Abstract
Members of the YABBY family of putative transcription factors participate in abaxial-adaxial identity determination in lateral organs in Arabidopsis (Arabidopsis thaliana). Two YABBY genes specifically expressed in reproductive structures, CRABS CLAW (CRC) and INNER NO OUTER (INO), have additional activities, with CRC promoting nectary development and carpel fusion, and INO responding to spatial regulation by SUPERMAN during ovule development. All YABBY coding regions, except YABBY5, were able to restore outer integument growth in ino-1 mutants when expressed from the INO promoter (PROINO). However, INO was the only YABBY family member that responded correctly to SUPERMAN to maintain the wild-type gynoapical-gynobasal asymmetry of the outer integument. By contrast, INO, FILAMENTOUS FLOWER, and YABBY3 failed to complement crc-1 when expressed from PROCRC. Roles of individual regions of CRC and INO in these effects were assessed using chimeric proteins with PROINO and PROCRC and the relatively constitutive cauliflower mosaic virus PRO35S. Regions of CRC were found to contribute additively to CRC-specific functions in nectary and carpel formation, with a nearly direct relationship between the amount of CRC included and the degree of complementation of crc-1. When combined with INO sequences, the central and carboxyl-terminal regions of CRC were individually sufficient to overcome inhibitory effects of SUPERMAN within the outer integument. Reproductive phenotypes resulting from constitutive expression were dependent on the nature of the central region with some contributions from the amino terminus. Thus, the YABBY family members have both unique and common functional capacities, and residues involved in differential activities are distributed throughout the protein sequences.
Functional characterization of plant transcription factors has been facilitated by genetic and transgenic analysis, and the activities of many such proteins have been clearly linked to diverse regulatory processes (Zhang, 2003). Transcription factors have been classified into families, such as the MADS and MYB families, based on sequence conservation of protein regions, most commonly DNA-binding domains (Riechmann, 2002). In many instances, members of the different families have become functionally diversified, resulting either from critical changes in protein coding sequence or from spatial and temporal changes in expression patterns. The distinct roles of the MADS-domain transcription factors AGAMOUS and APETELA3 in floral development represent an example of the former process (Riechmann et al., 1996; Riechmann and Meyerowitz, 1997), whereas the apparent interchangeability of the protein coding sequences of the MYB-domain proteins WEREWOLF and GLABARA1 exemplifies the latter process (Lee and Schiefelbein, 2001). Experimental interchange of protein coding sequences and regulatory regions has facilitated the understanding of both in vivo roles and mechanisms of functional divergence of such gene family members.
The YABBY family of putative transcription factors was identified in Arabidopsis (Arabidopsis thaliana; Bowman and Smyth, 1999; Sawa et al., 1999; Villanueva et al., 1999). The YABBY proteins have two conserved regions, a Cys-containing zinc finger motif, similar to ones functioning in a variety of macromolecular interactions (Takatsuji, 1998), and the YABBY region, which has similarity to a portion of the DNA-binding domain of the HMG family of transcription factors (Grosschedl et al., 1994). Based on both genetic and molecular evidence, members of the YABBY gene family function in the promotion of abaxial identity of lateral organs (Bowman, 2000). Some YABBY members share similar expression patterns; for example, FILAMENTOUS FLOWER (FIL) and YABBY3 (YAB3) are both expressed in most aerial lateral organs. Others, such as INNER NO OUTER (INO), have distinct endogenous expression domains, with this gene being limited to the outer integument of the ovule where no other YABBY gene expression has been detected. In addition to promoting abaxial cell types, CRABS CLAW (CRC), which has some expression domains not considered abaxial in nature, is also involved in the specification of the gynoecium and is required for nectary formation (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). Thus, perhaps aided by diverging expression patterns, some YABBY members may have acquired both specialized roles in polar identity and unique functional roles.
Asymmetric growth of the outer integument of ovules, mediated by interactions between INO and SUPERMAN (SUP; Meister et al., 2002), is another example of a specific role for a YABBY gene. Outer integument growth in wild-type ovules is mostly limited to the gynobasal (toward the base of the gynoecium) side of developing ovule primordia, correlated with confinement of INO expression to this region, and mutations in INO eliminate this growth. By contrast, sup mutant ovules exhibit outer integument growth on all sides of ovule primordia as a result of ectopic growth on the gynoapical side of the primordia. The ectopic growth is correlated with an expansion of INO expression to this region. Meister et al. (2002) demonstrated that both a PROINO:INO transgene and a PROINO:CRC transgene could restore outer integument growth in ino-1 mutants. However, while the PROINO:INO transgenic plants exhibited typical asymmetric outer integument growth, ovules of the PROINO:CRC transgenics phenocopied sup mutant ovules. SUP was thus hypothesized to maintain the endogenous INO expression pattern through attenuation of an INO autoregulatory circuit, with CRC being insensitive to the effects of SUP (Meister et al., 2002).
With diverged expression patterns and in vivo roles, we exploited the YABBY gene family as a model for mechanisms of functional divergence among plant transcription factors. Using both tissue-specific and constitutive promoters, we characterized the effects of expression of native or chimeric YABBY coding sequences on plant morphology. We demonstrate that FIL, YAB2, YAB3, and CRC could restore integument growth in ino-1 mutants but appear to be insensitive to the inhibitory effects of SUP. By contrast, INO, FIL, and YAB3 were unable to compensate for the loss of CRC in directing the proper development of both the nectaries and the gynoecium. Analysis of chimeric coding sequences suggested that each of three putative protein regions contributes to the observed functional differences in at least one of three expression domains examined and that the zinc finger motif may be dependent on an adjacent conserved sequence region for functionality in some expression domains. In addition to differences in expression domains, we speculate that differences in protein-protein interactions, due to changes in the coding regions, may make a significant contribution to functional divergence of the YABBY protein family.
RESULTS
INO Has a Unique Ability to Maintain Asymmetric Growth of the Outer Integument
Ovule development has been characterized previously through morphological, genetic, and molecular analyses (Gasser et al., 1998; Schneitz, 1999; Skinner et al., 2004). The Arabidopsis ovule initiates development as an apparently symmetrical primordium but is then partitioned along both proximal-distal and gynoapical-gynobasal (toward stigma and base of the gynoecium, respectively) axes, as evidenced by gene expression and growth patterns of the two integuments. Figure 1A shows a wild-type ovule where growth of the outer integument occurs in a gradient around the circumference of the ovule primordium, with maximal growth on the gynobasal side and essentially no growth on the gynoapical side. By contrast, while the outer integument in sup-5 ovules initiates on the gynobasal side, it subsequently develops from both sides of the ovule primordium (Fig. 1C; Gaiser et al., 1995). ino-1 ovules do not initiate an outer integument (Fig. 1B).
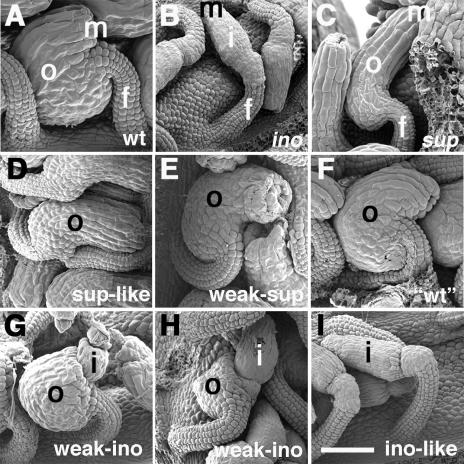
Figure 1.
Scanning electron micrographs of mature ovules from wild-type, mutant, and transgenic plants. In wild-type ovules (A), the outer integument initiates and grows from only the gynobasal side of the ovule primordium. In ino-1 ovules (B), the outer integument fails to initiate growth, whereas in sup-5 ovules (C), the outer integument grows from both the gynobasal and gynoapical sides of the ovule primordium. Outer integument growth in two phenotypic classes, sup-like (D) and weak-sup (E), resembled that of sup-5 ovules, but growth from the gynoapical side of the ovule primordium was reduced in the weak-sup class. Ovules that resembled wild type (F), exhibiting typical asymmetrical outer integument growth, were identified. Outer integument growth in the weak-ino class could vary from almost complete (G) to relatively rudimentary (H). In the ino-like class (I), outer integument growth did not occur. Representative ovule phenotypes were from plants containing the PROINO:YAB3 (D), PROINO:CRC (E), PROINO:INO (F and G), PROINO:FIL (H), and PROINO:YAB5 (I) transgenes. f, Funiculus; m, micropyle; i, inner integument; o, outer integument. Scale bar = 50 μm.
INO expression is limited to the site of outer integument initiation and the outermost (abaxial) cell layer of the developing outer integument (Villanueva et al., 1999; Balasubramanian and Schneitz, 2000). The identification of a genomic fragment (PROINO) sufficient to duplicate the endogenous expression of INO was described previously (Villanueva et al., 1999; Meister et al., 2002, 2004). Using PROINO, the ability of each YABBY member to complement the ino-1 mutant phenotype was assessed. Transgenic plants in this study exhibited a range of ovule morphologies. Ovules of transgenic plants were classified as wild type (complementing) if normal ovule development was fully restored (Fig. 1F) or “ino-like” if the ino-1 effects were not mitigated (Fig. 1I). Ovules resembling those of sup-5 were also identified and termed “sup-like” (Fig. 1D). In addition, two intermediate phenotypes were observed and referred to as “weak-sup” and “weak-ino.” In weak-sup ovules, integument growth from the gynoapical side of the ovule primordium was often less than observed in sup-5, but still greater than in wild type (Fig. 1E). Outer integument growth in weak-ino ovules resembled that of the ino-4 allele (Villanueva et al., 1999), where this structure partially covers but never fully envelops the inner integument yet still exhibits the characteristic asymmetric growth (Fig. 1, G and H).
Based on their phenotypic effects on ino-1 ovules, enumerated in Table I, the YABBY proteins partitioned into three statistically separable classes. As described previously (Villanueva et al., 1999; Meister et al., 2002), the PROINO:INO transgene fully complemented ino-1 and restored wild-type ovule development in the majority of transgenic lines (Table I). Approximately 20% showed only partial complementation and 14% were not complemented, consistent with variable activity of transgenes in independent transformants. In no case was ectopic integument growth observed on the gynoapical side of the ovules. Thus, INO efficiently restores integument growth and uniformly responds to spatial information provided by SUP. The INO coding region was the only native YABBY coding region with these properties.
Table I.
Transgenic complementation of ino-1
| Genotypea
|
PROINO Transgeneb
|
Ovule Phenotypec
|
Total Transgenic Lines
|
||||
|---|---|---|---|---|---|---|---|
| Sup-Like | Weak-Sup | Wild Type | Weak-Ino | Ino-Like | |||
| Wild type | –d | – | 10 | – | – | 10 | |
| ino-1/SUP | – | – | – | – | 8 | 8 | |
| INO/sup-5 | 7 | – | – | – | – | 7 | |
| YAB3 | 9 | 6 | – | – | – | 15 | |
| CRC | 7 | 13 | – | 4 | 1 | 25 | |
| FIL | – | 13 | 7 | 5 | 9 | 34 | |
| YAB2 | – | 3 | – | 10 | – | 13 | |
| INO | – | – | 28 | 8 | 6 | 42 | |
| YAB5 | – | – | – | – | 19 | 19 | |
| CcII | – | – | 9 | 13 | 2 | 24 | |
| CiII | – | – | – | 6 | 21 | 27 | |
| IiCC | 3 | 8 | 5 | 4 | 2 | 22 | |
| IcCC | 3 | 11 | 1 | 7 | 7 | 29 | |
| IiCI | – | 5 | 9 | 4 | 3 | 21 | |
| IcCI | – | 8 | – | 10 | 5 | 23 | |
| CcIC | 2 | 10 | 13 | 7 | 3 | 35 | |
| CiIC | 1 | 1 | 2 | 7 | 24 | 35 | |
| IiIC | – | 5 | 13 | – | – | 18 | |
| CcCI | 4 | 6 | 2 | 10 | 9 | 31 | |
Ovules were examined in wild-type, ino-1, or sup-5 mutant plants as designated. All transgenics were examined in an ino-1 mutant background.
Transgenes were constructed as transcriptional fusions of the identified coding sequence with PROINO.
Number of transgenic lines with the corresponding ovule phenotype, as described in the text and illustrated in Figure 1.
–, No plants of this class observed.
In contrast with INO, the majority of other Arabidopsis YABBY coding regions produced results similar to those observed for the PROINO:CRC construct. For CRC, YAB3, FIL, and YAB2, integument growth was at least partially restored, but the majority, or a significant fraction, of the transgenic lines also exhibited ectopic outer integument growth on the gynoapical side of the ovule primordia (Table I). Within this group, there was variation both in the ability to promote outer integument growth and for evidence of spatial regulation of this growth. PROINO:YAB3 and PROINO:CRC transgenics appeared to be most efficient at growth promotion and also had a higher frequency of ectopic integument growth from the gynoapical side of the ovule primordium relative to PROINO:FIL or PROINO:YAB2 transformants. Among these transgenes, only PROINO:FIL produced a significant fraction of plants (22%) with apparently wild-type ovules. PROINO:YAB5 was unable to support outer integument growth in any transformant analyzed and, thus, by itself represents a third class of transgene.
Chimeric Coding Sequences Evaluate ProteinDomain-Dependent Transgene Effects withinthe Outer Integument
Chimeric cDNAs, combining regions of INO and CRC coding sequences, were produced to identify protein regions responsible for the different effects of expression of these two coding regions on integument development. The chimeras represented permutations of three regions: the amino-terminal region (including the conserved zinc finger motif), the central variable region, and the carboxyl-terminal region (beginning at the start of the conserved YABBY region). Boundaries of the different regions are illustrated in Figure 2 and were placed at the junctions of the previously described conserved regions and the adjacent residues (Bowman and Smyth, 1999; Villanueva et al., 1999). Chimeric cDNAs and proteins are denoted by three capital letters, using I for INO sequence and C for CRC sequence, for each of the three regions. In addition, the seven residues adjacent to the final Cys of the zinc finger motif and within the previously described central variable region appear to be relatively conserved when considering the entire Arabidopsis YABBY family (Fig. 2). To assess the functional significance of this region, it was tested in the chimeras in linkage with either the amino-terminal or the central variable region. An additional lowercase letter in each protein designation indicates the source of this region in the chimeras enumerated in Table I.
Figure 2.
Alignment of Arabidopsis YABBY amino acid sequences. The zinc finger and YABBY regions, as previously defined (Siegfried et al., 1999), are indicated by solid and broken underlines, respectively. Residues conserved in at least five of the sequences are highlighted in gray. The seven conserved residues adjacent to the zinc finger motif are indicated with a double underline. Residues on either side of protein region boundaries that were used for chimeric proteins are in boldface type.
Chimeric coding regions were fused to PROINO and assessed in an ino-1 mutant background. As with PROINO-directed expression of the endogenous YABBY members, observed ovule phenotypes could be grouped into five classes: sup-like, weak-sup, wild type, weak-ino, and ino-like (Fig. 1). In general, we found that all chimeras were able to support some growth of the outer integument in at least a subset of transformants. Effects of the amino-terminal region exchange appeared to be sensitive to the conserved residues adjacent to the zinc finger motif. As evidenced by the CiII chimera (and the later described CiIC chimera), the inclusion of the INO sequence in this region in conjunction with the CRC amino terminus resulted in a severe reduction of the ability to promote integument growth. By contrast, the CcII coding region was able to support outer integument growth in most transformants. Since more than one-half of the CcII transformants had weak-ino ovules, this chimera was less effective than INO at promoting integument growth, but no transformants had weak-sup or sup-like integument growth, indicating a normal response to SUP effects. The INO amino-terminal region appeared to be less dependent on the nature of the adjacent sequence, as both the IcCC and IiCC chimeras were found to promote integument growth in a manner that was not statistically different from the effects of CRC (Table I).
Replacement of either the central variable or carboxyl-terminal regions of INO with CRC sequences produced transgenes that could promote growth of the outer integument from the gynoapical side of the ovule primordium. The IcCI chimera was the most effective; all PROINO:IcCI transgenics with substantial outer integument growth exhibited visible outer integument growth on the gynoapical side of the ovule. Both the IiCI and IiIC chimeras produced plants with either weak-sup or wild-type ovules. Uniquely, PROINO:IiIC transformants producing weak-ino or ino-like ovules were not identified. Of these three chimeras, both IcCI and IiIC produced effects that were statistically distinct from effects of INO. Although exchange of the variable or carboxyl-terminal regions of CRC resulted in the identification of plants containing wild-type ovules, which was not observed with the PROINO:CRC transgene, neither exchange eliminated the ability of the chimera to elicit growth of the outer integument from the gynoapical side of the ovule primordium in some transformants. However, due to the large percentage of wild-type ovules in transgenic plants, the CcIC chimera was statistically distinct from CRC, and CcCI was marginally distinct from CRC. Consistent with the apparent deleterious effects of the CiII amino-terminal exchange, the CiIC chimera appeared to be compromised in its ability to promote integument growth, producing mostly weak-ino or ino-like transformants.
INO Is Unable to Functionally Replace CRC in Gynoecium and Nectary Development
The Arabidopsis gynoecium initiates development as a continuous ring of tissue that is believed to represent two congenitally fused carpel primordia (Smyth et al., 1990; Alvarez and Smyth, 2002). An internal septum forms from outgrowths on the medial walls to divide the gynoecium into two chambers. The placentas, the sites of ovule development, are located at the boundaries of the septum with the ovary wall. The endogenous expression of CRC is limited in Arabidopsis to the developing gynoecium and the floral nectaries, which develop in a ring with glands at the abaxial base of each stamen (Baum et al., 2001). Within the gynoecium, CRC is localized to two domains, the carpel outer epidermis (the abaxial cell layer) and four internal domains of unknown function. crc mutant plants lack nectaries and the two carpels fail to fuse properly in the apical region of the gynoecium, as illustrated in Figure 3.
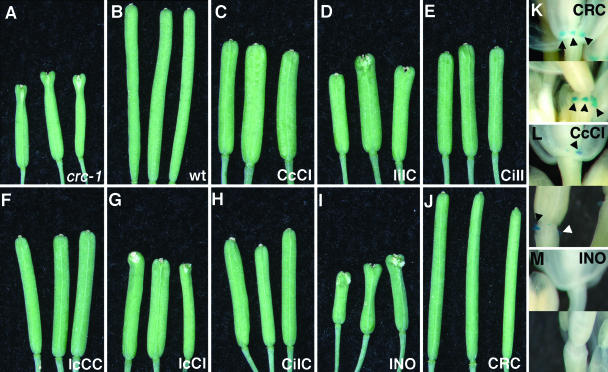
Figure 3.
Siliques of the Ler (wild type), crc-1 ET668, and PROCRC:CRC-INO transformants of crc-1 ET668. A, crc-1 ET668. B, Wild type. C, PROCRC:CcCI. D, PROCRC:IiIC. E, PROCRC:CiII. F, PROCRC:IcCC. G, PROCRC:IcCI. H, PROCRC:CiIC. I, PROCRC:INO. J, PROCRC:CRC. Chimeras with two CRC regions generally rescued carpel fusion but failed to rescue silique growth, with some having siliques broader than wild type. K, PROCRC:CRC fully complemented the crc-1 mutant with all nectaries, lateral and medial, present, as assayed by GUS staining in the nectary conferred by enhancer trap ET668 (arrowheads). L, Partial complementation, the development of lateral nectaries only, was observed using constructs that contained two of three CRC domains, e.g. PROCRC:CcCI. M, PROCRC:INO failed to complement crc-1, with no nectary development detected.
To assess the extent to which native YABBY proteins were able to complement crc-1 mutant plants, we used light microscopy to examine carpel defects and an enhancer trap line (ET668; Baum et al., 2001) that directs expression of the β-glucuronidase (GUS) coding sequence during nectary differentiation to assess nectary development. Using these assays, a PROCRC:CRC transgene was able to complement the crc-1 carpel and nectary defects as well as activate expression of the GUS reporter gene within the nectaries in almost all transformants examined (Fig. 3, J and K), as enumerated in Table II. In contrast with CRC, the PROCRC:INO transgene was unable to complement either mutant defect of crc-1 (Fig. 3, I and M; Table II). In the PROCRC:INO transformants, the carpels did not fuse in the apical region and GUS activity was not detected at the predicted site of nectary formation. In similar studies, the production of FIL or YAB3 regulated by PROCRC also failed to complement crc-1 mutant defects (Y. Eshed and J. Bowman, unpublished data).
Table II.
Transgenic complementation of crc-1
| Genotypea
|
PROCRC Transgeneb
|
Presence of Nectariesc
|
Apical Fusion of Pistild
|
||||
|---|---|---|---|---|---|---|---|
| + | ± | − | + | ± | − | ||
| Wild type | 20 | –e | – | 10 | – | – | |
| crc-1 ET668 | – | – | 20 | – | – | 10 | |
| CRC | 19 | – | 1 | 10 | – | – | |
| INO | – | – | 20 | – | – | 10 | |
| CiII | 2 | 3 | 15 | 4 | 4 | 2 | |
| IcCC | 12 | 5 | 3 | 7 | 2 | 1 | |
| IcCI | 4 | 4 | 12 | 2 | 2 | 5 | |
| CiIC | 16 | 1 | 3 | 10 | – | – | |
| IiIC | 1 | – | 19 | – | – | 10 | |
| CcCI | 12 | 3 | 5 | 9 | – | 1 | |
Genotype of plants examined in this study; all transgenics were genotypically crc-1 ET668.
Respective coding regions were expressed using PROCRC.
Presence of nectary tissue was determined by detectable GUS activity of the ET668 enhancer trap.
Fusion of the gynoecium apex was determined by light microscopy.
–, No plants of this class observed.
Chimeric Coding Sequences Assess ProteinDomain-Dependent Transgene Effects within the Gynoecium and Nectaries
The inability of INO to substitute for CRC within the nectaries and gynoecium contrasted with the ability of CRC to partially substitute for INO in outer integument development. To determine whether a specific protein region was responsible for this inactivity, INO-CRC chimeras were expressed under control of PROCRC. As with the PROCRC:INO transgene, the ability of the chimeras to complement the nectary and gynoecium defects of crc-1 was assayed in such mutants carrying the nectary-specific enhancer trap (Fig. 3; Table II). In some transformants, the transgene appeared to partially complement the nectary and/or carpel defects. For nectary development, this was evidenced by detectable GUS activity even when the nectaries were reduced in size or number of glands as compared to wild type. In regard to gynoecium development, partial complementation resulted in an incomplete fusion of the gynoecium apex and an overall length of the gynoecium intermediate between crc-1 and wild type (Fig. 3). The numbers of transgenic lines in each category (noncomplementation, partial complementation, or complete complementation of nectary of fusion defects) are enumerated in Table II.
Proteins including any two regions of CRC and only one region of INO (the IcCC, CiIC, or CcCI chimeras) appeared to have slightly less ability to complement the nectary defects of the crc-1 mutant relative to intact CRC (Table II), with CiIC most closely approximating the full CRC effect. Effects of these chimeras on closure of the gynoecium could not be statistically separated from effects of intact CRC (Table II; Supplemental Table III), but wild-type silique growth was not restored by any of the chimeras (Fig. 3). The relatively efficient complementation with the CiIC chimera shows that, in contrast to the observations for complementation of ino defects, the association between the amino-terminal region and adjacent conserved residues was not critical for complementation of crc defects. All three proteins containing only a single CRC region were significantly less active in complementation of crc-1 effects than were those with two CRC regions. A combination of the CRC carboxyl-terminal region with the other regions of INO was similar to INO in its inability to complement the nectary or gynoecium growth defects of crc-1. By contrast, replacement of either the amino-terminal region or the central diverged region of INO with the corresponding region of CRC led to a higher frequency of partial complementation of the crc-1 gynoecium defects than observed for INO, but still apparently less complementation than was observed for any of the chimeras containing two CRC regions. Overall, the results indicate that all three regions of CRC contribute to its unique ability to function in nectary and gynoecium development. The amino-terminal and central diverged regions play the most significant roles, with a lesser role for the carboxyl-terminal region. The positive contribution of the carboxyl-terminal region to CRC function was most apparent when examined in combination with one of the other two regions (compare results of CiII with CiIC and results of IcCI with IcCC in Table II).
Ectopic Expression of INO-CRC ChimericCoding Sequences
Common and distinct effects of ectopic expression of INO and CRC have been described previously (Eshed et al., 1999; Meister et al., 2002). Leaves of transgenic plants ectopically expressing either coding region were curled and could also be narrow and misshapen, apparently due to the production of abaxial cell types in adaxial domains, similar to the phenotypes observed when other YABBY gene family members were ectopically expressed (Sawa et al., 1999; Siegfried et al., 1999). Within the flower, ectopic CRC expression often resulted in gynoecia lacking a central cavity (the location of placental tissue and ovules; Eshed et al., 1999). By contrast, misexpression of INO often resulted in production of supernumerary organs in the outer three floral whorls and partial or complete loss of normal fourth whorl (gynoecium) tissue (Meister et al., 2002). In some transformants, ectopic expression of INO also resulted in the production of ovules with either ectopic outer integument growth, as in sup-5, or reduced or absent outer integuments similar to ino-4 and ino-1, respectively. However, the common reduction or loss of gynoecium tissues from ectopic expression of either INO or CRC precluded the analysis of ovule phenotypes in most transformants with floral defects. The differential effects of ectopic INO or CRC expression on flower development provided an alternative assay for functional analysis of the chimeric proteins.
The occurrence of vegetative and floral phenotypic effects in plants ectopically expressing the INO-CRC chimeras are illustrated and enumerated in Figure 4 and Table III, respectively. Among the proteins that included two regions of CRC, those containing the amino-terminal or carboxyl-terminal regions of INO (IiCC, IcCC, and CcCI) most closely approximated the activity of intact CRC and could not be statistically separated from this activity. The CcIC and CiIC proteins produced more divergent phenotypes, including the INO-like production of supernumerary floral organs (CcIC), but were still on the border of statistical separation from effects of CRC (Table III; Supplemental Table V). Thus, the central diverged region appears to have the largest role in effects of ectopic CRC expression on flower development. This contrasts with CRC complementation of crc-1 defects, where replacement of the central region with that of INO led to the smallest decrease in complementation.
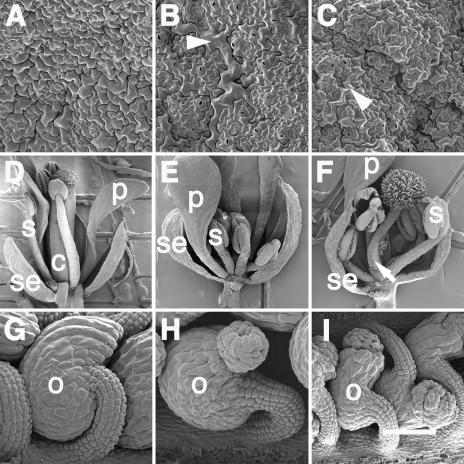
Figure 4.
Ectopic expression of INO-CRC chimeric proteins produced alterations to leaf, flower, and ovule morphology. The adaxial epidermis of wild-type leaves consists of cells that are approximately equivalent in size (A), whereas the abaxial epidermal layer (B) contains small cells interspersed with relatively large cells (arrowhead). The adaxial leaf epidermal layer of a transgenic plant (C) has characteristics of the abaxial epidermal layer, including the relatively large cells (arrowhead). Wild-type flowers consist of four concentric whorls of organs containing four sepals, four petals, six stamens, and a central gynoecium (sepal, petal, and two stamens removed to reveal gynoecium; D). Ectopic expression of INO-CRC chimeric proteins could produce flowers that either lacked a central gynoecium and contained supernumerary stamens (E) or had a gynoecium (arrow) without the central cavity (F). The outer integument of wild-type ovules (G), which initiates and develops asymmetrically, grows to fully envelop the inner integument and nucellus. Ectopic expression of INO-CRC chimeric proteins could produce ovules whose outer integument would initiate, as in wild-type (H) or sup-5 (I) plants, but not fully enclose the inner integument. Representative phenotypes were from plants containing the PRO35S:IiCI (C, H, and I), PRO35S:CcIC (E), or PRO35S:IiCC (F) transgenes. c, Carpel; o, outer integument; p, petal; se, sepal; s, stamen. Scale bars = 100 μm (A–C), 500 μm (D–F), and 50 μm (G–I).
Table III.
Phenotypic effects of ectopic expression of YABBY coding regions
|
PRO35S Transgenea
|
Wild Typeb
|
Weak-Sup Ovulesd
|
Weak-Ino Ovulesd
|
Curled Leavesc
|
Total Plants
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Supernumerary Floral Organse | Gynoecium Defectse | Weak-Sup Ovules | Weak-Ino Ovules | ||||||
| INO | –f | 1 | – | 9 | 9 | – | 1 | 8 | 28 |
| CRC | 24 | – | – | 6 | – | 3 | – | – | 33 |
| CcII | 8 | – | 7 | 7 | – | 5 | 20 | – | 47 |
| CiII | 19 | 18 | – | – | – | – | – | – | 37 |
| IiCC | 4 | – | – | 4 | – | – | – | – | 8 |
| IcCC | 3 | 1 | – | 1 | – | 3 | – | – | 8 |
| IiCI | 5 | – | 1 | 9 | 3 | 5 | 10 | 3 | 36 |
| IcCI | 7 | 3 | 1 | – | – | 11 | – | – | 22 |
| CcIC | 16 | 6 | – | 4 | 3 | – | – | – | 29 |
| CiIC | 8 | 2 | – | – | – | 8 | – | – | 18 |
| IiIC | 4 | – | – | 9 | 1 | – | 2 | – | 16 |
| CcCI | 2 | – | – | – | – | 2 | – | – | 4 |
Constructs utilizing the respective coding sequence driven by the PRO35S were transformed into wild-type plants.
Transgenics that lacked gross morphological defects in vegetative or reproductive tissues were classified as wild type.
Curled leaf phenotype was identified that resulted from alterations to leaf morphology.
Effects on outer integument morphology, resulting in either ectopic gynoapical growth (weak-sup) or overall reduced growth (weak-ino), were observed.
Transgene effects on floral morphology included supernumerary floral organs, most often stamens, and apparent loss of internal domains of the gynoecium.
–, No plants of this class observed.
None of the chimeras were able to duplicate the frequency of production of supernumerary floral organs observed for ectopic production of INO (Table III). Chimeras that did result in this relatively INO-specific effect included IiIC, IiCI, and CcIC. Statistical analysis (Supplemental Table V) indicated that the IiIC and IiCI chimeras were the closest to producing INO activity, indicating a likely importance of the amino-terminal region in this activity, with clear additive contributions from the other regions. The detrimental effects on function of combining the amino-terminal region of CRC with the adjacent conserved region of INO seen in the PROINO experiments were again observed, with the CiII chimera failing to elicit effects on vegetative or floral development in most transformants. By contrast, the CiIC chimera was able to elicit a phenotypic response. Thus, consistent with the positive effect seen in PROINO expression, the presence of the CRC carboxyl-terminal region appeared to mitigate the apparent detrimental effect of the CRC amino-terminal region in conjunction with the adjacent conserved region of INO. In summary, our results suggest that the central variable region, with contributions from either the amino-terminal region (in some instances) or the region adjacent to the zinc finger motif, is the most significant determinant of the INO- or CRC-specific phenotypic responses observed from ectopic expression.
DISCUSSION
Members of the YABBY gene family are hypothesized to function in the promotion of abaxial identity (Bowman, 2000; Bowman et al., 2002). Abaxial identity has been proposed as one component required for both proximal-distal and laminar extension of lateral organs (Waites and Hudson, 1995; McConnell and Barton, 1998). We observed that several YABBY members were sufficient to compensate for the loss of INO in promoting extension of the outer ovule integument when expressed using PROINO. As seen for other YABBY genes (Siegfried et al., 1999), INO and CRC were also able to promote abaxial epidermal identity in adaxial epidermal cells in vegetative tissues. These observations support a common functional capacity of the YABBY members in promotion of abaxial identity and the associated lateral organ growth. This also implies a common DNA-binding capacity for YABBY proteins in their functions in ovule and leaf development. However, this work indicates that YABBY family members involved in reproductive development have apparently evolved additional specialized functions.
INO is involved in an antagonistic relationship with SUP with respect to the regulation of PROINO-driven expression (Meister et al., 2002). This INO function was not reproduced by any other YABBY member. This observation further supports the hypothesis that the INO protein, and not PROINO alone, is critical in this interaction (Meister et al., 2002). CRC has multiple roles within the gynoecium, including postgenital carpel fusion and nectary formation (Alvarez and Smyth, 2002). We observed that INO could not compensate for the loss of CRC in either the nectaries or gynoecium, consistent with results from PROCRC-directed expression of other YABBY members, FIL and YAB3. This could imply an inability of INO and these other YABBY proteins to bind to specific transcriptional targets of CRC, despite the ability of CRC to apparently bind to YABBY targets involved in leaf and ovule development. The functional differentiation of YABBY proteins is also evidenced in ectopic expression experiments in which unique phenotypic responses in reproductive tissues could be correlated to various YABBY family members (Eshed et al., 1999; Siegfried et al., 1999; Meister et al., 2002). Therefore, our results support the hypothesis that the YABBY proteins have remained functionally similar in one regard, able to specify abaxial identity in nonendogenous domains, but that at least the two reproductive associated proteins, INO and CRC, have also significantly diverged to acquire novel, gene-specific functions.
Chimeric Proteins Indicate That Distributed Residues Participate in Differential Activities
Chimeric proteins have been used to evaluate regions responsible for differential function of members of other families of plant transcription factors. Serikawa and Zambryski (1997) found that functional differences between the KNAT1 and KNAT3 transcription factors were mainly in the ELK and homeodomain regions, indicating likely differences in DNA binding. By contrast, studies on some members of the MADS family of transcription factors indicated that the DNA-binding MADS domains were largely interchangeable between proteins, and that differences in interactions with other proteins were likely responsible for functional differentiation (Riechmann and Meyerowitz, 1997).
Our studies on chimeric INO-CRC proteins showed that there was a quantitative effect of the amount of CRC included in the chimeric protein on the protein's ability to complement crc-1. This additive effect was independent of which of the regions derived from CRC. When two CRC regions were included in the chimeric proteins, they commonly produced a degree of complementation close to that observed for the complete CRC protein, but still produced some transgenic plants that were deficient in nectary formation and carpel fusion. When a single CRC region was included in the chimeras, either the amino-terminal or central variable region in conjunction with the remaining INO protein was sufficient to partially complement crc-1, correcting the carpel phenotype more efficiently than the nectary defects.
For complementation of ino-1, most chimeric proteins could compensate for loss of outer integument growth, but only some responded correctly to the inhibitory effects of SUP. In contrast with the crc complementation analysis, replacement of either the variable or carboxyl-terminal regions with those of CRC resulted in relative insensitivity to SUP. Only the amino-terminal exchange (CcII) retained any ability to both promote integument growth and respond correctly to SUP, albeit at a reduced frequency. Consistently, replacement of any two regions of INO resulted in even more pronounced sup-like integument growth. In ectopic expression analysis, we demonstrated that exchange of either the CRC or INO variable region or the INO amino-terminal region was individually sufficient to alter the floral phenotype produced by the chimeric protein.
Taken together, our results imply that all three regions have roles in CRC- or INO-specific functional activity, but the relative contribution of each protein region was specific to the expression domain and function being complemented. This implies that all three regions of the protein participate in function-specific activities. In complementation of ino-1, the majority of INO-CRC chimeric proteins (and even other YABBY proteins) supported integument growth, indicating a common capacity to bind at least the majority of downstream targets of INO. However, since replacing any single region of INO with the corresponding region of CRC led to a decrease in the response to SUP, all three regions of INO must have some participation in this process, which likely depends on protein-protein interactions.
No other YABBY coding region could complement crc-1, and replacement of any two regions of CRC with the corresponding INO regions decreased the ability of the protein to carry out any function of CRC. This indicates that all three regions may participate in CRC-specific interactions with other proteins or with DNA, or a combination of these processes. These results indicate that all three regions of YABBY proteins participate in both DNA-binding and hypothesized protein-protein interactions necessary for YABBY function. Our results are consistent with the recent publication by Sieber et al. (2004) that showed that both the zinc finger motif region and the YABBY region were independently able to interact with NOZZLE/SPOROCYTELESS (NZZ/SPL), another nuclear protein essential for ovule development. Our results could not differentiate functional contributions of either the sequence conserved or nonconserved regions of the amino and carboxyl termini that could be facilitated by the creation and characterization of additional chimeric proteins.
We observed that the attachment of the CRC amino-terminal region with the immediately adjacent INO sequence (CiII) had a negative influence on protein activity when expressed from the PROINO or PRO35S regulatory regions. This effect was partially suppressed by replacing the INO region immediately adjacent to the putative zinc finger motif with that from CRC (CcII), even though the remainder of the central diverged region still derived from INO. The effect was also partially suppressed by exchanging the noncontiguous carboxyl-terminal region for that of CRC (CiIC). The combination of these two changes (CcIC) led to even greater activity that approached that of intact CRC in at least the PROINO expression domain. These results support two hypotheses. First, an independently functional zinc finger domain may extend outside the previously described conserved region, reminiscent of experiments demonstrating that residues adjacent to other zinc finger motifs were critical for functionality (Takatsuji, 1998). Second, since a nonsequential protein region is influencing activity, it is possible that the two conserved regions may form structure-stabilizing contacts within the folded protein or function in combination to produce protein-protein interacting interfaces for other proteins. Subtle contributions to functionality from the YABBY or zinc finger regions could then be supplied by supporting or interfering with the hypothesized protein-protein interactions. These models emphasize the requirement of employing in vivo functional assays, in addition to in vitro and in silico studies, to delineate YABBY protein domains that can be critically examined in further functional and structural studies.
A Model of YABBY Protein Function
The YABBY family members are hypothesized to function as DNA-binding transcriptional regulators. Although the YABBYs are thought to specify abaxial identity, differences in ectopic expression phenotypes and ability to complement ino or crc mutant defects in this and prior studies (Eshed et al., 1999; Siegfried et al., 1999; Meister et al., 2002) demonstrate that some members have functions outside this basic program. One possibility for these differences could be differences in binding-site specificity of some members. The inability of YAB5 or INO to complement ino or crc mutant defects, respectively, could be representative of such a divergence of DNA-binding consensus sequences. Since exchange of only the carboxyl-terminal region, including the putative DNA-binding YABBY region, in the PROCRC:IiIC transgene did not ameliorate this deficiency, it is possible that all three regions contribute to DNA-binding activity and that this activity is progressively reconstituted as the chimeric protein becomes more CRC like.
A second component of the functional differences of the YABBY members may be a differential ability to interact with distinct protein partners. Thus, DNA-binding activity may be similar for all of the chimeric proteins (as suggested by Kanaya et al. [2002] and by the ino-1 complementation experiments), but each region may make protein-specific contacts with interacting proteins necessary for manifestation of INO or CRC function. These contacts would then additively contribute to gene-specific function. This hypothesis is supported by our observation that amino acid differences throughout INO and CRC proteins, possibly extending outside the sequence conserved regions, appear to have significant effects in either flowers or ovules. This is also consistent with multiple regions of YABBY proteins being able to interact with NZZ/SPL (Sieber et al., 2004). However, the NZZ/SPL interactions cannot explain the INO-specific effects because both INO and YAB3 were able to interact with NZZ/SPL. Since a combination of the two proposed models is also a possibility, our results emphasize the importance of both sequence conserved and nonconserved regions within the YABBY proteins. While speculative, these hypotheses are directly testable once protein partners or DNA-binding sites for distinct YABBY family members have been identified.
MATERIALS AND METHODS
Construct Assembly
Chimeric Coding Sequences
All chimeric coding sequences were produced by a two-step PCR amplification process (Horton et al., 1990). The INO (pRJM23) and CRC (pRJM22) coding sequences (Meister et al., 2002) were used as templates for amplification of the amino-terminal (including the zinc finger motif), central variable, and carboxyl-terminal (including the YABBY region) coding regions. Complete cDNAs were cloned as BamHI/XbaI fragments into the same sites of pLITMUS28 (New England Biolabs, Beverly, MA), and the sequence was verified.
PROINO Constructs
The construction of a PROINO expression cassette (pRJM33), consisting of both the 5′ and 3′ regions flanking the INO genomic coding sequence, was described previously (Villanueva et al., 1999; Meister et al., 2002). These regions have been shown to be sufficient in combination to produce a pattern of expression that mimics the endogenous INO expression pattern and enables complementation of the ino-1 mutant phenotype (Villanueva et al., 1999; Meister et al., 2002). The protein coding regions of the remaining four YABBY family members were isolated by reverse transcription-PCR from cDNA (Siegfried et al., 1999; R. Khodosh and J. Bowman, unpublished data). All endogenous and chimeric YABBY coding regions were inserted into the BamHI/XbaI sites of pRJM33, replacing the INO protein coding sequence of that clone.
PROCRC Constructs
A PROCRC expression cassette comprising 4.2 kb 5′ of the CRC translation start site (this fragment drives reporter gene expression in a pattern that duplicates that of the endogenous gene; Lee et al., 2005), and the polyadenylation signal sequence of nopaline synthase (NOS3′) was used to drive expression of the endogenous and chimeric versions of CRC and INO. These constructs were analyzed for the extent to which they could complement crc-1 plants homozygous for the ET668 enhancer trap that exhibits GUS activity in differentiating nectaries (Baum et al., 2001).
Constitutive Expression Constructs
Protein coding sequences were transferred as BamHI/XbaI fragments into the BglII/XbaI sites of pMON999. pMON999 contained a modified cauliflower mosaic virus 35S promoter (PRO35S; Kay et al., 1987) and the NOS3′ sequence flanking a multiple cloning site.
Histochemical Staining and Microscopy
Lines containing the enhancer trap ET668 were stained for GUS activity as described by McConnell and Barton (1998), cleared in 70% ethanol, and observed using a Zeiss (Oberkochen, Germany) dissecting stereoscope. Scanning electron and light microscopy were performed as described previously (Broadhvest et al., 2000; Meister et al., 2002).
Plant Growth and Transformation
Transgenes for plant transformation were shuttled as NotI fragments into pMLBART (a gift from Kim Richardson, HortResearch, Auckland, New Zealand) and transferred into the Agrobacterium strain ASE by triparental matings (Figurski and Helinski, 1979; Fraley et al., 1985; Gleave, 1992). Wild-type Landsberg erecta (Ler), INO(COL)/ino-1(Ler), or crc-1 plants were grown in continuous light and transformed by the floral-dip method (Kranz and Kirchheim, 1987; Clough and Bent, 1998). Transformants were selected for phosphoinothricine (BASTA) resistance. Homozygous ino-1 plants were identified using a PCR-detectable COL/Ler polymorphism (Meister et al., 2002).
Sequence and Statistical Analysis
Sequences were aligned using ClustalX version 1.8 for Macintosh (Thompson et al., 1997). Pairwise evaluations of the statistical significance of differences in effects of chimeric transgenes were performed using Fisher's exact test in consultation with the University of California, Davis, Statistical Laboratory. An α-value that would represent the likelihood that two transgenes had similar phenotypic effects was determined. The Bonferroni adjustment (0.05/number of comparisons performed; http://home.clara.net/sisa/bonhlp.htm) was used in evaluating the statistical significance of the α-values.
Supplementary Material
Acknowledgments
We thank members of the Gasser and Bowman labs for helpful research discussions and Mitchell Watnik for statistical analysis. We appreciate the technical support of Christian Nelson, Louren Kotow, Peter Luu, Erin Goodson, and Chris Roxas.
This work was supported by a U.S. Department of Agriculture National Research Initiative Competitive Grant (2001–35304–09989 to C.S.G.); a grant from the Department of Energy, Division of Biosciences (DE–FG03–97ER20272 to J.L.B); a grant from the National Science Foundation (IBN 0077984 to J.L.B.); a National Science Foundation Plant Cell Biology Training Grant (to R.M.); and a University of California, Davis, Jastro-Shields Fellowship (to R.M.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055368.
References
- Alvarez J, Smyth DR (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR (2002) CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int J Plant Sci 163: 17–41 [Google Scholar]
- Balasubramanian S, Schneitz K (2000) NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 127: 4227–4238 [DOI] [PubMed] [Google Scholar]
- Baum SF, Eshed Y, Bowman JL (2001) The Arabidopsis nectary is an ABC-independent floral structure. Development 128: 4657–4667 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3: 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Broadhvest J, Baker SC, Gasser CS (2000) SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155: 895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76: 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick JS, Fink CL, Hoffmann NL, Sanders PR (1985) The SEV system: a new disarmed Ti plasmid vector for plant transformation. Biotechnology (N Y) 3: 629–635 [Google Scholar]
- Gaiser JC, Robinson-Beers K, Gasser CS (1995) The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser CS, Broadhvest J, Hauser BA (1998) Genetic analysis of ovule development. Annu Rev Plant Physiol Plant Mol Biol 49: 1–24 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10: 94–100 [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535 [PubMed] [Google Scholar]
- Kanaya E, Nakajima N, Okada K (2002) Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J Biol Chem 277: 11957–11964 [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Kranz AR, Kirchheim B (1987) Handling of Arabidopsis. In AR Kranz, ed, Arabidopsis Information Service, Vol 24: Genetic Resources in Arabidopsis. Arabidopsis Information Service, Frankfurt, pp 4.1.1–4.2.7
- Lee J-Y, Baum SF, Alvarez J, Patel A, Chitwood DH, Bowman JL (2005) Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128: 1539–1546 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton K (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- Meister RJ, Kotow LM, Gasser CS (2002) SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Meister RJ, Williams LA, Monfared MM, Gallagher TL, Kraft EA, Nelson CG, Gasser CS (2004) Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter region. Plant J 37: 426–438 [DOI] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomics overview. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, http://www.aspb.org/publications/arabidopsis/, p 46 [DOI] [PMC free article] [PubMed]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1997) Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol Biol Cell 8: 1243–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K (1999) The molecular and genetic control of ovule development. Curr Opin Plant Biol 2: 13–17 [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Zambryski PC (1997) Domain exchanges between KNAT3 and KNAT1 suggest specificity of the kn1-like homeodomains requires sequences outside of the third helix and N-terminal arm of the homeodomain. Plant J 11: 863–869 [DOI] [PubMed] [Google Scholar]
- Sieber P, Petrascheck M, Barberis A, Schneitz K (2004) Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiol 135: 2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews DN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 128: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Skinner DJ, Hill TA, Gasser CS (2004) Regulation of ovule development. Plant Cell 16: S32–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS (1999) INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Hudson A (1995) phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Zhang JZ (2003) Overexpression analysis of plant transcription factors. Curr Opin Plant Biol 6: 430–440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.