Abstract
Chlamydomonas reinhardtii is an excellent model system for plant biologists because of its ease of manipulation, facile genetics, and the ability to transform the nuclear, chloroplast, and mitochondrial genomes. Numerous forward genetics studies have been performed in Chlamydomonas, in many cases to elucidate the regulation of photosynthesis. One of the resultant challenges is moving from mutant phenotype to the gene mutation causing that phenotype. To date, complementation has been the primary method for gene cloning, but this is impractical in several situations, for example, when the complemented strain cannot be readily selected or in the case of recessive suppressors that restore photosynthesis. New tools, including a molecular map consisting of 506 markers and an 8X-draft nuclear genome sequence, are now available, making map-based cloning increasingly feasible. Here we discuss advances in map-based cloning developed using the strains mcd4 and mcd5, which carry recessive nuclear suppressors restoring photosynthesis to chloroplast mutants. Tools that have not been previously applied to Chlamydomonas, such as bulked segregant analysis and marker duplexing, are being implemented to increase the speed at which one can go from mutant phenotype to gene. In addition to assessing and applying current resources, we outline anticipated future developments in map-based cloning in the context of the newly extended Chlamydomonas genome initiative.
Sometimes called green yeast (Goodenough, 1992), the unicellular, eukaryotic green alga Chlamydomonas reinhardtii (hereafter called Chlamydomonas) is a venerable model system for plant biology as well as for cell motility. The tag green yeast refers to its haploid vegetative state, the existence of two mating types, and the general similarity in applicable genetic techniques. These aspects of Chlamydomonas biology have been previously reviewed (Rochaix, 1995).
Like many microorganisms, screening of Chlamydomonas strains for rare mutations is straightforward, since large numbers of cells can be plated on an appropriate selective medium, or nonswimmers, for example, can be selected from large numbers of swimming cells. At the same time, the ease of nuclear transformation in Chlamydomonas, coupled with the plant-like nonhomologous integration of transforming DNA, facilitates the creation of insertional mutant collections. Taken together, the assortment of techniques useable for Chlamydomonas indulges both the amateur and experienced geneticist, yielding sometimes overwhelming collections of mutant strains. In this report, we focus on mutants affecting photosynthesis, in keeping with the thrust of this journal, and the emphasis of the newly renewed and National Science Foundation-supported Chlamydomonas genome project (http://www.chlamy.org/). However, the map-based cloning tools described here are generally applicable to Chlamydomonas biology.
The use of Chlamydomonas to study the elaboration and regulation of the photosynthetic apparatus is long established and was recently reviewed (Grossman, 2000; Dent et al., 2001). Key to this is the ability to maintain nonphotosynthetic (PS−) mutants on acetate-containing media, as well as the ability to use replica plating ±acetate and/or chlorophyll fluorescence to identify such mutants (Bennoun and Béal, 1997; Niyogi et al., 1997). Furthermore, numerous photosynthetic (PS+) suppressors have been recovered from screening of PS− strains (e.g. Girard-Bascou et al., 1992; Levy et al., 1997; Bernd and Kohorn, 1998; Nickelsen, 2000; Esposito et al., 2001; Li et al., 2002).
Recovery of wild-type alleles of genes mutated in PS− strains has been successful, since both genomic complementation with selection on minimal medium or identification of DNA flanking an insertional mutant site are relatively straightforward although perhaps tedious methods (e.g. Gumpel et al., 1995; Boudreau et al., 2000; Vaistij et al., 2000; Auchincloss et al., 2002; Dauvillee et al., 2003). However, in cases where PS+ suppressors have been recovered from PS− strains or where a trait otherwise cannot be selected on minimal medium, recovery of the mutation requires other methods. In the case of PS+ suppressors, for example, a recessive suppressor would yield a PS− phenotype upon complementation, and even dominant suppressors such as mcd2, which suppresses the nuclear mcd1-2 mutation responsible for instability of the chloroplast petD mRNA (Esposito et al., 2001), require construction of a genomic library from the suppressed strain if they are to be cloned by complementation. Another example is the xanthophyll cycle mutant npq1, which is defective in nonphotochemical quenching (Niyogi et al., 1997). Although npq1 was generated in an insertional mutant population, its defect is not linked to the ARG7 insertional mutagen. In each of these cases, isolation of the gene of interest could be achieved through map-based cloning in a suitably developed system.
Map-based cloning relies on two basic principles, namely the existence of a genetic/physical map and the ability to generate progeny of sexual crosses that segregate for the trait of interest as well as phenotypic and/or molecular markers. In higher plants, such resources are most fully advanced in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), which, not coincidentally, have complete nuclear genome sequences. Furthermore, interfertile polymorphic ecotypes (Columbia and Landsberg erecta; and indica and japonica, respectively) have been utilized as sources of genetic variation to introduce into selected mutant backgrounds. Resources for Arabidopsis and rice have been extensively publicized and have been utilized in numerous examples of successful gene isolation (Chen et al., 2002; Garcia-Hernandez et al., 2002; Torjek et al., 2003); maize (Zea mays) is another subject of intensive efforts (Coe et al., 2002; Cone et al., 2002).
Here we present a case-based study to describe existing and projected resources for map-based cloning in C. reinhardtii. Mutations generated in this species can be mapped by crossing to the interfertile strain known as Chlamydomonas grossii, S1-D2 or its culture collection designation of CC-2290, which has a suitable profusion of sequence tagged sites (STS), cleavable amplified polymorphic sequence (CAPS), single nucleotide polymorphism (SNP), and RFLP markers (Gross et al., 1988; Vysotskaia et al., 2001; Grossman et al., 2003). Beginning with laborious RFLP-based mapping (Gross et al., 1988), Chlamydomonas mapping has moved toward a PCR-based method (Kathir et al., 2003) and now is poised to incorporate more high-throughput methods. This, in concert with an increasingly complete nuclear genome sequence (Grossman et al., 2003), provides the necessary tools for studies of all classes of mutations.
The nuclear mutants mcd4 and mcd5 were derived from strains petDLS2 and petDLS6, respectively, in which mutations engineered into the 5′-untranslated region of the chloroplast petD gene caused RNA instability and thus a PS− phenotype (Higgs et al., 1999). Both mcd4 and mcd5 are spontaneous PS+ mutants, do not carry a molecular tag, and genetic analysis (data not shown) showed them to be recessive. Thus, complementation of mcd4 or mcd5 with the wild-type genes would revert the PS+ phenotype to PS−, making genomic complementation an inappropriate approach. We therefore decided to map mcd4 and mcd5 by using available genomic resources and by developing new ones as opportunities or needs arose.
RESULTS AND DISCUSSION
A Composite Map of Molecular Markers
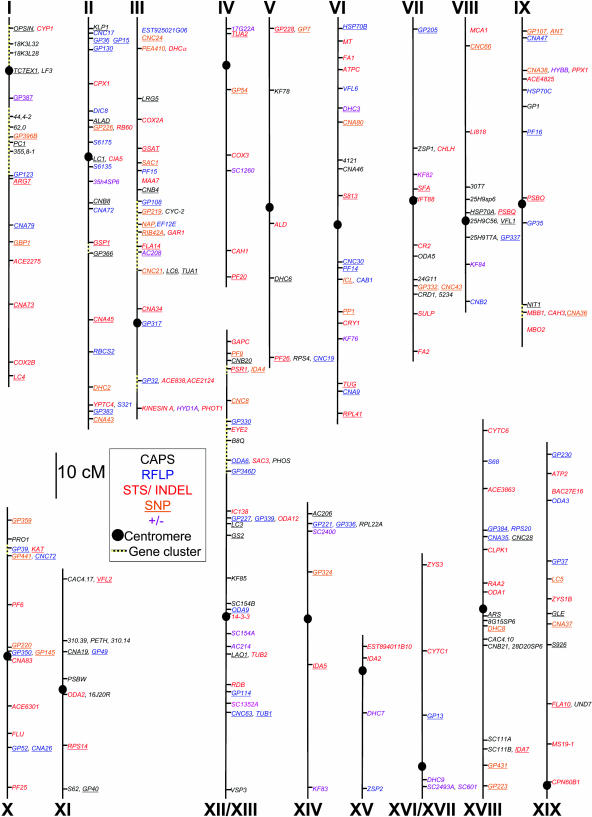
As noted above, Chlamydomonas molecular markers are derived from comparisons of laboratory strains and the interfertile polymorphic strain S1-D2. To date, 4 laboratories (see “Materials and Methods”) have generated at least 506 STS, insertion/deletion (InDel), CAPS, SNP, ± (a PCR fragment amplifiable in one polymorphic strain but not the other), and RFLP markers, whose respective utilities are discussed below. The combined molecular maps shown in Figure 1 contain 385 markers, representing 266 loci arranged on 17 linkage groups (LG), and differ from previously published maps (Kathir et al., 2003) by including markers generated by multiple laboratories. Fifteen markers could be assigned to an LG but not a specific location, whereas 107 are not displayed on the map because they are within marker-dense gene clusters or because precise marker order could not be determined due to insufficient data. Complete information on the markers is included as Supplemental Table I.
Figure 1.
Molecular map of C. reinhardtii. A total of 506 molecular markers have been arranged on the 17 LGs; 385 are shown here. Markers are color coded as to their type: CAPS markers are in black, RFLP markers are in blue, STS and InDel makers are in red, and ± markers are in purple. All SNP markers are underlined; those that can be assayed using an alternative method are indicated by a color other than orange. Broken lines indicate gene clusters, where only representative markers are shown; the remaining markers are in the supplemental data (Supplemental Table I).
Of the six types of markers mentioned above, five are PCR based (STS, InDel, CAPS, SNP, and ±), whereas RFLPs rely on DNA gel blots. STS markers can be generated when sequences of both strains are known. In Chlamydomonas, most STS markers have one primer conserved in both C. reinhardtii and S1-D2 and one that is specific for each strain. The three primers are used in a single reaction, generating products that differ in size for the C. reinhardtii or the S1-D2 allele. InDels have primers conserved in both strains but yield PCR products of different sizes due to insertions or deletions. Both STS and InDels can be assayed directly on an agarose gel to distinguish alleles. CAPS markers, on the other hand, require an additional step, since polymorphisms are revealed upon restriction enzyme digestion of the PCR product. SNPs are easy to find since only a single nucleotide difference is required, and on average 2.7 base substitutions were found in every 100 bp of sequence when S1-D2 and the laboratory strain were compared (Kathir et al., 2003). A disadvantage is that the reagents and technology for SNP detection are relatively expensive and some methods require special equipment. The last PCR-based marker shown on the map are ± markers. These amplify a PCR product from only one parent and thus have a higher error rate since failed PCR reactions would be scored as the nonamplifying parent, skewing recombination-based distance calculations. Use of RFLP markers is comparatively arduous, requiring isolation of genomic DNA and gel-blot hybridizations.
Because each laboratory whose markers are represented in Figure 1 used different mapping populations to determine either centromere linkage or recombination-based distances, such values could not be directly compared. Approximate distances were calculated based on the combined data and, where available, the genome sequence. Markers whose recombination data were not available were placed by alignment with the nuclear genome sequence, and their distances were calculated based on the assumption by Kathir et al. (2003) that 1 cM equals 100 kb. This estimate was based on the total number of cM in the genome and an approximate genome size of 108 bp. To determine the accuracy of this assumption, 23 marker pairs were examined for their actual kb to cM ratio. This ratio, not surprisingly, varied widely, from 0.860 kb/cM between Tpx and Fa1 on LG VI, to 511 kb/cM between Arg7 and GP123 on LG I. Varied rates of recombination are also seen in Arabidopsis, where the relationship between genetic and physical distance ranges, for example, from 30 kb/cM to >550 kb/cM on chromosome 4 (Schmidt et al., 1995). Due to this variance, most distances must be regarded as rough estimates and are annotated as such in the supplemental data (Supplemental Table II). Nonetheless, gene/marker order is expected to be accurate, especially where it has been confirmed by the genome sequence.
Mapping of Mcd4
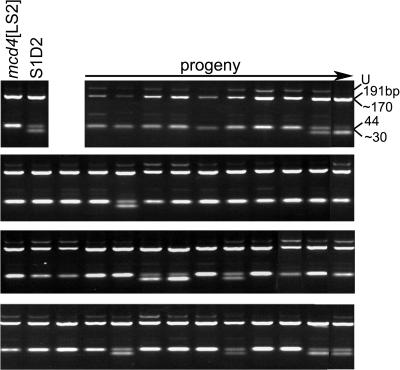
To develop a population for development and application of mapping methods, the mutant mcd4 [LS2] mt+ was crossed to S1-D2 mt−. By convention, the chloroplast genotype is given in brackets, except where it is wild type. The LS2 chloroplast genotype confers a PS− phenotype due to instability of the petD mRNA (Higgs et al., 1999), and this PS− phenotype is suppressed by the nuclear mutation mcd4. Thus, PS+ progeny of the cross carry the mutant mcd4 allele, whereas PS− progeny carry the S1-D2 wild-type allele Mcd4. To minimize analysis of duplicate recombination events, one PS+ progeny was chosen from each of 54 tetrads to create the mapping population for the mutant allele, mcd4. With a population of 54 progeny, a 17.7-cM (Kosambi units) resolution can be expected with a confidence interval of 95% (P > .05). At this resolution, 83% of the progeny should contain the mcd4 allele, although any marker giving greater than 70% mcd4 alleles was deemed significant by chi-squared analysis and the region examined further with additional markers. Sixty markers were scored for the percent of progeny that contained the mcd4 allele, resulting in an unambiguous assignment of mcd4 to LG II. The STS marker with the highest linkage was Gsp1, for which 42/53 (80%) progeny displayed the PCR product size associated with the mcd4 parent. Another tightly linked marker was the CAPS marker Dhc4, shown as an example in Figure 2. Since 41/52 (79%) of the progeny contained the mcd4 allele in this example, Mcd4 can be calculated as 22 cM distant from Dhc4. Linkage was also seen to Cia5 and CNA45, which based on PCR data are both 26 cM from Mcd4 (data not shown).
Figure 2.
Dhc4 is linked to mcd4. DNAs isolated from the mapping population described in the text were amplified with the Dhc4 CAPS primers (supplemental data) and digested with PstI. Digests were analyzed in a 3% agarose gel. The U indicates the undigested PCR product of 235 bp. Upon digestion, the mcd4 allele yields 191- and 44-bp products, whereas the S1-D2 (Mcd4) allele yields approximately 170-, 44-, and approximately 30-bp products. Forty-five out of 53 progeny tested are shown here, and the parental controls are at the left of the top row.
Bulked Segregant Analysis and Duplexing
To place markers at 20- to 30-cM intervals, as well as within 10 cM of the end of every linkage group, 57 to 72 markers would be needed to span the 1,107-cM genome. Markers placed at each end of the linkage group ensure adequate coverage in cases where the genetic maps are longer than the molecular maps, indicating potentially missing sequence. A subset of 67 markers, all of which are currently known (Supplemental Table I), could provide this coverage. Doing so with the 50 progeny required to achieve 20- to 30-cM coverage would entail 3,350 PCR reactions, a daunting number. To reduce the time and expense of mapping, bulked segregant analysis (BSA) and marker duplexing were evaluated. Although these techniques have been used in other systems, for example Arabidopsis (Lukowitz et al., 2000), they have not been tested systematically for Chlamydomonas. In BSA, DNAs from multiple segregating progeny are combined, and results from PCR-based markers are examined for significant bias from a roughly equal contribution from each parent.
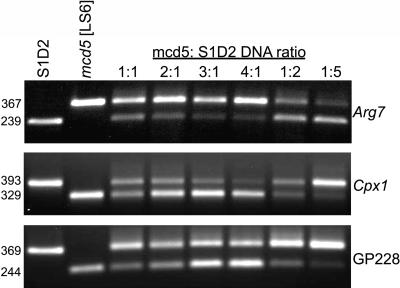
For our markers, BSA was evaluated by creating defined mixtures of mcd5 DNA and S1-D2 DNA, where the total DNA amount was held constant. The strain mcd5 is a laboratory (C. reinhardtii)-derived PS+ suppressor of a chloroplast mutation LS6 (Higgs et al., 1999) and analogous to mcd4. Forty STS markers have been examined for compatibility with BSA (Supplemental Table III), and three examples are shown in Figure 3. Examination of the reactions using 1:1 DNA ratios shows that the longer product stained more brightly in two cases, as would be expected for a roughly equal amplification of the two alleles. The relative prominence of products also changed as expected, although in some reactions one product or the other was stronger or weaker than anticipated. We have observed this variability for numerous primer sets and it is to some extent unavoidable. Importantly, however, even at the most extreme ratios (4-fold excess of mcd5 or 5-fold excess of S1-D2), the diluted allele was still visible. This suggests that BSA of five to six bulked progeny will still reveal a rare allele for these primer sets and that the overall banding pattern is to some degree indicative of the relative number of each allele. In particular, staining corresponding to the 1:1 ratio would be observed for unlinked markers to the gene of interest, which would avert the need to deconvolute the bulked DNAs for those particular markers. This technique was used successfully with much larger numbers of individuals from an Arabidopsis mapping population (Lukowitz et al., 2000), perhaps being facilitated by the lower [G+C] content and better sequence information.
Figure 3.
DNA of up to six progeny can be combined for BSA. The indicated ratios of mcd5:S1-D2 DNA were used to simulate bulked progeny. PCR products for the markers indicated at right were analyzed in 1% agarose gels, and product sizes in basepairs are shown at the left.
As we detail in the supplemental data (Supplemental Table III), 34 primer sets yielded good results with 4:1 or 1:5 mcd5:S1-D2 DNA ratios. Five primer sets failed to generate both parental bands at a 1:1 mcd5: S1-D2 ratio; thus, in these cases, individual progeny must be analyzed. The remaining primer sets worked for up to four progeny. Since some difficulties could be resolved with gradient PCR or other adjustments, it might be possible with further optimization to implement BSA for additional markers. Assuming that 34 primer sets can be used with 5 bulked progeny, 1 with 4 bulked progeny, and 5 with single samples, the 2,000 PCR reactions required for the analysis of 40 markers (the number we tested for BSA) with 50 progeny (a typical mapping population) would be reduced to only 603. A complete set of amplification instructions and actual results can be found at the Chlamydomonas genome web site (http://www.chlamy.org).
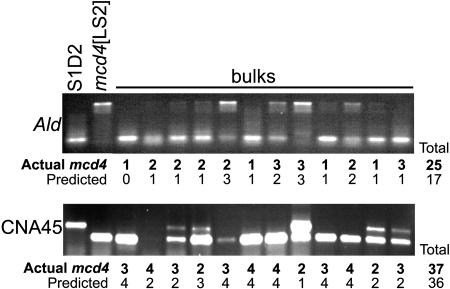
To determine whether BSA caused a loss of mapping resolution due to incorrect interpretation of gel band intensities, DNA from mcd4 mapping progeny were bulked in groups of four. Four markers were blind tested using the bulked samples, and results were compared to those using single progeny. Two such comparisons are shown in Figure 4. Previous BSA experiments (Arg7 and GP228 from Fig. 3 and others) showed that when bulks contain parental DNAs at equal concentrations, the products generated do not appear to have equal intensities. In light of this, results from the bulked progeny were compared among themselves to determine which of the five ratios each lane represented. Differences between predicted (deduced from BSA gels) and actual (known from single-progeny measurements) frequency of the mcd4 allele ranged from 4% to 15% of the total progeny, with the average being 9%. In principle, this uncertainty could prevent detection of markers >20 cM distant from a gene of interest, although this was not the case for CNA45, which is 26 cM away from mcd4 (Fig. 4). More experience scoring BSA, fewer progeny per PCR reaction, and/or the addition of a control lane with 1:1 ratio of mcd4/ S1-D2 would aid in reducing the margin of uncertainty to acceptable levels.
Figure 4.
BSA affects mapping resolution. DNAs from four randomly selected mcd4 progeny were bulked per reaction and analyzed for the markers indicated on the left, with separation of PCR products in 1% agarose gels. Products were scored for number of progeny that contained the mcd4 allele. Actual mcd4 allele frequencies were determined by scoring individual unbulked samples (data not shown). Bulks from 48 out of the 53 progeny tested are shown here.
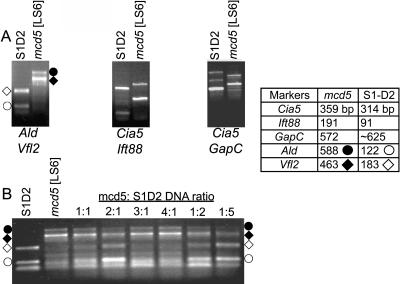
Combining two primer sets in one PCR reaction (duplexing) is a second method to reduce the amount of work needed for mapping. In choosing marker pairs for duplexing, annealing temperature and product size need to be considered. The four PCR products should be similar in size to avoid overly preferential amplification of the smaller bands, yet the products must be distinguishable by gel electrophoresis. Also, annealing temperatures should be close enough to prevent nonspecific amplification. Six paired markers were tested and four proved successful, as shown for three examples in Figure 5A. When duplexing and BSA were combined, all 4 parental bands were visible at DNA ratios of 1:2 or 2:1 mcd5:S1-D2 for Ald/Vfl2, whereas >3 DNAs did not offer sufficient sensitivity to detect the rare allele (Fig. 5B). These results suggest that, at least in some cases, combining duplexing and BSA holds promise for reducing labor while retaining resolution.
Figure 5.
Markers can be duplexed and combined with BSA. A, PCR products from 3 duplexed sets are shown, analyzed in 1% agarose gels. B, The Ald/Vfl2 duplexed primers were used in combination with BSA. PCR products were analyzed in a 1% agarose gel. The table at right shows expected product sizes for each marker. The symbols denoting the respective products from amplification of Ald or Vfl2 from mcd5 or S1-D2 are used to mark product positions on the relevant gels. S1-D2 generates a second, artifactual band with Ald primers that migrates at approximately 75 bp (below 122 bp S1-D2 product).
From Marker to Genome: Combining Mapping with Genome Data
The above data show the application of well-established molecular mapping technologies to Chlamydomonas. In plants, very high resolution mapping is needed prior to final identification of the gene of interest, because testing of numerous candidates by transformation is impractical. Since Chlamydomonas is easily transformed, the considerations are somewhat different. Nonetheless, 20 cM is far from adequate resolution to begin gene identification. In the case of mcd4, we combined phenotypic markers with custom-developed molecular markers based on comparative sequencing of mcd4 and S1-D2 in the region of interest.
Since mcd4 had been assigned to LG II, it was crossed to strains carrying two relevant phenotypic markers: pf12, a paralyzed flagella mutant (McVittie, 1972; Frey et al., 1997), and act1, a cycloheximide resistant mutant (Sager and Tsubo, 1961). Pf12 is tightly linked to the RFLP marker GP225, while Act1 is 3.5 cM from Pf12 (Harris, 1989). Progeny from a mcd4 [LS2] mt+ by pf12 mt− cross were scored for segregation of ability to swim and PS growth, and the results showed that pf12 and mcd4 were within 4 cM. In a cross using act1 as the mt− parent, act1 was estimated to be 9 cM from mcd4. The combined data from phenotypic and molecular markers yielded alternative positions between Act1 and CNA45, the latter an STS marker (Fig. 6), or close to pf12. This ambiguity exists because there is inconsistency between the phenotypic data and the mapping population data. This inconsistency may be caused by suppressed recombination between S1-D2 and the laboratory strain in this region. If this is the case, a high marker density or a larger mapping population will be needed to fine map mcd4.
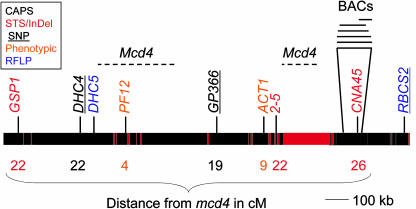
Figure 6.
Summary of mapping data showing the region of LG II surrounding mcd4. Available sequence from scaffolds 2 and 66 (genome version 2) is represented by the black bars, with red representing gaps in the sequence. The large gap is the estimated distance between scaffolds 2 (left) and 66 (right). Markers are color coded as shown in the box at top left. The potential positions of mcd4 are indicated by dotted lines. Distances between selected markers and mcd4 are given in centimorgans below the map. A BAC contig developed for the CNA45 region is shown to illustrate typical coverage where such contigs have been anchored.
Determining whether mcd4 is near Pf12 versus in the gap between scaffolds 2 and 66 awaits development of additional markers. act1 has not been cloned but has been reported to carry a mutation in a component of the 60S ribosome (Fleming et al., 1987). The Act1 gene has been tentatively placed at the end of scaffold 2 (Fig. 6) as allelic to the L10a gene, since L10a is the only 60S ribosomal protein encoded on either scaffold 2 or scaffold 66. Since a location on scaffold 2 is possible, especially given the very loose correlation between genetic and molecular distances, candidate gene searches were performed. Given that mcd4 suppresses an RNA instability phenotype and is recessive, one possibility was a loss-of-function in a chloroplast-targeted ribonuclease (for review, see Bollenbach et al., 2004); however, no such genes were found in these scaffolds. Another possibility was a tetratricopeptide repeat (TPR) protein; TPR motifs or a related motif, called pentatricopeptide repeats, are often found in posttranscriptional regulators of organelle gene expression, including in Chlamydomonas chloroplasts (Boudreau et al., 2000; Lurin et al., 2004; for review, see Small and Peeters, 2000). There are 4 TPR-containing predicted proteins encoded on scaffold 2, which remain in principle candidate genes. Inspection of the gene models does not predict chloroplast localization based on N-terminal sequence motifs; however, there are several reasons why this could be misleading (see “Future Perspectives”).
Generation of Additional Markers and Mapping Tools
Our initial mapping data placed Mcd4 into two possible locations. In both cases, it was desirable to generate bacterial artificial chromosome (BAC) contigs for eventual complementation, as well as additional markers. As a case study, we describe how BAC contigs can be extended using Chlamydomonas resources and our experience in generating site-specific markers.
Several BAC libraries have been constructed for Chlamydomonas, two of which are available through the Clemson Genomics Institute (http://www.genome.clemson.edu/groups/bac/). In addition, BAC contigs (http://www.biology.duke.edu/chlamy_genome/BAC/index.html) have been assembled for most of the STS and RFLP markers. In the case of mcd4, Gsp1 resides on scaffold 2 and CNA45 on scaffold 66. A 41-BAC contig exists for scaffold 2 covering approximately 1,000 kb (R. Nguyen, personal communication), and a smaller contig is linked to scaffold 66 and CNA45 (Fig. 6). An unknown amount of DNA separates scaffolds 2 and 66; such discontinuities in the genome sequence are represented by red bars in Figure 6. The distance between scaffolds would be 1,660 kb if 1 cM is equal to 100 kb in this region. As yet, no BAC contig has been assembled containing ends in both of them. Nonetheless, both scaffold 2 and CNA45 contigs have been extended using the genome to perform a virtual BAC walk. In this method, a scaffold end or segment is compared using BLAST to the BAC end database, and any BACs containing that sequence are identified. Then, the other end sequence associated with that BAC is used in BLAST searches with the genome sequence. BAC contigs can also be elongated by the traditional method of hybridization of single-copy BAC end probes to a filter representing the entire BAC library, followed by informatic analysis as described just above. This would be the preferred method if a BAC end lands in a hole between scaffolds (i.e. is not represented in available genome sequence). Eventually such contigs will rejoin the genome sequence, in effect bridging two scaffolds.
To place Mcd4 more accurately, additional markers were required. CAPS, STS, and SSR markers were all possible choices. CAPS markers can be made by converting existing RFLP markers, which entails generating primers flanking the polymorphic restriction site. This method was used to generate a CAPS marker from GP366, which has a PstI polymorphism. Eighty-one percent of the progeny contained the lab strain (mcd4) allele (Fig. 6). If RFLP markers are not available in the region of interest, one can use a brute-force method for any restriction site found in the nuclear genome sequence. This is because S1-D2 has 2.7 base substitutions/100 bp compared to the laboratory strain (Kathir et al., 2003), leading to an average of 1 useable marker/7 primer sets tested (for 6-base unique sequence restriction enzymes). When 15 primer sets were generated from scaffolds 2 and 66 using this concept, 5 sets produced a PCR product only for the C. reinhardtii allele, 2 sets yielded PCR products that differed in size between mcd4 and S1-D2 and could be used as InDel markers, while 3 contained polymorphisms when digested and could be used as CAPS markers. When the S1-D2 products from two nonpolymorphic markers were sequenced, one was converted to an STS marker after exhibiting enough sequence divergence to develop an S1-D2-specific primer. This demonstrates that the brute-force method can be a practical way to generate useful primer sets.
A second method for marker generation exploits the collection of S1-D2 expressed sequence tag (EST) sequences to create STS markers. Currently, 1,616 S1-D2 sequences have been deposited in the National Center for Biotechnology (NCBI) database, and additional sequencing is anticipated (see “Future Perspectives”). Because approximately 165,000 cDNA sequences from the laboratory strain are present in dbEST (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html), it is nearly always possible to align an S1-D2 EST with a corresponding laboratory strain sequence. Inspection of such alignments can directly suggest new markers based on sequence polymorphisms. If no polymorphisms exist within the aligned region, the 3′-UTRs, where more sequence divergence is inevitably found, should be inspected. This can be done by amplifying S1-D2 DNA with a primer from a conserved portion of the EST and a downstream primer generated from the C. reinhardtii sequence. Multiple downstream primers will likely need to be generated to identify one that will anneal with the S1-D2 sequence.
A third method of generating markers utilizes the ubiquitous repeat elements of the Chlamydomonas genome, which can be converted into short sequence repeat (SSR) markers to aid in fine mapping. The advantages of SSR markers are that they are available in all regions of the genome and are already annotated (repeat masker track). There are 370 SSRs on scaffold 66 alone, which on average is 1 SSR/1,443 bases of genome. Repeat motifs range from a single base repeat up to a 10-bp repeat. Primer sets encompassing the annotated SSRs can be generated from the genome sequence. These markers would require separation through a polyacrylamide gel or identification by capillary electrophoresis to distinguish differences of only one or two repeats between alleles. Once fine mapping is completed, gene isolation naturally becomes the focus. In general, complementation with BACs, plasmids, or cosmids would be used with either direct selection for phenotype or with cotransformation followed by screening. Because these strategies and their outcomes are highly case specific, they will not be discussed in detail here. An overview of the map-based cloning process from mutant to gene, as applied to Chlamydomonas, is shown in Figure 7. The ability to move quickly through this process is currently limited by the state of the nuclear genome assembly. As those resources improve, map-based cloning will progress rapidly in ease and popularity.
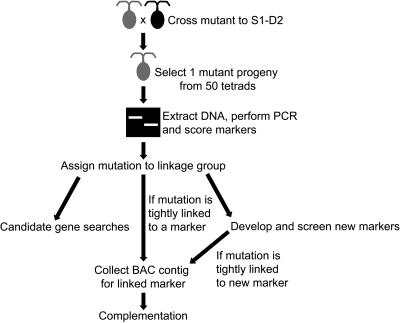
Figure 7.
Flow chart for map-based cloning in Chlamydomonas. The scheme begins with a cross between the strain carrying the mutation of interest and the appropriate mating type of S1-D2. In the second step, one clone showing the mutant phenotype is selected, although in principle the S1-D2 (nonmutant) allele could be mapped by selecting one wild-type clone per tetrad. The initial mapping phase identifies a linkage group and chromosome arm, and depending on location, phenotype, and linkage, several subsequent steps may ensue.
FUTURE PERSPECTIVES
New tools such as the updated molecular map (Fig. 1), genome sequences, and BAC contigs increase the attractiveness of Chlamydomonas. The molecular map currently contains 506 markers, far less than in Arabidopsis and rice, but new information is being generated to increase marker coverage (see below). DNA for PCR analysis is simple to generate; sufficient DNA can be prepared from cells picked up by a toothpick (see “Materials and Methods”). Once a linked marker has been identified, the genome sequence associated with that marker is accessible. Although the nuclear genome is not complete, 125 Mb has been sequenced and assembled into 3,211 scaffolds; roughly one-half of this is in 72 scaffolds of at least 504 kb in length. Genome finishing is under way, and completion in 2005 or 2006 is the current goal. Nuclear genome annotation has been performed on an automated basis and has been supplemented by manual annotation. This, combined with EST data displayed on the genome browser, allows for reasonably efficacious candidate gene searches.
Once a candidate is found, the gene sequence may need to be confirmed before complementation proceeds. This is particularly true for poorly annotated genes and/or those lacking EST support, or where sequence gaps are present. In addition, the gene prediction programs used have a significant error frequency, both in defining splice sites and in the recognition of mitochondrial and chloroplast transit peptides. Annotation has been aided by the combined EST and protein homolog data displayed on the genome browser, but only about 60% of the predicted genes have EST coverage and 42% have protein homologies (Li et al., 2003). These considerations, of course, are not unique to Chlamydomonas.
With existing tools, rough mapping in Chlamydomonas is straightforward. A kit of 67 PCR-based marker primers, allowing assignment of a mutant locus to a chromosome arm, is available to the community via the Stock Center. Using 50 progeny from a single cross and the marker kit, the entire genome can be screened at 20- to 30-cM resolution in less than 3 months. BSA and duplexing of markers can decrease the time and effort needed for mapping while reducing the cost. One can also anticipate the completion of the genome, facilitating candidate gene searches and determining which BACs should be used for complementation. Marker saturation will depend on two factors, namely additional sequence information from S1-D2 and high-throughput screening methods. Both of these are goals of the Chlamydomonas Genome Initiative in which our laboratory is participating. With respect to S1-D2 polymorphisms, immediate possibilities are to sequence BAC ends from a currently available library and to sequence much more deeply into the existing cDNA library. High-throughput methods for SNP markers are being examined, as such methods need to be adapted to the very GC-rich Chlamydomonas nuclear DNA. In the long run, this knowledge and associated technical advances will allow the mapping of interesting mutations to be a routine endeavor.
MATERIALS AND METHODS
Strains, Media, and Genetic Crosses
The Chlamydomonas reinhardtii laboratory strains mcd4 [LS2] mt+ or mcd5 [LS6] mt+ and S1-D2 mt− (CC-2290) were used as parents. Before crossing to S1-D2, backcrosses to a wild-type laboratory strain were conducted to ensure that the mcd4 and mcd5 phenotypes were caused by single, nuclear mutations. Then, mcd4 and mcd5 were crossed to S1-D2, and progeny were dissected as described previously (Levine and Ebersold, 1960). Progeny were tested for photoautotrophic growth by plating on minimal medium lacking acetate (Harris, 1989) and otherwise maintained on Tris-acetate phosphate medium (TAP; Gorman and Levine, 1965) under 23 h light and 1 h dark at 25°C. One PS+ progeny from each of 54 tetrads was used to create the mcd4 mapping population; the current mcd5 population is 38 individuals.
For crosses to phenotypic markers, mcd4 [LS2] mt+ was crossed to pf12 mt− (CC-610) or act1 mt− (CC-2953). Progeny were dissected and assayed for photosynthesis and swimming ability (pf12 cross) or cycloheximide resistance (act1 cross). For the pf12 cross, swimming ability was determined by light microscopy at 100× magnification. The act1 genotype was determined by growth on TAP medium containing 10 ng/mL cycloheximide. Genetic distance was determined as described (Harris, 1989).
DNA Preparation and PCR Conditions
Total DNA from the mapping population was prepared using a protocol adapted from Steve Pollock (http://www.biology.duke.edu/chlamy/methods/quick_pcr.html). A toothpick of cells from a TAP plate was resuspended in 50 μL of 10 mm NaEDTA in a 1.5-mL microfuge tube. The tube was vortexed and incubated at 100°C for 5 min. Then, the tube was centrifuged at 12,000 rpm for 1 min. The supernatant was retained as the DNA sample, and its concentration was measured on a spectrophotometer, then diluted to 20 ng/μL.
Markers used were reported previously or developed in the Dutcher, Mets, or Stern lab, and are listed in Supplemental Table I (Kathir et al., 2003; Torjek et al., 2003). PCR conditions were 8.5% glycerol, 0.83% formamide, 7 μL of GoTaq buffer (Promega, Madison, WI), 1 μL of each primer (10 mm stock), 0.5 μL of 10 mm dNTPs, 20 ng of Chlamydomonas DNA, and 0.25 μL of GoTaq (Promega) in a final volume of 30 μL. The PCR program began with denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 1 min, annealing for 1 min at primer-specific temperatures (see Supplemental Table I) and 72°C for 1 min. A final extension was performed at 72°C for 10 min. PCR products were analyzed in 1% or 3% agarose gels and visualized using ethidium bromide.
For BSA, the mcd5 and S1-D2 DNAs were combined so that total amount of DNA in the PCR reaction remained at 20 ng. Ratios ranged from 4:1 to 1:5 mcd5:S1-D2. PCR reactions were performed as specified above. For duplexing reactions, markers were chosen that had sizes that were distinguishable in a 3% agarose gel and had similar annealing temperatures. When the total DNA amount was not held constant, reactions containing DNA from usually >3 progeny did not yield any PCR products. Thus, the total DNA amount is a key parameter.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank Carolyn Silflow for her advice on mapping and Rachel Nguyen and Nancy Haas for BAC contig and marker information. Laurens Mets and Susan Dutcher graciously provided markers for mapping. Thanks also to the Stern lab for their help and support, especially Tom Bollenbach for valuable advice.
This work was supported by the National Science Foundation (grant nos. MCB–0235878 and MCB–0091020 to D.B.S.). M.T. was supported in part by a summer undergraduate research fellowship from the Plant Genome Research Outreach Program at Boyce Thompson Institute/Cornell University.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.054221.
References
- Auchincloss AH, Zerges W, Perron K, Girard-Bascou J, Rochaix JD (2002) Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J Cell Biol 157: 953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P, Béal D (1997) Screening algal mutant colonies with altered thylakoid electrochemical gradient through fluorescence and delayed luminescence digital imaging. Photosynth Res 51: 161–165 [Google Scholar]
- Bernd KK, Kohorn BD (1998) Tip loci: six Chlamydomonas nuclear suppressors that permit the translocation of proteins with mutant thylakoid signal sequences. Genetics 149: 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Schuster G, Stern DB (2004) Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog Nucleic Acid Res Mol Biol 78: 305–337 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix J-D (2000) The Nac2 gene of Chlamydomonas reinhardtii encodes a chloroplast TPR protein involved in psbD mRNA stability, processing and/or translation. EMBO J 19: 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S, et al (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E, Cone K, McMullen M, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H, et al (2002) Access to the maize genome: an integrated physical and genetic map. Plant Physiol 128: 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, McMullen MD, Bi IV, Davis GL, Yim YS, Gardiner JM, Polacco ML, Sanchez-Villeda H, Fang Z, Schroeder SG, et al (2002) Genetic, physical, and informatics resources for maize: on the road to an integrated map. Plant Physiol 130: 1598–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillee D, Stampacchia O, Girard-Bascou J, Rochaix JD (2003) Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J 22: 6378–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Han M, Niyogi KK (2001) Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii. Trends Plant Sci 6: 364–371 [DOI] [PubMed] [Google Scholar]
- Esposito D, Higgs DC, Drager RG, Stern DB, Girard-Bascou J (2001) A nucleus-encoded suppressor defines a new factor which can promote petD mRNA stability in the chloroplast of Chlamydomonas reinhardtii. Curr Genet 39: 40–48 [DOI] [PubMed] [Google Scholar]
- Fleming GH, Boynton JE, Gillham NW (1987) The cytoplasmic ribosomes of Chlamydomonas reinhardtii: characterization of antibiotic sensitivity and cycloheximide-resistant mutants. Mol Gen Genet 210: 419–428 [DOI] [PubMed] [Google Scholar]
- Frey E, Brokaw CJ, Omoto CK (1997) Reactivation at low ATP distinguishes among classes of paralyzed flagella mutants. Cell Motil Cytoskeleton 38: 91–99 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, et al (2002) TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics 2: 239–253 [DOI] [PubMed] [Google Scholar]
- Girard-Bascou J, Pierre Y, Drapier D (1992) A nuclear mutation affects the synthesis of the chloroplast psbA gene product in Chlamydomonas reinhardtii. Curr Genet 22: 47–52 [DOI] [PubMed] [Google Scholar]
- Goodenough UW (1992) Green yeast. Cell 70: 533–538 [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardii. Proc Natl Acad Sci USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CH, Ranum LPW, Lefebvre PA (1988) Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr Genet 13: 503–508 [DOI] [PubMed] [Google Scholar]
- Grossman AR (2000) Chlamydomonas reinhardtii and photosynthesis: genetics to genomics. Curr Opin Plant Biol 3: 132–137 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Harris E, Hauser C, Lefebvre P, Martinez D, Rokhsar D, Shrager J, Silflow C, Stern D, Vallon O, et al (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2: 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumpel NJ, Ralley L, Girard-Bascou J, Wollman FA, Nugent JH, Purton S (1995) Nuclear mutants of Chlamydomonas reinhardtii defective in the biogenesis of the cytochrome b6f complex. Plant Mol Biol 29: 921–932 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego [DOI] [PubMed]
- Higgs DC, Shapiro RS, Kindle KL, Stern DB (1999) Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol Cell Biol 19: 8479–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P, LaVoie M, Brazelton WJ, Haas NA, Lefebvre PA, Silflow CD (2003) Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell 2: 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, Ebersold WT (1960) The genetics and cytology of Chlamydomonas. Annu Rev Microbiol 14: 197–216 [DOI] [PubMed] [Google Scholar]
- Levy H, Kindle KL, Stern DB (1997) A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas chloroplasts. Plant Cell 9: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Holloway SP, Lee J, Herrin DL (2002) Nuclear genes that promote splicing of group I introns in the chloroplast 23S rRNA and psbA genes in Chlamydomonas reinhardtii. Plant J 32: 467–480 [DOI] [PubMed] [Google Scholar]
- Li JB, Lin S, Jia H, Wu H, Roe BA, Kulp D, Stormo GD, Dutcher SK (2003) Analysis of Chlamydomonas reinhardtii genome structure using large-scale sequencing of regions on linkage groups I and III. J Eukaryot Microbiol 50: 145–155 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVittie AC (1972) Flagellum mutants of Chlamydomonas reinhardtii. J Gen Microbiol 71: 525–540 [DOI] [PubMed] [Google Scholar]
- Nickelsen J (2000) Mutations at three different nuclear loci of Chlamydomonas suppress a defect in chloroplast psbD mRNA accumulation. Curr Genet 37: 136–142 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Bjorkman O, Grossman AR (1997) Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD (1995) Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet 29: 209–230 [DOI] [PubMed] [Google Scholar]
- Sager R, Tsubo Y (1961) Genetic analysis of streptomycin resistance and dependence in Chlamydomonas. Z Vererbungsl 92: 430–438 [DOI] [PubMed] [Google Scholar]
- Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C (1995) Physical map and organization of Arabidopsis thaliana chromosome 4. Science 270: 480–483 [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Torjek O, Berger D, Meyer RC, Mussig C, Schmid KJ, Rosleff Sorensen T, Weisshaar B, Mitchell-Olds T, Altmann T (2003) Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J 36: 122–140 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD (2000) Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 97: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vysotskaia VS, Curtis DE, Voinov AV, Kathir P, Silflow CD, Lefebvre PA (2001) Development and characterization of genome-wide single nucleotide polymorphism markers in the green alga Chlamydomonas reinhardtii. Plant Physiol 127: 386–389 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.