [DOI] [PubMed]
- Kroger N, Deutzmann R, Sumper M (1999) Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 286: 1129–1132 [DOI] [PubMed] [Google Scholar]
- Lane TW, Morel FMM (2000) A biological function for cadmium in marine diatoms. Proc Natl Acad Sci USA 97: 4627–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res 28: 3657–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaud MF, Lichtle C, Apt K, Martin W, Cerff R (2000) Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol Biol Evol 17: 213–223 [DOI] [PubMed] [Google Scholar]
- Maheswari U, Montsant A, Goll J, Krishnasamy S, Rajyashri KR, Patell VM, Bowler C (2005) The Diatom EST Database. Nucleic Acids Res 33: D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He SQ, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31: D383–D387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Yoshida Y, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Medlin LK, Kooistra W, Gersonde R, Wellbrock U (1996) Evolution of the diatoms (Bacillariophyta). 2. Nuclear-encoded small-subunit rRNA sequence comparisons confirm a paraphyletic origin for the centric diatoms. Mol Biol Evol 13: 67–75 [DOI] [PubMed] [Google Scholar]
- Medlin LK, Kooistra WCHF, Sam M (2000) A review of the evolution of the diatoms: a total approach using molecules, morphology and geology. In A Witkowski, J Sieminska, eds, The Origin and Early Evolution of the Diatoms: Fossil, Molecular and Biogeographical Approaches. W. Szafer Institute of Botany, Polish Academy of Sciences, Kracow, Poland, pp 13–34
- Morel FMM, Reinfelder JR, Roberts SB, Chamberlain CP, Lee JG, Yee D (1994) Zinc and carbon co-limitation of marine phytoplankton. Nature 369: 740–742 [Google Scholar]
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B (1995) Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles 9: 359–372 [Google Scholar]
- Nowrousian M, Kuck U, Loser K, Weltring KM (2000) The fungal acl1 and acl2 genes encode two polypeptides with homology to the N- and C-terminal parts of the animal ATP citrate lyase polypeptide. Curr Genet 37: 189–193 [DOI] [PubMed] [Google Scholar]
- Patron NJ, Rogers MB, Keeling PJ (2004) Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot Cell 3: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen N, Sumper M, Kroger N (2003) Biosilica formation in diatoms: characterization of native silaffin-2 and its role in silica morphogenesis. Proc Natl Acad Sci USA 100: 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D, Ratledge C (2000) Compartmentation of ATP: citrate lyase in plants. Plant Physiol 122: 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C (2002) Regulation of lipid accumulation in oleaginous micro-organisms. Biochem Soc Trans 30: 1047–1050 [DOI] [PubMed] [Google Scholar]
- Raven JA, Waite AM (2004) The evolution of silicification in diatoms: inescapable sinking and sinking as escape? New Phytol 162: 45–61 [Google Scholar]
- Reinfelder JR, Kraepiel AML, Morel FMM (2000) Unicellular C-4 photosynthesis in a marine diatom. Nature 407: 996–999 [DOI] [PubMed] [Google Scholar]
- Rogers M, Keeling PJ (2004) Lateral transfer and recompartmentalization of Calvin cycle enzymes of plants and algae. J Mol Evol 58: 367–375 [DOI] [PubMed] [Google Scholar]
- Round FE, Crawford RM, Mann DG (1990) The Diatoms: Biology and Morphology of the Genera. Cambridge University Press, London
- Satoh D, Hiraoka Y, Colman B, Matsuda Y (2001) Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol 126: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S, Carels N, Falciatore A, Chiusano ML, Bowler C (2002) Genome properties of the diatom Phaeodactylum tricornutum. Plant Physiol 129: 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang CW, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR (2003) Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol 131: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KS, Jakubzick C, Whittam TS, Ferry JG (1999) Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci USA 96: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Dewachter R (1994) Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Veldhuis MJW, Cucci TL, Sieracki ME (1997) Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J Phycol 33: 527–541 [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D (2002) The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA 99: 15507–15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Bin JC, McKay DJ, Snyder FF (1999) Cloning and characterization of human guanine deaminase: purification and partial amino acid sequence of the mouse protein. J Biol Chem 274: 8175–8180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.