Abstract
Transgenic tomato (Solanum lycopersicum) plants expressing a fragment of the mitochondrial malate dehydrogenase gene in the antisense orientation and exhibiting reduced activity of this isoform of malate dehydrogenase show enhanced photosynthetic activity and aerial growth under atmospheric conditions (360 ppm CO2). In comparison to wild-type plants, carbon dioxide assimilation rates and total plant dry matter were up to 11% and 19% enhanced in the transgenics, when assessed on a whole-plant basis. Accumulation of carbohydrates and redox-related compounds such as ascorbate was also markedly elevated in the transgenics. Also increased in the transgenic plants was the capacity to use l-galactono-lactone, the terminal precursor of ascorbate biosynthesis, as a respiratory substrate. Experiments in which ascorbate was fed to isolated leaf discs also resulted in increased rates of photosynthesis providing strong indication for an ascorbate-mediated link between the energy-generating processes of respiration and photosynthesis. This report thus shows that the repression of this mitochondrially localized enzyme improves both carbon assimilation and aerial growth in a crop species.
Improving plant productivity is now of unprecedented importance given the twin problems of environmental deterioration and the world population explosion (Miyagawa et al., 2001). One of the major determinants of crop growth and yield is believed to be photosynthetic carbon metabolism (Galtier et al., 1993; Sweetlove et al., 1998; Miyagawa et al., 2001); however, to date very few molecular approaches aimed at improving the efficiency of these pathways have proven successful. Those strategies that did work generally involved expression of cyanobacterial enzymes in higher plants. The expression of Fru-1,6/sedoheptulose 1,7-bisphosphatase (Miyagawa et al., 2001) in tomato (Solanum lycopersicum); ictB, a gene involved in HCO3− accumulation; tobacco (Nicotiana tabacum); and Arabidopsis (Arabidopsis thaliana; Lieman-Hurwitz et al., 2003) resulted in increased photosynthetic rates and growth under normal and limiting carbon dioxide concentrations, respectively. A recent report showed that overexpression of the gene otsA from Escherichia coli, encoding trehalose phosphate synthase, resulted in elevated rates of photosynthesis and growth in tobacco leaves (Pellny et al., 2004). Interestingly, however, none of the successful strategies described above has been the result of alterations in the levels of the endogenous plant proteins.
As part of an ongoing project to determine the function of the TCA cycle in the illuminated leaf, we previously comprehensively phenotyped the tomato wild species (Solanum pennellii) mutant Aco1, which exhibits a deficiency in expression of one of the two isoforms of aconitase present in the tomato (Tanksley et al., 1992; Carrari et al., 2003). Biochemical analyses of the Aco1 mutant revealed that it exhibited a decreased flux through the TCA cycle and decreased levels of TCA cycle intermediates but was characterized by elevated adenylate levels and an increased rate of carbon dioxide assimilation. In addition, although it must be taken into account that S. pennellii is a green-fruited species bearing very small fruits (Schauer et al., 2005), these plants were characterized by a dramatically increased fruit weight. These data are in close agreement with those from other researchers (Raghavendra and Padmasree, 2003) in hinting that the importance of the respiratory pathways in photosynthetic metabolism is greater than once imagined.
In this study we turn our attention to the analysis of the importance of the mitochondrial malate dehydrogenase (mMDH) for photosynthetic metabolism. The enzyme MDH catalyses the reversible reduction of oxaloacetate to malate and is important in multiple metabolic pathways. Higher plants contain multiple forms of MDH that differ in coenzyme specificity and subcellular localization (Gietl et al., 1996; Miller et al., 1998). Chloroplasts contain an NADP-dependent MDH that plays an important role in balancing redox equivalents between the cytosol and the stroma. Plants also contain NAD-dependent MDHs, which are found in the mitochondria as a component of the Krebs cycle, in the cytosol and peroxisome where they function in malate-Asp shuttles and glyoxysomes wherein they function in β-oxidation (Gietl et al., 1996; Miller et al., 1998). The mMDH is believed to be important not only in oxidizing NADH but also as a component of the malate/Asp shuttle for exchange of substrate and reducing equivalent across the mitochondrial membrane (Gietl, 1992; Scheibe, 2004). This exchange is of particular importance during photorespiration wherein it is responsible for the export of malate from the mitochondria. Although a range of biochemical and physiological analyses has illustrated the importance of this enzyme in photosynthetic metabolism, these hypotheses have yet to be tested in a molecular manner. Here we describe the generation and characterization of transgenic plants exhibiting decreased expression of the mMDH. The results obtained are discussed in the context of the regulation of energy metabolism in the illuminated leaf.
RESULTS
Cloning of a cDNA-Encoding mMDH from Tomato
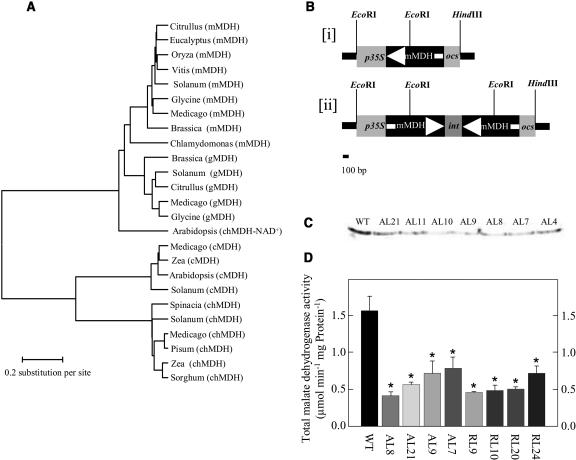
Searching tomato expressed sequence tag collections (Van der Hoeven et al., 2003) facilitated the isolation of the full-length 1,041-bp mMDH clone cLEI112D19 (subsequently named SlmMDH; GenBank accesssion no. AY725474). Sequence analysis of the tomato MDH revealed an open reading frame of 346 amino acids. Comparison with functionally characterized MDHs revealed relatively high identity with the mitochondrial isoforms and a much lower homology with plastidial, cytosolic, and glycoxysomal isoforms (Fig. 1A). SlmMDH bears characteristics of a mitochondrial transit peptide sequence, indicating a mitochondrial location for the protein encoded by the SlmMDH cDNA. These in silico predictions were further corroborated by promoter-green fluorescent protein fusion studies (C. Studart-Rodriguez, F. Carrari, and A.R. Fernie, unpublished data). Analysis of mRNA northern blots using the SlmMDH cDNA as a probe indicates constitutive expression of the gene, with the transcript present at approximately equivalent levels in young and mature leaves, stems, roots, and fruits. In addition, the transcript is ubiquitous during fruit development (data not shown).
Figure 1.
Characterization and expression of tomato mMDH. A, Dendrogram of plant MDH sequences. MDH sequences were aligned using the ClustalW alignment program (Higgins and Sharp, 1988). Organellar targeting sequences were removed from the aligned sequences. The taxonomic names of the species and the accession numbers of the sequences used in the phylogenetic analysis are: Solanum (S. lycopersicum), mMDH, AY725474; Glycine (G. max), mMDH, AF068687; Chlamydomonas (C. reinhardtii), mMDH, AAA84971.1; Medicago (M. sativa), mMDH, AF020271; Brassica (B. napus), mMDH, X89451; Vitis (V. vinifera), mMDH, AF195869; Citrullus (C. lanatus), mMDH, P17783; Eucalyptus (E. gunnii), mMDH, X78800; Oryza (O. sativa) mMDH, AF444195; Solanum (S. lycopersicum), gMDH, AY725476; Brassica (B. napus), gMDH, 1050435; Medicago (M. sativa) gMDH, AF020270; Glycine (G. max), gMDH, P37228; Citrullus (C. lanatus), gMDH, P19446; Arabidopsis (Arabidopsis thaliana), chMDH-NAD dependent, Y13987; Solanum (S. lycopersicum), chMDH, AY725477; Zea (Z. mays), chMDH, X16084; Pisum (P. sativum), chMDH, X74507; Medicago (M. sativa), chMDH, AF020269; Sorghum (S. bicolor), chMDH, 100755; Spinacia (S. oleracea), chMDH, X84020; Solanum (S. lycopersicum), cMDH, AY725475; Medicago (M. sativa), cMDH, AF020272; Zea (Z. mays), cMDH, 2286153; Arabidopsis, cMDH, 2341034. B, Construction of a chimeric gene for the expression of tomato mMDH antisense RNA (subsection i) or RNA interference (RNAi; subsection ii) consisting of a 514-bp fragment encoding the CaMV 35S promoter and a 961-bp (antisense) or two 961-bp tandem fragments separated by a stem loop (RNAi; ocs terminator). C, Western-blot analysis of leaves of transgenic plants with altered expression of mMDH as compared to wild type (WT). D, Total MDH activity determined in 6-week-old leaves taken from fully expanded source leaves of transgenic plants with altered expression of mMDH as compared to wild type.
Transgenic Plants Show an Elevated Growth Rate But Not an Accelerated Developmental Phenotype
A 996-bp fragment of the cDNA-encoding mMDH was cloned either in the antisense orientation into the transformation vector pBINAR between the cauliflower mosaic virus (CaMV) promoter and the ocs terminator (Fig. 1B, subsection i) or using the RNAi design (Fig. 1B, subsection ii). We then transferred 60 transgenic tomato plants obtained by Agrobacterium tumefaciens-mediated transformation to the greenhouse.
Screening of the lines for a reduction of total cellular MDH activity yielded eight lines that exhibited a significant reduction in activity as determined by activity gel assays (data not shown). To demonstrate that this reduction was a consequence of a specific reduction in the mitochondrial isoform we carried out northern (data not shown) and western (Fig. 1C) analyses in addition to direct assay of total MDH activity (Fig. 1D). These preliminary studies suggested that the antisense, but not the RNAi, transformants were suitable for further study since in these lines the reduction in MDH expression was confined to the mitochondrial isoform. Subcellular fractionation studies on samples from a second harvest (Table I), provided further evidence, in addition to that provided by zymogram and northern analysis, that the reduction in activity was confined to the mitochondria. It should be noted that, although the absolute values for total MDH activity are variable between the harvests, the relative degree of inhibition in the antisense lines is conserved between the harvests.
Table I.
Enzyme activities in antisense mMDH transgenic lines
Activities were determined in 6-week-old fully expanded source leaves. Data presented are mean ± se of determinations on six individual plants per line. MDH activities determined in different fractions of Percoll gradients. Data represent the mean of measurements from different fractions (subcellular fractionation was carried out on only four plants per line, samples were pooled to ensure high mitochondrial yield, activities were calculated with respect to the level of contamination as assessed by marker enzyme assays). Values in bold type were determined by the t test to be significantly different (P < 0.05) from the wild type. Dashes represent not measured.
| Enzyme Activity
|
|||||
|---|---|---|---|---|---|
| Wild Type | AL7 | AL8 | AL9 | AL21 | |
| nmol mg protein−1 min−1 | |||||
| Total MDH | 3,129 ± 388 | 1,697 ± 292 | 2,054 ± 104 | 1,465 ± 332 | 2,367 ± 55 |
| mMDH | 1,329 | – | 647 | – | 699 |
| Rubisco (initial) | 43.9 ± 8.0 | 73.9 ± 13.0 | 46.0 ± 7.9 | 40.47 ± 7.1 | 57.2 ± 19.3 |
| Rubisco (total) | 50.3 ± 6.2 | 83.0 ± 10.8 | 56.1 ± 9.4 | 65.7 ± 30.1 | 79.3 ± 26.0 |
| UGPasea | 109 ± 24 | 91.1 ± 9.1 | 84.6 ± 12.2 | 157 ± 36 | 108 ± 19.6 |
| PFK | 155 ± 28 | 313 ± 122 | 150 ± 17.6 | 400 ± 98 | 286 ± 29 |
| Pyruvate kinase | 106 ± 43 | 122 ± 19.7 | 86.7 ± 21.0 | 143 ± 35 | 105 ± 21 |
| Phosphoglucomutase | 57.1 ± 19.9 | 82.3 ± 18.0 | 64.1 ± 8.7 | 79.0 ± 22 | 75 ± 18 |
| Transaldolase | 1.0 ± 0.2 | 1.0 ± 0.3 | 1.6 ± 0.1 | 0.9 ± 0.3 | 2.3 ± 0.7 |
| Phosphoglycerate kinase | 169 ± 21 | 251 ± 37 | 177 ± 18 | 216 ± 60 | 200 ± 31 |
| NADP-GAPDHb | 5.5 ± 1.7 | 28.4 ± 6.0 | 10.1 ± 3.4 | 19.5 ± 6.2 | 8.4 ± 2.3 |
| GLDH | 107 ± 46 | 49 ± 10 | 80 ± 19 | 191 ± 47 | 114 ± 44 |
UGPase, UDPGlc pyrophosphorylase.
NADP-GAPDH, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase.
Analysis of the maximal catalytic activities of other important enzymes of photosynthetic carbohydrate metabolism revealed no changes in important enzymes of photosynthesis (Rubisco and Calvin cycle enzymes) but a mild increase in the glycolytic enzyme phosphofructokinase (PFK). In addition, no significant differences were observed in the maximal catalytic activity of l-galactono-lactone dehydrogenase (GLDH), the terminal step of ascorbic acid biosynthesis in plants (Wheeler et al., 1998); however, this activity was highly variable both across and within genotypes.
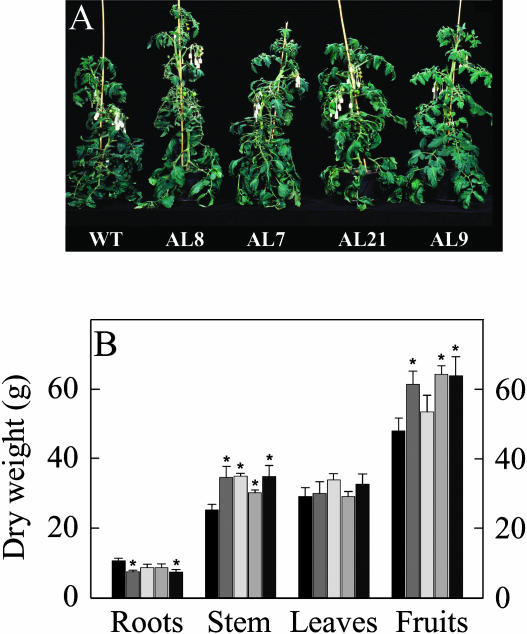
When we grew the transgenic plants in the greenhouse side by side with wild-type controls, a clear increase in the growth of the aerial parts of the transformants was observed (Fig. 2A). Close examination of the transgenic plants revealed that they were significantly taller and appeared to have marginally bigger fruit; however, there were no difference in leaf formation or onset of senescence. These visual observations were supported by experimental measurements (Fig. 2B). Furthermore, there was a tendency of earlier flowering in the transformants (significant in the cases of lines AL7 and AL8). While the total leaf mass did not vary across the genotypes, analysis of root biomass revealed that this was somewhat restricted in the transformants (Fig. 2B). When taken together these findings imply an improved harvest index. Assessment of pigment content of leaves revealed that the photosynthetic pigment content was largely unaltered with the exception of small decreases in violaxanthin and significant increases in the level of antheraxanthin (data not shown).
Figure 2.
Growth phenotype of antisense mMDH tomato plants. Transgenic plants showed an enhanced aerial biomass with respect to the wild type. A, Photograph showing representative plants after 7 weeks growth. B, Biomass (in grams dry weight) of various plant organs on plant maturity. Wild type, black bar; AL7, very dark gray bar; AL8, dark gray bar; AL9, light gray bar; and AL21, very light gray bar.
Reduction in mMDH Activity Results in an Increased Chloroplastic-Electron Transport Rate and Enhanced Photosynthesis
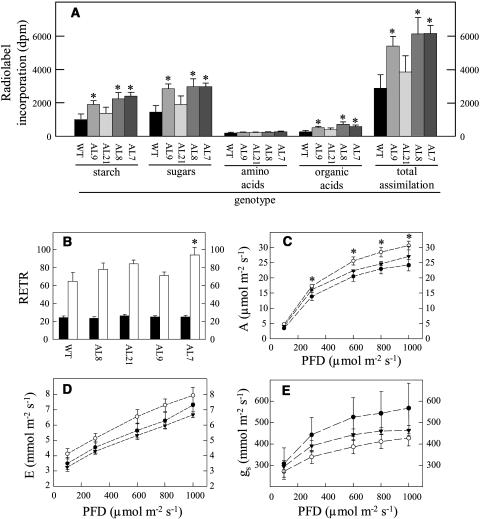
Given the increase in aerial yield in the transformants, we next analyzed whether they exhibited altered photosynthetic rates. Firstly, we studied the metabolism of 14CO2 by leaf discs excised from the wild-type and transformant plants. Notably the assimilation rate was markedly increased in plants exhibiting inhibition of MDH (Fig. 3A). Furthermore, this increased carbon fixation was coupled with an increase in the accumulation of [14C]Suc, [14C]-starch, and [14C]-organic acids. When these data are expressed as a percentage of 14CO2 metabolised, decreased mMDH activity led to a decreased partitioning into organic and amino acids, implying a reduced respiration rate in these plants.
Figure 3.
Effect of decreased mMDH activity on photosynthesis. A, Photosynthetic assimilation and carbon partitioning at the onset of illumination. Leaf discs were cut from six separate untransformed potato plants and four independent transgenic lines at the end of the night and illuminated at 250 μmol photosynthetically active radiation m−2 s−1 in an oxygen electrode chamber containing air saturated with 14CO2. After 30 min the leaf discs were extracted and fractionated. B, In vivo fluorescence emission was measured as an indicator of the ETR by use of a PAM fluorometer at PFDs of 250 (black bars) and 700 μmol m−2 s−1 (white bars). C, Assimilation rate. D, Transpiration rate. E, Stomatal conductance. The lines used were: wild type, black circles; AL21, white circles; and AL7, black triangles. Values are presented as mean ± se of determinations on six individual plants per line; an asterisk indicates values that were determined by the t test to be significantly different (P < 0.05) from the wild type.
As a second experiment, fluorescence emission was measured in vivo using a pulse amplitude modulation (PAM) fluorimeter in order to calculate relative electron transport rates (ETRs). When exposed to higher irradiance (photon flux density [PFD] of 700 μmol m−2 s−1) the mMDH plants exhibited elevated ETRs (significantly so in the case of line AL7; Fig. 3B). Finally, gas exchange was measured directly in a subset of the transformants under PFDs that ranged from 100 to 1,000 μmol m−2 s−1 (Fig. 3, C–E). The transformants exhibited assimilation rates that were higher than the wild type under all conditions except the lowest irradiance (significantly so in the case of AL21; Fig. 3C). Similar results were obtained from a second harvest. Interestingly, when data from both harvests are plotted together a clear negative correlation becomes apparent between the activity of MDH and the rate of photosynthesis (data not shown). Analysis of other parameters of gas exchange revealed that the transformants also exhibited minor changes in transpiration rate at all PFD (Fig. 3D) and that the stomatal conductance is noticeably (yet not significantly) lower in the transgenics. The lower stomatal conductance, paradoxically, suggests increased resistance to carbon dioxide uptake (Fig. 3E) and, in keeping with this, the internal carbon dioxide concentration inside the stomatal cavity was estimated to be lower (data not shown).
Photosynthetic Carbon Metabolism in the mMDH Transformants
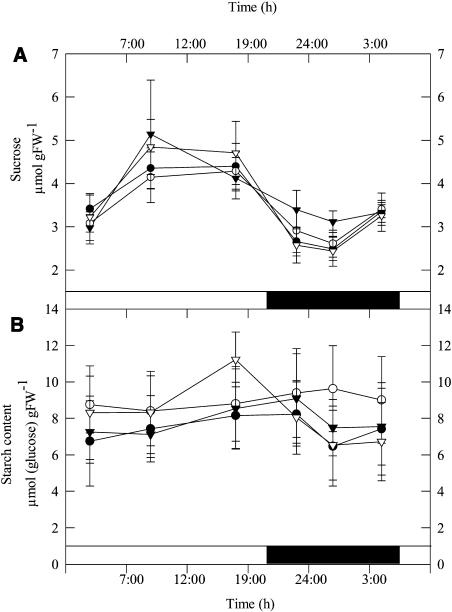
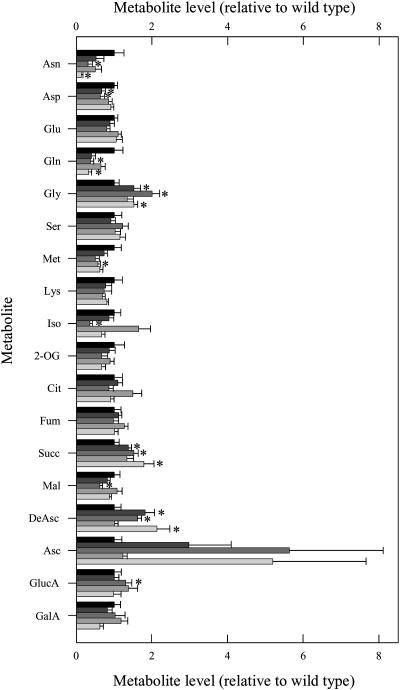
Analysis of the carbohydrate content of leaves from 6-week-old plants during a diurnal cycle revealed that the transformants were characterized by small, but nonsignificant, increases in Suc and starch contents (Fig. 4) and significant increases in Glc and Fru (data not shown). In all cases metabolite contents were in a similar range to those reported previously for tomato (e.g., Galtier et al., 1993). We next decided to extend this study to major primary pathways of plant photosynthetic metabolism by utilizing an established gas chromatography-mass spectrometry (GC-MS) protocol for metabolic profiling (Fernie et al., 2004a). These studies revealed a decrease in levels of Asn and Gln (despite no consistent change in the levels of Asp or Glu) alongside a decrease in Met content in the transgenic lines (Fig. 5). In addition, trends of increased Gly and succinate content were observable in the transformants; however, the levels of the other TCA cycle intermediates were largely unaffected. When other organic acids were analyzed, the cell wall constituents galacturonic and GlcUA showed no consistent pattern of change, but ascorbate and dehydroascorbate increased by up to 5.7-fold in the transgenic lines (significantly in the case of dehydroascorbate but notably not in line AL9).
Figure 4.
Diurnal changes in Suc (A) and starch (B) content in leaves of 7-week-old transgenic and wild-type tomato plants. At each time point, samples were taken from mature source leaves, and the data presented are mean ± se of determinations on six individual plants per line. The lines used were: wild type, black circles; AL7, white circles; AL8, black triangles; and AL21, white triangles. Black bars indicate the dark period; white bars indicate the light period.
Figure 5.
Relative metabolite content of fully expanded leaves from 8-week-old plants of the antisense mMDH lines. Metabolites were determined as described in “Materials and Methods.” The full data set from these metabolic profiling studies can be accessed at our Web site: www.mpimp-golm.mpg.de/fernie. Data are normalized with respect to the mean response calculated for the wild type (to allow statistical assessment, individual samples from this set of plants were normalized in the same way). Values are presented as mean ± se of determinations on six individual plants per line; asterisk indicates values that were determined by the t test to be significantly different (P < 0.05) from the wild type. Wild type, black bar; AL7, very dark gray bar; AL8, dark gray bar; AL9, light gray bar; and AL21, very light gray bar.
Since this method does not readily allow the determination of phosphorylated intermediates, which are important diagnostic markers for alterations in photosynthesis (Stitt, 1997) we assayed these by means of cycling assays, regular linear spectrophotometric assays, and HPLC methods (Table II). There were no significant differences in either the individual pools or the total phosphoester levels in the transformants, and furthermore, little change in the levels of nucleotides. The one exception to this statement is the fact that line AL9 displayed a decreased level of UTP and consequently a decreased level of total uridinylates. Interestingly, there was, however, a tendential decrease in the level of inorganic phosphate in the transgenics (significant in lines AL8 and AL9) and a consequent increase in the deduced phosphoester to inorganic phosphate ratio.
Table II.
Phosphorylated intermediates in antisense mMDH transgenic lines
Metabolite levels were determined in 6-week-old fully expanded source leaves. Samples used were harvested at exactly the same time as those for enzyme determinations presented in Table I. Data presented are mean ± se of determinations on six individual plants per line. Values in bold type were determined by the t test to be significantly different (P < 0.05) from the wild type.
| Metabolite Level
|
|||||
|---|---|---|---|---|---|
| Wild Type | AL7 | AL8 | AL9 | AL21 | |
| nmol gFW−1 | |||||
| Glc-6-P | 94 ± 11 | 101 ± 16 | 102 ± 13 | 95 ± 18 | 120 ± 23 |
| Fru-6-P | 28 ± 4 | 23 ± 4 | 29 ± 2 | 25 ± 1 | 32 ± 4 |
| Glc-1-P | 17 ± 2 | 18 ± 2 | 16 ± 1 | 18 ± 1 | 21 ± 2 |
| 3-Phosphoglycerate | 220 ± 28 | 249 ± 37 | 281 ± 22 | 222 ± 17 | 276 ± 36 |
| Inorganic phosphate | 3.7 ± 0.8 | 3.1 ± 0.9 | 1.4 ± 0.2 | 1.8 ± 0.2 | 2.3 ± 0.6 |
| ATP | 36 ± 5 | 29 ± 8 | 30 ± 6 | 32 ± 5 | 31 ± 6 |
| ADP | 13 ± 2 | 9 ± 2 | 10 ± 3 | 13 ± 3 | 12 ± 2 |
| UTP | 13 ± 1 | 9 ± 3 | 7 ± 2 | 8 ± 1 | 8 ± 2 |
| UDP | 10 ± 1 | 8 ± 1 | 7 ± 2 | 9 ± 2 | 7 ± 1 |
| UDPGlc | 82 ± 7 | 82 ± 10 | 67 ± 6 | 69 ± 4 | 75 ± 7 |
| Σ hexose phosphates | 139 ± 14 | 143 ± 16 | 147 ± 14 | 119 ± 7 | 173 ± 25 |
| Σ phosphoesters | 359 ± 41 | 391 ± 49 | 418 ± 23 | 339 ± 27 | 449 ± 52 |
| Σ adenylates | 49 ± 7 | 38 ± 10 | 40 ± 9 | 45 ± 7 | 43 ± 8 |
| Σ uridinylates | 105 ± 7 | 99 ± 13 | 82 ± 10 | 85 ± 3 | 89 ± 8 |
| ATP/ADP | 3.0 ± 0.2 | 3.6 ± 0.9 | 3.6 ± 0.5 | 2.7 ± 0.3 | 2.4 ± 0.3 |
| UTP/UDP | 1.3 ± 0.2 | 1.1 ± 0.2 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.2 |
| Phosphoester/Pi | 121 ± 27 | 251 ± 121 | 375 ± 73 | 212 ± 13 | 328 ± 132 |
Inhibition in mMDH Activity Results in Reduced Rates of Respiration
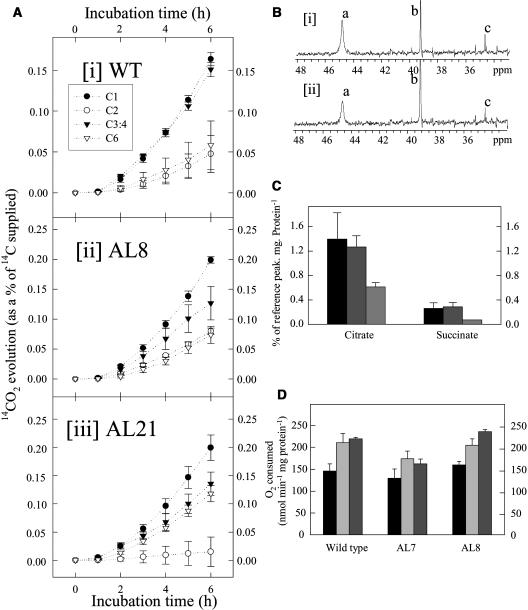
While some of the results presented above suggest a decreased rate of respiration, they do not provide direct evidence in support of this. We therefore next directly evaluated the rate of respiration in the transformants. For this purpose we adopted two strategies. First, we recorded the evolution of 14CO2 following incubation of leaf discs in positionally labeled 14C-Glc molecules, and second, we used a combination of conventional respiratory and NMR studies of isolated mitochondria to assess flux through the TCA cycle. In the first experiment, we incubated leaf discs taken from plants in the light and supplied these with [1-14C], [2-14C], [3:4-14C], or [6-14C]Glc over a period of 6 h. During this time, we collected the 14CO2 evolved at hourly intervals. Carbon dioxide can be released from the C1 position by the action of enzymes that are not associated with mitochondrial respiration, but carbon dioxide evolution from the C3:4 positions of Glc cannot (ap Rees and Beevers, 1960). Thus, the ratio of carbon dioxide evolution from C1 to C3:4 positions of Glc provides an indication of the relative rate of the TCA cycle with respect to other processes of carbohydrate oxidation. When the relative 14CO2 release of the transgenic and wild-type lines is compared for the various fed substrates, an interesting pattern emerges (Fig. 6A). The rate of 14CO2 evolution is always highest in leaves incubated in [1-14C]Glc; however, the absolute rate of carbon dioxide evolution from the C1 position of the transgenic lines is far in excess of that observed in the wild type. In contrast, the release from the C3:4 positions is much lower in the transgenic lines (both in relative and absolute terms) than in the wild type. Thus, these data reveal that a lower proportion of carbohydrate oxidation is carried out by the TCA cycle in the transgenic lines.
Figure 6.
Respiratory parameters in leaves and isolated fruit mitochondria of the antisense mMDH lines. A, 14CO2 evolution from isolated leaf discs in the light. Leaf discs were taken from 10-week-old-plants and were incubated in 10 mm MES-KOH, pH 6.5, supplemented with [1-14C]-, [2-14C]-, [3:4-14C]-, or [6-14C]Glc (at an activity concentration of 2.32 KBq mL−1 and specific activity of 58, 55, 54, and 56 mCi mmol−1, respectively). The 14CO2 liberated was captured (in hourly intervals) in a KOH trap and the amount of radiolabel released was subsequently quantified by liquid scintillation counting. Values are presented as mean ± se of determinations on six individual plants per line. Note the scale on the y axis is different in the plots of the transgenic and wild-type data. B, Proton-decoupled 13C NMR spectra of tomato mitochondria isolated from (subsection i) wild type and (subsection ii) line 8 MDH antisense breaker fruits. The spectra were recorded over a 4-h incubation period under state III respiration in the presence of 10 mm [3- 13C]pyruvate, 2.1 mm citrate, 0.02 mm isocitrate, 1.3 mm succinate, 0.02 mm fumarate, and 0.6 mm malate. a, Citrate; b, DMSO; c, succinate. C, Average amount of [4-13C]citrate and [2-13C]succinate production in the first hour by wild type (black bar), line 7 MDH antisense (light gray bar), and line 8 MDH antisense (dark gray bar) mitochondria as measured by 13C NMR (n = 4 except line 8 where n = 2 for citrate measurements and n = 1 for the succinate measurement). D, The rate of oxygen consumption of isolated tomato mitochondria in the presence of 10 mm pyruvate, 10 mm malate, 0.1 mm TPP, and 1 mm ADP (black bar); 1 mm NADH (light gray bar); or 1 mm NADH and 2 mm l-galactono-lactone (dark gray bar). Values are presented as mean ± se of determinations on four individual plants per line; an asterisk indicates values that were determined by the t test to be significantly different (P < 0.05) from NADH alone.
Carbon dioxide evolution from the C2 and C6 positions was relatively similar across the genotypes with the exception that evolution from the C6 position was somewhat higher, in both lines tested, and that from the C2 position was lower, in line AL21, than that observed in wild type.
Isolation of suitable quantities of leaf mitochondria for respiratory measurements was not feasible, so for the second experiment we isolated mitochondria from breaker fruits obtained from the wild type and two transgenic lines (AL7 and AL8). The mitochondria were incubated with [3-13C]pyruvate under simulated cytosolic conditions (described in detail in Smith et al., 2004), and representative 13C NMR spectra from line AL8 and the wild type can be seen in Figure 6B. The citrate and succinate signal intensities are lower in the spectra from the transgenic line, and a quantitative analysis confirms that there was a significant decrease in the [4-13C]citrate signal in line 8 (Fig. 6C). Surprisingly, the rate of oxygen consumption in the presence of malate and pyruvate showed no difference between the wild type and transgenic lines (Fig. 6D). However, the rate of oxygen consumption in the presence of l-galactono-lactone and NADH was significantly increased in line AL8 (Fig. 4D). The lack of change in oxygen consumption could, however, potentially be explained by the fact that the inhibition of mitochondrial activity was not as dramatic as that observed in the leaf (data not shown).
The Assimilation Rate Is Elevated following the Incubation of Wild-Type Leaf Discs on High Concentrations of Ascorbate
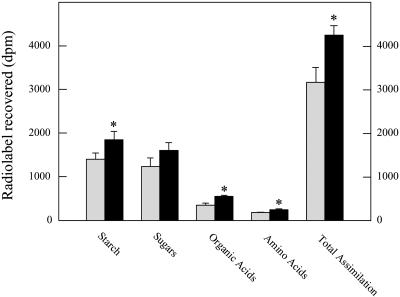
To summarize, the transgenic lines are characterized by a decreased rate of flux through the TCA cycle, a decreased rate of respiration, an increase in the cellular level of ascorbate, and an enhanced photosynthetic rate. To ascertain if the level of ascorbate has any functional significance with respect to the rate of respiration, we next incubated leaf discs isolated from wild-type plants in 50 mm ascorbate in the dark for a period of 2 h before illuminating them and measuring the assimilation of 14CO2 (Fig. 7). We chose this concentration of ascorbate since it is similar to that estimated to be present in the chloroplast (Smirnoff, 2000). Intriguingly, ascorbate feeding led to an increase in assimilation of approximately 34% (when this experiment was repeated with a range of concentrations, a similar result was observed; however, there was no enhanced assimilation rate at concentrations lower than 50 mm). This elevated assimilation rate was coupled to increased incorporation into starch, organic acids, and amino acids with the increase most prominent in the case of organic acids.
Figure 7.
Effect of ascorbate loading on photosynthesis. Photosynthetic assimilation and carbon partitioning at the onset of illumination. Leaf discs were cut from six separate untransformed tomato plants at the end of the night and incubated in the dark for 2 h in incubation medium (10 mm MOPS buffer, pH 6.0) prior to illumination at 250 μmol photosynthetically active radiation m−2 s−1 in an oxygen electrode chamber containing air saturated with 14CO2. After 30 min, the leaf discs were extracted and fractionated. Gray bars indicate leaf discs incubated in the absence and black bars in the presence of 50 mm ascorbate; an asterisk indicates values that were determined by the t test to be significantly different (P < 0.05) between experimental conditions.
DISCUSSION
The ability of plants to convert solar energy into biomass is of immense importance since the very survival of both photoauxotrophic and nonphotoautotrophic organisms depends on this process. Although the process of domestication of crop species has lead to huge increases in productivity, this has taken many generations and has come at the cost of narrowing the base of genetic diversity (Tanksley and McCouch, 1997; Zamir, 2001) and has generally occurred without an improvement in leaf photosynthesis (Richards, 2000). Recently, a number of biotechnology-based approaches utilizing transgenesis have been attempted. However, it is important to note that the majority of reports claiming enhanced photosynthetic rates are very much conditional; for example, the overexpression of maize (Zea mays) sucrose-phosphate synthase in tomato resulted in an increase rate of photosynthesis under saturating carbon dioxide and light but not in air (Galtier et al., 1993), whereas in contrast, plants expressing the cyanobacterial ictB gene displayed higher rates of photosynthesis only under limiting conditions (Lieman-Hurwitz et al., 2003). The mMDH antisense plants generated here were similarly characterized by elevated rates of photosynthesis over a range of light intensities; however, it seems most unlikely that a conserved mechanism underlies these observations. In both previous studies, either the initial or the total Rubisco activity was elevated in lines exhibiting enhanced assimilation, but this was not the case for the mMDH antisense plants. The reason for the increased assimilation in the AL lines is also not apparent when looking at metabolite levels that are usually considered to be diagnostic for the regulation of photosynthesis (Stitt, 1997), since, for example, the phosphate levels are repressed, which would be expected, paradoxically, to limit photosynthesis. However, the fact that the levels of carbohydrates do not accumulate dramatically in the AL lines, although they exhibit elevated shoot and fruit growth, may facilitate the high rates of photosynthesis observed.
Thus far we have discussed the elevated photosynthetic rates only in terms of the classically considered mechanisms of photosynthetic regulation rather than concentrating on the primary metabolic lesion that these plants display. That the TCA cycle could have a direct role in the regulation of the rate of photosynthesis is in keeping with current reevaluation of the role of the TCA cycle in the illuminated leaf (Ragahvendra and Padmasree, 2003; Fernie et al., 2004b). In this study, we utilized a novel method to demonstrate that flux through the Krebs cycle was compromised in the transformants. The transgenic tomato lines generated here display a similar phenotype to that which we previously characterized in the Aco1 mutant of the wild tomato species (Carrari et al., 2003) in that they exhibit a higher rate of photosynthesis and an enhanced fruit yield but depressed root growth. However, unlike the Aco1 mutant, the mMDH antisense plants were not characterized by dramatic elevations in leaf carbohydrate or adenylate content. These data do not contradict our previous postulate that the increase in photosynthesis is a compensation for the reduction in energy production by respiration; however, they do not support the hypothesis that this reflected a shift in cellular carbon balance. These results are somewhat at odds with the conventional understanding of the operation of the mMDH in the light, which has previously been suggested to be in the direction of malate production to supply malate required for important cytosolic and glyoxysomal reactions that are integral to photosynthesis (Fernie et al., 2004b). Data presented here show that if the mMDH does contribute to malate production, its contribution is not essential. These data thus suggest that it would be very interesting to test the importance for malate production in the light of the other, variously localized, isoforms of MDH. Although it remains possible that the mMDH operates in the direction of malate production in the light, the cumulative data presented here suggest that, at least in the tomato leaf, its operation in the reverse direction is more likely.
Surveying the levels of a wide range of primary metabolites suggested that these transgenic plants exhibit reduced ammonium assimilation and an increase in photorespiration. They also revealed a somewhat surprising trend in increasing succinate with decreasing mMDH activity; however, no consistent changes were observed in the levels of other Krebs cycle intermediates. Given that the evaluation of subcellular metabolite concentrations is not yet of high enough resolution to determine intracellular organic acids, the exact reason for this pattern of change remains unclear. By far the most striking feature of the metabolite data was, however, the dramatic increase in ascorbate observed in the transgenics. Intriguingly, reanalysis of GC-MS spectra revealed that the Aco1 mutant also displayed a dramatically elevated ascorbate level (F. Carrari and A.R. Fernie, unpublished data). That ascorbate levels increase following a restriction of flux through the Krebs cycle is fascinating, especially in light of recent reports suggesting that the terminal enzyme of ascorbate biosynthesis GLDH is coupled to the cytochrome pathway (Bartoli et al., 2000; Millar et al., 2003). Despite this fact that we could not determine a difference in the maximal catalytic activity of this enzyme in the transgenics, our results obtained in isolated fruit mitochondria from these lines are consistent with an elevated capacity of the transgenics for the synthesis of ascorbate from l-galactono-lactone (at least in the case of line AL8).
The increase in ascorbate in the transformants, while interesting in its own right, may additionally be pivotal in the elevation of photosynthetic rate observed in these plants. Much is known concerning the importance of ascorbate within photosynthesis. Ascorbate acts in the Mehler peroxidase reaction with ascorbate peroxidase to regulate the redox state of photosynthetic electron carriers and as a cofactor for violaxanthin deepoxidase, an enzyme involved in xanthophyll cycle-mediated photoprotection (Smirnoff and Wheeler, 2000; Danna et al., 2003). The ascorbate-redox state has also recently been shown to control guard cell signaling and stomatal movement (Chen and Gallie, 2004) as well as the expression levels of both nuclear and chloroplastic components of the photosynthetic apparatus (Smirnoff, 2000; Kiddle et al., 2003; Pastori et al., 2003). An additional function of ascorbate is that of cofactor for prolyl hydroxylase, an enzyme that posttranslationally hydroxylates Pro residues in cell wall Hyp-rich glycoproteins required for cell division and expansion (Smirnoff and Wheeler, 2000). Thus, when these functions are considered together with the phenotype described for the antisense mMDH plants, it is apparent that ascorbate represents a strong link between the major pathways of energy metabolism within the illuminated leaf. While the exact mechanism by which this is achieved is unclear from this study, the fact that the increased photosynthetic rate can be phenocopied by incubation of leaf discs in ascorbate gives support to our view that it is ascorbate dependent. Moreover, this experiment demonstrates that the change in photosynthesis can occur within a short period of time, suggesting that changes in gene expression are not a prerequisite for ascorbate-mediated elevation of photosynthesis. Alongside the recent demonstration that several enzymes of the Krebs cycle are redox regulated (Balmer et al., 2004), this finding makes it tempting to speculate that the pathways of energy metabolism are tightly regulated by redox factors including the cellular levels of ascorbate. It is interesting to note that the increase in ascorbate recorded here is of a similar magnitude to those previously achieved on the overexpression of enzymes directly involved in ascorbate biosynthesis (Jain and Nessler, 2000; Agius et al., 2003). Thus, this modification could prove to be an alternative to the costly chemical production of ascorbate, which is the single most important specialty chemical manufactured in the world (Hancock and Viola, 2002), in addition to a novel way to improve photosynthesis and growth in crop species.
MATERIALS AND METHODS
Materials
Tomato (Solanum lycopersicum) L. cv Moneymaker was obtained from Meyer Beck (Berlin). Plants were handled as described in the literature (Carrari et al., 2003). All chemicals and enzymes used in this study were obtained from Roche Diagnostics (Mannheim, Germany), with the exception of radiolabeled sodium bicarbonate and d-[1-14C]-, d-[3:4-14C]-, and d-[6-14C]Glc, which were from Amersham International (Braunschweig, Germany); d-[2-14C]Glc was from American Radiolabeled Chemicals (St. Louis); and the [3-13C]pyruvate was from Aldrich Chemical (Milwaukee, WI).
cDNA Cloning and Expression
The 996-bp fragment of the mitochondrial SlMDH was cloned in antisense orientation or using an RNAi approach into the vector pBINAR (Liu et al., 1999) between the CaMV 35S promoter and the ocs terminator. This construct was introduced into plants by an Agrobacterium-mediated transformation protocol, and plants were selected and maintained as described in the literature (Tauberger et al., 2000). Initial screening of 58 lines (26 antisense and 32 RNAi) was carried out using a combination of total enzyme activity determinations and activity gels. These screens allowed the selection of eight lines, which were taken to the next generation. Total MDH activity, MDH protein content, and expression of the various isoforms of MDH were confirmed in the second harvest of these lines after which four lines were chosen for detailed physiological and biochemical analyses.
Northern-Blot Analysis
Total RNA was isolated using the commercially available Trizol kit (Gibco BRL, Karlsruhe, Germany) according to the manufacturer's suggestions for the extraction from plant material. Hybridization using standard conditions was carried out using the expressed sequence tags for the various isoforms of MDH obtained from Clemson University (Clemson, SC) collection.
Immunodetection of MDH Protein and Gel Stains of MDH Activities
Western analysis of MDH protein was carried out on either crude protein extract or mitochondrial protein extract, exactly as previously described (Gietl et al., 1996) using antibodies kindly provided by the authors. Activity gels were performed on 20 and 40 μg of crude protein extract from 2 different plants, using the protocol described by Shaw and Prasad (1970).
Analysis of Enzyme Activities
Enzyme extracts were prepared as described previously (Tauberger et al., 2000). All activities were determined as described by Carrari et al. (2003) with the exception of PFK and phosphoglucomutase activities, which were determined as defined in Fernie et al. (2001a), and MDH, which were determined as defined in Jenner et al. (2001). GLDH activity was determined as described by Bartoli et al. (2000). The mitochondrial activity of MDH was determined by applying the same method to mitochondrial fractions following the protocol for isolation described in Sweetlove et al. (2002), while those for the plastid, peroxisome, and cytosol were estimated in comparison to marker enzymes for these compartments. The purity of the mitochondrial preparations was confirmed by measurement of the cytochrome c oxidase (Neuberger, 1985) and pyrophosphate-dependent phosphofructokinase (Fernie et al., 2001b), which serve as markers for the mitochondria and cytosol, respectively. Modifications in the activities of peroxisomal and plastidial isoforms of MDH were assessed by comparison to the levels of catalase (Aebi, 1984) and alkaline pyrophosphatase (Farre et al., 2001a), respectively.
Determination of Metabolite Levels
Leaf samples were taken at the time point indicated, immediately frozen in liquid nitrogen, and stored at −80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of the appropriate extraction buffer. The levels of starch, Suc, Fru, and Glc in the leaf tissue were determined exactly as described previously (Fernie et al., 2001a). Levels of glycolytic intermediates, nucleotides, and nucleosides were measured as described in Gibon et al. (2002), while phosphate was determined using the protocol described by Sharkey and Vanderveer (1989). The levels of all other metabolites was quantified by GC-MS exactly following the protocol described by Roessner et al. (2001) with the exception that the peak identification was optimized to tomato tissues (Roessner-Tunali et al., 2003).
Photosynthetic Pigment Determination
The determination of the levels of chlorophylls a and b, β-carotene, lutein, neoxanthin, violaxanthin, antheraxanthin, and zeaxanthin were performed in acetone extracts (Thayer and Björkmann, 1990).
Measurements of Photosynthetic Parameters
The 14C-labeling pattern of Suc, starch, and other cellular constituents was performed by illuminating leaf discs (10-mm diameter) in a leaf-disc oxygen electrode (Hansatech, Kings Lynn, Norfolk, UK) in saturating 14CO2 at a PFD of 250 μmol m−2 s−1 of photosynthetically active radiation at 20°C for 30 min, and subsequent fractionation was performed exactly as detailed by Lytovchenko et al. (2002). Fluorescence emission was measured in vivo using a PAM fluorometer (Walz, Effeltrich, Germany) on 9-week-old plants maintained at fixed irradiance (250 and 700 μmol photons m−2 s−1) for 30 min prior to measurement of chlorophyll fluorescence yield and relative ETR calculated using the WinControl software package (Walz). Gas-exchange measurements were performed in a special custom-designed open system (Lytovchenko et al., 2002).
Measurement of Respiratory Parameters
Dark respiration was measured using the same gas exchange system as defined above. Estimations of the TCA cycle flux on the basis of 14CO2 evolution were carried out following incubation of isolated leaf discs in 10 mm MES-KOH, pH 6.5, containing 2.32 KBq mL−1 of [1-14C]-, [2-14C]-, [3:4-14C]-, or [6-14C]Glc. 14CO2 evolved was trapped in KOH and quantified by liquid scintillation counting. The results were interpreted following ap Rees and Beevers (1960). Estimations of the TCA cycle flux were also performed in isolated mitochondria. For this purpose, mitochondria were isolated from fruit following the method of Holtzapffel et al. (2002) and rapidly frozen and stored at −80°C prior to NMR analysis described below.
13C NMR Spectroscopy
Proton-decoupled 13C NMR spectra of low-density mitochondrial suspensions were recorded at 150.9 MHz on a Varian Unity Inova 600 spectrometer (Palo Alto, CA) using a 10-mm diameter broadband probehead. The suspension was oxygenated with an air-lift (Fox et al., 1989), and NMR spectra were recorded in 15-min blocks over a period of 4 h using acquisition conditions similar to those described previously (Aubert et al., 2001). Mitochondria were suspended at a density of 200 μg mitochondrial protein/milliliter in a buffer containing 0.2 m mannitol, 0.1 m MOPS, 5 mm MgCl2, 0.1% w/v bovine serum albumin, and 20 mm KH2PO4 in 10% D20, corrected to pH 7.2 with KOH. The metabolism of 10 mm [3-13C]pyruvate was observed in the presence of 20 mm Glc, 2.1 mm citrate, 1.3 mm succinate, 0.6 mm malate, 0.3 mm NAD+, 0.2 mm ADP, 0.1 mm thiamine pyrophosphate, 0.02 mm fumarate, 0.02 mm isocitrate, and 0.1 unit hexokinase. Glc and hexokinase were included to regenerate ADP (Aubert et al., 2001) and the TCA cycle acids to mimic the likely cytosolic composition (Farre et al., 2001b).
Statistical Analysis
The t tests were performed using the algorithm embedded into Microsoft Excel (Microsoft, Seattle). The term significant is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with the t test.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY725474 (mMDH), AY725475 (cMDH), AY725476 (gMDH), and AY725477 (chMDH).
Acknowledgments
We are very grateful to Dr. Joachim Fisahn (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for help in organization of gas-exchange measurements and discussion of the results and are indebted to Helga Kulka (same institute) for excellent care of the plants. We are also most grateful to Christine Gietl (Technische Universität Munich, Germany) for the generous gift of the MDH antibodies.
This work was supported by the Max-Planck-Gesellschaft (to F.C., A.N.-N., A.L., and A.R.F.), by Conselho Nacional de Desenvolvimento Científico e Tecnológico (A.N.-N.), by Deutscher Akademischer Austauschdienst (A.N.-N.), and by the United Kingdom Biotechnology and Biological Sciences Research Council (A.M.O.S., R.G.R., and L.J.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055566.
References
- Aebi H (1984) Catalase in vitro. Methods Enzymol 105: 121–126 [DOI] [PubMed] [Google Scholar]
- Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat Biotechnol 21: 177–181 [DOI] [PubMed] [Google Scholar]
- ap Rees T, Beevers H (1960) Pathways of glucose dissimilation in carrot slices. Plant Physiol 35: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Douce R, Gout E, Ratcliffe RG, Roberts JKM (2001) Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by 13C and 31P nuclear magnetic resonance. J Exp Bot 52: 37–45 [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schuurmann P, Droux M, et al (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH (2000) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gallie DR (2004) The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16: 1143–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna CH, Bartoli CG, Sacco F, Ingala LR, Santa-Maria GE, Guiamet JJ, Ugalde RA (2003) Thylakoid bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiol 132: 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre EM, Bachmann A, Willmitzer L, Trethewey RN (2001. a) Acceleration of potato tuber sprouting by the expression of a bacterial pyrophosphatase. Nat Biotechnol 19: 268–272 [DOI] [PubMed] [Google Scholar]
- Farre EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001. b) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids and sugar alcohols in potato tubers using a nonaquous fractionation method. Plant Physiol 127: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004. b) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport chain. Curr Opin Plant Biol 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roessner U, Trethewey RN, Willmitzer L (2001. a) The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta 213: 418–426 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001. b) Fructose 2,6-bisphosphate activates pyrophosphate: fructose 6 phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky A, Willmitzer L (2004. a) Metabolic profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Fox GG, Ratcliffe RG, Southon TE (1989) Airlift systems for in vivo NMR spectroscopy of plant tissues. J Magn Reson 82: 360–366 [Google Scholar]
- Galtier N, Foyer CH, Huber J, Voelker TA, Huber SC (1993) Effects of elevated sucrose phosphate synthase activity on photosynthesis, assimilate partitioning, and growth in tomato (Lycoperscion esculentum var UC82B). Plant Physiol 101: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel cycling system. Plant J 30: 221–233 [DOI] [PubMed] [Google Scholar]
- Gietl C (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim Biophys Acta 1100: 217–234 [DOI] [PubMed] [Google Scholar]
- Gietl C, Seidel C, Svendsen I (1996) Plant glyoxysomal but not mitochondrial malate dehydrogenase can fold without chaperone assistance. Biochim Biophys Acta 1274: 48–58 [DOI] [PubMed] [Google Scholar]
- Hancock RD, Viola R (2002) Biotechnological approaches for L-ascorbic acid production. Trends Biotechnol 20: 299–305 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244 [DOI] [PubMed] [Google Scholar]
- Holtzapffel RC, Finnegan PM, Millar AH, Badger MR, Day DA (2002) Mitochondrial protein expression in tomato fruit during on-vine ripening and cold storage. Funct Plant Biol 29: 827–834 [DOI] [PubMed] [Google Scholar]
- Jain AK, Nessler CL (2000) Metabolic engineering of an alternative pathway for ascorbate biosynthesis in plants. Mol Breed 6: 73–78 [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KM, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, Foyer CH (2003) Effects of leaf ascorbate content on defence and photosynthesis gene expression in Arabidopsis thaliana. Antioxid Redox Signal 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Lieman-Hurwitz J, Rachmilevitch S, Mittler R, Marcus Y, Kaplan A (2003) Enhanced photosynthesis and growth of transgenic plants that express ictB, a gene involved in HCO3- accumulation in cyanobacteria. Plant Biotechnol J 1: 43–50 [DOI] [PubMed] [Google Scholar]
- Liu XJ, Prat S, Willmitzer L, Frommer WB (1999) Cis-regulatory elements directing tuber specific and sucrose inducible expression of a chimeric class I patatin promoter-GUS gene fusion. Mol Gen Genet 223: 101–106 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A, Sweetlove LJ, Pauly M, Fernie AR (2002) The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Driscoll BT, Gregerson RG, Gantt JS, Vance CP (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J 15: 173–184 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19: 965–969 [DOI] [PubMed] [Google Scholar]
- Neuberger M (1985) Preparation of plant mitochondria, criteria for assessment of mitochondrial integrity and impurity, survival in vitro. In R Douce, D Day, eds, Higher Plant Cell Respiration. Springer-Verlag, Berlin, pp 7–24
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Janovic S, Verrier PJ, Graham N, Foyer CH (2003) Leaf vitamin C contents modulate plant defence transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellny TK, Ghannoum O, Conroy JP, Schluepmann H, Smeekens S, Andralojc J, Krause KP, Goddijn O, Paul MJ (2004) Genetic modification of photosynthesis with E. coli genes for trehalose synthesis. Plant Biotechnol J 2: 71–82 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Richards RA (2000) Selectable traits to increase crop photosynthesis and yield of grain crops. J Exp Bot 51: 447–458 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR (2005) Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicon complex. J Exp Bot 56: 297–307 [DOI] [PubMed] [Google Scholar]
- Scheibe R (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Vanderveer PJ (1989) Stromal phosphate concentration is low during feedback limited photosynthesis. Plant Physiol 91: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CR, Prasad R (1970) Starch gel electrophoresis of enzymes: a compilation of recipes. Biochem Genet 4: 297–320 [DOI] [PubMed] [Google Scholar]
- Smirnoff N (2000) Ascorbate biosynthesis and function in photoprotection. Philos Trans R Soc Lond B Biol Sci 355: 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. CRC Crit Rev Plant Sci 19: 267–290 [DOI] [PubMed] [Google Scholar]
- Smith AMO, Ratcliffe RG, Sweetlove LJ (2004) Activation and function of plant mitochondrial uncoupling protein. J Biol Chem 279: 51944–51952 [DOI] [PubMed] [Google Scholar]
- Stitt M (1997) The flux of carbon between the chloroplast and the cytoplasm. In DT Dennis, DH Turpin, DD Lefebre, DB Layzell, eds, Plant Metabolism. Longman, Harlow, UK, pp 382–400
- Sweetlove LJ, Heazelwood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Kossmann J, Riesmeier JW, Trethewey RN, Hill SA (1998) The control of source to sink carbon flux during tuber development in potato. Plant J 15: 697–706 [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vincente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose 6-phosphate. Plant J 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Thayer SS, Björkmann O (1990) Leaf xanthophyll content and composition in sun and shade determined by HPLC. Photosynth Res 23: 331–343 [DOI] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2003) Deductions about the number, organization and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393: 365–368 [DOI] [PubMed] [Google Scholar]
- Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2: 983–989 [DOI] [PubMed] [Google Scholar]