Abstract
Sphingolipids are a major component of membrane lipids and their metabolite sphingosine-1-phosphate (S1P) is a potent lipid mediator in animal cells. Recently, we have shown that the enzyme responsible for S1P production, sphingosine kinase (SphK), is stimulated by the phytohormone abscisic acid in guard cells of Arabidopsis (Arabidopsis thaliana) and that S1P is effective in regulating guard cell turgor. We have now characterized SphK from Arabidopsis leaves. SphK activity was mainly associated with the membrane fraction and phosphorylated predominantly the Δ4-unsaturated long-chain sphingoid bases sphingosine (Sph) and 4,8-sphingadienine, and to a lesser extent, the saturated long-chain sphingoid bases dihydrosphingosine and phytosphingosine (Phyto-Sph). 4-Hydroxy-8-sphingenine, which is a major sphingoid base in complex glycosphingolipids from Arabidopsis leaves, was a relatively poor substrate compared with the corresponding saturated Phyto-Sph. In contrast, mammalian SphK1 efficiently phosphorylated Sph, dihydrosphingosine, and 4,8-sphingadienine, but not the 4-hydroxylated long-chain bases Phyto-Sph and 4-hydroxy-8-sphingenine. Surface dilution kinetic analysis of Arabidopsis SphK with Sph presented in mixed Triton X-100 micelles indicated that SphK associates with the micellar surface and then with the substrate presented on the surface. In addition, measurements of SphK activity under different assay conditions combined with phylogenetic analysis suggest that multiple isoforms of SphK may be expressed in Arabidopsis. Importantly, we found that phytosphingosine-1-phosphate, similar to S1P, regulates stomatal apertures and that its action is impaired in guard cells of Arabidopsis plants harboring T-DNA null mutations in the sole prototypical G-protein α-subunit gene, GPA1.
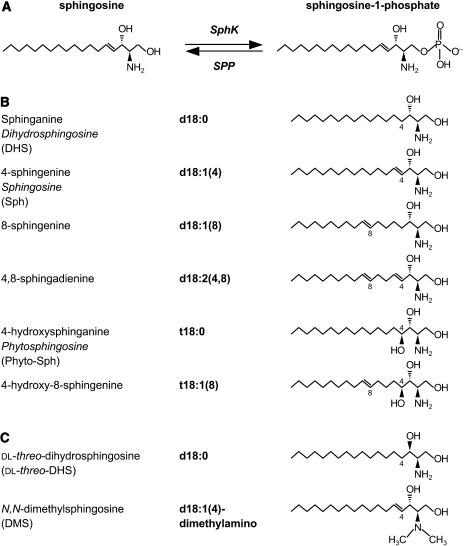
Sphingolipids are ubiquitous components of cellular membranes in eukaryotic cells and in a few bacteria (Lynch and Dunn, 2004). They are composed of a long-chain sphingoid base having one amide-linked fatty acyl chain and a polar head group. In mammals, the prevalent long-chain base is sphingosine (Sph, d18:1) with a chain length of 18 carbon atoms and an E-double bond between carbons 4 and 5, whereas in the yeast Saccharomyces cerevisiae, the predominant long-chain base is phytosphingosine (Phyto-Sph, t18:0), a saturated and 4-hydroxylated form of Sph (Fig. 1, A and B). In plant sphingolipids, the long-chain-base moieties are mostly the 8-E and 8-Z-isomers of 4,8-sphingadienine (d18:2) and 4-hydroxy-8-sphingenine (t18:1), although Phyto-Sph and dihydrosphingosine (DHS, d18:0) are the predominant free long-chain bases (Abbas et al., 1994; Imai et al., 2000; Sperling and Heinz, 2003; Wright et al., 2003; Fig. 1B).
Figure 1.
Reaction catalyzed by SphK and structural diversity of plant sphingoid bases. A, SphK, using ATP as the phosphate donor, catalyzes the phosphorylation of Sph to produce S1P. S1P phosphatase (SPP) regenerates Sph from S1P. B, Structures of common C18 long-chain bases from plants. Names and shorthand designations are given for each long-chain base. For certain long-chain bases, the common name is also given in italics. The naturally occurring dihydroxy (d) and trihydoxy (t) long-chain bases have D-erythro and D-ribo configurations, respectively. The double bond in the Δ4 position of d18:1(4) and d18:2 (4,8) is primarily in the E (trans) configuration, whereas the double bond in the Δ8 position of d18:1(8), d18:2(4,8), and t18:1(8) may be in the Z (cis) or E (trans) configuration. C, Structures of SphK inhibitors.
Sphingosine-1-phosphate (S1P) has received renewed attention recently because it regulates many biological processes in mammals through its interactions with a family of specific cell surface G-protein-coupled receptors (GPCRs) and also serves as an intracellular second messenger in eukaryotes to regulate Ca2+ homeostasis, cell growth, and survival (Spiegel and Milstien, 2003). In plants, only recently has S1P been identified and its role in plant cell signaling been investigated (Ng et al., 2001; Coursol et al., 2003; Pandey and Assmann, 2004). S1P regulates guard cell behavior via Ca2+ mobilization (Ng et al., 2001), inhibition of plasma membrane inwardly rectifying K+ channels, and stimulation of slow anion channels (Coursol et al., 2003). The action of S1P on ion channels is impaired in guard cells of Arabidopsis (Arabidopsis thaliana) plants harboring T-DNA null mutations in the sole prototypical G-protein α-subunit gene, GPA1, suggesting that heterotrimeric G-proteins are downstream targets for S1P in plants, as in mammals (Coursol et al., 2003). Interestingly, recent evidence shows that GCR1, one putative GPCR in Arabidopsis, can directly interact with GPA1 and negatively controls S1P regulation of stomatal apertures (Pandey and Assmann, 2004).
In eukaryotes, cellular levels of S1P are ultimately regulated by the balance between synthesis from Sph via sphingosine kinase (SphK; Fig. 1A) and degradation by S1P lyase or phosphohydrolases specific for S1P (Le Stunff et al., 2004). Two different isotypes of SphK (SphK1 and SphK2) have previously been cloned and characterized in mammals (Kohama et al., 1998; Liu et al., 2000; Melendez et al., 2000; Nava et al., 2000; Pitson et al., 2000; Fukuda et al., 2003). Although similar in amino acid sequence, SphK1 and SphK2 have different substrate specificities, kinetic properties, tissue distribution, and opposite actions in apoptosis and proliferation (Olivera et al., 1999, 2003; Igarashi et al., 2003; Inagaki et al., 2003; Liu et al., 2003). Recently, two putative Drosophila SphK genes, Sk1 and Sk2, have been characterized (Herr et al., 2004). They both show greater resemblance to mammalian SphK2 than to SphK1, suggesting that SphK2 is the more primitive of the two mammalian isoenzymes. A null mutant of Sk2 demonstrated elevated long-chain-base levels, impaired flight performance, and diminished fecundity (Herr et al., 2004). S. cerevisiae also has two SphKs, referred to as long-chain sphingoid base kinase 4 (LCB4) and 5 (LCB5; Nagiec et al., 1998). Although LCB4 and LCB5 have similar substrate specificities (Nagiec et al., 1998) and function redundantly in heat stress-induced cell cycle arrest (Jenkins and Hannun, 2001), they have different subcellular localizations (Hait et al., 2002; Funato et al., 2003) that could be related to specific functions. Indeed, recent studies have shown a specific role of LCB4 in cell growth inhibition and ceramide synthesis from exogenous sphingoid base (Kim et al., 2000; Funato et al., 2003). In contrast, LCB5 appears to play a role in heat stress resistance during induced thermotolerance (Ferguson-Yankey et al., 2002).
Until recently, no specific demonstration of SphK activity was reported in plants, although related kinases able to phosphorylate DHS were reported in Zea mays and Arabidopsis (Crowther and Lynch, 1997; Nishiura et al., 2000). We recently showed that SphK activity is present in Arabidopsis and that the enzyme activity can be stimulated by the plant hormone abscisic acid (ABA) in guard cell and mesophyll cell protoplasts (Coursol et al., 2003). Although that study provided evidence for S1P formation in Arabidopsis, much remains to be learned about plant SphKs. In this study, we characterized SphK activity in Arabidopsis leaves and examined its intracellular distribution. We found that Phyto-Sph, the predominant free long-chain base in plants, is a substrate for Arabidopsis SphK. In addition, our results reveal that phytosphingosine-1-phosphate (Phyto-S1P) is an effective regulator of guard cell aperture that requires the presence of functional GPA1.
RESULTS
Characterization of SphK from Arabidopsis Leaves
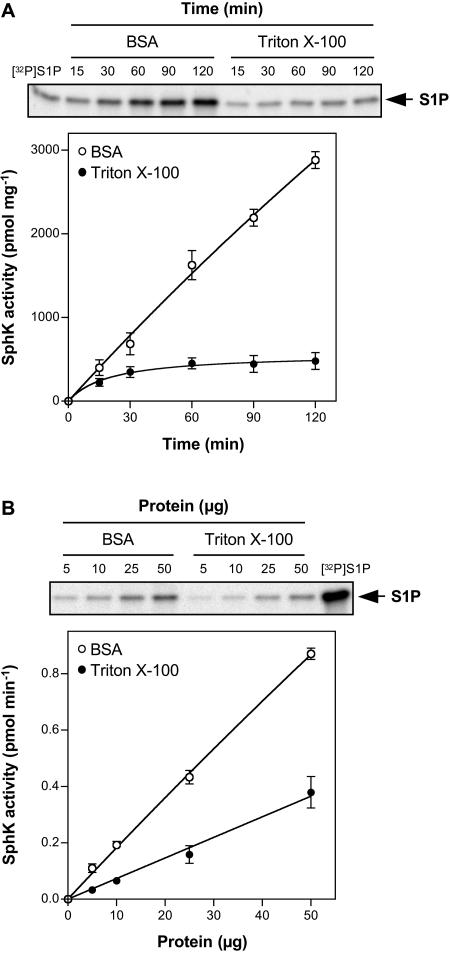
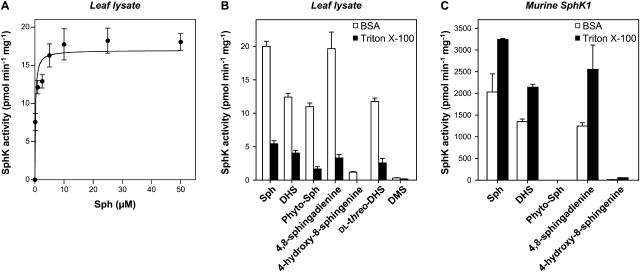
SphK activity in Arabidopsis leaf lysates was measured with [γ-32P]ATP and Sph as substrates by quantifying the formation of [32P]S1P. Because Sph is highly lipophilic, for in vitro SphK assays it is usually presented either in micellar form with nonionic detergents, such as Triton X-100, or as a complex with bovine serum albumin (BSA; Liu et al., 2000). It was previously shown that mammalian SphK1 and SphK2 are both active when Sph is added in BSA. However, when Sph is presented in Triton X-100, SphK1 activity is stimulated, while SphK2 is inactive (Liu et al., 2003). Thus, differential substrate presentation can be used to qualitatively distinguish between SphK1 and SphK2 activities in tissues or cells that express both isoforms (Billich et al., 2003; Paugh et al., 2003). Arabidopsis SphK activity was assayed at 37°C to allow direct comparison with the properties of mammalian SphKs. When Sph was presented to the leaf extracts as Triton X-100 mixed micelles, a 32P-labeled lipid that comigrated on thin-layer chromatography (TLC) with 32P-labeled S1P standard was produced (Fig. 2A). S1P formation was only linear for a short period of time and plateaued after 30 min. Sph consumption did not exceed 1%, suggesting that the plateau in S1P formation in Triton X-100 mixed micelles was not due to substrate depletion. In contrast, when Sph was presented as a BSA complex, the rate of S1P formation was much greater and nearly constant for at least 120 min (Fig. 2A). Additionally, S1P formation was proportional to the amount of protein added over the range of 5 to 50 μg when Sph was presented either in Triton X-100 mixed micelles or as a BSA complex (Fig. 2B). High KCl concentrations inhibit mammalian SphK1 but stimulate SphK2 when Sph is added as a BSA complex (Liu et al., 2000). We found that Arabidopsis SphK activity was increased by 56% when Sph was added as a BSA complex and the KCl concentrations were raised from 0 to 0.5 m KCl (data not shown).
Figure 2.
Dependence of Arabidopsis SphK activity on time and protein amount. SphK activity in leaf lysates was measured with Sph (50 μm) added either as a BSA complex or as Triton X-100 mixed micelles. A, Leaf lysates (25 μg of protein) were incubated for the indicated times. B, The indicated amounts of leaf lysate protein were incubated for 30 min. When linearity and saturation were implied, a straight line and a saturation curve were fitted, respectively; R2 = 0.9966 and 0.9912 for BSA and Triton X-100, respectively, in A; R2 = 0.9986 and 0.9909 for BSA and Triton X-100, respectively, in B. Data are means ± se of three independent experiments. Note that, when error bars are not indicated, the se is less than the size of the symbols. Top images show representative TLCs demonstrating the formation of [32P]S1P with increasing time (A) or amounts (B) of protein. The phosphorylated product of Sph produced by murine SphK1 was used as an authentic [32P]S1P standard.
Kinetic Characterization of Arabidopsis SphK
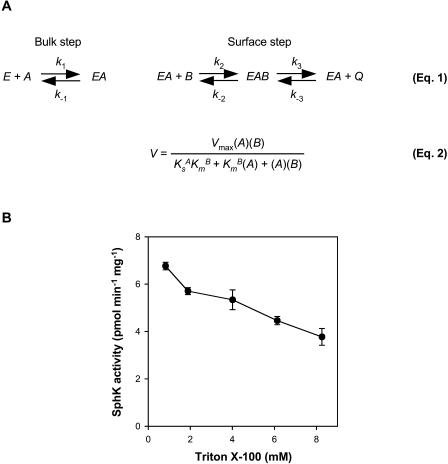
To study the kinetic behavior of Arabidopsis SphK in leaf lysates, we first used the Triton X-100 mixed-micelle assay system, following the surface dilution model of enzyme kinetics (Carman et al., 1995). This model takes into account both two-dimensional surface interaction and three-dimensional bulk interaction between the enzyme and the lipid substrate (Fig. 3A). Such consideration is critical in determining the kinetic parameters of these enzymes, because lipid-dependent enzymes, such as SphK, catalyze reactions at a water-lipid interface (Carman et al., 1995). Triton X-100 is one of the most frequently employed detergents for surface dilution kinetics because it has been shown to form uniform mixed micelles with a variety of lipid molecules, including phosphatidylcholine, phosphatidylinositol, and long-chain sphingoid bases (Carman et al., 1995). Our data (Fig. 3B) show that Arabidopsis SphK activity decreased when the surface concentration of Sph in the mixed micelle was diluted by the addition of Triton X-100 while the bulk concentration of Sph was kept the same. Therefore, the enzyme activity exhibited typical surface dilution effects in the presence of increasing concentrations of Triton X-100 in mixed micelles.
Figure 3.
Principle of the surface dilution model. A, The principle of the surface dilution model is presented in Equation 1 (Carman et al., 1995). According to this model, the action of the enzyme E consists of two consecutive steps. First, the enzyme interacts noncatalytically with the surface of detergent/lipid mixed micelles A. Subsequently, the enzyme mixed-micelle complex EA binds to the individual lipid substrate B presented on the surface, which leads to the conversion of substrate to product Q. The first association depends on the bulk concentration of both E and A, whereas the second step depends on the surface concentrations of EA and B. A is defined as the sum of the molar concentrations of detergent plus lipid substrate, and B is the mole fraction of lipid substrate in the mixed micelle. Equation 2 is the rate expression for the surface dilution kinetic model (Carman et al., 1995). Three kinetic parameters, Vmax, KsA, and KmB, can be determined from this equation. Vmax is the true Vmax when both the bulk concentration and the surface concentration of the lipid substrate approach infinity. KsA, which equals k−1/k1 and is expressed in bulk concentration terms, is the dissociation constant describing the interaction of the enzyme with the mixed micelles in the first binding step. KmB, equal to (k−2 + k3)/k2, defines the interfacial Km for the second binding step and is expressed in surface concentration units, such as mole fraction. B, SphK activity in leaf lysates measured with increasing Triton X-100 molar concentrations in the mixed micelles. The molar concentration of Sph was held constant at 50 μm. Data are means ± se of five independent experiments.
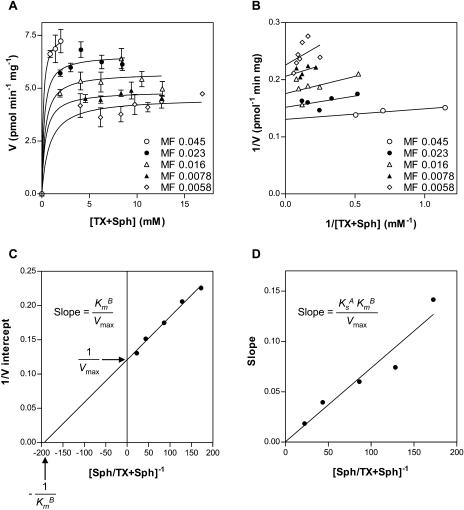
To determine the surface dilution kinetic parameters, SphK activity was measured as a function of the sum of the molar concentrations of Triton X-100 plus Sph (A of Fig. 3A, Eq. 2) at a series of mole fractions of Sph (B of Fig. 3A, Eq. 2). The activity was dependent on the sum of molar concentrations of Triton X-100 plus Sph at each surface concentration (Fig. 4A). As the surface concentration of Sph in the mixed micelles decreased, the apparent Vmax decreased. A double reciprocal plot of the data in Figure 4A indicated that Arabidopsis SphK exhibits saturation kinetics when the bulk concentration of Triton X-100 plus Sph is varied at each fixed mole fraction of Sph (Fig. 4B). According to Equation 2 (Fig. 3A), the replot of the 1/V intercept from Figure 4B versus 1/B should be linear; the intercept of the 1/V intercept axis is equal to 1/Vmax, and the intercept of the 1/B axis is equal to −1/KmB (Carman et al., 1995). A replot of the data was indeed linear (Fig. 4C), and the Vmax and KmB were 8.3 pmol min−1 mg−1 and 0.0052 mol fraction, respectively (Table I). Equation 2 also predicts that a replot of the slopes from Figure 4B versus 1/B should be linear and cross the origin, and the slope of the replot is equal to KsAKmB/Vmax (Carman et al., 1995). Figure 4D shows that such a replot was indeed linear and passed through the origin. By using the slope of the line in Figure 4C, the dissociation constant KsA for the Triton X-100 mixed micelles was calculated to be 1.2 mm (Table I).
Figure 4.
Activity of Arabidopsis SphK toward Sph in mixed micelles with Triton X-100 according to the surface binding model. A, SphK activity in leaf lysates was measured as a function of the sum of the molar concentrations of Triton X-100 (TX) plus Sph at set mole fractions (MF) of Sph. Data are means ± se of five independent experiments. V refers to picomoles of Sph hydrolyzed per minute per milligram of enzyme. B, Reciprocal plot of the data in A. C, Replot of 1/V intercepts obtained in B versus the reciprocal of the mole fraction of Sph; R2 = 0.9896. D, Replot of slopes obtained in B versus the reciprocal of the mole fraction of Sph; R2 = 0.9210. The lines drawn in A and B were a result of a least-squares analysis of the individual data sets using Equation 2. They are statistically different [P(F4,21 > 36.75) < 0.00001].
Table I.
Kinetic parameters of Arabidopsis SphK
Phosphorylation of Sph presented in Triton X-100 micelles was measured as a function of the substrate's bulk concentration at set surface concentrations of Sph in leaf lysates. Kinetic parameters were calculated from Figure 4, C and D, according to Equation 2 of the surface dilution kinetic model depicted in Figure 3A. Apparent KsA (mm) is the dissociation constant describing the interaction of the enzyme with the mixed micelles. Apparent Vmax (picomoles per minute per milligram) is the true Vmax at an infinite mole fraction and an infinite bulk concentration of Sph. Apparent KmB (mole fraction) is the interfacial Km. Phosphorylation of Sph presented as a BSA complex was measured at various substrate concentrations in lysate, cytosolic, and membrane fractions from leaves. Apparent Vmax (picomoles per minute per milligram) and Km (μm) values were determined from the data depicted in Figures 5A and 6, A and B, using GraphPad Prism by a weighted nonlinear regression fit to the equation V = Vmax A/(Km + A), where V is the initial rate and A is the substrate concentration.
| Fraction | Vmax(app) | Km(app) | Ks(app)A | Km(app)B | |
|---|---|---|---|---|---|
| Triton X-100 | Lysate | 8.3 | - | 1.2 | 0.0052 |
| BSA | Lysate | 17.3 | 0.18 | - | - |
| BSA | Cytosol | (2.4)a | (3)a | - | - |
| BSA | Membrane | 49.6 | 0.94 | - | - |
Values in parentheses are estimates because saturation of the enzyme with Sph was not reached.
We next investigated the catalytic properties of Arabidopsis SphK toward Sph complexed to BSA in leaf lysates. SphK activity exhibited saturation kinetics and the data fit well to the Michaelis-Menten equation (Fig. 5A). Surprisingly, the Km(app) was 0.18 μm, which is much lower than the value of 3.4 μm previously reported for SphK2 (Liu et al., 2000; Table I).
Figure 5.
Substrate specificity of Arabidopsis SphK activity. A, SphK activity was measured in leaf lysates with the indicated concentrations of Sph added as a BSA complex. Data were fitted to the Michaelis-Menten equation as described in “Materials and Methods”; R2 = 0.9296. Data are means ± se of three independent experiments. B and C, Phosphorylation of various long-chain bases (50 μm) added either as a BSA complex or as Triton X-100 mixed micelles was measured in lysates from leaves (B) and HEK 293 cells overexpressing SphK1 (C). Data are means ± se of five independent experiments.
Substrate Specificity of Arabidopsis SphK Activity
We next determined the substrate specificity of Arabidopsis SphK activity (Fig. 5B). The data obtained with leaf lysates indicate a relative rank order of substrate selectivity of Sph > DHS ≥ 4,8-sphingadienine ≥ dl-threo-dihydrosphingosine (dl-threo-DHS) ≫ Phyto-Sph for Triton X-100 mixed micelles and Sph = 4,8-sphingadienine > DHS ≥ dl-threo-DHS ≥ Phyto-Sph ⋙ 4-hydroxy-8-sphingenine for BSA complexes (Fig. 5B). In agreement with our previous results (Coursol et al., 2003), the unnatural isomer of DHS, dl-threo-DHS (Fig. 1C), a potent inhibitor of mammalian SphK (Kohama et al., 1998), was also phosphorylated, but to a lesser extent than Sph, whereas N,N-dimethylsphingosine (DMS; Fig. 1C), another potent inhibitor of mammalian (Edsall et al., 1998) and Arabidopsis (Coursol et al., 2003) SphKs, was not phosphorylated regardless of the method of substrate presentation (Fig. 5B).
The data show that Sph, a low-abundance, naturally occurring long-chain base in Arabidopsis leaves (Sperling et al., 1998), was the best substrate under Triton X-100 assay conditions (P < 0.05, Student's t test). Under BSA assay conditions, Sph and 4,8-sphingadienine, which is structurally similar to Sph except for the presence of an additional double bond between carbons 8 and 9 (Fig. 1B), were the favored substrates. Importantly, our results also show that a plant SphK can utilize Phyto-Sph, a free long-chain base that is enriched in plants (Abbas et al., 1994; Wright et al., 2003), as a substrate. 4-Hydroxy-8-sphingenine, a major long-chain base in glucosylceramide (GlcCer) of Arabidopsis leaves (Imai et al., 2000) that is structurally similar to Phyto-Sph except for the presence of an additional double bond between carbons 8 and 9 (Fig. 1B), was phosphorylated by Arabidopsis SphK in BSA, albeit much less efficiently than Sph (6%), and was not phosphorylated in Triton X-100.
SphK1 shows a preference for Sph over DHS (Kohama et al., 1998; Melendez et al., 2000; Nava et al., 2000; Pitson et al., 2000) just as does Arabidopsis SphK, whereas SphK2 shows a preference for DHS compared to Sph (Liu et al., 2000). Therefore, we were interested to determine whether SphK1 was also able to phosphorylate the plant long-chain bases 4,8-sphingadienine and 4-hydroxy-8-sphingenine. We found that 4,8-sphingadienine was phosphorylated by mammalian SphK1 as efficiently as Sph in Triton X-100 mixed micelles (P = 0.3, Student's t test; Fig. 5C). In contrast, mammalian SphK1 did not have significant activity toward 4-hydroxy-8-sphingenine regardless of the method of substrate presentation (Fig. 5C).
Subcellular Distribution of Arabidopsis SphK Activity
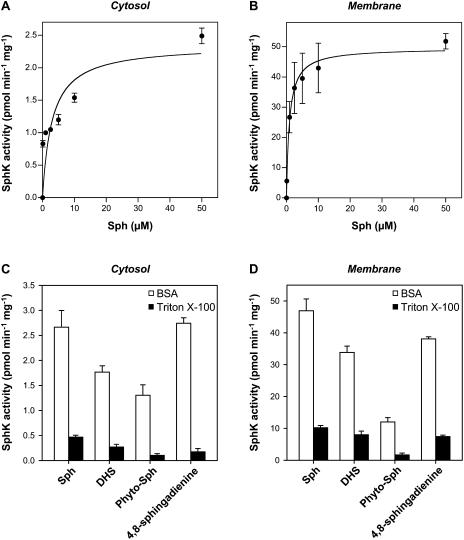
To further characterize the Arabidopsis SphK activity, we examined the subcellular distribution of the enzyme activity. Approximately 75% of the total leaf lysate SphK activity was membrane associated, as determined either in Triton X-100 or BSA (data not shown). Membrane-associated SphK had a 20-fold higher specific activity than cytosolic SphK regardless of the method of substrate presentation (Fig. 6, A and B; Table I). Additionally, SphK activity in both cytosolic and membrane fractions was higher in BSA than in Triton X-100 mixed micelles (Fig. 6, C and D). With Sph complexed to BSA as substrate, typical Michaelis-Menten kinetics were observed for Arabidopsis SphK in both cytosolic and membrane fractions (Fig. 6, A and B). The Km(app) of the membrane fraction was 0.94 μm, which is much lower than the value of 3 μm found for the cytosolic fraction (Table I).
Figure 6.
Distribution of SphK activity in Arabidopsis leaves. A and B, SphK activity was measured in cytosolic (A) and membrane (B) fractions from leaves with the indicated concentrations of Sph added as a BSA complex. Data were fitted to the Michaelis-Menten equation as described in “Materials and Methods”; R2 = 0.7189 and 0.9902 for A and B, respectively. C and D, Phosphorylation of various long-chain bases (50 μm) added either as a BSA complex or as Triton X-100 mixed micelles was measured in cytosolic (C) and membrane (D) fractions from leaves. Data are means ± se of three independent experiments. Note change in scale between A and C, and B and D. When error bars are not indicated, the se is less than the size of the symbols.
We next compared the substrate specificity of SphK activity in cytosolic and membrane fractions (Fig. 6, C and D) with that of leaf lysates (Fig. 5B). The natural long-chain bases Sph, DHS, Phyto-Sph, and 4,8-sphingadienine were chosen for the analysis because they were significantly phosphorylated by SphK in leaf lysates (Fig. 5B). The data obtained indicate a relative rank order of substrate selectivity that is comparable to that obtained with leaf lysates. Nevertheless, in a comparison of the cytosolic and membrane fractions, the cytosolic fraction shows a greater relative preference for Sph compared to the other substrates in Triton X-100 (Fig. 6C). Additionally, in BSA the cytosolic fraction shows a 2-fold greater relative preference for Phyto-Sph compared to the membrane fraction (Fig. 6D).
Responses of Arabidopsis Guard Cells to Phyto-S1P
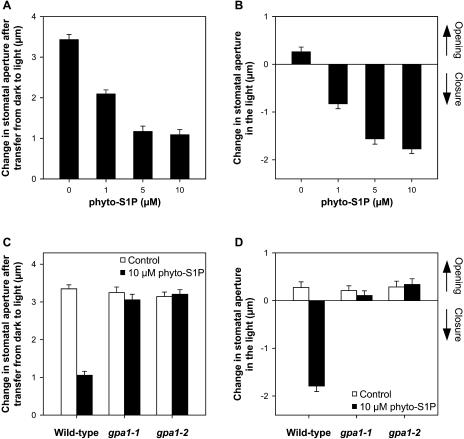
Stomata form pores on leaf surfaces that regulate uptake of CO2 for photosynthesis and loss of water vapor during transpiration (Hetherington and Woodward, 2003). During drought, ABA plays an essential role by triggering rapid, osmotically driven water loss from the pair of guard cells that surround each stomatal pore (Schroeder et al., 2001). This results in guard cell shrinkage, stomatal closure, and consequent reductions in water loss. Recently, we showed that S1P is involved in ABA inhibition of stomatal opening and promotion of stomatal closure in Arabidopsis, affecting the activities of plasma membrane inward K+ channels and slow anion channels (Coursol et al., 2003). Because Phyto-Sph is the predominant free sphingoid base present in plants (Abbas et al., 1994) and is an effective substrate for Arabidopsis SphK (Fig. 5), it was of interest to test the ability of its phosphorylated form, Phyto-S1P, to modulate Arabidopsis guard cell turgor. Exogenous Phyto-S1P (1–10 μm) inhibited the light-induced opening of closed stomata (Fig. 7A). In addition, Phyto-S1P (1–10 μm) promoted closure of open stomata (Fig. 7B). Phyto-S1P concentrations above 10 μm did not have any further effects on stomatal apertures (data not shown). Together, these results provide evidence that Phyto-S1P, similarly to S1P, is competent to regulate stomatal apertures in Arabidopsis.
Figure 7.
Effects of Phyto-S1P on stomatal aperture in wild-type and G-protein α-subunit null mutant gpa1-1 and gpa1-2 plants. A and C, Apertures after a 2.5-h pretreatment in the dark and a further 2-h light treatment with or without Phyto-S1P. Changes in stomatal aperture reflect final minus initial (3.15 ± 0.15 μm) aperture in the dark. B and D, Apertures after a 2.5-h light pretreatment and a further 3-h light treatment with or without Phyto-S1P. Changes in stomatal aperture reflect final minus initial (6.09 ± 0.07 μm) aperture. Data are means ± se of 3 independent experiments; n = 40 apertures/experiment. Pairwise comparisons are simultaneously significant at P ≤ 0.05 by Scheffe's method.
In animal cells, extracellular actions of S1P via GPCRs are well established (Spiegel and Milstien, 2003). Recently, we used two independent Arabidopsis T-DNA knockout lines, gpa1-1 and gpa1-2 (Ullah et al., 2001; Wang et al., 2001), to show that the sole canonical plant G-protein α-subunit, GPA1, is required for S1P to regulate guard cell ion channel activities and stomatal apertures (Coursol et al., 2003). Therefore, we next examined whether Phyto-S1P signals in guard cells are also transduced via GPA1. In contrast to the response of wild-type plants, exogenous Phyto-S1P neither inhibited stomatal opening (Fig. 7C) nor promoted stomatal closure (Fig. 7D) in gpa1-1 or gpa1-2 null mutant plants. These results suggest that GPA1 mediates both S1P and Phyto-S1P signaling in Arabidopsis guard cells.
DISCUSSION
Although S1P recently has been implicated in plant cell signaling (Ng et al., 2001; Coursol et al., 2003; Pandey and Assmann, 2004), plant SphKs have not yet been well defined. In this report, we characterized the enzymatic properties of SphK from Arabidopsis leaves. We found that presentation of Sph as Triton mixed micelles gives much lower activity than presentation of the substrate as a complex with BSA (Fig. 2). Results from the surface dilution kinetics suggest that Arabidopsis SphK binds to Triton mixed micelles with the same affinity regardless of the micellar composition (Fig. 4). Thus, the initial binding step (Fig. 3A) is likely to occur through nonspecific hydrophobic interactions with the micelle surface. The Km(app)B of 0.0052 mol fraction is comparable to the Km(app) of 0.002 and 0.0035 mol fraction previously reported for yeast and rat brain SphK, respectively (Buehrer and Bell, 1992; Lanterman and Saba, 1998; Table I). In striking contrast, the Km(app) value obtained with Sph complexed to BSA as substrate is much lower than the value previously determined for SphK2 (Liu et al., 2000; Table I), suggesting that Arabidopsis SphK has a very high affinity for Sph complexed to BSA. These data suggest that Arabidopsis SphK has unique catalytic properties compared to mammalian SphKs.
Arabidopsis SphK demonstrated activity with Sph, DHS, an intermediate in the de novo synthesis pathway in eukaryotes (Merrill, 2002), and Phyto-Sph, which is mainly derived from the hydroxylation of free DHS (Wright et al., 2003) and is a high-abundance, naturally occurring free long-chain base in plants (Abbas et al., 1994; Fig. 5B). Although Sph is a quantitatively minor free long-chain base in plants (Imai et al., 2000), it was the best substrate for Arabidopsis SphK (Fig. 5B). These data demonstrate that specificity of Arabidopsis SphK for long-chain bases is distinct from that of mammalian SphKs. Indeed, SphK1 shows a preference for Sph over DHS, as does Arabidopsis SphK, but does not phosphorylate Phyto-Sph or dl-threo-DHS (Kohama et al., 1998; Melendez et al., 2000; Nava et al., 2000; Pitson et al., 2000). By contrast, SphK2 shows a greater preference for DHS compared to Sph, but is able to phosphorylate Phyto-Sph and dl-threo-DHS (Liu et al., 2000). Our data also reveal that Arabidopsis SphK has unique substrate specificity compared to the related Z. mays DHS kinase, which, based on competition experiments, phosphorylates Sph and DHS but does not phosphorylate Phyto-Sph and dl-threo-DHS (Crowther and Lynch, 1997).
In animal cells, Sph is the predominant long-chain-base moiety. It is derived solely from breakdown of complex sphingolipids through the endocytic/salvage pathway, allowing the cells to minimize the de novo synthesis of DHS (Gillard et al., 1998). In contrast, in plants, 8-sphingenine, 4,8-sphingadienine, and 4-hydroxy-8-sphingenine are the predominant long-chain-base moieties in complex sphingolipids (Imai et al., 2000; Lynch and Dunn, 2004). These long-chain bases are produced through the action of Δ4- and Δ8-desaturases, which introduce a double bond between carbons 4 and 5 and carbons 8 and 9, respectively, in DHS and dihydroceramide (Sperling et al., 1998; Ternes et al., 2002). We found that 4,8-sphingadienine and, to a lesser extent, 4-hydroxy-8-sphingenine, but not 8-sphingenine (data not shown), were phosphorylated by Arabidopsis SphK (Fig. 5B). Surprisingly, 4,8-sphingadienine, which is virtually absent in mammalian systems (Renkonen, 1970), was also phosphorylated by mammalian SphK1 (Fig. 5C). In contrast, 4-hydroxy-8-sphingenine was not a substrate for mammalian SphK1 and was a poor substrate for Arabidopsis SphK (Fig. 5C). Therefore, the presence of a Δ8-double bond alone or associated with a hydroxyl group at position 4 on the acyl chain of the long-chain base drastically reduced catalytic efficiency of Arabidopsis SphK, suggesting that this enzyme is regioselective. Interestingly, the Δ8-double bond had no effect on Arabidopsis SphK activity when present together with the Δ4-double bond in 4,8-sphingadienine. Collectively, these results suggest that plant SphK and mammalian SphK1 have marked preferences for long-chain bases with a Δ4-double bond. Interestingly, studies using mammalian cells and guard cells showed that dihydrosphingosine-1-phosphate, a phosphorylated long-chain base lacking the Δ4-double bond, was not effective in altering apoptosis (Cuvillier et al., 1996), Ca2+ mobilization (Ng et al., 2001), and stomatal apertures (Ng et al., 2001; Coursol et al., 2003), suggesting the importance in signaling of modification of the fourth carbon.
In contrast to mammalian SphK1, which is mainly cytosolic (Kohama et al., 1998), Arabidopsis SphK activity is predominantly associated with membranes (Fig. 6; Table I). Membrane localization was also observed for the DHS kinase from Z. mays (Crowther and Lynch, 1997). The hidden Markov-based TMHMM software and the SignalP program predict that two of the three candidate Arabidopsis SphK proteins (discussed in detail below) lack transmembrane-spanning domains and signal anchors. However, SignalP did predict the presence of a signal peptide in the third candidate NP_193885 (data not shown). Data from S. cerevisiae, where membrane association of SphK activity is reported (Nagiec et al., 1998; Olivera et al., 1998; Hait et al., 2002; Funato et al., 2003) in the absence of membrane localization signals in LCB4 and LCB5 (Nagiec et al., 1998), indicate that such signals are not required for membrane localization of these lipid kinases. Recently, we showed that the phytohormone ABA stimulated the phosphorylation of exogenously supplied Sph in leaf and guard cell lysates of Arabidopsis (Coursol et al., 2003). Given the membrane localization of the majority of Arabidopsis SphK activity, it is reasonable to assume that Arabidopsis SphK activity is regulated by the availability of substrate rather than by translocation to membranes. However, because 25% of Arabidopsis SphK activity is cytosolic, it is possible that this SphK might be translocated from the cytosol to membranes upon stimulation of guard cells by ABA. Consistent with this notion, recent studies have shown that various agonists can induce translocation of mammalian SphK1 to membranes (Kleuser et al., 2001; Johnson et al., 2002; Melendez and Khaw, 2002; Young et al., 2003), resulting in spatial and temporal regulation of S1P formation.
It is conceivable that the cytosolic Arabidopsis SphK may be a different isozyme than the membrane bound SphK. Several lines of evidence suggest that multiple isoforms of SphKs, which exhibit different substrate specificities and subcellular localizations than SphK1 and SphK2, exist in mammals (Banno et al., 1998; Gijsbers et al., 2001; Fukuda et al., 2003). Indeed, membrane bound SphKs with high activity toward Phyto-Sph have been detected in human platelets (Banno et al., 1998) and in rat tissues (Gijsbers et al., 2001; Fukuda et al., 2003). This activity was not attributable to either SphK1 or SphK2 because it was not depleted by anti-SphK1 and anti-SphK2 antibodies (Fukuda et al., 2003). Therefore, it was of interest to compare the enzymatic properties of the cytosolic and membrane bound SphK from Arabidopsis leaves. Although little SphK activity was detected in the cytosolic fraction of Arabidopsis leaves, the kinetic behavior of the cytosolic SphK (Fig. 6A) was found to differ markedly from that of the total (Fig. 5A) and membrane bound enzyme (Fig. 6B). Thus, the Km(app) of the cytosolic SphK was 3 μm, which is about 3- and 17-fold higher than the Km(app) of the total and membrane bound SphKs, respectively (Table I). Moreover, there were differences in the substrate preference of the lysate, cytosolic, and membrane SphKs from Arabidopsis leaves. The cytosolic fraction showed a greater relative preference for Sph compared to DHS, Phyto-Sph, and 4,8-sphingadienine in Triton X-100 (Fig. 6C), and the membrane fraction showed a lower relative preference for Phyto-Sph in BSA (Fig. 6D). Collectively, these results suggest that SphK activity may be catalyzed by multiple isoforms in Arabidopsis.
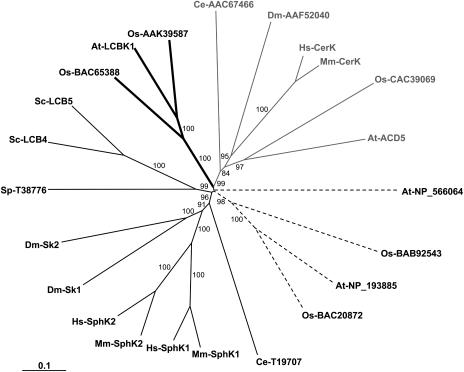
To assess whether multiple SphK genes may exist in the Arabidopsis genome, we analyzed the public databases for the presence of putative SphK coding sequences. Three gene loci were detected that could potentially code for SphK proteins (At2g46090, At4g21540, and At5g23450, which encode NP_566064, NP_193885, and At-LCBK1, respectively; Worrall et al., 2003). The predicted amino acid sequences of these three genes contain the five conserved domains (C1–C5) previously identified in SphKs (Le Stunff et al., 2004), suggesting that they might be SphKs. This interpretation should, however, be tempered by the fact that substrate specificity cannot be deduced from protein sequence homology. The recent cloning of mammalian ceramide kinase (CerK; Sugiura et al., 2002) and Arabidopsis ACD5, recently identified as a plant CerK (Liang et al., 2003), are good examples. Although mammalian and Arabidopsis CerKs contain the five conserved catalytic domains previously identified in the SphKs (Le Stunff et al., 2004), they do not catalyze the phosphorylation of Sph but instead are specific for ceramide (Sugiura et al., 2002; Liang et al., 2003). Nevertheless, phylogenetic analysis clearly indicates that the three candidate proteins fall into three major clusters that are distinguishable from the cluster CerKs (Fig. 8). The two putative Arabidopsis SphK proteins, NP_193885 and At-LCBK1, fall into 2 plant clusters that are distinguishable from both CerK and metazoan SphK cluster enzymes, while At-NP_566064 has no clear relationship with any cluster of the phylogenetic tree (Fig. 8). To date, only At-LCBK1 can unequivocally be designated as a long-chain-base kinase in Arabidopsis since recombinant At-LCBK1 has been shown to phosphorylate DHS (Nishiura et al., 2000).
Figure 8.
Phylogenetic tree of SphKs and CerKs. Deduced amino acid sequences were aligned using ClustalW multiple alignment and visually refined using BioEdit Sequence Alignment Editor 4.8.8. The phylogenetic tree was generated by the neighbor-joining method using the PAUP 4.0b10 package. Branch lengths are scaled proportional to genetic distance. Bootstrap values (percent) were obtained with 1,000 replicates and those exceeding 50% are indicated above the supported branches. Sequences from the following organisms were used: At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. Known SphKs and CerKs are as follows: At-LCBK1 (accession no. NP_568432), At-ACD5 (accession no. AAQ62904), Dm-Sk1 (accession no. AAF48045), Dm-Sk2 (accession no. AAF47706), Hs-SphK1 (accession no. AAF73423), Hs-SphK2 (accession no. AAF74124), Hs-CerK (accession no. BAC01154), Mm-SphK1 (accession no. AAC61697), Mm-SphK2 (accession no. AAF74125), Mm-CerK (accession no. BAC01155), Sc-LCB4 (accession no. NP_014814), Sc-LCB5 (accession no. NP_013361). Accession numbers of putative SphKs, CerKs, and long-chain-base kinases are indicated in the dendrogram. Thin shading, thick shading, gray, and dotted lines refer to Cluster SphKs, long-chain-base kinases, CerKs, and putative long-chain-base kinases, respectively.
In Arabidopsis guard cells, S1P seems to function upstream of the sole prototypical G-protein α-subunit GPA1 because gpa1 knockout plants are insensitive to S1P inhibition of stomatal opening and promotion of stomatal closure as well as to S1P inhibition of inwardly rectifying K+ channels and activation of slow anion channels (Coursol et al., 2003). In support of positioning S1P upstream of GPA1, basal levels of SphK activity were found to be similar in leaf and guard cell lysates of wild-type and gpa1 null mutant plants (Coursol et al., 2003; data not shown). Here, we show that Phyto-S1P, similarly to S1P, inhibits stomatal opening and promotes stomatal closure in Arabidopsis and that its action is impaired in guard cells of gpa1 knockout plants. This observation suggests a biological role of Phyto-S1P in plant cell signaling. The high abundance of free Phyto-Sph in plants indicates that Phyto-Sph may not be a rate-limiting substrate for Arabidopsis SphK. However, further experiments are needed to determine the relative importance of Phyto-S1P and S1P as signaling molecules in guard cells. It would also be worthwhile to investigate whether the ABA response is mediated by Phyto-S1P in guard cells. Meanwhile, our results also have broader implications for Phyto-S1P signaling events in plants. GPA1 regulates not only guard cell function (Wang et al., 2001; Coursol et al., 2003) but also plant cell division (Ullah et al., 2001; Perfus-Barbeoch et al., 2004). In this regard, it is well established that Phyto-S1P regulates cell proliferation in yeast (Skrzypek et al., 1999; Kim et al., 2000). Our finding that GPA1 may be implicated in Phyto-S1P action in guard cells thus opens the exciting possibility that Phyto-S1P may also play a role in other G-protein-mediated processes in plants. Interestingly, it has been reported recently that Phyto-S1P is a high-affinity ligand for the mammalian S1P4 GPCR (Candelore et al., 2002). Therefore, our data raise the intriguing possibility that a functional relationship between Phyto-S1P and heterotrimeric G-proteins is evolutionarily conserved between animals and plants.
In conclusion, this study shows that Phyto-S1P, similar to S1P, is active in guard cell signaling, supporting a role for phosphorylated long-chain sphingoid bases in modulating physiological processes in plants. In addition, enzymatic analyses presented here provide the biochemical basis for future molecular characterization of Arabidopsis SphKs. Analysis of which of the three putative SphK genes encode true SphKs and characterization of which of these enzyme(s) encode the Phyto-S1P-related kinase activities we have identified will provide the next step in clarifying the roles of SphKs in plant metabolism and signaling.
MATERIALS AND METHODS
Materials
[γ-32P]ATP (3,000 Ci/mmol) was purchased from Amersham Biosciences (Buckinghamshire, UK); Sph, DHS, dl-threo-DHS, DMS, and Phyto-Sph were obtained from Biomol Research Laboratory (Plymouth Meeting, PA); and Phyto-S1P from Avanti Polar Lipids (Alabaster, AL). Stock solutions were prepared in ethanol (Sph, DHS, dl-threo-DHS, DMS, Phyto-Sph) or in chloroform (Phyto-S1P). Prior to use, Phyto-S1P was dried under a stream of nitrogen and then resuspended by sonication in water. Soybean lecithin powder was from Archer Daniels Midland (Decatur, IL). Bradford protein assay reagent was from Bio-Rad (Hercules, CA). Leupeptin, aprotinin, and pepstatin were obtained as a complete EDTA-free protease inhibitor cocktail from Roche (Mannheim, Germany). Triton X-100, β-glycerophosphate, BSA, phenylmethylsulfonyl fluoride, insoluble polyvinyl pyrrolidone, and sand were purchased from Sigma (St. Louis). Silica gel G60 TLC plates were from Merck (Darmstadt, Germany).
Preparation of Cytosolic and Membrane Fractions from Arabidopsis Leaves
Plants of Arabidopsis thaliana L. ecotype Wassilewskija were grown as previously described (Pandey et al., 2002). Leaves were harvested from 4- to 6-week-old plants and homogenized in SphK buffer (0.2 m Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm 2-mercaptoethanol, 10% [v/v] glycerol, 0.5 mm 4-deoxypyridoxine, 1 mm NaVO3, 15 mm NaF, 40 mm β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, 10 μg/mL each of leupeptin, aprotinin, pepstatin, and soybean trypsin inhibitor) containing 0.6% (w/v) insoluble polyvinyl pyrrolidone and sand (−50 + 70 mesh). Homogenates were then centrifuged at 10,000g for 10 min. Resultant supernatants were centrifuged at 100,000g for 30 min to obtain cytosol and total membrane fractions. Pellets were resuspended in SphK buffer (see below). Protein concentrations were determined using the Bradford reagent, with BSA as the standard. All extraction and purification procedures were carried out at 4°C. All fractions were frozen in liquid nitrogen and stored at −80°C.
Isolation of 4-Hydroxy-8-Sphingenine and 4,8-Sphingadienine from Soybean
The predominant long-chain sphingoid bases of plant sphingolipids are isomers of 4-hydroxy-8-sphingenine (t18:1) and 4,8-sphingadienine (d18:2), which are not commercially available. Accordingly, these were prepared from soybean GlcCer essentially as described previously (Cahoon and Lynch, 1991; Sullards et al., 2000). Briefly, crude soybean lecithin powder was subjected to mild alkaline hydrolysis in 20 volumes (w/v) 0.6 n NaOH in methanol. Equal volumes of chloroform and water were added to separate phases. The GlcCer recovered in the chloroform phase was precipitated with cold acetone and hydrolyzed with 2 mL of 0.5 n HCl in methanol at 65°C overnight. The liberated long-chain bases were extracted by adding equal volumes of chloroform and 0.6 n NaOH and separated by TLC on silica gel G60 using chloroform:methanol:acetic acid:water (85:15:15:3; v/v/v/v). The long-chain bases were extracted from the silica gel with 3 mL of methanol:chloroform:0.1 n NH4OH (10:5:1; v/v/v) and analyzed by HPLC as described previously (Merrill et al., 1988; Wright et al., 2003). Two major fractions were collected: one contained >94% 4-hydroxy-8-E-sphingenine; the other fraction contained 8-E- (64%) and 8-Z- (32%) isomers of 4,8-sphingadienine. Only minor amounts of other long-chain bases were present in these fractions (see Supplemental Fig. 1).
Preparation of Triton X-100/Sph Mixed Micelles
Sph in ethanol was transferred to a siliconized glass tube and dried under a stream of nitrogen. Triton X-100-Sph mixed micelles were prepared by adding Triton X-100 to the dried Sph and vortexing. The total Sph concentration in Triton X-100-Sph mixed micelles did not exceed 0.15 mol fraction to ensure that the structure of the micelles was not disrupted (Furneisen and Carman, 2000). The mole fraction of Sph in mixed micelles was calculated according to the formula [Sph (mole fraction)] = [Sph (mm)]/([Sph (mm)] + [Triton X-100 (free) (mm)]), where [Triton X-100 (free) (mm)] = [Triton X-100 (total) (mm)] − critical micelle concentration of Triton X-100 (0.24 mm).
Assay of SphK Activity
SphK activity was measured essentially as previously described (Olivera et al., 1998). In brief, cell extracts were incubated at 37°C in 200 μL SphK buffer in the presence of 50 μm Sph added in micellar form with 0.25% (v/v) Triton X-100, and [γ-32P]ATP (10 μCi, 1 mm) in 10 mm MgCl2. In some experiments, Sph prepared as a complex with 4 mg/mL BSA (final concentration of 0.2 mg/mL) was added in the presence of 200 mm KCl. Reactions were stopped by addition of 800 μL chloroform:methanol:concentrated HCl (100:200:1; v/v/v). Chloroform (250 μL) and 2 m KCl (250 μL) were then added sequentially to generate a two-phase system. The labeled lipids in the organic phases were separated by TLC with chloroform:acetone:methanol:acetic acid:water (10:4:3:2:1; v/v/v/v) and visualized and quantified with a phosphorimager (Molecular Dynamics, Sunnyvale, CA). SphK activity was expressed as picomoles of S1P formed per minute and per milligram of protein. Michaelis-Menten plots were performed using GraphPad Prism 3.02 (GraphPad Software, San Diego).
Stomatal Aperture Measurements
Stomatal aperture measurements were conducted essentially as described (Wang et al., 2001). Briefly, rosette leaves from plants 4 to 6 weeks old were harvested in darkness at the end of the night period. To determine stomatal opening, leaves were placed cuticle side up in 4.5-cm-diameter plastic petri dishes containing 10 mm KCl, 7.5 mm potassium iminodiacetate, 10 mm MES-KOH, pH 6.15, at 20°C for 2 h in the dark to ensure closed stomata. Baseline stomatal aperture was determined by measuring 10 apertures from 4 different paradermal sections of abaxial epidermis with an optical micrometer. Leaves were then incubated at 20°C for 2 h under illumination at 200 μmol m−2 s−1 in the absence or presence of Phyto-S1P. Stomatal aperture measurements (10 apertures from 4 different paradermal sections) were then made for each treatment. To induce open stomata whose closing responses could be assessed, leaves were placed cuticle side up in petri dishes containing 20 mm KCl, 1 mm CaCl2, 5 mm MES-KOH, pH 6.15, at 20°C for 2 h under illumination at 200 μmol m−2 s−1. Leaves were then incubated at 20°C for a further 3 h in the light in the absence or presence of Phyto-S1P. Stomatal apertures were measured as described for stomatal opening experiments prior to and following a 3-h incubation in the light.
Sequence Alignment and Phylogenetic Analysis
Amino acid sequences of open reading frames were initially aligned using ClustalW (Thompson et al., 1994) with the BioEdit Sequence Alignment Editor 4.8.8 (Hall, 1999). The alignments were then visually refined. Only regions of unambiguous alignments were used in the subsequent phylogenetic analysis as previously described (Sugiura et al., 2002). The N-terminal portions of the proteins were omitted because of the difficulty of confidently assessing primary homology among these sequences. After the N-terminal regions, the sequences of SphKs and CerKs encompass five conserved domains recently identified in all SphKs and CerKs (Sugiura et al., 2002) and share high degrees of similarity (Sugiura et al., 2002). These regions were therefore included in the phylogenetic analyses that were performed using the neighbor-joining method with the PAUP 4.0b10 package (PPC AltiVec). A dendrogram was constructed using TreeView 1.5.2 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requester.
Supplementary Material
Acknowledgments
We thank Ana Olivera and Sheldon Milstien for technical advice, Catherine Damerval for guiding us through the phylogenetic analysis, Jean Coursol for statistical analyses, and Carl K.-Y. Ng for critically reading the manuscript. We also thank three anonymous reviewers for comments that improved the manuscript.
This work was supported by the Institut National de la Recherche Agronomique (grant to S.C.), by the National Science Foundation (grant nos. MCB–0312864 to D.V.L. and MCB–0209694 to S.M.A.), by the U.S. Department of Agriculture (grant nos. 01–35304–09898 to S.G. and 01–35304–09916 to S.M.A.), and by the National Cancer Institute (grant no. CA61774 to S.S.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055806.
References
- Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, Sessions AE, Wang E, Merrill AH Jr, Riley RT (1994) Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol 106: 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno Y, Kato M, Hara A, Nozawa Y (1998) Evidence for the presence of multiple forms of Sph kinase in human platelets. Biochem J 335: 301–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billich A, Bornancin F, Dévay P, Mechtcheriakova D, Urtz N, Baumruker T (2003) Phosphorylation of the immunomodulatory drug FTY720 by sphingosine kinases. J Biol Chem 278: 47408–47415 [DOI] [PubMed] [Google Scholar]
- Buehrer BM, Bell RM (1992) Inhibition of sphingosine kinase in vitro and in platelets. Implications for signal transduction pathways. J Biol Chem 267: 3154–3159 [PubMed] [Google Scholar]
- Cahoon EB, Lynch DV (1991) Analysis of glucocerebrosides of rye (Secale cereale L. cv. Puma) leaf and plasma membrane. Plant Physiol 95: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelore MR, Wright MJ, Tota LM, Milligan J, Shei GJ, Bergstrom JD, Mandala SM (2002) Phytosphingosine 1-phosphate: a high affinity ligand for the S1P4/Edg-6 receptor. Biochem Biophys Res Commun 297: 600–606 [DOI] [PubMed] [Google Scholar]
- Carman GM, Deems RA, Dennis EA (1995) Lipid signaling enzymes and surface dilution kinetics. J Biol Chem 270: 18711–18714 [DOI] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Crowther GJ, Lynch DV (1997) Characterization of sphinganine kinase activity in corn shoot microsomes. Arch Biochem Biophys 337: 284–290 [DOI] [PubMed] [Google Scholar]
- Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind JS, Spiegel S (1996) Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381: 800–803 [DOI] [PubMed] [Google Scholar]
- Edsall LC, Van Brocklyn JR, Cuvillier O, Kleuser B, Spiegel S (1998) N,N-Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry 37: 12892–12898 [DOI] [PubMed] [Google Scholar]
- Ferguson-Yankey SR, Skrzypek MS, Lester RL, Dickson RC (2002) Mutant analysis reveals complex regulation of sphingolipid long chain base phosphates and long chain bases during heat stress in yeast. Yeast 19: 573–586 [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Kihara A, Igarashi Y (2003) Distribution of sphingosine kinase activity in mouse tissues: contribution of SPHK1. Biochem Biophys Res Commun 309: 155–160 [DOI] [PubMed] [Google Scholar]
- Funato K, Lombardi R, Vallée B, Riezman H (2003) Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J Biol Chem 278: 7325–7334 [DOI] [PubMed] [Google Scholar]
- Furneisen JM, Carman GM (2000) Enzymological properties of the LPP1-encoded lipid phosphatase from Saccharomyces cerevisiae. Biochim Biophys Acta 1484: 71–82 [DOI] [PubMed] [Google Scholar]
- Gijsbers S, Van der Hoeven G, Van Veldhoven PP (2001) Subcellular study of sphingoid base phosphorylation in rat tissues: evidence for multiple sphingosine kinases. Biochim Biophys Acta 1532: 37–50 [DOI] [PubMed] [Google Scholar]
- Gillard BK, Clement RG, Marcus DM (1998) Variations among cell lines in the synthesis of sphingolipids in de novo and recycling pathways. Glycobiology 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Hait NC, Fujita K, Lester RL, Dickson RC (2002) Lcb4p sphingoid base kinase localizes to the Golgi and late endosomes. FEBS Lett 532: 97–102 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98 [Google Scholar]
- Herr DR, Fyrst H, Creason MB, Phan VH, Saba JD, Harris GL (2004) Characterization of the Drosophila sphingosine kinases and requirement for Sk2 in normal reproductive function. J Biol Chem 279: 12685–12694 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura SI (2003) Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J Biol Chem 278: 46832–46839 [DOI] [PubMed] [Google Scholar]
- Imai H, Morimoto Y, Tamura K (2000) Sphingoid base composition of monoglucosylceramide in Brassicaceae. J Plant Physiol 157: 453–456 [Google Scholar]
- Inagaki Y, Li PY, Wada A, Mitsutake S, Igarashi Y (2003) Identification of functional nuclear export sequences in human sphingosine kinase 1. Biochem Biophys Res Commun 311: 168–173 [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Hannun YA (2001) Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J Biol Chem 276: 8574–8581 [DOI] [PubMed] [Google Scholar]
- Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM (2002) PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA). J Biol Chem 277: 35257–35262 [DOI] [PubMed] [Google Scholar]
- Kim S, Fyrst H, Saba J (2000) Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleuser B, Maceyka M, Milstien S, Spiegel S (2001) Stimulation of nuclear sphingosine kinase activity by platelet-derived growth factor. FEBS Lett 503: 85–90 [DOI] [PubMed] [Google Scholar]
- Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S (1998) Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem 273: 23722–23728 [DOI] [PubMed] [Google Scholar]
- Lanterman MM, Saba JD (1998) Characterization of sphingosine kinase (SK) activity in Saccharomyces cerevisiae and isolation of SK-deficient mutants. Biochem J 332: 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Stunff H, Milstien S, Spiegel S (2004) Generation and metabolism of bioactive sphingosine-1-phosphate. J Cell Biochem 92: 882–899 [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17: 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, Spiegel S (2000) Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem 275: 19513–19520 [DOI] [PubMed] [Google Scholar]
- Liu H, Toman RE, Goparaju S, Maceyka M, Nava VE, Sankala H, Payne SG, Bektas M, Ishii I, Chun J, et al (2003) Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. J Biol Chem 278: 40330–40336 [DOI] [PubMed] [Google Scholar]
- Lynch DV, Dunn TM (2004) An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol 161: 677–702 [DOI] [PubMed] [Google Scholar]
- Melendez AJ, Carlos-Dias E, Gosink M, Allen JM, Takacs L (2000) Human sphingosine kinase: molecular cloning, functional characterization and tissue distribution. Gene 251: 19–26 [DOI] [PubMed] [Google Scholar]
- Melendez AJ, Khaw AK (2002) Dichotomy of Ca2+ signals triggered by different phospholipid pathways in antigen stimulation of human mast cells. J Biol Chem 277: 17255–17262 [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr (2002) De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J Biol Chem 277: 25843–25846 [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr, Wang E, Mullins RE, Jamison WCL, Nimkar S, Liotta DC (1988) Quantitation of free sphingosine in liver by high-performance liquid chromatography. Anal Biochem 171: 373–381 [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Skrzypek M, Nagiec EE, Lester RL, Dickson RC (1998) The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J Biol Chem 273: 19437–19442 [DOI] [PubMed] [Google Scholar]
- Nava VE, Lacana E, Poulton S, Liu H, Sugiura M, Kono K, Milstien S, Kohama T, Spiegel S (2000) Functional characterization of human sphingosine kinase-1. FEBS Lett 473: 81–84 [DOI] [PubMed] [Google Scholar]
- Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM (2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410: 596–599 [DOI] [PubMed] [Google Scholar]
- Nishiura H, Tamura K, Morimoto Y, Imai H (2000) Characterization of sphingolipid long-chain base kinase in Arabidopsis thaliana. Biochem Soc Trans 28: 747–748 [PubMed] [Google Scholar]
- Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, Spiegel S (1999) Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol 147: 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera A, Kohama T, Tu Z, Milstien S, Spiegel S (1998) Purification and characterization of rat kidney sphingosine kinase. J Biol Chem 273: 12576–12583 [DOI] [PubMed] [Google Scholar]
- Olivera A, Rosenfeldt HM, Bektas M, Wang F, Ishii I, Chun J, Milstien S, Spiegel S (2003) Sphingosine kinase type 1 induces G12/13-mediated stress fiber formation, yet promotes growth and survival independent of G protein-coupled receptors. J Biol Chem 278: 46452–46460 [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Wang XQ, Coursol S, Assmann SM (2002) Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol 153: 517–526 [DOI] [PubMed] [Google Scholar]
- Paugh SW, Payne SG, Barbour SE, Milstien S, Spiegel S (2003) The immunosuppressant FTY720 is phosphorylated by sphingosine kinase type 2. FEBS Lett 554: 189–193 [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7: 719–731 [DOI] [PubMed] [Google Scholar]
- Pitson ST, d'Andrea RJ, Vandeleur L, Moeretti PAB, Xia P, Gamble JR, Vadas MA, Wattenberg BW (2000) Human sphingosine kinase: purification, molecular cloning and characterization of the native and recombinant enzymes. Biochem J 350: 429–441 [PMC free article] [PubMed] [Google Scholar]
- Renkonen O (1970) Presence of sphingadienine and trans-monoenoic fatty acids in ceramide monohexosides of human plasma. Biochim Biophys Acta 210: 190–192 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC (1999) Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol 181: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Heinz E (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632: 1–15 [DOI] [PubMed] [Google Scholar]
- Sperling P, Zähringer U, Heinz E (1998) A sphingolipid desaturase from higher plants. Identification of a new cytochrome b5 fusion protein. J Biol Chem 273: 28590–28596 [DOI] [PubMed] [Google Scholar]
- Spiegel S, Milstien S (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 4: 397–407 [DOI] [PubMed] [Google Scholar]
- Sugiura M, Kono K, Liu H, Shimizugawa T, Minekura H, Spiegel S, Kohama T (2002) Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J Biol Chem 277: 23294–23300 [DOI] [PubMed] [Google Scholar]
- Sullards MC, Lynch DV, Merrill AH Jr, Adams J (2000) Structure determination of soybean and wheat glucosylceramides by tandem mass spectrometry. J Mass Spectrom 35: 347–353 [DOI] [PubMed] [Google Scholar]
- Ternes P, Franke S, Zahringer U, Sperling P, Heinz E (2002) Identification and characterization of a sphingolipid Δ4-desaturase family. J Biol Chem 277: 25512–25518 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Worrall D, Ng CK, Hetherington AM (2003) Sphingolipids, new players in plant signaling. Trends Plant Sci 8: 317–320 [DOI] [PubMed] [Google Scholar]
- Wright BS, Snow JW, O'Brien TC, Lynch DV (2003) Synthesis of 4-hydroxysphinganine and characterization of sphinganine hydroxylase activity in corn. Arch Biochem Biophys 415: 184–192 [DOI] [PubMed] [Google Scholar]
- Young KW, Willets JM, Parkinson MJ, Bartlett P, Spiegel S, Nahorski SR, Challiss RA (2003) Ca2+/calmodulin-dependent translocation of sphingosine kinase: role in plasma membrane relocation but not activation. Cell Calcium 33: 119–128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.