Abstract
The PCR is an extremely powerful method for detecting microorganisms. However, its full potential as a rapid detection method is limited by the inhibition of the thermostable DNA polymerase from Thermus aquaticus by many components found in complex biological samples. In this study, we have compared the effects of known PCR-inhibiting samples on nine thermostable DNA polymerases. Samples of blood, cheese, feces, and meat, as well as various ions, were added to PCR mixtures containing various thermostable DNA polymerases. The nucleic acid amplification capacity of the nine polymerases, under buffer conditions recommended by the manufacturers, was evaluated by using a PCR-based detection method for Listeria monocytogenes in the presence of purified template DNA and different concentrations of PCR inhibitors. The AmpliTaq Gold and the Taq DNA polymerases from Thermus aquaticus were totally inhibited in the presence of 0.004% (vol/vol) blood in the PCR mixture, while the HotTub, Pwo, rTth, and Tfl DNA polymerases were able to amplify DNA in the presence of 20% (vol/vol) blood without reduced amplification sensitivity. The DNA polymerase from Thermotoga maritima (Ultma) was found to be the most susceptible to PCR inhibitors present in cheese, feces, and meat samples. When the inhibitory effect of K and Na ions was tested on the nine polymerases, HotTub from Thermus flavus and rTth from Thermus thermophilus were the most resistant. Thus, the PCR-inhibiting effect of various components in biological samples can, to some extent, be eliminated by the use of the appropriate thermostable DNA polymerase.
The usefulness of PCR-based detection of microorganisms in complex biological samples, such as clinical, environmental, and food samples, is limited in part by the presence of substances that inhibit the PCR or reduce the amplification efficiency. The PCR inhibitors may act through one or more of the following mechanisms (34): (i) interference with the cell lysis step, (ii) degradation or capture of the nucleic acids, or (iii) inactivation of the thermostable DNA polymerase.
A number of components have been reported to be PCR inhibitors, namely, bile salts and complex polysaccharides in feces (17, 22), heme in blood (2), humic substances in soil (30), proteinases in milk (25), and urea in urine (11). Much effort is being devoted to the development of sample preparation methods to overcome the problem of PCR inhibitors (for reviews, see references 13 and 14). Different techniques are being employed to reduce the effect of PCR inhibitors and/or to separate the microorganisms from the PCR inhibitors. For example, aqueous two phase-systems (15), boiling (23), density gradient centrifugation (18), dilution (5), DNA extraction methods (12), enrichment media (31), filtration (6), and immunological techniques (8) have been used to facilitate PCR. The thermostable DNA polymerase is perhaps the most important target site of PCR-inhibiting substances. The most widely used polymerase in PCR-based methods for the detection of microorganisms is Taq DNA polymerase from Thermus aquaticus. The polymerase can be degraded by proteinases (25), denatured by phenol (10) or detergents (27), and inhibited by blocking of the active site by the inhibitor, which is the effect of heme (2).
In a study by Wiedbrauk et al. (32), it was noted that both Tfl and Tth DNA polymerases were more resistant to aqueous and vitreous fluids of the eye than the polymerases Taq, Tli, and Stoffel fragment. The aim of this study was to systematically investigate the capacity of nine commercially available thermostable DNA polymerases to amplify DNA in the presence of certain complex biological samples and in the presence of certain ions. The samples are all known to reduce the amplification capacity of Taq DNA polymerase (13). We used a PCR-based detection assay for Listeria monocytogenes to study the effect of increasing the concentration of purified template DNA and of adding different concentrations of the inhibitory samples. Increasing the concentration of target DNA may be useful in overcoming the effect of inhibitors (interfering with DNA and/or binding reversibly to the DNA-binding domain of the polymerase), while adding various concentrations of the inhibitory sample may be useful in evaluating the effect of the inhibitory samples on the amplification capacity of the nine polymerases.
MATERIALS AND METHODS
Bacterial strain and DNA extraction.
DNA of L. monocytogenes 167 vet (which was obtained from the Meat Research Institute, Kävlinge, Sweden) was used as the target DNA in this study. Extraction of L. monocytogenes DNA was performed in accordance with a standard technique described by Sambrook et al. (28), modified by the addition of 30 U of mutanolysin (Sigma Chemical Co., St. Louis, Mo.) per ml to the lysis solution. The concentration of DNA was measured spectrophotometrically (28), and the size of the L. monocytogenes genomic DNA is estimated to be 3,150 kb (21).
PCR assay and incubation conditions.
The PCR assay was carried out as previously described by Lantz et al. (15). Briefly, the PCR assay was performed with the primer ru8 [5′-AAGGAGGTGATCCA(G/A)CCGCA(G/C)(G/C)TTC-3′], which is complementary to the universal region of the 16S rRNA gene (26), and the primer LM2 (5′-CCTTTGACCACTCTGGAGACAGAGC-3′), which is complementary to the L. monocytogenes-specific region of the 16S rRNA gene (15). The 553-bp PCR product was visualized by 1.3% agarose gel electrophoresis containing ethidium bromide (28). The gel was analyzed by using a gel documentation system (Bio-Rad Laboratories, Hercules, Calif.). The results were scored as follows: +, strong intensity of PCR product on agarose gel; ±, weak intensity of PCR product on agarose gel; −, no visible PCR product on agarose gel. Each of the PCR results given here was obtained from at least two independent PCR amplifications.
The volume of the PCR mixture was 25 μl. All of the PCR mixtures contained a 0.5 mM concentration of each of the primers ru8 and LM2 and a 0.2 mM concentration of each of the deoxyribonucleoside triphosphates. Reaction buffers for the polymerases, as specified by the manufacturers, were as follows: the PCR buffer of AmpliTaq Gold DNA polymerase (Thermus flavus) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, and 0.75 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.); the PCR buffer of the Expand High Fidelity (Expand HF) PCR system (which is a mixture of two DNA polymerases, Taq [Thermus aquaticus] and Pwo [Pyrococcus woesei]) contained 1× Expand HF buffer, 1.5 mM MgCl2, and 1.33 U of Expand DNA polymerase (Boehringer Mannheim GmbH, Mannheim, Germany); the PCR buffer of HotTub DNA polymerase (Thermus ubiquatous) contained 50 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, 2.5 mM MgCl2, and 0.75 U of HotTub DNA polymerase (Amersham Life Science, Cleveland, Ohio); the PCR buffer of Pwo DNA polymerase (Pyrococcus woesei) contained 10 mM Tris-HCl (pH 8.85; 20°C), 25 mM KCl, 5 mM (NH4)2SO4, 1.5 mM MgCl2, and 1.25 U of Pwo DNA polymerase (Boehringer Mannheim); the PCR buffer of rTth DNA polymerase (Thermus thermophilus) contained 5% (vol/vol) glycerol, 10 mM Tris-HCl (pH 8.3), 0.1 M KCl, 0.05% (wt/vol) Tween 20, 0.75 mM EGTA, 2.5 mM MgCl2, and 1.25 U of rTth DNA polymerase (Perkin-Elmer Cetus); the PCR buffer of Taq DNA polymerase (Thermus aquaticus) contained 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3; 20°C), and 0.75 U of Taq DNA polymerase (Boehringer Mannheim); the PCR buffer of Tfl DNA polymerase (Thermus flavus) contained 20 mM Tris-Acetate (pH 9.0), 10 mM (NH4)2SO4, 75 mM potassium acetate, 0.05% Tween 20, 2.5 mM MgSO4, and 0.5 U of Tfl DNA polymerase (Promega Corporation, Madison, Wis.); the PCR buffer of Tli DNA polymerase (Thermus litoralis) contained 10 mM Tris-HCl (pH 9.0; 25°C), 0.1% Triton X-100, 50 mM KCl, 2.5 mM MgCl2, and 0.3 U of Tli DNA polymerase (Promega); the PCR buffer of Ultma DNA polymerase (Thermotoga maritima) contained 10 mM Tris-HCl (pH 8.8; room temperature), 10 mM KCl, 0.002% (vol/vol) Tween 20, 2.5 mM MgCl2, and 0.75 U of Ultma DNA polymerase (Perkin-Elmer Cetus). The reaction mixtures were subjected to 30 amplification cycles consisting of heat denaturation at 94°C for 40 s, primer annealing at 53°C for 40 s, and DNA extension at 72°C for 40 s. Finally, the mixtures were maintained at 72°C for 7 min for the final extension of DNA. These incubation conditions were the same for all amplification reaction mixtures, except those containing AmpliTaq Gold, since this polymerase requires a hot start (95°C for 10 min). Incubation was carried out in a model 2400 thermal cycler (Perkin-Elmer Cetus).
PCR-inhibitory samples.
The blood sample used was drawn from a healthy person into 10-ml evacuated blood-collecting tubes containing 0.1 ml of 0.47 M EDTA (Terumo Europe N. V., Leuven, Belgium). The Danish Blue Castello soft cheese (henceforth referred to as cheese) was diluted 10-fold in physiological saline solution and homogenized for 2 min in a stomacher (Lab-Blender 400; Steward Laboratory, London, United Kingdom). The fecal sample was obtained from a healthy person, diluted 10-fold in physiological saline solution, and homogenized for 2 min in the stomacher. The minced pork meat (henceforth referred to as meat) was diluted 10-fold in physiological saline solution and homogenized for 2 min in the stomacher. Each of the PCR-inhibitory samples was poured into a sterile 1.5-ml Eppendorf tube and stored at −20°C. The frozen blood and cheese, feces, and meat homogenates were thawed at room temperature, mixed with a vortex mixer, and allowed to stand for 5 min to allow large particles to settle before they were added directly or after dilution to the reaction mixtures of the nine polymerases.
The effect of target DNA concentration on sensitivity in the presence of PCR-inhibitory samples was studied by adding different concentrations of L. monocytogenes DNA (1 μg to 10 fg per reaction tube) to reaction mixtures containing 20% (vol/vol) blood, 20% (vol/vol) cheese homogenate, 20 or 0.4% (vol/vol) fecal homogenate, and 20 or 0.2% (vol/vol) meat homogenate.
To study the effect of different concentrations of the PCR-inhibitory samples on the amplification capacity of the nine polymerases, different blood concentrations (20, 2, 0.2, 0.04, 0.02, 0.004, and 0.002%) and different cheese, feces, and meat homogenate concentrations (20, 2, 0.2, 0.1, 0.07, 0.05, and 0.04%) were added to PCR mixtures of the nine polymerases containing 1 ng of L. monocytogenes DNA per reaction tube.
Ion solutions.
Different concentrations of CaCl2 · 2H2O, KCl, MgCl2 · 6H2O, and NaCl (Merck, Darmstadt, Germany) were prepared, as sterile solutions, to study their effect on the amplification capacity of the nine polymerases. The ion concentrations were 1, 5, 10, 20, 30, 40, and 50 mM CaCl2; 10, 50, 100, 200, 300, 400, 500, and 750 mM KCl; 25, 50, 75, 100, and 150 mM MgCl2; and 50, 100, 200, 300, 400, 500, and 750 mM NaCl. Five microliters of the different ion concentrations were added to the PCR mixtures of the nine polymerases containing 1 ng of L. monocytogenes DNA per reaction tube.
RESULTS AND DISCUSSION
Amplification capacity of the nine thermostable DNA polymerases.
The DNA amplification capacities of nine thermostable DNA polymerases were compared in accordance with a PCR-based assay previously designed to detect L. monocytogenes (15). To assess the detection limit for L. monocytogenes DNA in PCR-inhibitory samples relative to the target DNA in the absence of the sample (i.e., in pure water), various amounts of DNA were added to the PCR mixtures containing different polymerases (Table 1). Six of the nine polymerases detected 0.1 ng of DNA. The detection limits for the polymerases Expand, Tli, and Ultma were 1 pg, 10 pg, and 1 ng per reaction tube, respectively.
TABLE 1.
Inhibitory effects of blood, cheese, feces, and meat on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | Samples | PCR resultsa for mixtures containing indicated amt (g) of L. monocytogenes DNA/reaction tube
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 10−6 | 10−7 | 10−8 | 10−9 | 10−10 | 10−11 | 10−12 | 10−13 | ||
| AmpliTaq Gold | 20% Blood | −− | −− | −− | −− | [−−] | −− | −− | −− |
| 20% Cheese | ++ | ++ | +± | +− | [−−] | −− | −− | −− | |
| 0.4% Feces | ++ | −− | −− | −− | [−−] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | +± | −− | [−−] | −− | −− | −− | |
| Expand High Fidelity | 20% Blood | ++ | ±− | −− | −− | −− | −− | [−−] | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | ++ | +− | [−−] | −− | |
| 0.4% Feces | ++ | −− | −− | −− | −− | −− | [−−] | −− | |
| 0.2% Meat | ++ | ++ | +± | −− | −− | −− | [−−] | −− | |
| HotTub | 20% Blood | ++ | ++ | ++ | ++ | [+±] | −− | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | [+±] | −− | −− | −− | |
| 0.4% Feces | ++ | −− | −− | −− | [−−] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | ++ | −− | [−−] | −− | −− | −− | |
| Pwo | 20% Blood | ++ | ++ | ++ | ++ | [+±] | −− | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ±± | [−−] | −− | −− | −− | |
| 0.4% Feces | ++ | ++ | ++ | ++ | [+−] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | ±− | −− | [−−] | −− | −− | −− | |
| rTth | 20% Blood | ++ | ++ | ++ | ++ | [++] | −− | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | [+±] | −− | −− | −− | |
| 0.4% Feces | ++ | ++ | ++ | ++ | [++] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | ++ | −− | [−−] | −− | −− | −− | |
| Taq | 20% Blood | −− | −− | −− | −− | [−−] | −− | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | [±±] | −− | −− | −− | |
| 0.4% Feces | ++ | −− | −− | −− | [−−] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | +± | −− | [−−] | −− | −− | −− | |
| Tfl | 20% Blood | ++ | ++ | ++ | ++ | [±±] | −− | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | [+±] | −− | −− | −− | |
| 0.4% Feces | ±− | −− | −− | −− | [−−] | −− | −− | −− | |
| 0.2% Meat | ++ | ++ | ++ | +± | [−−] | −− | −− | −− | |
| Tli | 20% Blood | ++ | ++ | ++ | ++ | −− | [−−] | −− | −− |
| 20% Cheese | ++ | ++ | ++ | ++ | ++ | [±±] | −− | −− | |
| 0.4% Feces | ++ | −− | −− | −− | −− | [−−] | −− | −− | |
| 0.2% Meat | ++ | ++ | ++ | −− | −− | [−−] | −− | −− | |
| Ultma | 20% Blood | −− | −− | −− | [−−] | −− | −− | −− | −− |
| 20% Cheese | ++ | ++ | −− | [−−] | −− | −− | −− | −− | |
| 0.4% Feces | ±− | −− | −− | [−−] | −− | −− | −− | −− | |
| 0.2% Meat | ++ | +− | −− | [−−] | −− | −− | −− | −− | |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band; brackets indicate that results are for L. monocytogenes DNA in pure water instead of the sample in the PCR. The amounts of DNA indicated are per reaction tube.
Percentage (vol/vol) of blood or homogenate of cheese, feces, or meat in the PCR mixtures containing different amounts of L. monocytogenes DNA.
When blood was added to the PCR mixtures, it was found to be highly inhibitory to the DNA polymerases from Thermus aquaticus (AmpliTaq Gold and Taq) and Thermotoga maritima (Ultma) (Table 1). The addition of 20% (vol/vol) blood was completely inhibitory to these polymerases, even after the concentration of L. monocytogenes DNA was increased to 1 μg per reaction tube. However, the polymerases Expand, HotTub, Pwo, rTth, Tfl, and Tli successfully amplified the selected region of the 16S rRNA gene in the presence of 20% (vol/vol) blood. The amplification sensitivities of four of these six polymerases were not affected by the presence or absence of blood in the PCR, while the sensitivity of Expand was decreased by 5 log units and the sensitivity of Tli was decreased by 2 log units in the presence of 20% (vol/vol) blood. Akane et al. (2) found that the heme compound of blood is an inhibitor for AmpliTaq DNA polymerase, and they showed that this inhibition was not due to irreversible inactivation of the polymerase. Heme has been suggested to form a stable complex with the DNA polymerase, thereby preventing formation, and also causing dissociation, of the DNA polymerase complex (4).
The addition of 20% (vol/vol) cheese homogenate decreased the sensitivity of the polymerases AmpliTaq Gold, Expand, and Pwo by 1 log unit and the sensitivity of the polymerase Ultma by 2 log units, while the detection sensitivities of the other polymerases in the presence of 20% (vol/vol) cheese homogenate were the same as for pure water solutions of DNA (Table 1). When 20% (vol/vol) fecal homogenate or 20% (vol/vol) meat homogenate was added to reaction mixtures of the nine polymerases containing different concentrations of L. monocytogenes DNA (1 μg to 0.1 pg per reaction tube), the nine polymerases were completely inhibited (data not shown). Decreasing the fecal concentration to 0.4% (vol/vol) resulted in a marked difference between the polymerases (Table 1), so that 0.4% (vol/vol) fecal homogenate was found not to be inhibitory to the polymerases Pwo, from Pyrococcus woesei, and rTth, from Thermus thermophilus, and they exhibited the same sensitivity as for pure water solutions of DNA. On the other hand, the other polymerases had a sensitivity to 1 μg of L. monocytogenes DNA per reaction tube in the presence of 0.4% (vol/vol) fecal homogenate. Decreasing the meat homogenate concentration to 0.2% (vol/vol) gave rise to a small difference between the sensitivities of the polymerases (Table 1). The polymerase Tfl, from Thermus flavus, was slightly inhibited by the addition of 0.2% (vol/vol) meat homogenate, such that it had a sensitivity 1 log unit less than pure water solutions of DNA, while the polymerase Ultma required an increase in the concentration of L. monocytogenes DNA to 0.1 μg per reaction tube to overcome the inhibitory effect of 0.2% (vol/vol) meat homogenate. The rest of the polymerases exhibited a sensitivity to 10 ng of L. monocytogenes DNA per reaction. These results agree with previous studies in which both fecal and meat homogenates were found to be highly inhibitory to AmpliTaq and Taq DNA polymerases (17, 19, 33, 35).
Effect of different concentrations of blood, cheese, feces, and meat on the DNA polymerases.
The polymerases HotTub, Tfl, and Tli amplified the selected region of L. monocytogenes DNA in the presence of all tested concentrations of blood (Table 2). Although the polymerases Pwo and rTth were not inhibited in the presence of 20% (vol/vol) blood, and the polymerases Expand and Ultma were not inhibited in the presence of 2% (vol/vol) blood, a PCR-inhibitory span was observed for the polymerases Pwo (0.02%), rTth (2 to 0.02%), and Ultma (0.2 to 0.02%). A similar range of PCR inhibition was noted when we studied the effect of adding different concentrations of culture medium containing 28.6% whole blood to PCR mixtures containing Taq DNA polymerase and Streptococcus pneumoniae DNA (1). The addition of 0.5 μg of bovine serum albumin counteracted the inhibitory effect of the blood culture at various concentrations. A possible explanation of this may be the ability of bovine serum albumin to bind heme and/or serve as a competitive target for proteinases. On the other hand, the concentration of blood had to be reduced to as little as 0.002% (vol/vol) for the polymerases AmpliTaq Gold and Taq to regain their amplification capacity in the presence of 1 ng of L. monocytogenes DNA (Table 2). In a study by Mercier et al. (20), it was found that the addition of 4% (vol/vol) blood to the reaction mixture was totally inhibitory to Taq DNA polymerase, whereas PCR amplification took place in the presence of 1 or 2% (vol/vol) blood. Panaccio and Lew (24), however, found that the addition of 1% (vol/vol) blood was totally inhibitory to Taq DNA polymerase, while they succeeded in amplifying a target sequence in the presence of up to 4% (vol/vol) blood by using Tth DNA polymerase. A possible explanation of these conflicting results for the effect of blood on Taq DNA polymerase may be the use of different target DNA concentrations and PCR conditions.
TABLE 2.
Inhibitory effects of different blood concentrations on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | PCR resultsa with bloodb concn (vol/vol) of:
|
||||||
|---|---|---|---|---|---|---|---|
| 20% | 2% | 0.2% | 0.04% | 0.02% | 0.004% | 0.002% | |
| AmpliTaq Gold | −− | −− | −− | −− | −− | −− | ±+ |
| Expand High Fidelity | −− | ++ | ++ | ++ | ±± | −+ | ++ |
| HotTub | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Pwo | ±+ | ++ | ++ | +− | −− | ++ | ++ |
| rTth | ++ | −− | −− | −− | −− | ++ | ++ |
| Taq | −− | −− | −− | −− | −− | −− | ++ |
| Tfl | ++ | ++ | −+ | ++ | ++ | ++ | ++ |
| Tli | ++ | ++ | ++ | ++ | ++ | −± | −± |
| Ultma | −− | ±± | −− | −− | −− | −± | −± |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band.
Percentage (vol/vol) of blood in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
When different concentrations of the cheese homogenate were added to PCR mixtures of the nine polymerases containing 1 ng of L. monocytogenes DNA (Table 3), eight of the nine polymerases were not inhibited by the cheese homogenate, whereas the amplification capacity of the polymerase Ultma was totally inhibited at concentrations of cheese homogenate over 0.2% (vol/vol). The generally low level of inhibition by cheese may be due to the choice of the particular cheese used. In a previous study, variations in PCR inhibition were observed for different soft cheeses (16).
TABLE 3.
Inhibitory effects of different concentrations of cheese, feces, and meat on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | Sampleb | PCR resultsa with sample concn of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 20% | 2% | 0.2% | 0.1% | 0.07% | 0.05% | 0.04% | ||
| AmpliTaq Gold | Cheese | −+ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | ++ | ++ | ++ | ++ | ++ | |
| Meat | −− | −− | −− | −+ | −+ | ++ | ++ | |
| Expand High Fidelity | Cheese | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | ++ | ++ | ++ | ++ | ++ | |
| Meat | −− | −− | ±+ | ++ | ++ | ++ | ++ | |
| HotTub | Cheese | ±+ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | −− | ++ | ++ | ++ | ++ | |
| Meat | −− | −− | −− | ±+ | ±+ | ++ | ++ | |
| Pwo | Cheese | ±+ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −+ | ++ | ++ | ++ | ++ | ++ | |
| Meat | −− | −− | −− | ++ | ++ | ++ | ++ | |
| rTth | Cheese | ±+ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | ++ | ++ | ++ | ++ | ++ | |
| Meat | −− | −− | −± | ++ | ++ | ++ | ++ | |
| Taq | Cheese | ±+ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | −− | ±+ | ++ | ++ | ++ | |
| Meat | −− | −− | −− | ±+ | ++ | ++ | ++ | |
| Tfl | Cheese | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | −− | −± | ++ | ++ | ++ | |
| Meat | −− | −− | −± | −+ | ++ | ++ | ++ | |
| Tli | Cheese | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | −− | −− | −+ | ++ | ++ | |
| Meat | −− | −− | −− | ±+ | ++ | ++ | ++ | |
| Ultma | Cheese | −− | −− | ++ | ++ | ++ | ++ | ++ |
| Feces | −− | −− | −− | −± | −+ | ++ | ++ | |
| Meat | −− | −− | −− | −− | −− | −− | −− | |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band.
Percentage (vol/vol) of homogenates of cheese, feces, and meat in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
The concentration of fecal homogenate had to be reduced to 0.2% (vol/vol) to detect the PCR product in both replicates for the polymerases AmpliTaq Gold, Expand, Pwo, and rTth (Table 3). The amplification capacity of the polymerases HotTub, Taq, Tfl, and Ultma was restored after the fecal homogenate concentration was reduced to 0.1% (vol/vol). The polymerase most sensitive to fecal homogenate was Tli, which required a decrease in the concentration of fecal homogenate to 0.07% (vol/vol) to restore its amplification capacity. In the case of the meat homogenate, a concentration as low as 0.04% (vol/vol) was inhibitory to the polymerase Ultma (Table 3). On the other hand, the amplification capacities of the polymerases Expand, rTth, and Tfl were restored when the concentration of meat homogenate was decreased to 0.2% (vol/vol). The PCR product was detected with the remaining polymerases in tubes containing meat homogenate at concentrations of, but not higher than, 0.1% (vol/vol).
Effect of trypsin inhibitor on rTth inhibitors in blood.
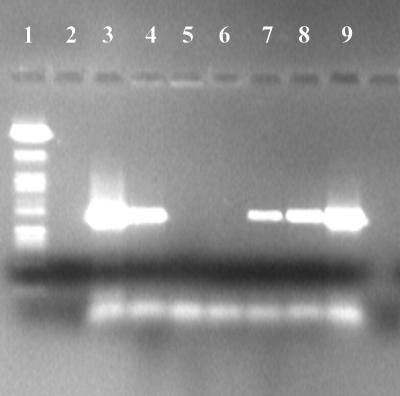
To investigate whether a trypsin inhibitor relieves inhibition of rTth DNA polymerase in PCR mixtures with 2 to 0.02% (vol/vol) blood concentrations (see Table 2), 5 μg of lima bean trypsin inhibitor (Sigma Chemical Co.) was added to PCR mixtures containing different concentrations of blood and 1 ng of L. monocytogenes DNA. The addition of trypsin inhibitor decreased the range of inhibitory blood concentrations relative to those shown in Table 2. However, blood concentrations of 0.2 and 0.04% (vol/vol) were still inhibitory to the rTth DNA polymerase (Fig. 1). Powell et al. (25) have also shown that the addition of proteinase inhibitors to the reaction mixtures can partially counteract the effect of PCR inhibitors in milk. They studied the detection of L. monocytogenes DNA in PCR mixtures with various concentrations of milk and found the PCR to be inhibited by the milk in the concentration range of 0.5 to 0.01% (vol/vol). They suggested that a proteinase which inhibits the PCR by degrading the DNA polymerase may be present in milk.
FIG. 1.
Relief of rTth inhibition due to blood by the addition of 5 μg of lima bean trypsin inhibitor in a PCR mixture containing 1 ng of L. monocytogenes DNA and blood at different concentrations. Lanes: 1, 100-bp molecular weight marker (Boehringer Mannheim); 2, negative control; 3, positive control; 4, 20% blood; 5, 2% blood; 6, 0.2% blood; 7, 0.04% blood; 8, 0.02% blood; 9, 0.004% blood.
Effect of ions on the nine thermostable DNA polymerases.
Different concentrations of ions were added to reaction mixtures containing 1 ng of L. monocytogenes DNA to study their effect on the amplification capacity of the nine polymerases (Tables 4 to 7). The inhibitory effect of divalent ions (Ca and Mg) was more pronounced than that of monovalent ions (K and Na), with Ca being the most inhibitory. The polymerase most sensitive to K, Mg, and Na was Ultma from Thermatoga maritima, whereas the polymerases AmpliTaq Gold and Taq were the most sensitive to Ca. On the other hand, the polymerases from Thermus flavus (HotTub) and Thermus thermophilus (rTth) were the most resistant to the monovalent ions, while the polymerase HotTub was the most resistant to Ca, which can be related to the low ionic content of HotTub buffer. The levels of resistance of the polymerase Taq to K, Mg, and Na were higher than that of the polymerase AmpliTaq Gold, although they are derived from the same organism. The resistance pattern of the polymerase Expand, which is a mixture of two polymerases (Pwo and Taq), to K, Mg, and Na was more similar to that of Taq than to that of Pwo. However, the polymerase Expand was more resistant to Ca ions than were the polymerases Taq and Pwo. It has previously been found that the addition of K ions at concentrations higher than 75 mM was completely inhibitory to amplification of DNA by AmpliTaq (9), and increasing the concentration of Mg ions to 15 mM has been found to inhibit the ability of Taq DNA polymerase to detect herpes simplex virus (32). The highly inhibitory effect of the Ca ion may be due to its ability to compete with the Mg ion. Ca ion concentrations above 3 mM have previously been found to be inhibitory to the amplification capacity of AmpliTaq DNA polymerase. This inhibitory effect was reversed by increasing the Mg ion concentration in the reaction mixture (3). Also, it has previously been shown that variations in the performance of DNA polymerases in coamplification PCR were salt dependent (7).
TABLE 4.
Effect of Ca ions on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | PCR resultsa with CaCl2 concn (mM)b of:
|
||||||
|---|---|---|---|---|---|---|---|
| 0.2 | 1.0 | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 | |
| AmpliTaq Gold | ++ | −− | −− | −− | −− | −− | −− |
| Expand High Fidelity | ++ | ++ | ++ | −− | −− | −− | −− |
| HotTub | ++ | ++ | ++ | ++ | −− | −− | −− |
| Pwo | ++ | +− | −− | −− | −− | −− | −− |
| rTth | ++ | ++ | −− | −− | −− | −− | −− |
| Taq | ++ | −− | −− | −− | −− | −− | −− |
| Tfl | ++ | ++ | +− | −− | −− | −− | −− |
| Tli | ++ | ++ | −− | −− | −− | −− | −− |
| Ultma | ++ | ++ | −− | −− | −− | −− | −− |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band.
Concentrations of CaCl2 in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
TABLE 7.
Effect of Na ions on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | PCR resultsa with NaCl concn (mM)b of:
|
||||||
|---|---|---|---|---|---|---|---|
| 10 | 20 | 40 | 60 | 80 | 100 | 150 | |
| AmpliTaq Gold | ++ | ++ | −− | −− | −− | −− | −− |
| Expand High Fidelity | ++ | ++ | ++ | ++ | −− | −− | −− |
| HotTub | ++ | ++ | ++ | ++ | ++ | −− | −− |
| Pwo | ++ | ++ | ++ | −− | −− | −− | −− |
| rTth | ++ | ++ | ++ | ++ | ++ | ±− | −− |
| Taq | ++ | ++ | ++ | ++ | −− | −− | −− |
| Tfl | ++ | ++ | ++ | ++ | −− | −− | −− |
| Tli | ++ | ++ | ++ | ++ | −− | −− | −− |
| Ultma | ++ | ++ | −− | −− | −− | −− | −− |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band.
Concentrations of NaCl in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
Conclusions.
The inhibition of Taq DNA polymerase amplification by the components of biological samples can be reduced or eliminated by the use of an appropriate thermostable DNA polymerase. A number of DNA polymerases are now commercially available. These polymerases exhibit different properties with regard to resistance to various components in clinical, environmental, and food samples. By selecting an appropriate polymerase, it is possible to more efficiently amplify nucleic acid in the presence of biological material without the need for extensive sample processing prior to PCR.
Based on the results given in Tables 1 and 2, eight of the nine polymerases can be divided into three groups: (i) the blood-sensitive polymerases (AmpliTaq Gold, Taq, and Ultma), i.e., those whose amplification capacities were not restorable by either increasing the template DNA concentration (Table 1) nor decreasing the blood concentration (Table 2); (ii) the blood-resistant polymerases (HotTub, Tfl, and Tli), which amplified the template DNA in the presence of all tested blood concentrations; and (iii) the feces-resistant polymerases (Pwo and rTth), which can amplify DNA in the presence of 20% blood but have a range of inhibition around the blood concentration of 0.02%. The polymerase Expand, which is a mixture of Pwo and Taq, has a resistance pattern to the inhibitory samples that is different from the other eight enzymes.
We have shown that components in biological samples can differentially inhibit different polymerases in the PCR, oftentimes by several orders of magnitude. However, many intrinsic factors may also play a role in the resistance of the thermostable DNA polymerases to PCR inhibition, for instance, the difference in enzyme purification techniques, whether the polymerase is native or recombinant, and the buffer content. Taq DNA polymerase from different commercial sources has previously been reported to be inhibited differentially by humic substances in soil extracts (29). Although not examined in this study, the DNA extraction protocol and the presence of trace levels of extraction reagents in the purified DNA can also affect extraction efficiency and increase PCR inhibition, thereby affecting the sensitivity of detection. The polymerase Tth has been found by Katcher and Schwartz (10) to maintain both DNA- and RNA-dependent DNA polymerase activities in the presence of 5% (vol/vol) phenol, while a trace amount of phenol was found to be inhibitory to Taq DNA polymerase.
TABLE 5.
Effect of K ions on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | K ion concn (mM) in the supplied buffer | PCR resultsa with extra KCl concn (mM)b of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 10 | 20 | 40 | 60 | 80 | 100 | 150 | ||
| AmpliTaq Gold | 50 | ++ | ++ | ++ | ±± | −− | −− | −− | −− |
| Expand High Fidelity | Unknown | ++ | ++ | ++ | ++ | ++ | −− | −− | −− |
| HotTub | 0 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | −− |
| Pwo | 25 | ++ | ++ | ++ | ++ | −− | −− | −− | −− |
| rTth | 100 | ++ | ++ | ++ | ++ | ++ | ++ | ±− | −− |
| Taq | 50 | ++ | ++ | ++ | ++ | ++ | −− | −− | −− |
| Tfl | 75c | ++ | ++ | ++ | ++ | ++ | +− | −− | −− |
| Tli | 50 | ++ | ++ | ++ | ++ | ++ | +− | −− | −− |
| Ultma | 10 | ++ | ++ | −− | −− | −− | −− | −− | −− |
Two independent PCR results. Symbols: band of good intensity; ±, band of low intensity; −, no band.
Concentrations of KCl in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
The buffer contained potassium acetate instead of KCl.
TABLE 6.
Effect of Mg ions on the amplification capacities of nine thermostable DNA polymerases
| DNA polymerase | Mg ion concn (mM) in the supplied buffer | PCR resultsa with extra MgCl2 concn (mM)b of:
|
||||
|---|---|---|---|---|---|---|
| 5.0 | 10.0 | 15.0 | 20.0 | 30.0 | ||
| AmpliTaq Gold | 2.5 | ++ | +− | +− | −− | −− |
| Expand High Fidelity | 1.5 | ++ | ++ | ++ | +± | −− |
| HotTub | 2.5 | ++ | ++ | ±± | −− | −− |
| Pwo | 1.5c | ++ | ++ | ++ | ±− | −− |
| rTth | 2.5 | ++ | ++ | ±± | ±− | −− |
| Taq | 1.5 | ++ | ++ | ++ | +± | −− |
| Tfl | 2.5c | ++ | ±− | −− | −− | −− |
| Tli | 2.5 | ++ | ++ | ++ | −− | −− |
| Ultma | 2.5 | −− | −− | −− | −− | −− |
Two independent PCR results. Symbols: +, band of good intensity; ±, band of low intensity; −, no band.
Concentrations of MgCl2 in the PCR mixtures containing 1 ng of L. monocytogenes DNA.
The buffer contained MgSO4 instead of MgCl2.
ACKNOWLEDGMENTS
We thank the following companies for donating the DNA polymerases used in this study: Amersham Life Science, Boehringer Mannheim, Perkin-Elmer Cetus, and Promega.
This work was supported by a grant from the Swedish Council for Forestry and Agricultural Research.
REFERENCES
- 1.Abu Al-Soud W, Lantz P-G, Bäckman A, Olcén P, Rådström P. A sample preparation which facilitates detection of bacteria in blood cultures by the polymerase chain reaction. J Microbiol Methods. 1998;32:217–224. [Google Scholar]
- 2.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from blood strains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 3.Bickley J, Short J K, McDowell D G, Parkes H C. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153–158. doi: 10.1111/j.1472-765x.1996.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 4.Byrnes J J, Downey K M, Esserman L, So A G. Mechanism of hemin inhibition of erythroid cytoplasmic DNA polymerase. Biochemistry. 1975;14:796–799. doi: 10.1021/bi00675a023. [DOI] [PubMed] [Google Scholar]
- 5.Chernesky M A, Jang D, Sellors L, Luinstra K, Chong S, Castriciano S, Mahony J B. Urinary inhibitors of polymerase chain reaction and ligase chain reaction and testing of multiple specimens may contribute to lower assay sensitivities for diagnosing Chlamydia trachomatis infected women. Mol Cell Probes. 1997;11:243–249. doi: 10.1006/mcpr.1997.0109. [DOI] [PubMed] [Google Scholar]
- 6.DiMichele L J, Lewis M J. Rapid, species-specific detection of lactic acid bacteria from beer using the polymerase chain reaction. J Am Brew Chem Soc. 1993;51:63–66. [Google Scholar]
- 7.Favre N, Rudin W. Salt-dependent performance variation of DNA polymerases in co-amplification PCR. BioTechniques. 1996;21:28–30. doi: 10.2144/96211bm04. [DOI] [PubMed] [Google Scholar]
- 8.Fluit A C, Terensma R, Visser M J C, Aarsman C J M, Poppelier M J J G, Keller B H I, Klapwijk P, Verhoef J. Detection of Listeria monocytogenes in cheese with the magnetic immuno-polymerase chain reaction assay. Appl Environ Microbiol. 1993;59:1289–1293. doi: 10.1128/aem.59.5.1289-1293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Innis M A, Myambo K B, Gelfand D H, Brow M A D. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katcher H L, Schwartz I. A distinctive property of Tth DNA polymerase: enzymatic amplification in the presence of phenol. BioTechniques. 1994;16:84–92. [PubMed] [Google Scholar]
- 11.Khan G, Kangro H O, Coates P J, Heath R B. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J Clin Pathol. 1991;44:360–365. doi: 10.1136/jcp.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein A, Barsuk R, Dagan S, Nusbaum O, Shouval D, Galun E. Comparison of methods for extraction of nucleic acids from hemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J Clin Microbiol. 1997;35:1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lantz P-G. Ph.D. thesis. Lund, Sweden: Lund University; 1998. [Google Scholar]
- 14.Lantz P-G, Hahn-Hägerdal B, Rådström P. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci Technol. 1994;5:384–389. [Google Scholar]
- 15.Lantz P-G, Tjerneld F, Borch E, Hahn-Hägerdal B, Rådström P. Enhanced sensitivity in PCR detection of Listeria monocytogenes in soft cheese through use of an aqueous two-phase system as a sample preparation method. Appl Environ Microbiol. 1994;60:3416–3418. doi: 10.1128/aem.60.9.3416-3418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lantz P-G, Tjerneld F, Hahn-Hägerdal B, Rådström P. Use of aqueous two-phase system in sample preparation for polymerase chain reaction-based detection of microorganisms. J Chromatogr B. 1996;680:165–170. doi: 10.1016/0378-4347(95)00496-3. [DOI] [PubMed] [Google Scholar]
- 17.Lantz P-G, Matsson M, Wadström T, Rådström P. Removal of PCR inhibitors from human fecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods. 1997;28:159–167. [Google Scholar]
- 18.Lindqvist R, Norling B, Lambertz S T. A rapid sample preparation method for detection of food pathogens based on buoyant density centrifugation. Appl Microbiol. 1997;24:306–310. doi: 10.1046/j.1472-765x.1997.00069.x. [DOI] [PubMed] [Google Scholar]
- 19.Marsiglia M L, Ikuta N, Fonseca A K, Schuch D T, Hötzel I, Ozaki L S, Marques E K, Lunge V R. Development of a combined selective enrichment method and polymerase chain reaction (PCR) assay for sensitive detection of Salmonella in food samples. J Microbiol Biotechnol. 1997;13:649–654. [Google Scholar]
- 20.Mercier B, Gaucher C, Feugeas O, Mazurier C. Direct PCR from whole blood, without DNA extraction. Nucleic Acids Res. 1990;18:5908. doi: 10.1093/nar/18.19.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel E, Cossart P. Physical map of the Listeria monocytogenes chromosome. J Bacteriol. 1992;174:7098–7103. doi: 10.1128/jb.174.22.7098-7103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monteiro L, Bonnemaison D, Vekris A, Petry K G, Bonnet J, Vidal R, Cabrita J, Megraud F A. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olcén P, Lantz P-G, Bäckman A, Rådström P. Rapid diagnosis of bacterial meningitis by a seminested PCR strategy. Scand J Infect Dis. 1995;27:537–539. doi: 10.3109/00365549509047063. [DOI] [PubMed] [Google Scholar]
- 24.Panaccio M, Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991;19:1151. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell H A, Gooding C M, Garrett S D, Lund B M, McKee R A. Proteinase inhibition of the detection of Listeria monocytogenes in milk using the polymerase chain reaction. Lett Appl Microbiol. 1994;18:59–61. [Google Scholar]
- 26.Rådström P, Bäckman A, Qian N, Kragsbjerg P, Påhlson C, Olcén P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossen L, Nørskov P, Holmstrømet K, Rasmussen O F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Tebbe C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai Y-L, Olson B H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernars K, Delfgou E, Soentoro P S, Notermans S. Successful approach for detection of low numbers of enterotoxigenic Escherichia coli in minced meat by using the polymerase chain reaction. Appl Environ Microbiol. 1991;57:1914–1919. doi: 10.1128/aem.57.7.1914-1919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedbrauk D L, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal speciments for detection of group A retroviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witham P K, Yamashiro C T, Livak K J, Batt C A. A PCR-based assay for detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl Environ Microbiol. 1996;62:1347–1353. doi: 10.1128/aem.62.4.1347-1353.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]