Abstract
In grasses, residues homologous to residues Ile-1,781 and Ile-2,041 in the carboxyl-transferase (CT) domain of the chloroplastic acetyl-coenzyme A (CoA) carboxylase (ACCase) from the grass weed black-grass (Alopecurus myosuroides [Huds.]) are critical determinants for sensitivity to two classes of ACCase inhibitors, aryloxyphenoxypropionates (APPs) and cyclohexanediones. Using natural mutants of black-grass, we demonstrated through a molecular, biological, and biochemical approach that residues Trp-2,027, Asp-2,078, and Gly-2,096 are also involved in sensitivity to ACCase inhibitors. In addition, residues Trp-2,027 and Asp-2,078 are very likely involved in CT activity. Using three-dimensional modeling, we found that the side chains of the five residues are adjacent, located at the surface of the inside of the cavity of the CT active site, in the vicinity of the binding site for APPs. Residues 1,781 and 2,078 are involved in sensitivity to both APPs and cyclohexanediones, whereas residues 2,027, 2,041, and 2,096 are involved in sensitivity to APPs only. This suggests that the binding sites for these two classes of compounds are overlapping, although distinct. Comparison of three-dimensional models for black-grass wild-type and mutant CTs and for CTs from organisms with contrasted sensitivity to ACCase inhibitors suggested that inhibitors fitting into the cavity of the CT active site of the chloroplastic ACCase from grasses to reach their active sites may be tight. The three-dimensional shape of this cavity is thus likely of high importance for the efficacy of ACCase inhibitors.
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) is a biotinylated enzyme that catalyzes the carboxylation of acetyl-CoA to produce malonyl-CoA. This is a two-step, reversible reaction, consisting of the ATP-dependent carboxylation of the biotin group on the carboxyl carrier domain by the biotin-carboxylase activity, followed by the transfer of the carboxyl group from biotin to acetyl-CoA by the carboxyl-transferase (CT) activity (for review, see Nikolau et al., 2003). In eukaryotes and prokaryotes, ACCase is a key enzyme in fatty acid biosynthesis (Harwood, 1988). In plants, ACCase activity is not only involved in primary fatty acid biosynthesis, which is located in chloroplasts and mitochondria (Focke et al., 2003; Nikolau et al., 2003), but also in the formation of long-chain fatty acids and flavonoids and in malonylation, which occurs in the cytosol (Sasaki et al., 1995; Post-Beittenmiller, 1996). Two ACCase isoforms are thus found in plant cytosol and organites, respectively (Sasaki et al., 1995; Konishi et al., 1996), with the chloroplastic isoform accounting for more than 80% of the total ACCase activity (Herbert et al., 1996). The cytosolic ACCase isoform in all plants studied so far is a homodimeric enzyme, as it is in all eukaryotes (Nikolau et al., 2003). Each monomer carries all ACCase functional domains on a single polypeptide. Homomeric ACCases thus share a high degree of conservation between eukaryotes (Nikolau et al., 2003). In most plant species, the chloroplastic ACCase is a heteromeric enzyme consisting of three nuclear-encoded subunits and one subunit encoded by a chloroplastic gene (Konishi et al., 1996). However, in the Poaceae (grasses), the chloroplastic ACCase is a homodimeric enzyme (Konishi et al., 1996) encoded by a nuclear gene distinct from that coding for the cytosolic ACCase isoform (Gornicki et al., 1994, 1997; Podkowinski et al., 1996).
Aryloxyphenoxypropionates (APPs) and cyclohexanediones (CHDs) are two chemical classes of molecules that selectively inhibit homomeric, chloroplastic ACCase from grasses (Rendina et al., 1990; Burton et al., 1991), which makes them postemergent herbicides used worldwide to control grass weeds. APPs and CHDs are reversible (Gronwald, 1991), mutually exclusive (Rendina et al., 1990) inhibitors of the CT reaction. Heteromeric, chloroplastic ACCase is insensitive to CHDs and APPs (Alban et al., 1994). The highly different structure of heteromeric ACCase likely accounts for this insensitivity. Surprisingly, homomeric, cytosolic ACCases are less sensitive by several orders of magnitude than the rather similar homomeric, chloroplastic ACCases from the Poaceae (e.g. Egli et al., 1993; Herbert et al., 1996; Evenson et al., 1997; Joachimiak et al., 1997; Price et al., 2003). Although the structure of the yeast (Saccharomyces cerevisiae) ACCase CT domain in complex with APPs has been solved (Zhang et al., 2004), the molecular bases for those differences in sensitivity to APPs and CHDs remain largely unknown.
To identify the determinants for sensitivity to ACCase-inhibiting herbicides, grass mutants with altered sensitivity due to mutation(s) within the target enzyme are of great interest. Since their introduction to world agriculture in the 1980s, APPs and CHDs have been widely used to control a variety of grass weeds. As a consequence, weed plants that are resistant to these herbicides have been reported in 33 species so far, with altered ACCase being a widespread resistance mechanism (see the International Survey of Herbicide Resistant Weeds homepage at www.weedscience.com). One of the best-studied weeds is black-grass (Alopecurus myosuroides [Huds.]), a major grass weed in winter crops in Europe. Analysis of resistant ACCase mutants in black-grass revealed that an Ile-1,781-Leu substitution confers resistance to some, but not all, APPs and CHDs, and that an Ile-2,041-Asn substitution confers resistance to APPs but not to CHDs (Délye et al., 2002a, 2002b, 2003). Both substitutions are located within the CT domain of homomeric, chloroplastic ACCase. Similar findings were obtained in mutant plants from the grass weeds Lolium rigidum (Gaud.), Setaria viridis (L. Beauv.), and Avena fatua (L.) for the Ile-to-Leu substitution (Zhang and Devine, 2000; Zagnitko et al., 2001; Christoffers et al., 2002; Délye et al., 2002b, 2002c), and from L. rigidum for the Ile-to-Asn substitution (Délye et al., 2003). Herein we demonstrate that three other residues, located within black-grass ACCase CT domain at positions 2,027, 2,078, and 2,096 are also crucial for sensitivity to APP and/or CHD inhibitors. Using protein modeling, we discuss the structural consequences of the five amino acid substitutions listed above for ACCase-inhibitor interactions.
RESULTS
In the following, the reference sequence for ACCase is EMBL accession AJ310767 (black-grass chloroplastic ACCase). Unless otherwise stated, all nucleotide and amino acid positions referred to hereafter correspond to those in this sequence.
Sensitivity to APPs and CHDs and Amino Acid Substitutions within the ACCase CT Domain
Among the 16 black-grass seedlings resistant to ACCase inhibitors that were used for sequencing experiments, 11 contained 2 identical ACCase alleles. A total of 21 sequences were thus obtained for analysis. Here we only considered the black-grass ACCase coding sequence for analysis. After discarding noncoding intron and 3′ sequences, the 21 sequences were aligned. A total of 25 single-nucleotide polymorphisms (SNPs), consisting of 21 synonymous and 4 nonsynonymous changes, were identified. The four nonsynonymous changes were a Trp-2,027-Cys, an Arg-2,078-Gly, a Gly-2,096-Ala, and a Lys-2,264-Arg substitution. The 21 sequences comprised a total of 10 haplotypes, 7 of which contained nonsynonymous SNPs. Eight of the 10 haplotypes found herein were not observed in a previous study (Délye et al., 2003; EMBL accession ALIGN_000483). They displayed one nonsynonymous and six synonymous SNPs that were not found previously. While the nonsynonymous substitution at position 2,027 was never reported, each of the amino acid replacements at positions 2,078 and 2,096 was previously observed in a single haplotype present in one haloxyfop-resistant, in one fenoxaprop-resistant, and in one fenoxaprop-sensitive seedling, respectively (Délye et al., 2003). Amino acid substitution at position 2,264 was previously observed in four haplotypes mostly found in herbicide-sensitive seedlings (Délye et al., 2003).
The three seedlings in population 00-016 consisted of one clodinafop-resistant and two fenoxaprop-resistant seedlings. They contained four haplotypes. Two haplotypes displayed a Trp-to-Cys change caused by a G-to-T transversion at the first position in codon 2,027. Each seedling contained at least one Cys-2,027 haplotype. One haplotype present in a single seedling contained a Lys-to-Arg change caused by a G-to-C transversion at the second position in codon 2,264 that is located outside the ACCase CT domain. Allele-specific PCR assays enabling the identification of the codons present at positions 2,027, 2,078, and 2,096 were used to genotype a total of 500 seedlings from populations 00-016 and from the derived, purified population Cys-2027-F1 (Table I). The presence of Leu-1,781 ACCase alleles that confer resistance to cycloxydim and fenoxaprop and of Asn-2,041 ACCase alleles that confer resistance to the three APPs used was also assessed in these populations using previously described allele-specific PCR assays (Délye et al., 2002a, 2003). Cys-2,027 ACCase alleles exclusively occurred in about 60% of the seedlings in population 00-016. All seedlings containing one or two Cys-2,027 allele(s) were resistant to the three APPs used and sensitive to the two CHDs. Association of the presence of Cys-2,027 allele(s) with resistance to APPs was supported by Fisher's exact test (Table I). Results from the purified population Cys-2,027-F1 further supported this association, with 100% of the seedlings in this population being resistant to the three APPs and sensitive to the two CHDs.
Table I.
Herbicide sensitivity of and mutant chloroplastic ACCase alleles detected in seedlings from four field and two purified black-grass populations
| Population
|
Mutant ACCase Allele(s) Detected
|
CHD Herbicides
|
APP Herbicides
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cycloxydim
|
Clethodim
|
Fenoxaprop
|
Clodinafop
|
Haloxyfop
|
|||||||
| Ra | Sa | R | S | R | S | R | S | R | S | ||
| 00-017b | None | 0 | 50 | 0 | 50 | 0 | 50 | 0 | 50 | 0 | 50 |
| 00-016 | None | 0 | 20 | 0 | 18 | 12 | 7 | 0 | 21 | 0 | 17 |
| Cys-2,027c | 0 | 30 | 0 | 32 | 31 | 0 | 29 | 0 | 33 | 0 | |
| P-value (Cys-2,027)d | 1.0 | 1.0 | 5.10−4 | 0.0 | 0.0 | ||||||
| 00-049 | None | 0 | 3 | 0 | 4 | 0 | 2 | 1 | 1 | 0 | 3 |
| Leu-1,781c | 32 | 0 | 0 | 30 | 32 | 0 | 18 | 13 | 0 | 33 | |
| Gly-2,078c | 4 | 0 | 6 | 0 | 7 | 0 | 5 | 0 | 5 | 0 | |
| Leu-1,781 + Gly-2,078f | 11 | 0 | 10 | 0 | 9 | 0 | 12 | 0 | 9 | 0 | |
| P-value (Gly-2,078)d | 0.0286 | 2.10−4 | 0.0278 | 0.1053 | 0.0015 | ||||||
| 00-099 | None | 0 | 5 | 0 | 4 | 0 | 6 | 0 | 6 | 0 | 5 |
| Leu-1,781c | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | |
| Asn-2,041c | 0 | 5 | 0 | 6 | 4 | 0 | 6 | 0 | 6 | 0 | |
| Ala-2,096c | 0 | 29 | 0 | 31 | 28 + 3e | 0 | 28 + 2e | 0 | 26 + 2e | 0 | |
| Leu-1,781 + Asn-2,041f | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| Leu-1,781 + Ala-2,096f | 2 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 0 | |
| Asn-2,041 + Ala-2,096f | 0 | 7 | 0 | 6 | 7 | 0 | 5 | 0 | 7 | 0 | |
| P-value (Ala-2,096)d | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | ||||||
| Cys-2,027-F1b,g | Cys-2,027h | 0 | 50 | 0 | 50 | 50 | 0 | 50 | 0 | 50 | 0 |
| Gly-2,078-F1b,g | Gly-2,078h | 50 | 0 | 50 | 0 | 50 | 0 | 50 | 0 | 50 | 0 |
| Ala-2,096-F1b,g | Ala-2,096h | 0 | 50 | 0 | 50 | 50 | 0 | 50 | 0 | 50 | 0 |
S, Sensitive; R, resistant.
Population used for ACCase enzyme assay.
One or two allele(s) mutated at the same codon were detected.
Fisher's exact test for the association of the presence of the mutant ACCase allele with resistance to each herbicide.
Resistant plants with a green first leaf of less than 1 cm in length.
A single copy of each mutant allele was detected.
Purified population (see “Materials and Methods”).
Two alleles mutated at the same codon were detected.
The 10 seedlings in population 00-049 consisted of 1 clodinafop-resistant, 6 cycloxydim-resistant, and 3 fenoxaprop-resistant seedlings. They contained three haplotypes, one of which displayed an Asp-to-Gly change caused by an A-to-G transition at the first position in codon 2,078. This haplotype was present in all 10 seedlings. No other nonsynonymous mutation was recorded among the 10 seedlings. A total of 500 seedlings from populations 00-049 and from the derived purified population Gly-2,078-F1 were genotyped as above (Table I). Gly-2,078 ACCase alleles exclusively occurred in about 30% of the seedlings in population 00-049. However, 65% of the seedlings containing a Gly-2,078 allele also contained a Leu-1,781 allele (Table I). The few seedlings containing only one or two Gly-2,078 allele(s) were all resistant across the range of ACCase inhibitors tested. Although Leu-1,781 ACCase alleles did not confer resistance to clethodim, clodinafop, and haloxyfop, all seedlings containing one Leu-1,781 and one Gly-2,078 allele were resistant to these inhibitors (Table I). Association of the presence of Gly-2,078 allele(s) with resistance to all herbicides but clodinafop was supported by Fisher's exact test (Table I). Results from the purified population Gly-2,078-F1 supported this association, with 100% of the seedlings in this population being resistant to each of the five herbicides used, including clodinafop.
The three seedlings in population 00-099 consisted of one fenoxaprop-resistant and two clodinafop-resistant seedlings. They contained three haplotypes, all displaying a Gly-to-Ala change caused by a G-to-C transversion at the second position in codon 2,096. No other nonsynonymous mutation was recorded among the three seedlings. A total of 500 seedlings from populations 00-099 and from the derived, purified population Ala-2,096-F1 were genotyped as above (Table I). Ala-2,096 ACCase alleles exclusively occurred in about 75% of the seedlings in population 00-099. This population also contained Leu-1,781 and Asn-2,041 alleles. Most seedlings containing Ala-2,096 alleles did not contain another mutant allele (Table I). These seedlings were always sensitive to the two CHDs used. Most seedlings containing Ala-2,096 ACCase alleles were resistant to the three APPs. However, several such seedlings were first considered sensitive to fenoxaprop (three seedlings), clodinafop (two seedlings), or haloxyfop (two seedlings) because their first leaf was less than 1 cm in length (Letouzé and Gasquez, 1999). Herbicide bioassay with the three APPs was thus repeated for populations 00-099 and 00-017. Closer observation prior to genotyping revealed that the first leaf of seedlings containing Ala-2,096 ACCase allele(s) was always fully green and alive, even when it was less than 1 cm in length. In contrast, the first leaf of sensitive seedlings not containing mutant ACCase alleles in population 00-017 and 00-099 was always softened and bleached. We consequently considered that seedlings with a green first leaf of less than 1 cm were herbicide resistant (Table I). All seedlings containing one or two Ala-2,096 ACCase allele(s) were thus sensitive to the two CHDs used and resistant to the three APPs. Association of the presence of Ala-2,096 allele(s) with resistance to APPs was supported by Fisher's exact test (Table I). Results from the purified population Ala-2,096-F1 further supported this association, with 100% of the seedlings in this population being resistant to the three APPs and sensitive to the two CHDs (Table I).
As previously (Délye et al., 2003), we found that seedlings not containing any of the five mutant ACCase alleles identified to date were resistant to fenoxaprop (12 seedlings) or clodinafop (one seedling; Table I). This was consistent with a previous demonstration that other mechanisms, such as enhanced herbicide metabolism, may also account for resistance to ACCase inhibitors in black-grass (e.g. Letouzé and Gasquez, 2003).
ACCase Inhibition by APPs and CHDs
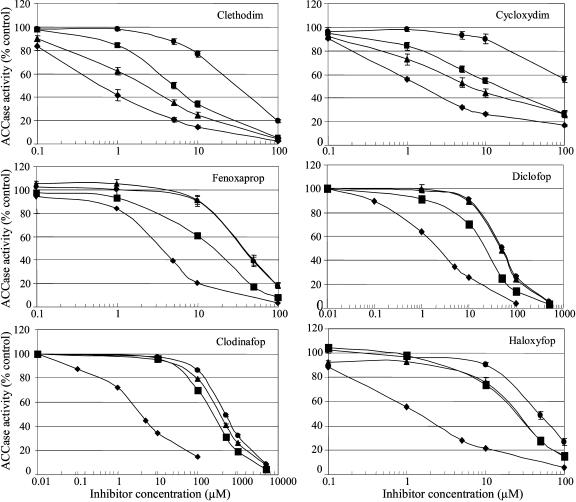
ACCase activity measured in vitro without the presence of inhibitors was always 2 times lower in extracts from the resistant black-grass-purified populations, Cys-2,027-F1 and Gly-2,078-F1, than in extracts from the sensitive population, 00-017. It was also moderately lower in extracts from the resistant population, Ala-2,096-F1, than in extracts from population 00-017 (data not shown). The action of four APPs and two CHDs upon enzymatic activity of wild-type Cys-2,027, Gly-2,078, and Ala-2,096 ACCases is shown in Figure 1. A high level of resistance to the four APPs assayed is observed for all three mutant ACCase alleles (Fig. 1). Herbicide doses required to inhibit 50% ACCase activity (I50) values were 10 to 125 times higher than those obtained for the wild-type ACCase (Table II). Cys-2,027 and Ala-2,096 ACCase alleles displayed inhibition patterns similar to that of the wild-type ACCase allele for the two CHDs assayed (Fig. 1), although the I50 values obtained were slightly higher for both mutant alleles (Table II). In contrast, the Gly-2,078 ACCase allele displayed a high level of resistance to the two CHDs (Fig. 1), with I50 values for clethodim and cycloxydim 36 and 83.5 times higher than those obtained for the wild-type ACCase allele, respectively (Table II). These results fully supported the association of the Trp-2,027-Cys and Gly-2,096-Ala substitutions with resistance to APPs, and of the Asp-2,078-Gly substitution with resistance to all APPs and CHDs assayed found using allele-specific PCR in black-grass (Table I). We thus concluded that Trp-2,027-Cys and Gly-2,096-Ala substitutions confer resistance to APPs but not to CHDs, while the Asp-2,078-Gly substitution confers resistance to all APPs and CHDs assayed.
Figure 1.
Inhibition of ACCase activity of wild-type (population 00-017; ♦), Cys-2,027 (population Cys-2,027-F1; ▴), Gly-2,078 (population Gly-2,078-F1; •), and Ala-2,096 (population Ala-2,096-F1; ▪) ACCase alleles by clethodim and cycloxydim (CHDs) and by fenoxaprop, diclofop, clodinafop, and haloxyfop (APPs). Averages of two independent experiments are shown with error bars. ACCase activity is expressed as a percentage of ACCase activity without inhibitor for each allele.
Table II.
I50 values obtained for wild-type, Cys-2,027, Gly-2,078, and Ala-2,096 ACCase alleles
Values are mean ± se of two independent experiments.
| Inhibitor
|
00-017 (S) | Cys-2,027-F1 (R)
|
Gly-2,078-F1 (R)
|
Ala-2,096-F1 (R)
|
|||
|---|---|---|---|---|---|---|---|
| I50 | I50 | R:S I50 Ratio | I50 | R:S I50 Ratio | I50 | R:S I50 Ratio | |
| μm | μm | μm | μm | ||||
| Cycloxydim (CHD) | 1.6 ± 0.1 | 7.8 ± 2.9 | 5.0 | 133.8 ± 6.7 | 83.5 | 14.7 ± 2.5 | 9.0 |
| Clethodim (CHD) | 0.8 ± 0.2 | 2.2 ± 0.4 | 3.0 | 29.0 ± 4.6 | 36.0 | 5.2 ± 0.5 | 6.5 |
| Fenoxaprop (APP) | 0.7 ± 0.2 | 37.0 ± 4.8 | 53.0 | 37.1 ± 3.7 | 53.0 | 14.3 ± 1.0 | 20.5 |
| Clodinafop (APP) | 4.0 ± 1.1 | 361.8 ± 25.9 | 90.5 | 501.4 ± 50.7 | 125.5 | 230.1 ± 2.3 | 57.5 |
| Haloxyfop (APP) | 1.4 ± 0.1 | 26.6 ± 3.0 | 19.0 | 48.7 ± 4.6 | 35.0 | 22.2 ± 0.1 | 16.0 |
| Diclofop (APP) | 2.2 ± 0.5 | 46.9 ± 3.5 | 21.5 | 50.1 ± 2.3 | 23.0 | 21.8 ± 0.2 | 10.0 |
Black-Grass ACCase Modeling
For a detailed evaluation of the effects of amino acid substitutions at positions 1,781, 2,027, 2,041, 2,078, and 2,096 in black-grass ACCase upon herbicide binding, we reconstructed two types of three-dimensional models of homodimeric ACCase. The first type used a model built into a 2.5-Å density map obtained by electron crystallography of the yeast free ACCase CT domain (Protein Data Bank [PDB] accession 1UYT; Zhang et al., 2004) as a template. The second type used a model built into a 2.8-Å density map obtained by electron crystallography of the yeast ACCase CT domain in complex with the APP haloxyfop (PDB accession 1UYS; Zhang et al., 2004) as a template. Alignments of the primary amino acid sequences of chains B and C in PDB accessions 1UYT and 1UYS with black-grass ACCase CT domain (residues Leu-1,639 to Leu-2,204 in EMBL accession AJ310767) were visually checked for inconsistency. The sequence identity between the black-grass CT domain and the template sequences was 55.3%, which was sufficient to achieve a satisfactory model (Schwede et al., 2003). In the following, B or C after an amino acid position number refers to the residue included in the ACCase CT monomer modeled after the B or C chain in PDB accession 1UYT or 1UYS, respectively.
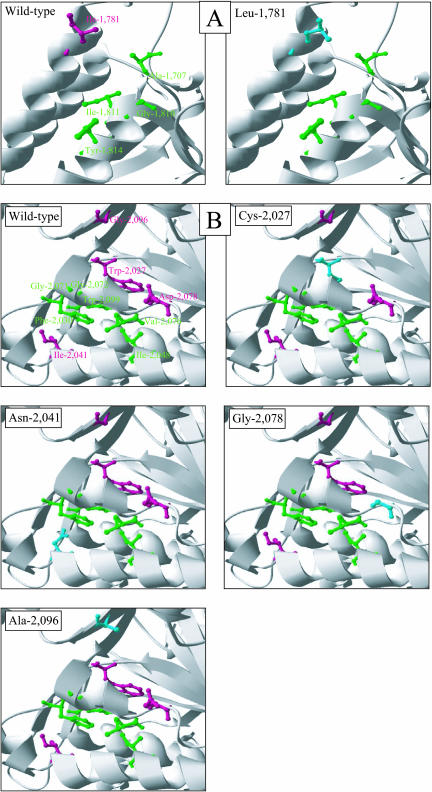
The free ACCase CT dimer consisted of the head-to-tail arrangement of two monomers. The two active sites of the CT dimer were located in cavities at the interface of the dimer, with each monomer equally contributing to the active-site shape (Zhang et al., 2003). Because of the symmetry of the ACCase CT dimer, we will consider only one of the two active sites in the following. In the yeast CT domain, the binding site for the APPs haloxyfop and diclofop is located within the cavity of the active site. APP molecules interact with the side chains of residues Ala-1,627C, Leu-1,705C, Gly-1,734C, Ile-1,735C, Tyr-1,738C, Trp-1,924B, Phe-1,956C, Val-1,967C, Ile-1,974C, Gly-1,997C, Gly-1,998C, and Val-2,002C (Zhang et al., 2004), which are homologous to residues Ala-1,707C, Ile-1,781C, Gly-1,810C, Ile-1,811C, Tyr-1,814C, Trp-1,999B, Phe-2,030C, Ile-2,041C, Ile-2,048C, Gly-2,071C, Gly-2,072C, and Val-2,076C, respectively, in black-grass ACCase. Aligning the 31 full CT sequences available in the databases showed that all but three of the residues in the binding site for APPs were absolutely conserved. Residue 1,999 was either Trp or Leu (1 sequence out of 31). Only residues 1,781 (Ile/Leu) and 2,041 (Ile/Val/Met) that are involved in sensitivity to APPs and/or CHDs (Zhang and Devine, 2000; Zagnitko et al., 2001; Christoffers et al., 2002; Délye et al., 2002b, 2002c, 2003) showed appreciable variation between the available CT sequences. Because of the dimer structure of the CT domain, the side chains in residues 1,781C, 2,027B, 2,041B, 2,078B, and 2,096B were adjacent in the models for black-grass. All five residues were located at the dimer interface, deep inside the active-site cavity (Fig. 2). Residues Ile-1,781C and Ile-2,041B were homologous to residues Leu-1,705C and Val-1,967B, respectively, which belong to the binding site of APPs in yeast CT (Zhang et al., 2004). The side chains in residues Trp-2,027B and Asp-2,078B were located close to those in residues Val-2,076B and Trp-1,999B, respectively, in the binding site for APPs, while the side chain in residue Gly-2,096B was more remote (Fig. 2B).
Figure 2.
Ribbon diagram views of the inside of the CT active-site cavity in a three-dimensional model for black-grass chloroplastic ACCase. Side-chain colors: Green, APP-binding site; magenta, variable amino acid, residue present in wild-type black-grass ACCase; blue, variable amino acid, residue present in mutant black-grass ACCase. Hydrogen atoms and side chains not discussed in the text are not shown for clarity. A, Views of the inside of the active-site cavity part formed by monomer C in Ile-1,781 and Leu-1,781 ACCase alleles. Monomer B is not shown. B, Views of the inside of the active-site cavity part formed by monomer B in wild-type ACCase allele after a 180° rotation with respect to the views of monomer C. Monomer C is not shown.
The structural changes caused in the free ACCase CT domain by amino acid replacements at positions 1,781, 2,027, 2,041, 2,078, and 2,096 are illustrated in Figure 2. The Ile-1,781C-Leu change resulted in the aliphatic side chain that is oriented toward the inside of the CT active site in the Ile-1,781C residue being replaced with an aliphatic side chain protruding toward the opening of the cavity in residue Leu-1,781C (Fig. 2A). In yeast CT, a Leu residue is present at position 1,781. Its side chain favors herbicide binding via van der Waals interactions with the methyl group in APPs (Zhang et al., 2004). This is the case in the model for CT-herbicide complexes computed for the black-grass mutant, Leu-1,781 ACCase, but not in that computed for the wild type, Ile-1,781 ACCase. The change at position 2,027B caused the replacement of a mostly buried aromatic cycle in the model for the Trp-2,027 ACCase with a buried thiol group in the model for the mutant, Cys-2,027 ACCase (Fig. 2B). The change at position 2,078 caused the carboxyl group that strongly protruded within the active-site cavity of CT in the model for the Asp-2,078 ACCase to be fully removed in the model for the mutant, Gly-2,078 ACCase (Fig. 2B). The Gly-2,096B-Ala substitution resulted in the occurrence of an extra methyl group protruding within the active-site cavity of CT in the model for the mutant, Ala-2,096 ACCase (Fig. 2B). None of these three changes directly interfered with herbicide binding in models for black-grass CT-herbicide complexes (data not shown).
The substitution at position 2,041 caused the hydrophobic, aliphatic side chain that protruded toward the opening of the CT active-site cavity in the model for Ile-2,041 ACCase to be replaced with a polar side chain containing an amide group oriented toward the inside of the cavity in the model for the mutant, Asn-2,041 ACCase (Fig. 2B). In the model for black-grass Asn-2041B ACCase in complex with APPs, this change caused a major clash that was not compatible with herbicide binding (data not shown). The clash involved residue Phe-2,030B that is crucial for herbicide binding in yeast CT (Zhang et al., 2004) and caused a heavy twisting of a CT section encompassing residues 2,035B to 2,042B.
Comparison of ACCase CT Domain Three-Dimensional Structures
Herein we assessed black-grass ACCase sensitivity to APPs and CHDs using both a whole-seedling-based bioassay and an ACCase enzyme assay. The most suitable way of comparing ACCase sensitivity to inhibitors between different organisms is by an enzyme assay. In the literature, there are a total of 10 organisms, including 5 grasses, 1 dicotyledonous plant, 1 alga, 1 fungus, 1 mammal, and 1 protozoan, for which both enzyme sensitivity studies and CT protein sequences are available (Table III). From enzyme studies, it appeared that the chloroplastic, homomeric ACCase from grasses is highly sensitive to APPs and CHDs, while cytosolic ACCases are far less sensitive (i.e. tolerant) to these compounds (Table III). For each ACCase isoform in Table III, three-dimensional models were reconstructed as described above. No obvious differences could be observed between species when comparing models for CT-herbicide complexes, which may be due to models being computed from yeast-tolerant ACCase. We therefore focused upon models for free CT domains reconstructed after PDB accession 1UYT to search for differences in the three-dimensional shape of the active-site cavity that may interfere with herbicides accessing their binding site. Two models corresponding to the plastidic and cytosolic isoforms were thus built for black-grass, wheat (Triticum aestivum), and Toxoplasma gondii. One model was built for each of the other seven organisms. No ACCase sequence from pea was available in the databases. However, because a very high conservation was observed between the CT sequences from the two legume plants available in the databases, i.e. alfalfa (Medicago sativa) and soybean (Glycine max; 92.9% amino acid identity), we used the sequence from alfalfa as a model for pea ACCase.
Table III.
Sensitivity to ACCase inhibitors of and amino acids present at specific positions in ACCase from 10 eukaryotic organisms
I50 values correspond to APP or CHD concentrations inhibiting 50% ACCase activity.
| Organism
|
Accession No.
|
Range of I50 Values
|
Amino Acid Present at Position
|
References for I50 Values
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APPs | CHDs | 1,781 | 2,027 | 2,037 | 2,041 | 2,078 | 2,079 | 2,080 | 2,088 | 2,096 | |||
| μm | |||||||||||||
| Black-grass cpa | AJ310767 | 0.06–4.0 | 0.7–9.0 | Ile | Trp | Leu | Ile | Asp | Ser | Lys | Cys | Gly | Price et al. (2004); Menéndez and De Prado (1999); This work |
| Rye-grass cpa | AF359515, AJ519781 | 0.2–0.6 | 4.2 | .b | . | . | . | . | . | . | . | . | Evenson et al. (1997); Christopher and Holtum (1998); Kuk et al. (2000) |
| Wheat cpa | AF029895 | 0.2–2.0 | <10.0 | . | . | . | . | . | . | . | Phe | . | Gornicki and Haselkorn (1993); Konishi and Sasaki (1994); Joachimiak et al. (1997) |
| Maize cpa | U19183 | 0.04–10.0 | 10.0 | . | . | . | . | . | . | . | . | . | Egli et al. (1993); Herbert et al. (1996); Price et al. (2003) |
| Green foxtail cpa | AF294805 | 0.08–4.4 | 0.3–7.7 | . | . | . | . | . | . | . | . | . | Marles et al. (1993); Shukla et al. (1997) |
| Black-grass cya | AJ632096 | 35.0 | NAc | Leu | . | . | . | . | . | . | Met | . | Menéndez and De Prado (1999) |
| Wheat cya | U39321 | 250.0 | 250.0–>500.0 | Leu | . | . | . | . | . | . | Met | . | Joachimiak et al. (1997) |
| Pea cya | L25042d | 7.0–150.0 | >1,000.0 | Leu | . | . | . | . | . | Arg | Met | . | Alban et al. (1994); Konishi and Sasaki (1994); Herbert et al. (1996); Christopher and Holtum (1998) |
| C. cryptica | L20784 | >100.0 | 23.0 | Leu | . | Met | . | . | Pro | Thr | Met | . | Roessler (1990) |
| T. gondii cpa | AF157612 | 5.0 | >400.0 | Leu | . | Met | . | . | Pro | Thr | Met | . | Jelenska et al. (2002) |
| T. gondii cya | AF330145 | >400.0 | >400.0 | Leu | . | Met | . | . | . | Arg | Met | . | Jelenska et al. (2002) |
| Yeast | Z71631 | >500.0 | >500.0 | Leu | . | Met | Val | . | Pro | Thr | Met | Ala | Joachimiak et al. (1997) |
| Rat (β isoform) | AB004329 | NAc | 18.0–>100.0 | Leu | . | Met | Met | . | . | Ser | Met | . | Seng et al. (2003) |
cp, Plastidic; cy, cytosolic ACCase isoform.
Dots, Residues identical to those in black-grass chloroplastic ACCase
NA, None available.
Sequence from alfalfa (see text).
To assess the variability of the residues identified as potentially important for the shape of the active-site cavity, all 31 complete sequences of homomeric ACCase CT domains available in databases were aligned (data not shown). The three-dimensional shape of the active-site cavity was highly conserved between organisms (data not shown). In particular, all three-dimensional models for chloroplastic ACCases exhibited near-identical active-site three-dimensional structures (data not shown). The only structural difference between ACCase isoforms sensitive or tolerant to herbicides that could be identified in the region of the active site modeled after chain C in PDB accession 1UYT was due to the presence of a Leu-1,781 residue in all sequences but those from chloroplastic ACCases from grasses that contained an Ile-1,781 residue (Table III; see Fig. 2 for an illustration).
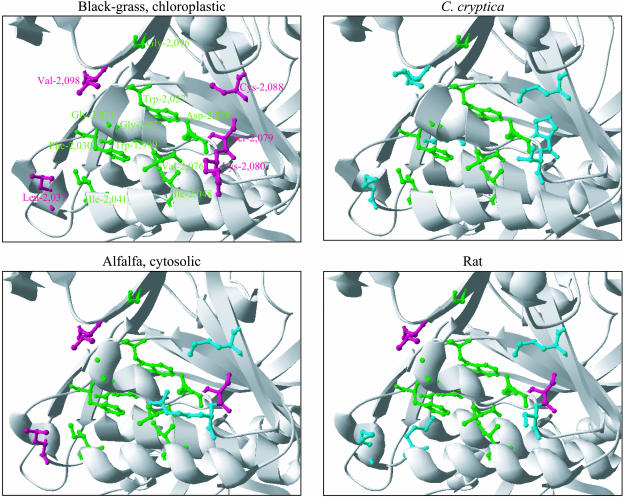
Five variable side chains, belonging to residues 2,037, 2,041, 2,079, 2,080, and 2,088, were observed in the region of the active site modeled after chain B in PDB accession 1UYT. Figure 3 portrays representative models. Residue Leu-2,037, the side chain of which was located near residue 2,041 in the herbicide-binding site, was conserved between all known CT sequences from higher plants. This residue was Met in all other known sequences. Residue 2,041 in the binding site for APPs was Ile in all known sequences from higher plants. It was either Ile or Val in all other sequences but that from rat (Met). The side chains in residues at positions 2,079B, 2,080B, and 2,088B were located close to that in residue Asp-2,078B. Consensus at position 2,079 between the 31 known CT sequences was Ser/Pro/Ala. Among the 10 organisms used in the modeling study, a Ser residue is found in all CT sequences from higher plants, in the sequence from rat, and in that from T. gondii cytosolic ACCase (Table III), while the sequences for the other organisms contained a Pro-2,079 residue. The residue at position 2,080 was variable enough between the 31 CT sequences, with consensus at this position being Lys/Arg/Thr/Ser/Gln/Ala/Leu. A Lys-2,080 residue occurred in all CT sequences from grasses (Table III), while the sequences from other organisms contained an Arg, a Thr, or a Ser residue at this position (Table III). Residue at position 2,088 was fairly conserved, being most frequently Cys or Met. A Phe-2,088 or a Thr-2,088 residue was found in 1 out of the 31 available CT sequences. All chloroplastic homomeric ACCases but that from wheat (Phe-2,088) contained a Cys-2,088 residue (Table III). The sequences from the five other ACCases in Table III contained a Met-2,088B residue.
Figure 3.
Ribbon diagram views of the CT active-site cavity on monomer B in three-dimensional models for black-grass chloroplastic, C. cryptica, alfalfa cytosolic, and rat (D) ACCases. Side-chain colors: Green, APP-binding site and variable amino acid identified in black-grass ACCase; magenta, amino acid variable between organisms; blue, amino acid variable between organisms, residue differing from that present in black-grass at this position (see Table III). Hydrogen atoms and side chains not discussed in the text are not shown for clarity. Monomer C is not shown.
The main differences in three-dimensional structure observed between models for ACCase isoforms sensitive or tolerant to APPs and CHDs resulted from the combination of residues present at positions 1,781C, 2,037B, 2,041B, 2,079B, 2,080B, and 2,088B in the various CT sequences (see Fig. 3 for an illustration). These combinations altered the distribution of polar groups at the surface of the bottom of the cavity of the CT active site, the level of obstruction of the opening of the cavity by hydrophobic chains, and the shape of the cavity within or at the vicinity of the binding site for APPs.
DISCUSSION
APPs and CHDs have long been known to interfere at the level of the ACCase CT domain (Rendina et al., 1990). Only recently has the three-dimensional structure of the ACCase CT domain in complex with the APPs haloxyfop and diclofop been determined (Zhang et al., 2004). This structure, however, has been determined from yeast ACCase, which is tolerant to APPs and CHDs (Joachimiak et al., 1997). Yeast ACCase contains a Leu and a Val residue at positions homologous to Ile-1,781 and Ile-2,041, respectively, in black-grass wild-type ACCase (Table III). In grasses, the Ile-1,781-Leu change confers resistance to diclofop but not to haloxyfop, while the Ile-2,041-Val substitution may confer resistance to haloxyfop (Délye et al., 2002a, 2002b, 2003). Furthermore, the yeast CT sequence, like the black-grass mutant Ala-2,096 ACCase that is resistant to APPs, contains an Ala residue at a position homologous to Gly-2,096 in black-grass wild-type ACCase (Table III). Because of this, the details of the interaction of sensitive, chloroplastic ACCases from grasses with APP inhibitors cannot be fully understood from yeast models. No data are available regarding the binding mode of CHD inhibitors to ACCase. Previous studies with grass weed mutants with altered sensitivity to ACCase inhibitors identified two Ile residues at positions 1,781 and 2,041 within the CT domain of chloroplastic, homomeric ACCase that are critical for interaction with APP and/or CHD herbicides (Zhang and Devine, 2000; Zagnitko et al., 2001; Christoffers et al., 2002; Délye et al., 2002a, 2002b, 2002c, 2003). Herein, we found that three more residues (Trp-2,027, Asp-2,078, and Gly-2,096) are also associated with sensitivity to ACCase inhibitors in black-grass. ACCase enzyme assays and three-dimensional model analyses clearly show that these three residues are direct causes for altered sensitivity to ACCase inhibitors. This conclusion is further strengthened by the preliminary report of Trp-2,027-Cys and Asp-2,078-Gly substitutions as candidate herbicide mutations in wild oat (Christoffers et al., 2000).
Residue Trp-2,027 is absolutely conserved in all 31 homomeric ACCase sequences known so far. Residue Gly-2,096 is conserved in all ACCase sequences but those from yeasts (Ala-2,096, as in a black-grass mutant, resistant ACCase). Residues Trp-2,027 and Gly-2,096, like residue Ile-2,041, are both involved in sensitivity to the four APPs, but not to the two CHDs assayed (Tables I and II; Délye et al., 2003). In contrast, residue Asp-2,078 that is conserved in 30 out of the 31 known ACCase sequences (Val in the sequence from soybean) is the only one identified so far that is involved in sensitivity to all APPs and CHDs assayed (Tables I and II). In particular, the Asp-2,078-Gly substitution is the first reported mutation, to our knowledge, conferring a high level of resistance to the highly effective CHD clethodim (Table II). APPs and CHDs are reversible, competitive inhibitors of acetyl-CoA binding to the CT active site (Herbert et al., 1996; Zhang et al., 2003). The molecules of APPs and CHDs consist of a hydrophobic carbon skeleton with polar substituents (developed formulas are available at www.weedscience.com). Their bioactivity is limited by rather strict structural requirements (Webb et al., 2000; Turner and Pernich, 2002). The structure-activity relationship and the mode of binding to ACCase are poorly understood for CHDs (Webb et al., 2000). Because residues 1,781 and 2,078 are involved in sensitivity to both APPs and CHDs, and because residues 2,027, 2,041, and 2,096 are involved in sensitivity to APPs only, it is clear that binding sites for both inhibitor classes are largely overlapping, but not identical, in grasses. The side chain in residue Asp-2,078B is located at the bottom of the cavity of the CT active site, suggesting that CHDs, like APPs, bind deep inside the CT active-site cavity to inhibit ACCase CT activity.
From the three-dimensional models of black-grass CT-herbicide complexes, only the Ile-2,041B-Asn substitution directly interfered with herbicide binding, very likely because of a change in the conformation of the APP-binding site (Fig. 2B). The Gly-2,096B-Ala substitution very likely did not interfere with APP binding per se. Occurrence of an extra methyl group protruding within the cavity of the CT active site would either hamper access of APP molecules to their binding site and/or interfere with the allosteric changes in CT that are necessary for proper APP binding (Zhang et al., 2004). In the models reconstructed from yeast CT-herbicide complexes, the Trp-2,027B-Cys and the Asp-2,078B-Gly substitutions, which both occur at the bottom of the cavity of the CT active site, did not interfere with APP binding. However, they resulted in a number of small allosteric changes in the three-dimensional shape of the cavity that may not accurately be modeled from yeast. The heavily reduced activity observed for Cys-2,027 and Gly-2,078 ACCases supports the hypothesis of alteration of the shape of the active site, suggesting that changes at positions 2,027 and 2,078 might interfere with herbicide action by hampering access to their binding site. The reason why the Ile-1,781C-Leu substitution causes resistance to some APPs and CHDs cannot be directly deduced from three-dimensional models derived from the structure of yeast CT-APP complexes. In yeast, the residue homologous to residue Ile-1,781 is a Leu residue, the side chain of which contributes to increasing the stability of the CT-APP complex through van der Waals interactions with the R-methyl group in APPs (Zhang et al., 2004). These van der Waals interactions do not exist in black-grass wild-type Ile-1,781 ACCase. From the yeast model, the Ile-1,781C-Leu substitution should result either in no alteration in sensitivity to haloxyfop and diclofop or in increased sensitivity of black-grass ACCase to both compounds. In fact, Leu-1,781 ACCase is sensitive to haloxyfop and resistant to diclofop (Délye et al., 2002b), thus illustrating the limits of using the three-dimensional structure of the yeast CT-APP complex as a template to model the black-grass CT-APP complex.
In the three-dimensional models for the five grass species analyzed (Table III), the molecular surface of the region around the APP-binding site is absolutely conserved. However, substantial variation in I50 values are observed within the APP and CHD inhibitor classes, even within a given grass species (Tables II and III). This, and the heavy structural requirements for herbicidal activity observed for APPs and CHDs (Webb et al., 2000; Turner and Pernich, 2002), suggests that herbicide fitting into the cavity of the chloroplastic ACCase CT domain active site from grasses to reach their binding site(s) is extremely tight, and that some herbicide molecules fit better than others in the active-site cavity. Tight fitting is also likely the reason why such a subtle change as the Ile-1,781-Leu substitution, which may be considered as highly conservative, has a drastic effect upon the sensitivity of ACCase to the APPs fenoxaprop and diclofop and to the CHD cycloxydim, while it does not significantly alter sensitivity to the APPs clodinafop and haloxyfop, nor to the CHD clethodim (Zhang and Devine, 2000; Zagnitko et al., 2001; Christoffers et al., 2002; Délye et al., 2002a, 2002b, 2002c). Until the details of the binding of each inhibitor molecule within the cavity of the chloroplastic ACCase CT active site from grasses are fully understood, the cross-resistance pattern associated with specific structural changes will remain unpredictable.
All three-dimensional models for the ACCase CT domain from ACCase isoforms tolerant to APPs and CHDs (Table III) display both obstructing, hydrophobic side chains at the opening of the active-site cavity, differences in the shape of the bottom of this cavity, and differences in the distribution of polar groups at the bottom of the cavity when compared to three-dimensional models for chloroplastic ACCases from grasses (Fig. 3). Considering a similar binding mode for APPs and CHDs, these differences likely reduce the ability of the inhibitor molecules to reach their respective binding sites, thus making these ACCase isoforms tolerant to APPs and CHDs. Differences at the level of the binding site for APPs itself, because of variation at positions 1,781 in all tolerant ACCases, and at positions 2,096 and/or 2,041 in yeast and rat ACCases, are likely an additional reason for tolerance to ACCase inhibitors. In this respect, it can be proposed that the considerable differences in sensitivity to the APP inhibitors haloxyfop and clodinafop observed between the ACCase from the alga Cyclotella cryptica and the apicoplast ACCase from the protozoan T. gondii (Table III; Roessler, 1990; Jelenska et al., 2002), which display identical residues at the nine positions discussed above (Table III), might be attributed to the presence of a Leu-1,999 residue and/or an Ile-2,098 residue in the sequence from C. cryptica. All 30 other ACCase sequences known so far display a Trp-1,999 and a Val-2,098 residue. The Trp-1,999 residue belongs to the binding site for APPs. In three-dimensional models, the side chain in residue Ile-2,098B protrudes close to the opening of the active-site cavity, possibly impeding the access of APPs (Fig. 3).
Homomeric ACCase isoforms sensitive (chloroplastic) or tolerant (cytosolic) to APPs or CHDs have been shown to have similar catalytic properties (Herbert et al., 1996; Price et al., 2003). Mutant, chloroplastic ACCases with an Ile-1,781-Leu substitution did not display altered catalytic activity (Marles et al., 1993; Shukla et al., 1997). Similarly, mutant chloroplastic ACCases with an Ile-2,041-Asn or a Gly-2,096-Ala substitution displayed an activity close to that of wild-type ACCase (Délye et al., 2003). Residues 1,781, 2,041, and 2,096 are all located close to the opening of the cavity of CT active site, in the hydrophobic part of the cavity (Fig. 2). Unaltered activity of ACCase alleles mutated at these positions suggests that these residues do not play a major role in CT activity. In contrast, mutant chloroplastic ACCases with a Trp-2,027-Cys or an Asp-2,078-Gly substitution displayed a strongly reduced ACCase activity (about one-half the activity of the wild-type enzyme). This suggests that these residues that are located in the polar area at the bottom of the CT active-site cavity are involved in CT catalytic activity. This is consistent with the polar acetyl-thioester group in acetyl-CoA being inserted inside the cavity (Zhang et al., 2003, 2004), and with residues Trp-2,027 and Asp-2,078 being conserved in 31 and 30 out of the 31 known ACCase CT sequences, respectively.
This raises the question of whether there is a fitness penalty associated with black-grass mutant chloroplastic ACCase alleles. All black-grass mutant ACCase alleles discussed herein have been selected for in black-grass field populations by two APPs, fenoxaprop and clodinafop. As discussed before, the cross-resistance pattern associated with a given mutation is unpredictable. The cross-resistance to other APPs and/or CHDs observed for each mutant allele is thus a collateral effect of the selection. Black-grass plants containing mutant ACCase alleles are capable of surviving and spreading in the field under agronomic conditions. It may be that reduced ACCase activity does not significantly hamper the fitness of the whole plant. In this respect, a reduction of 70% in ACCase activity was shown to enable normal growth in yeast (Zagnitko et al., 2001), which suggests that ACCase activity might be overabundant enough in an organism to tolerate a significant reduction in enzyme efficiency without deleterious consequences. Black-grass populations, such as populations 00-049 or 00-099 that contain several mutant ACCase alleles (Table I), are potentially of interest for conducting in situ fitness cost studies. Allele-specific PCR assays developed in this work and in previous studies (Délye et al., 2002b, 2003) would then be of great use in assessing the evolution of the respective allele frequencies.
This work raised the number of residues within the black-grass homomeric ACCase CT domain that proved to be involved in sensitivity to APPs and/or CHDs to five. Among these residues, polar residues Trp-2,027 and Asp-2,078 also likely play a role in the CT reaction. Although the cavity of the CT active site is constituted almost equally by two distinct regions of the CT domain, we found that one proved determinant for herbicide sensitivity in grasses (residue 1,781) occurred in a region of highly conserved three-dimensional structure, while the other four were clustered in a more variable region encompassing amino acid positions 2,027 to 2,096. It is possible that additional, yet unknown, mutations exist there that confer altered sensitivity to ACCase inhibitors. Characterizing field or laboratory mutants at these positions will certainly improve our understanding of the mechanism of both the CT reaction and the inhibitory action of APPs and CHDs.
MATERIALS AND METHODS
Plant Material and Chloroplastic ACCase CT Domain Sequencing
We investigated three black-grass (Alopecurus myosuroides [Huds.]) populations (Table I) collected in France in 2000. Preliminary herbicide and genotyping assays revealed that, in these populations, the frequency of herbicide-resistant plants was considerably higher than the sum of frequencies of plants containing Leu-1,781 or Asn-2,041 ACCase alleles, which we expected was due to the presence of novel resistant ACCase allele(s). Resistance to 3 APPs (fenoxaprop, clodinafop, and haloxyfop) and 2 CHDs (cycloxydim and clethodim) was assessed using 50 seedlings per population and per herbicide as described elsewhere (Letouzé and Gasquez, 1999). The concentrations discriminating herbicide-resistant from herbicide-sensitive seedlings were 6 μm for clethodim and as described for other herbicides (Délye et al., 2002b). All seedlings were subsequently genotyped using previously described allele-specific PCR assays enabling the identification of amino acids present at positions 1,781 (Délye et al., 2002a) and 2,041 (Délye et al., 2003).
A total of 16 herbicide-resistant seedlings not containing Leu-1,781 or Asn-2,041 ACCase alleles were selected for sequencing experiments. They consisted of one clethodim-resistant, six cycloxydim-resistant, and three fenoxaprop-resistant seedlings from population 00-049, one clodinafop-resistant and two fenoxaprop-resistant seedlings from population 00-016, and one fenoxaprop-resistant and two clodinafop-resistant seedlings from population 00-099. DNA extraction and PCR amplification of a DNA fragment, including nucleotide positions 4,368 to 7,329 in the black-grass chloroplastic ACCase coding sequence (EMBL accession AJ310767; Délye et al., 2002a) with a proofreading polymerase, was as described (Délye et al., 2003). The amplified DNA fragment encompassed the entire CT domain of black-grass ACCase. The amplicon obtained from at least five independent PCRs pooled was purified using a Nucleospin Extract kit (Macherey-Nagel, Duren, Germany) and directly sequenced on both strands using gene-specific primers. When seedlings contained two different ACCase alleles, the amplicon was cloned in plasmid pDrive (Qiagen, Valencia, CA), and three different DNA inserts were sequenced for each ACCase allele. All differences observed after aligning the sequences were visually rechecked from chromatograms. Sequence assembling and alignments were performed using BioEdit software (Hall, 1999) and Multalin software (Corpet, 1988), respectively.
Bidirectional Allele-Specific PCR
Two four-primer bidirectional allele-specific PCR assays (Délye et al., 2002c) were designed to identify the codons present at positions 2,078 and 2,096. Primers VG2078-For: GCCTGGGTCGTGATTGG and VD2078-Rev: CGATCTGGGTTTATCTTGCTAT were designed to specifically prime ACCase sequences containing G or A at nucleotide position 6,389, respectively. Primers VA2096-For: CTGAGAGGACTGCAAAGGC and VG2096-Rev: CCTTGAGGTTCGAGAACATTAC were designed to specifically prime ACCase sequences containing C or G at nucleotide position 6,443, respectively. Primers VG2078-For and VD2078-Rev were used together with primers ACVII11: CTGCAAACATTGGTGGACCTCTTCCTATTAC and ACVII11R: CAGTCGGTGCTTCCTGCTGCAGCTG at a final concentration of 0.2 μm for each of the four primers. Primers VA2096-For and VG2096-Rev were used together with primers ACVII11 and ACVII11R at a final concentration of 0.2 μm for each of the four primers but VG2096-Rev (0.4 μm). Primers were designed to generate up to three distinct sizes of amplicons in a PCR, depending on the ACCase alleles present within one plant. Primer pair ACVII11/ACVII11R yielded a 1,082-bp fragment. Primer pairs ACVII11/VD2078-Rev and VG2078-For/ACVII11R yielded a 596- and a 524-bp fragment, respectively. Primer pairs ACVII11/VG2096-Rev and VA2096-For/ACVII11R yielded a 650- and a 472-bp fragment, respectively.
Development of a four-primer bidirectional allele-specific PCR was not possible for codon 2,027, possibly because of DNA secondary structure. We thus used two three-primer PCR assays for genotyping at codon 2,027. Primers VC2027-For: TCTGTTCATACTTGCTAACTGT and VW2027-For: CTGTTCATACTTGCTAACTGG were designed to specifically prime ACCase sequences containing T or G at nucleotide position 6,237, respectively. Each primer was used together with primers ACVII11 and ACVII11R at a final concentration of 0.2 μm for each of the three primers. Up to two distinct sizes of amplicons were expected in a PCR, depending on the ACCase alleles present within one plant. Primer pairs VC2027-For/ACVII11R and VW2027-For/ACVII11R yielded a 681- and a 680-bp fragment, respectively.
All PCR mixes were as described (Délye et al., 2002a). For all primer combinations, the cycling program consisted of one denaturation step of 30 s at 95°C, followed by 37 cycles of 10 s at 95°C, 15 s at 62°C, and 30 s at 72°C.
Inhibition of Black-Grass ACCase Activity by APPs and CHDs
For enzyme studies, three purified populations were obtained by enabling cross-pollination between six black-grass plants that each contained two ACCase alleles with the same mutant codon. Each of the Cys-2,027-F1, Gly-2,078-F1, and Ala-2,096-F1 populations was thus derived from six plants containing two Cys-2,027, two Gly-2,078, or two Ala-2,096 ACCase alleles, respectively. The plants used for pairings were selected from populations 00-016 (Cys-2,027 allele), 00-049 (Gly-2,078 allele), and 00-099 (Ala-2,096 allele), respectively, using the allele-specific PCR assays described above. Sequencing the ACCase CT domain in these 18 plants showed no other amino acid change when compared to the CT domain from sensitive plants.
Populations 00-017, Cys-2,027-F1, Gly-2,078-F1, and Ala-2,096-F1 (Table I) were used for ACCase assays. Population 00-017 is a field-sensitive population where no mutant ACCase alleles could be detected. ACCase extraction, enzyme assay, and determination of APP and CHD inhibitory action were performed as described (Shukla et al., 1997) from three to four leaf seedlings. Assays were performed twice for each inhibitor concentration. The data were fitted to the logistic sigmoidal, three-parameter model of Sigmaplot software (SPSS Science, Chicago), and the concentrations inhibiting 50% of ACCase activity (I50 values) were computed as the x0 from the equation for each inhibitor.
Protein Modeling
Homodimeric ACCase CT models were computed using the Swiss-model server (Schwede et al., 2003). For all CT sequences studied, each monomer was modeled separately using the three-dimensional structure of the free CT domain from yeast (Saccharomyces cerevisiae) as a template (PDB accession 1UYT; Zhang et al., 2003). To assess the consequences of the amino acid substitutions we identified in the black-grass CT domain upon the herbicide-binding site, mutant and wild-type black-grass CT domains were also modeled using the three-dimensional structure of the yeast CT domain in complex with the APP haloxyfop as a template (PDB accession 1UYS; Zhang et al., 2004). CT dimers were visualized using Deepview 3.7 (Schwede et al., 2003).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ310767 and AJ632096.
This work was supported by the Département Santé des Plantes et Environnement of the Institut National de la Recherche Agronomique.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046144.
References
- Alban C, Baldet P, Douce R (1994) Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem J 300: 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JD, Gronwald JW, Keith RA, Somers DA, Gegenbach BG, Wyse DL (1991) Kinetics of inhibition of acetyl-coenzyme A carboxylase by sethoxydim and haloxyfop. Pestic Biochem Physiol 39: 100–109 [DOI] [PubMed] [Google Scholar]
- Christoffers MJ, Berg ML, Messersmith CG (2000) Analysis of acetyl-CoA carboxylase gene sequences from fenoxaprop-P-resistant wild oat biotypes (abstract no. 121). Proc North Cent Weed Sci Soc 55: 67 [Google Scholar]
- Christoffers MJ, Berg ML, Messersmith CG (2002) An isoleucine to leucine mutation in acetyl-CoA carboxylase confers herbicide resistance in wild oat. Genome 45: 1049–1056 [DOI] [PubMed] [Google Scholar]
- Christopher JT, Holtum JAM (1998) The dicotyledonous species Erodium moschatum (L) L'Hér. ex. Aiton is sensitive to haloxyfop herbicide due to herbicide-sensitive acetyl-coenzyme A carboxylase. Planta 207: 275–279 [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C, Calmès É, Matéjicek A (2002. a) SNP markers for black-grass (Alopecurus myosuroides Huds.) genotypes resistant to acetyl CoA carboxylase-inhibiting herbicides. Theor Appl Genet 104: 1114–1120 [DOI] [PubMed] [Google Scholar]
- Délye C, Matéjicek A, Gasquez J (2002. b) PCR-based detection of resistance to acetyl-CoA carboxylase-inhibiting herbicides in black-grass (Alopecurus myosuroides Huds) and ryegrass (Lolium rigidum Gaud). Pest Manag Sci 58: 474–478 [DOI] [PubMed] [Google Scholar]
- Délye C, Wang T, Darmency H (2002. c) An isoleucine-leucine substitution in chloroplastic acetyl-Co A carboxylase from green foxtail (Setaria viridis L. Beauv.) is responsible for resistance to the cyclohexanedione herbicide sethoxydim. Planta 214: 421–427 [DOI] [PubMed] [Google Scholar]
- Délye C, Zhang X-Q, Chalopin C, Michel S, Powles SB (2003) An isoleucine residue within the carboxyl-transferase domain of multidomain acetyl-CoA carboxylase is a major determinant of sensitivity to aryloxyphenoxypropionate but not to cyclohexanedione inhibitors. Plant Physiol 132: 1716–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli MA, Gegenbach BG, Gronwald JW, Somers DA, Wyse DL (1993) Characterization of maize acetyl-coenzyme A carboxylase. Plant Physiol 101: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson KJ, Gronwald JW, Wyse DL (1997) Isoforms of acetyl-coenzyme A carboxylase in Lolium multiflorum. Plant Physiol Biochem 35: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focke M, Gieringer E, Schwan S, Jänsch L, Binder S, Braun H-P (2003) Fatty acid biosynthesis in mitochondria of grasses: malonyl-coenzyme A is generated by mitochondrial-localized acetyl-coenzyme A carboxylase. Plant Physiol 133: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R (1997) Plastid-localised acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc Natl Acad Sci USA 94: 14179–14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P, Haselkorn R (1993) Wheat acetyl-CoA carboxylase. Plant Mol Biol 22: 547–552 [DOI] [PubMed] [Google Scholar]
- Gornicki P, Podkowinski J, Scappino LA, DiMaio J, Ward E, Haselkorn R (1994) Wheat acetyl-coenzyme A carboxylase: cDNA and protein structure. Proc Natl Acad Sci USA 91: 6860–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald JW (1991) Lipid biosynthesis inhibitors. Weed Sci 39: 435–449 [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Harwood JL (1988) Fatty acid metabolism. Annu Rev Plant Physiol 39: 101–138 [Google Scholar]
- Herbert D, Price LJ, Alban C, Dehaye L, Job D, Cole DJ, Pallett KE, Harwood JL (1996) Kinetic studies on two isoforms of acetyl-CoA carboxylase from maize leaves. Biochem J 318: 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Sirikhachornkit A, Haselkorn R, Gornicki P (2002) The carboxyltransferase activity of the apicoplast acetyl-CoA carboxylase of Toxoplasma gondii is the target of aryloxyphenoxypropionate inhibitors. J Biol Chem 277: 23208–23215 [DOI] [PubMed] [Google Scholar]
- Joachimiak M, Tevzadze G, Podkowinski J, Haselkorn R, Gornicki P (1997) Wheat cytosolic acetyl-CoA carboxylase complements an ACC1 null mutation in yeast. Proc Natl Acad Sci USA 94: 9990–9995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y (1994) Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance towards herbicides. Proc Natl Acad Sci USA 91: 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y (1996) Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37: 117–122 [DOI] [PubMed] [Google Scholar]
- Kuk Y-I, Burgos NR, Talbert RE (2000) Cross- and multiple resistance of diclofop-resistant Lolium spp. Weed Sci 48: 412–419 [Google Scholar]
- Letouzé A, Gasquez J (1999) A rapid reliable test for screening aryloxyphenoxypropionic acid resistance within Alopecurus myosuroides and Lolium spp populations. Weed Res 39: 37–48 [Google Scholar]
- Letouzé A, Gasquez J (2003) Enhanced activity of several herbicide-degrading enzymes: a suggested mechanism responsible for multiple resistance in black-grass (Alopecurus myosuroides Huds.). Agronomie (Paris) 23: 601–608 [Google Scholar]
- Marles MAS, Devine MD, Hall JC (1993) Herbicide resistance in Setaria viridis conferred by a less sensitive form of acetyl coenzyme A carboxylase. Pestic Biochem Physiol 46: 7–14 [Google Scholar]
- Menéndez J, De Prado R (1999) Characterization of two acetyl-CoA carboxylase isoforms in diclofop-methyl-resistant and -susceptible biotypes of Alopecurus myosuroides. Pestic Biochem Physiol 65: 82–89 [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414: 211–222 [DOI] [PubMed] [Google Scholar]
- Podkowinski J, Sroga GE, Haselkorn R, Gornicki P (1996) Structure of a gene encoding a cytosolic acetyl-CoA carboxylase of hexaploid wheat. Proc Natl Acad Sci USA 93: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D (1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 405–430 [DOI] [PubMed] [Google Scholar]
- Price LJ, Herbert D, Cole DJ, Harwood JL (2003) Use of plant cell cultures to study graminicide effects on lipid metabolism. Phytochemistry 63: 533–541 [DOI] [PubMed] [Google Scholar]
- Price LJ, Moss SR, Cole DJ, Harwood JL (2004) Graminicide resistance in a black-grass (Alopecurus myosuroides) population correlates with insensitivity of acetyl-CoA carboxylase. Plant Cell Environ 27: 15–26 [Google Scholar]
- Rendina AR, Craig-Kennard AC, Beaudoin JD, Breen MK (1990) Inhibition of acetyl-coenzyme A carboxylase by two classes of grass-selective herbicides. J Agric Food Chem 38: 1282–1287 [Google Scholar]
- Roessler PG (1990) Purification and characterization of acetyl-CoA carboxylase from the diatom Cyclotella cryptica. Plant Physiol 92: 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y (1995) The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol 108: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seng TW, Skillman TR, Yang N, Hammond C (2003) Cyclohexanedione herbicides are inhibitors of rat heart acetyl-CoA carboxylase. Bioorg Med Chem Lett 13: 3237–3242 [DOI] [PubMed] [Google Scholar]
- Shukla A, Leach GE, Devine MD (1997) High-level resistance to sethoxydim conferred by an alteration in the target enzyme, acetyl-CoA carboxylase, in Setaria faberi and Setaria viridis. Plant Physiol Biochem 35: 803–807 [Google Scholar]
- Turner JA, Pernich DJ (2002) Origin of enantiomeric selectivity in the aryloxyphenoxypropionic acid class of herbicidal acetyl coenzyme A carboxylase (ACCase) inhibitors. J Agric Food Chem 50: 4554–4566 [DOI] [PubMed] [Google Scholar]
- Webb SR, Durst GL, Pernich D, Hall JC (2000) Interaction of cyclohexanediones with acetyl coenzyme-A carboxylase and an artificial target-site antibody mimic: a comparative molecular field analysis. J Agric Food Chem 48: 2506–2511 [DOI] [PubMed] [Google Scholar]
- Zagnitko O, Jelenska J, Tevzadze G, Haselkorn R, Gornicki P (2001) An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors. Proc Natl Acad Sci USA 98: 6617–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tweel B, Tong L (2004) Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA 101: 5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang Z, Shen Y, Tong L (2003) Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 299: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Devine MD (2000) A possible point mutation of plastidic ACCase gene conferring resistance to sethoxydim in green foxtail (Setaria viridis) (abstract no. 81). Weed Sci Soc Am 40: 33 [Google Scholar]