Abstract
In many species, salt sensitivity is associated with the accumulation of sodium (Na+) in photosynthetic tissues. Na+ uptake to leaves involves a series of transport steps and so far very few candidate genes have been implicated in the control of these processes. In this study, Na+ transport was compared in two varieties of durum wheat (Triticum turgidum) L. subsp. durum known to differ in salt tolerance and Na+ accumulation; the relatively salt tolerant landrace line 149 and the salt sensitive cultivar Tamaroi. Genetic studies indicated that these genotypes differed at two major loci controlling leaf blade Na+ accumulation (R. Munns, G.J. Rebetzke, S. Husain, R.A. James, R.A. Hare [2003] Aust J Agric Res 54: 627–635). The physiological traits determined by these genetic differences were investigated using measurements of unidirectional 22Na+ transport and net Na+ accumulation. The major differences in Na+ transport between the genotypes were (1) the rate of transfer from the root to the shoot (xylem loading), which was much lower in the salt tolerant genotype, and (2) the capacity of the leaf sheath to extract and sequester Na+ as it entered the leaf. The genotypes did not differ significantly in unidirectional root uptake of Na+ and there was no evidence for recirculation of Na+ from shoots to roots. It is likely that xylem loading and leaf sheath sequestration are separate genetic traits that interact to control leaf blade Na+.
Salinity imposes both ionic and osmotic stresses on plants. In monocots, salt tolerance is typically associated with the ability to exclude sodium (Na+) from the photosynthetic tissues of the shoot (Flowers and Yeo, 1988; Yeo et al., 1990; Tester and Davenport, 2003). In hexaploid (AABBDD) bread wheat (Triticum aestivum), the ability to maintain low shoot levels of Na+ is associated with the Kna1 locus on the 4D chromosome (Gorham et al., 1987; Dubcovsky et al., 1996). Tetraploid (AABB) durum wheat (Triticum turgidum) L. subsp. durum lacks the D genome and is more salt sensitive than bread wheat and accumulates higher levels of Na+ in the shoot (Gorham et al., 1987, 1990). Within durum wheat varieties, the osmotic effects of salinity cause rapid and persistent growth inhibition and depression of grain yield and show little genotypic variation (James et al., 2002; Munns and James, 2003). In contrast, the ion-specific effects of Na+ accumulation on growth and leaf senescence begin to appear only after several weeks of salt treatment and show substantial genotypic variation. Therefore, differences in salt tolerance between durum wheats are generally correlated with Na+ exclusion from leaves (Husain et al., 2003; Munns and James, 2003). A screen of 64 modern cultivars and ancient landraces identified several landraces with leaf blade Na+ levels comparable to those of bread wheats, indicating the presence of Na+ exclusion traits within the A or B genomes (Munns et al., 2000). To identify the genetic bases of Na+ exclusion in durum wheat, crosses were made between the relatively salt tolerant landrace line 149 (with low leaf blade Na+) and a salt sensitive commercial variety Tamaroi (with high leaf blade Na+). Genetic analysis showed that the low leaf blade Na+ phenotype was associated with two dominant loci of major effect and that these loci were interactive (epistatic) rather than additive (Munns et al., 2003). These loci were designated Nax1 and Nax2 (Na+ exclusion loci). Recently, a molecular marker linked to Nax1 was identified and proven to facilitate the rapid transfer of this trait into commercial varieties of durum wheat (Lindsay et al., 2004).
The aim of this study was to determine the physiological correlates of the genetic control of leaf Na+ in durum wheats to facilitate screening for novel traits and to enhance understanding of the control of Na+ transport within grasses. The screening method previously used to identify plants differing in Na+ exclusion involved sampling of the leaf 3 blade 10 d after emergence of the leaf (Munns and James, 2003; Fig. 1). This method was nondestructive and Na+ measurements correlated well with salt sensitivity assayed by biomass reduction; however, it provided little information about the processes leading to low leaf blade Na+. In this study, plants were destructively harvested to determine Na+ allocation with the whole plant, and both net and unidirectional movements of Na+ were analyzed. Na+ transport was divided into a number of processes: (1) initial influx of Na+ into root cells; (2) Na+ efflux from root cells to the external solution; (3) Na+ transfer into the xylem for transport to the shoot; (4) removal of Na+ from the xylem in the shoot, before entry into leaf blades; and (5) recirculation of Na+ from leaf blades to other tissues including the roots.
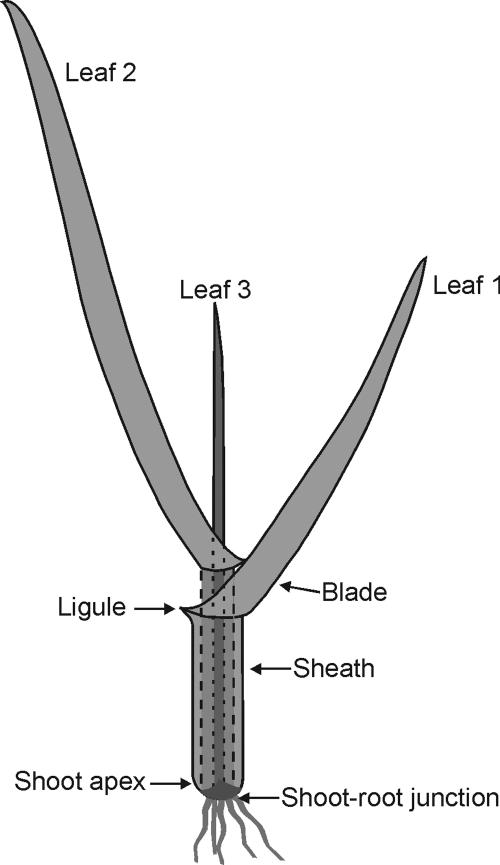
Figure 1.
Plant at the three-leaf stage that was used for most experiments. Leaves are clearly divided into sheath and blades by the ligule. The sheath of leaf 1 is about 40 mm in length. The stem of the seedling is comprised of leaf sheaths, the shoot apex, and leaf meristems. The coleoptile was dead at this stage and was removed before measurements.
Unidirectional influx of Na+ into roots of nonhalophytes is very rapid and may exceed net uptake into the plant by at least 10-fold (Tester and Davenport, 2003). Thus, a very rapid efflux of Na+ from roots must occur to control net rates of influx. Na+ that has entered the root can be effluxed, stored in root vacuoles, or transported to the xylem. Once loaded into the xylem, Na+ may be withdrawn at various points along the vascular pathway, into mature root cells, or into cells at the root shoot junction or leaf sheath (Fig. 1) before reaching the leaf blade. Na+ that reaches the leaf blade may be stored or recirculated in the phloem to other tissues including the roots. We have assumed in this description that Na+ is transported to leaf cells primarily via the xylem, although in immature leaves recirculation from source leaves could be the major pathway of Na+ uptake. Using radioactively labeled 22Na+, we tested which of these unidirectional transport processes contributed to differences in net Na+ accumulation in the shoot and leaf blades of the two genotypes.
RESULTS
The Salt Tolerant Genotype Excludes Na+ from the Shoot and Sequesters Na+ in Leaf Sheaths
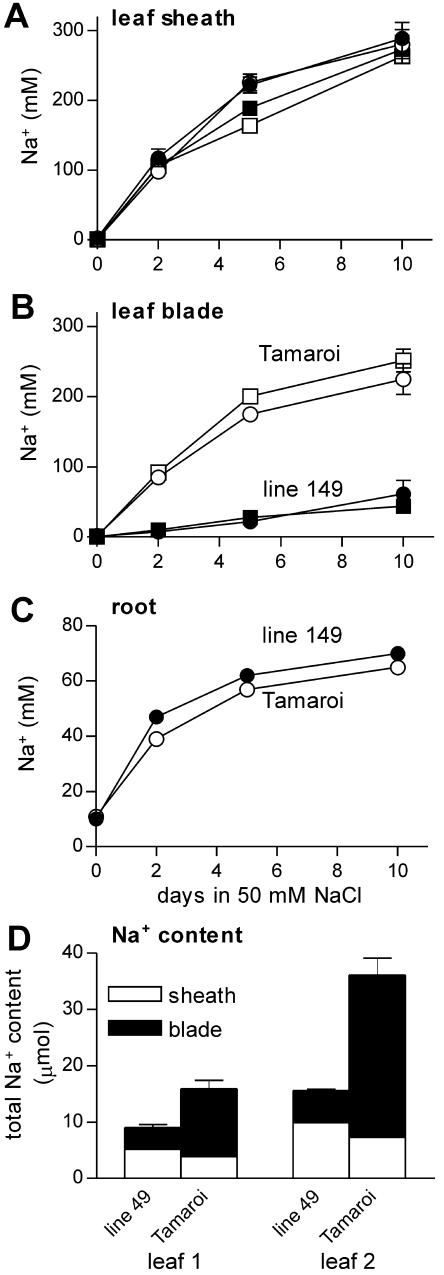
Measurement of net Na+ accumulation over 10 d indicated 2 major differences in Na+ transport between the salt tolerant line 149 and salt sensitive Tamaroi. The salt tolerant plants accumulated less Na+ in the shoot and also accumulated the major proportion of shoot Na+ in leaf sheaths. The difference in rate of net whole plant accumulation between the genotypes was less than 2-fold (Table I). Both genotypes accumulated Na+ to similar levels in the root, and therefore the relatively small difference in total uptake produced a larger (3-fold) difference in the rate of Na+ accumulation in the shoot (Table I). Within individual leaves, Tamaroi accumulated Na+ to similar concentrations in leaf sheaths compared with line 149 but accumulated much higher concentrations in the leaf blades (Fig. 2). On a Na+ content basis, the genotypes differed only 2-fold in leaf accumulation of Na+ by day 10 but differed 3- to 5-fold in Na+ content of the sensitive blade tissue, due to large differences in the proportion of leaf Na+ that was stored in the sheath (Fig. 2D). The experiment indicated that rates of Na+ accumulation declined in all tissues over 10 d and that the roots approached a steady state of Na+ concentration by around day 5 (Fig. 2C). Therefore, plants for radioactive experiments were pretreated for a minimum of 5 d to ensure that root Na+ concentrations were near equilibrium.
Table I.
Genotypic differences in net Na+ uptake
Net Na+ uptake, Na+ transport to the shoot (shoot Na+ uptake), and root Na+ uptake in salt tolerant line 149 and salt sensitive Tamaroi, calculated over a 5- to 10-d period in half-strength modified Hoagland plus 50 mm NaCl and 2 mm CaCl2. Values are means ± sem (n = 4).
| Na+ Uptake Parameter | Line 149 | Tamaroi |
|---|---|---|
| μmol g FW−1(root) d−1 | ||
| Net Na+ uptake (whole plant) | 17.3 ± 0.4 | 26.3 ± 2.9 |
| Shoot Na+ uptake | 7.1 ± 0.5 | 17.3 ± 1.5 |
| Root Na+ uptake | 10.2 ± 0.5 | 9.0 ± 0.5 |
Figure 2.
Increase in Na+ concentrations in the leaf sheaths (A) and leaf blades (B) of leaf 1 (squares) and leaf 2 (circles), and roots (C), of salt tolerant line 149 (black symbols) and salt sensitive Tamaroi (white symbols) during 10 d exposure to 50 mm NaCl and 2 mm CaCl2 in half-strength modified Hoagland. D, Na+ content of leaves 1 and 2 after 10 d. Data represent mean ± sem (n = 4).
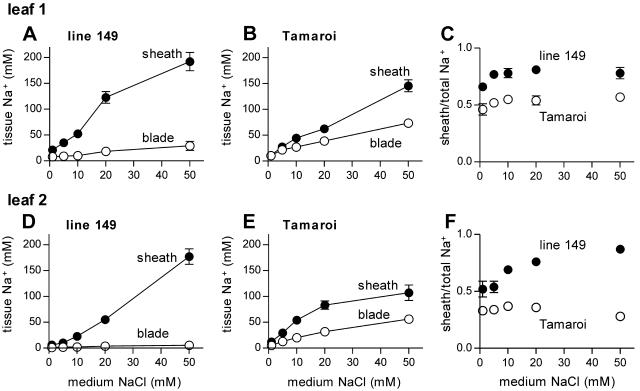
Storage of Na+ in the leaf sheaths was investigated further by measuring Na+ content of the leaf sheath and leaf blade of leaves 1 and 2 after 5 d of exposure to different levels of NaCl. In this experiment, the 2 genotypes accumulated Na+ to similar concentrations in the leaf sheath except at the highest Na+ concentration (50 mm), where the salt tolerant line 149 accumulated a substantially higher Na+ concentration than Tamaroi with no evidence of saturation of storage capacity (Fig. 3). While it is possible that both genotypes have a similar capacity to store Na+ in the sheath (i.e. they can accumulate Na+ to similar concentrations), they may differ in the ability of the sheath cells to extract Na+ from the xylem stream. This possibility was supported by genotypic differences in the proportion of total leaf Na+ content that was stored in the sheath (Fig. 3, C and F). Line 149 was able to extract up to 80% of total leaf Na+ for storage in the sheath, and this capacity appeared to increase with increasing NaCl, at least in the younger leaf (Fig. 3F). The salt sensitive Tamaroi could store up to 50% of leaf Na+ in the sheath in the oldest leaf but only around 30% in the younger leaf, with little change in response to external NaCl levels (Fig. 3F).
Figure 3.
Na+ content of leaves 1 (A–C) and 2 (D–F) after 5 d exposure to different concentrations of NaCl (all solutions contained half-strength modified Hoagland and CaCl2 to maintain a Na+:Ca2+ ratio below 15:1). A, B, D, and E, Na+ concentrations in the leaf blade (white symbols) and sheath (black symbols). C and F, Ratio of sheath Na+ content to total leaf Na+ (blade + sheath). Data represent mean ± sem (n = 4).
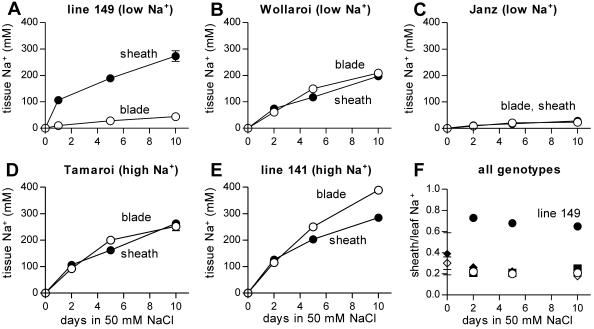
The existence of a distinct sheath Na+ storage mechanism in line 149 was supported by a wider genotypic comparison of allocation of Na+ between sheath and blade of leaf 2, using 2 other varieties of durum wheat with low (Wollaroi) and high (line 141) leaf blade Na+ as well as a hexaploid bread wheat, cv Janz (with low leaf blade Na+ typical of bread wheats; Fig. 4). With the exception of line 149, the genotypes all allocated approximately 25% of leaf Na+ to the sheath, resulting in an almost 1:1 concentration ratio between leaf blade and sheath (because the sheath comprises approximately 25% of total leaf tissue; Fig. 4, B–F). In contrast, line 149 allocated approximately 70% of leaf Na+ to the sheath, resulting in a much lower blade Na+ concentration compared to the sheath (Fig. 4, A and F). These results suggest that line 149 possesses some additional mechanism for maintenance of low blade Na+, involving efficient withdrawal of leaf Na+ into the sheath.
Figure 4.
Leaf sheath and blade Na+ partitioning in four durum varieties and a bread wheat (C) differing in leaf blade Na+ (low versus high). A to E, Increase in Na+ concentrations in the leaf sheaths (black symbols) and leaf blades (white symbols) of leaf 2 during 10 d exposure to 50 mm NaCl and 2 mm CaCl2 in half-strength modified Hoagland. F, Ratio of leaf sheath Na+ content to total leaf Na+ content in line 149 (black circles), Wollaroi (black diamonds), Janz (black squares), Tamaroi (white circles), and line 141 (white diamonds). Data represent mean ± sem (n = 4).
The Genotypes Differ in Unidirectional Uptake of 22Na+ to the Shoot
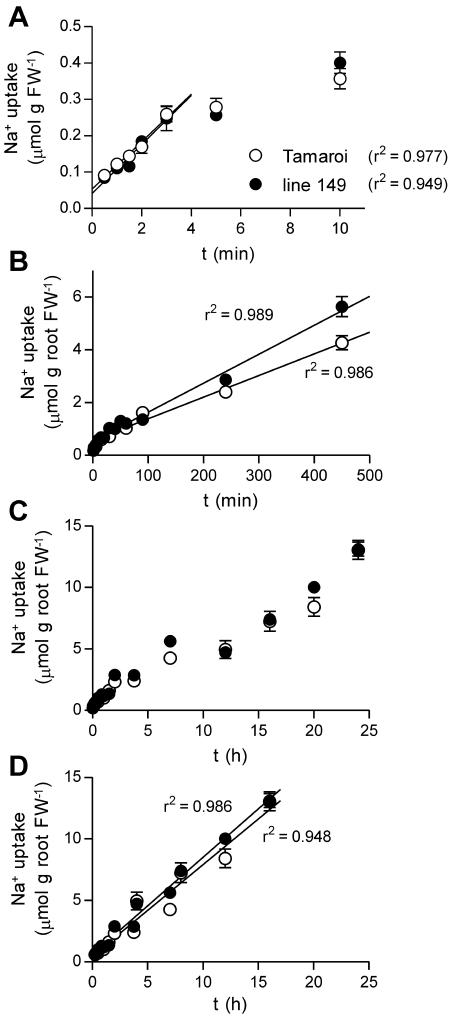
22Na+ accumulation in roots was rapid over the first 5 min then slowed to a steady rate (Fig. 5). Shoot Na+ accumulation became measurable after 30 min (Fig. 6). These data could be described by a classic three compartment flux model in which the root cytoplasm, vacuole, and shoot exchanged Na+ and the Na+ content of the root compartments was at steady state (Supplemental Fig. 1). According to the model, 22Na+ accumulates in the root cytoplasm first. Initially, the labeled Na+ is a small proportion of the cytoplasmic Na+, so there is negligible efflux of 22Na+ and therefore the rate of uptake represents unidirectional influx (the first 3 min of uptake). As the cytoplasmic Na+ pool becomes labeled, some of the 22Na+ is effluxed (to the solution and to the shoot) and therefore the rate of accumulation in the root slows. Once the cytoplasmic 22Na+ has equilibrated with the external solution, net accumulation in the root becomes linear and represents vacuolar influx (until the vacuolar Na+ pool becomes labeled and there is significant efflux of 22Na+ from the vacuole, causing eventual saturation and cessation of root 22Na+ accumulation). Shoot 22Na+ accumulates very slowly initially because root Na+ pools are only partially labeled. As the root Na+ pools contributing to shoot uptake become fully labeled, shoot 22Na+ accumulation reaches a steady rate, representing the rate of unidirectional transfer to the shoot. This rate may be permanent (varying with transpiration but always unidirectional from root to shoot) or may slow due to recirculation from shoot to root once shoot Na+ pools become significantly labeled with 22Na+.
Figure 5.
Root Na+ uptake measured as root 22Na+ accumulation in wheat seedlings pretreated for 5 d in 25 mm NaCl and 2 mm CaCl2 (+ modified half-strength Hoagland). Root uptake measured over 0 to 10 min (A); 15 to 450 min (B); 0.3 to 24 h (C); 0.3 to 24 h with 8-h-dark period subtracted (D). Uptake rates were estimated by fitting linear regressions using least squares method; results are presented in Table II. Black symbols represent salt tolerant line 149; white symbols represent salt sensitive Tamaroi; mean ± sem (n = 4).
Figure 6.
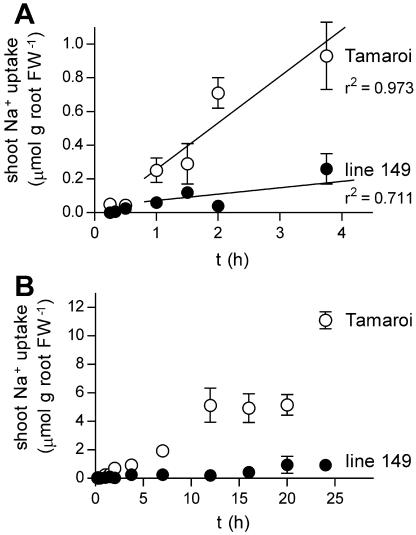
Shoot Na+ uptake measured as shoot 22Na+ accumulation per root FW in wheat seedlings pretreated for 5 d in 25 mm NaCl and 2 mm CaCl2 in modified half-strength Hoagland. A, Shoot uptake over 4 h showing delay in initial uptake. B, Shoot uptake over 24 h. Black symbols represent salt tolerant line 149; white symbols represent salt sensitive Tamaroi; mean ± sem (n = 4).
Unidirectional influx was estimated from the first linear phase of 22Na+ uptake (0–3 min; Fig. 5A). Na+ influx rates did not differ significantly between genotypes (Table II). Vacuolar uptake was estimated from the second linear phase of root 22Na+ accumulation (0.25–7 h; Fig. 5B) and was slightly slower in Tamaroi, although the difference diminished over 24 h (but was also evident in the efflux data described in Fig. 7; Table II). Tonoplast influx continued for up to 24 h; however, the pattern of uptake was not linear over the period but included a slow phase between 8 and 12 h (Fig. 5C). When the 8-h period of darkness was subtracted from the longer time points (all time points over 8 h included an 8-h-dark period) then root uptake appeared linear over the period of the experiment (Fig. 5D; Table II). It is not clear why this would occur in roots, and a slightly different pattern of uptake was observed in shoots (Fig. 6B). The rates of uptake calculated by linear fits to dark subtracted data over 24 h are higher than those calculated from uptake before the dark period (Table II) and the values from the earlier part of the timecourse (0.25–7 h) are likely to provide more accurate estimates of tonoplast influx. Salt sensitive Tamaroi exhibited a slightly lower rate of tonoplast influx by both measures. Vacuolar loading was slow (an influx of 0.7 μmol h−1 g fresh weight [FW]−1 into a vacuolar compartment of perhaps 50 mm Na+, equivalent to 40.5 μmol g FW−1, assuming a vacuolar volume of 90% and root FW:DW of 10, would result in exchange of less than one-half the vacuolar Na+ within 24 h) and therefore the relatively constant rate of tonoplast influx over 24 h probably represents slow loading of an equilibrated vacuolar pool rather than a rise in net root vacuolar Na+ concentration.
Table II.
Genotypic differences in influx, efflux, and unidirectional shoot uptake
Unidirectional transport across the plasma membrane and tonoplast of root cells, measured as 22Na+ uptake or efflux, and shoot uptake. Influx data represent slope of linear regressions estimated using least squares method and 95% confidence intervals (shoot uptake estimated between 1–8 h). Efflux data are presented as absolute rates and also as the proportion of the cytoplasmic (for plasma membrane) or vacuolar (for tonoplast) Na+ that was effluxed per minute (the relative rates).
| Line 149 (Salt Tolerant) | Tamaroi (Salt Sensitive) | |
|---|---|---|
| Plasma membrane influx (nmol g FW−1 min−1) | 65.3 (46.8–83.9) | 67.4 (38.8–96.1) |
| Tonoplast influx (nmol g FW−1 min−1) | ||
| 15 to 450 min | 11.0 (9.9–12.0) | 8.2 (6.9–9.5) |
| 0.3 to 24 h (dark subtracted) | 13.2 (12.1–14.2) | 12.4 (10.0–14.8) |
| Shoot uptake (nmol g root FW−1 min−1) | 0.6 (−0.1–1.4) | 4.6 (3.2–6.0) |
| Root efflux (nmol g FW−1 min−1) | ||
| Plasma membrane | 335 | 108 |
| Tonoplast | 5.2 | 3.5 |
| Relative efflux (min−1) | ||
| Plasma membrane efflux (min−1) | 0.05907 | 0.07261 |
| Tonoplast efflux (min−1) | 0.00122 | 0.00071 |
Figure 7.
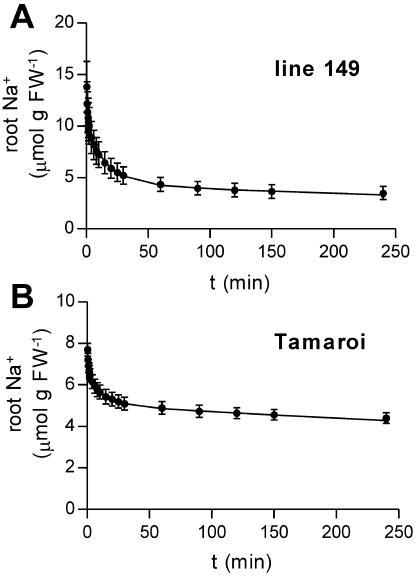
Efflux of 22Na+ from excised roots expressed as decrease in tissue 22Na+ after transfer to nonradioactive solution (black circles). Data were fitted with a double exponential equation (lines) to estimate plasma membrane and tonoplast efflux rates. Half-times for exchange of the cytoplasm (k1) and tonoplast (k2) in min−1 were 0.060 and 0.0012 (line 149) and 0.0726 and 0.0007 (Tamaroi), respectively. Initial contents of the cytoplasm (Qc) and vacuole (Qv) in nmol g FW−1 were 5,676 and 4,224 (line 149) and 1,494 and 4,956 (Tamaroi), respectively. Data are presented as root Na+ (calculated using the specific activity of the loading solution) and are equivalent to 22Na+ remaining in the tissue. A, Salt tolerant line 149. B, Salt sensitive Tamaroi. Data represent mean ± sem (n = 5).
The fact that the two genotypes had similar total root Na+ contents and rates of influx into the root means that the root specific activity of 22Na+ (22Na+ per mole Na+) was similar in both. The specific activity of Na+ translocated to the shoot depends on the specific activity of the root Na+ pool(s) from which it is derived and will provide an underestimate of the true rate of Na+ transfer to the shoot until the specific activity of translocated Na+ matches that of the external solution. The similarity in root Na+ concentration and 22Na+ accumulation between the genotypes means that the rate of appearance of 22Na+ into the shoot is likely to provide a fair reflection of the relative rate of unidirectional influx of total Na+ from the root to the shoot.
The genotypes showed similar time courses of uptake to the shoot but large differences in the rate of transfer to the shoot (Fig. 6). There was very little uptake to the shoot before 30 min, reflecting the need for root Na+ to become labeled before xylem loading (Fig. 6A). Tamaroi showed rapid uptake to the shoot over 24 h compared with line 149 (Fig. 6B). Uptake over 24 h was not linear and showed a reduction in uptake during the dark period. However, this period of reduced shoot uptake did not match the period of reduced root uptake but followed with a lag of approximately 4 h. Subtraction of the dark period of the experiment produced a linear pattern of shoot uptake in line 49 but not in Tamaroi (data not shown). Because of the different effects of the dark period in the two lines, only the earlier time points (1–8 h) were used to calculate rates of transfer to the shoot (Fig. 6A; Table II). Using these estimates, the genotypes varied nearly 8-fold in the rate of transfer to the shoot, a difference more than sufficient to explain the 3-fold difference in net shoot Na+ accumulation (Table I). The genotypes do not differ in transpiration rates (Rivelli et al., 2002) so a difference in the rate of bulk flow of Na+ through the xylem cannot explain the large genotypic difference in Na+ translocation. There was no apparent increase in the rate of transport to the shoot over time after 1 h, suggesting that the specific activity of translocated 22Na+ was unchanging over this period (and also likely to be the same as the external solution, implying that shoot Na+ uptake was not underestimated). In neither genotype was there evidence for a reduction in the rate of uptake over time that would indicate the possibility of recirculation of shoot 22Na+.
The Genotypes Are Unlikely to Differ in Na+ Efflux across the Root Plasma Membrane
Na+ efflux from excised roots was analyzed using a two compartment model that gave estimates of plasma membrane and tonoplast efflux rates and the 22Na+ content of the cytoplasmic and vacuolar compartments. However, the data did not strictly satisfy the criteria for a model of efflux from two serial, independent compartments. Departures from ideal fits were observed at both earlier and later time points, possibly due to, respectively, contamination by efflux from the wall and changes in the physiological state of the roots. Manual fitting produced a close fit of the data points over the middle 70% to 80% of each time series, with deliberate departure allowed between fitted and experimental data at the earlier and later time points (Fig. 7). Estimates of efflux derived from these fits indicated a higher rate of plasma membrane Na+ efflux in line 149 (Table II). However, in this experiment, the genotypes differed markedly in tissue 22Na+ content at the beginning of the efflux period (evident in the genotypic difference in total content at t = 0 in Fig. 7), in contrast to other experiments where root uptake did not differ more than 10% between the genotypes (Table I; Fig. 5). The analysis of the efflux data indicated that line 149 had a much higher cytoplasmic Na+ content, and this accounted for its higher efflux rate. When the difference in Na+ content was taken into account, the relative rates of efflux across the plasma membrane (the proportion of cytoplasmic Na+ effluxed per minute) were very similar in both genotypes, indicating that they had similar capacity for Na+ efflux (Table II). It is possible that line 149 does maintain higher root cytoplasmic Na+ concentrations than Tamaroi; however, this would be difficult to reconcile with the data on influx into roots of intact plants, which showed similar rates of cytoplasmic and vacuolar loading (indicating similar Na+ contents in these compartments in both genotypes). Therefore, the absolute efflux values must be treated with caution. The relative efflux values describe the capacity of the membrane transport systems (independent of substrate supply) and indicate much smaller genotypic differences. Relative tonoplast efflux was more rapid in the salt tolerant line 149 (Fig. 7). Genotypic differences in the measures of relative efflux are consistent with the unidirectional influx experiment, which showed no difference in influx and a small difference in vacuolar influx rates between the genotypes (Table II). Assuming that root vacuolar Na+ concentration was at steady state, a higher vacuolar influx would require a higher efflux to maintain equilibrium.
The Salt Tolerant Line 149 Extracts Na+ More Efficiently into the Leaf Sheath
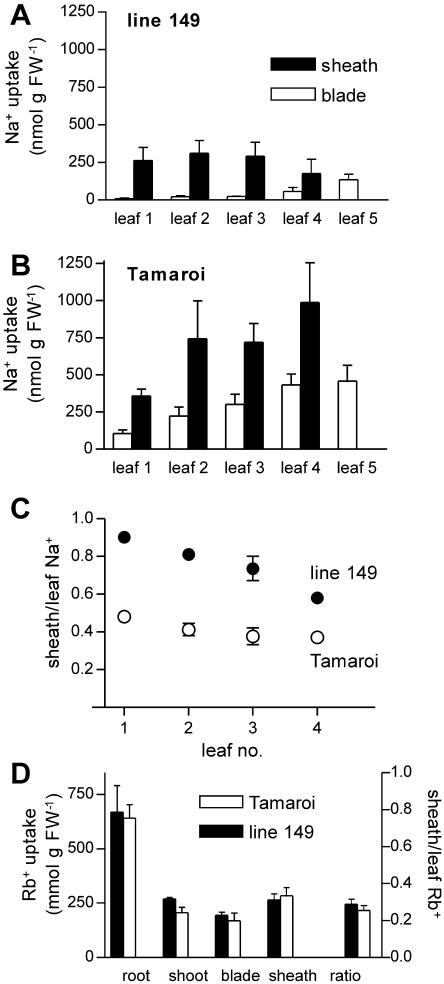
22Na+ uptake into leaves over 2 h represents unidirectional transfer into the leaf (as indicated by the rapid rise in shoot 22Na+ over the first 2 h of shoot uptake; Fig. 6A). Both genotypes accumulated more 22Na+ in the sheath than the blade on a concentration basis (Fig. 8, A and B). Salt sensitive Tamaroi accumulated much more 22Na+ than line 149 in the leaf as a whole and most markedly in the leaf blades (Fig. 8B). However, Tamaroi also accumulated more 22Na+ (on a content as well as a concentration basis) in the sheaths than did line 149, suggesting that the capacity for Na+ uptake into the sheath was as high (in fact higher) in Tamaroi as in line 149. The main genotypic difference in sheath storage was in the proportion of Na+ entering the leaf that was withdrawn into the sheath. In line 149, 72% to 97% of xylem 22Na+ was withdrawn into the leaf sheaths, compared with 36% to 50% in Tamaroi (Fig. 8C). This suggests higher affinity for Na+ uptake by sheath cells in line 149. These ratios of uptake from the xylem (Fig. 8C) match the ratios of net Na+ distribution within the leaf (Fig. 3, C and F), suggesting that the mechanism of sheath sequestration relies on extraction of Na+ from the xylem stream (rather than subsequent recirculation from the blade to the sheath).
Figure 8.
Unidirectional uptake of 22Na+ (A–C) and 86Rb+ (D) into leaf blades and sheaths after 2 h root exposure to radioactively labeled solution (all solutions contained 25 mm NaCl and 2 mm CaCl2 in half-strength modified Hoagland with K+ replaced with Rb+ in D). Uptake is expressed as the concentration of Na+ or Rb+ per leaf organ (nmol g FW−1) and as the ratio of sheath to total leaf uptake (measured as nanomole content). A to C, Uptake of Na+ into salt tolerant line 149 (A and C) and salt sensitive Tamaroi (B and C). D, Uptake of Rb+ into roots, total shoot, and sheath and blade of leaf 1, and ratio of sheath to total leaf Rb+ in leaf 1. Data represent mean ± sem (n = 5 [A–C]; n = 6 [D]).
To investigate whether the preferential accumulation of Na+ in leaf sheaths in line 149 represented a general solute accumulation mechanism, we measured 86Rb+ uptake into shoot tissues. 86Rb+ is used as an analog of K+ in transport studies because it has a much longer radioactive half-life than 42K+ and is transported with apparently similar properties by many K+ transport systems. After 2 h uptake, 86Rb+ had accumulated to high levels in leaves, with slightly more 86Rb+ in leaf sheaths than blades but with no evidence for genotypic differences in uptake or sequestration (Fig. 8D). Interestingly, the ratio of sheath to blade 86Rb+ accumulation in both genotypes was around 30%, close to the ratio of Na+ accumulation observed in Tamaroi and other wheat varieties (Figs. 4F and 8C).
Na+ Retranslocation Did Not Contribute to Control of Leaf Blade Na+
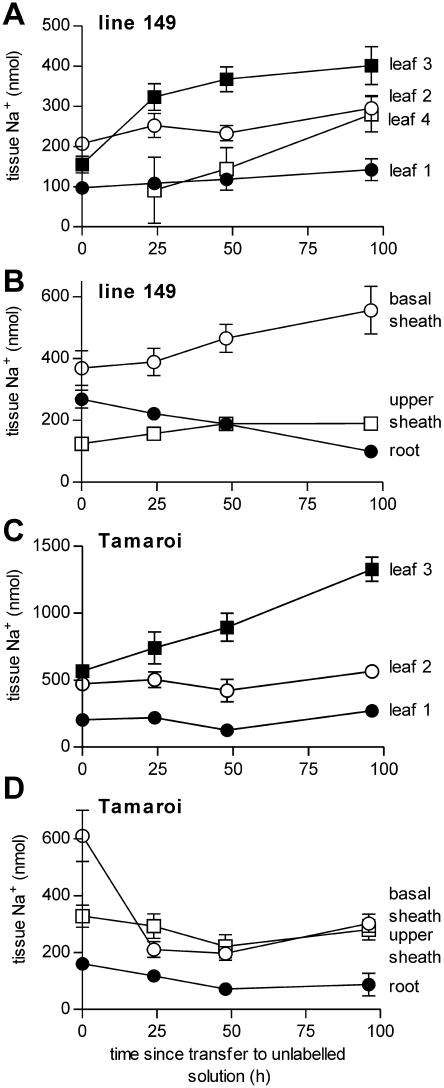
Plants were loaded with 22Na+ for 24 h before transfer to identical nonradioactive solution for a maximum of 4 d (Fig. 9). During the loading phase, root Na+ pools became highly labeled with 22Na+ and therefore some transfer of 22Na+ from root to shoot continued after transfer of roots to nonradioactive solution, complicating measurement of recirculation. 22Na+ content of the youngest leaves (leaves 3 and 4) rose over the course of the experiment in both genotypes as did the 22Na+ content of the basal sheath (comprising the lower parts of the sheaths of all leaves) in line 149 (Fig. 9, A and C). Over 96 h, the roots showed significant declines in 22Na+ content in both genotypes (Fig. 9, B and D). The salt sensitive Tamaroi also showed a reduction in 22Na+ content of the basal sheath (Fig. 9D). This could reflect retranslocation from the basal sheath to the roots. Alternatively, it could indicate transfer from the basal sheath to the youngest leaf (although the timecourse of increase in leaf 3 22Na+ was much more rapid than the decline in basal sheath 22Na+) or the fact that the unemerged leaves comprise a portion of the sheath tissue harvested, so that their emergence would remove 22Na+ from the sheath. The rest of the shoot tissues showed no consistent changes in 22Na+ content in either genotype. There was therefore no evidence for recirculation of Na+ from shoot to root (except possibly in the lower part of the leaf sheaths in the salt sensitive Tamaroi). Therefore, recirculation is unlikely to contribute to the differences in leaf Na+ between the genotypes.
Figure 9.
Recirculation of Na+ from shoot to root measured as decrease in tissue 22Na+ after transfer to nonradioactive solution. Plants were loaded with 22Na+ for 24 h then transferred to unlabeled solution for up to 4 d. A and B, Salt tolerant line 149. C and D, Salt sensitive Tamaroi. Note that root data are divided by 10. Data are presented as tissue Na+ and are equivalent to tissue 22Na+ content. Data represent mean ± sem (n = 8).
DISCUSSION
We compared two durum wheat genotypes in which differences in leaf blade Na+ (an indicator of Na+ sensitivity) had been shown to be associated with two main genetic loci. We found two major physiological differences between the genotypes (xylem loading/retrieval of Na+ and leaf sheath sequestration of Na+) that could account in combination for the differences in leaf blade Na+.
Root Uptake and Whole Plant Na+ Accumulation
The genotypes varied almost 2-fold in net uptake into the whole plant (Table I) and this must result from differences in net uptake via the roots (i.e. the sum of unidirectional influx and efflux across the plasma membrane of root cells). However, the differences in root influx and relative efflux (efflux capacity) between genotypes were small and could not explain a 2-fold difference in net uptake (Table II; as discussed in the “Results” section, relative efflux measures were considered more reliable than absolute values). The apparent absence of genotypic differences in influx and (relative) efflux is probably because net uptake was so low relative to unidirectional influx that small differences in efflux would cause large relative differences in net accumulation rates. In conditions where root Na+ concentration is at steady state, net uptake consists only of net shoot Na+ accumulation. In both genotypes, root Na+ influx was very high relative to shoot uptake (Table II). The rate of Na+ efflux required to balance influx with net uptake was calculated as the difference between influx and net uptake. For line 149, efflux = 65.3 − 0.6 = 64.7 nmol g FW−1 min−1 = 99.0% of influx. For Tamaroi, efflux = 67.4 − 4.6 = 62.8 nmol g FW−1 min−1 = 93.2% of influx. The difference in efflux between genotypes (64.7 versus 62.8 nmol g FW−1 min−1) would be almost impossible to detect but is sufficient to cause a 7-fold difference in net uptake (0.6 versus 4.6 nmol g FW−1 min−1). A genotypic difference in influx or efflux could reflect genetic differences in abundance, selectivity, or control of plasma membrane cation transporters. However, it is more likely that the difference in net uptake arises mainly from a difference in the net rate of xylem loading of Na+ for transfer to the shoot. A difference in leak to the shoot would have only a small effect on root Na+ pools (because transfer to the shoot occurs at a very low rate relative to root plasma membrane exchange with the external solution) and therefore would cause a large change in total and shoot Na+ content but only a slight change in root influx or efflux rates. The absence of genotypic differences in root Na+ influx has been observed in other species of wheat (Nevo et al., 1992; Davenport et al., 1997) and suggests that Na+ uptake pathways have other conserved physiological functions. Very high rates of Na+ influx relative to net uptake are typical of glycophytic species including Arabidopsis (Arabidopsis thaliana), and it remains to be tested whether the high rates of Na+ efflux required to balance influx impose an energetic burden on plants in saline conditions (Essah et al., 2003; Tester and Davenport, 2003). The extent to which manipulation of Na+ influx or efflux could affect translocation of Na+ to the shoot remains an open question.
Root vacuolar influx and efflux were slightly more rapid in the salt tolerant line 149. This implies that Na+ cycles across the root tonoplast more rapidly in line 149 than in Tamaroi. It is not clear whether this would have any effect on root Na+ content and transfer to the shoot. Overexpression of the Arabidopsis tonoplast Na+(K+)/H+ antiporter AtNHX1 (that would increase Na+ influx into the vacuole) improved Na+ tolerance without increasing Na+ content of transgenic wheat plants (Xue et al., 2004), suggesting that enhanced capacity to retain Na+ in vacuoles could reduce cytoplasmic Na+ toxicity. However, it is difficult to see any advantage to rapid cycling of Na+ across the tonoplast (Tester and Davenport, 2003).
Na+ Translocation to the Shoot
The genotypes did not differ in the timecourse of 22Na+ transfer to the shoot (i.e. the time taken for 22Na+ to appear in measurable amounts in the shoot after exposure of roots to 22Na+), suggesting that Na+ was loaded into the xylem from similar pools of root Na+ in both genotypes. However, the salt sensitive Tamaroi transferred Na+ to the shoot at a much higher rate than line 149 and this difference was sufficient to account for the difference in net shoot Na+ content between the genotypes (Table I; Fig. 6). Because the kinetics of root influx and vacuolar accumulation were very similar in the two genotypes, the difference in rate of transfer to the shoot is likely to be due to a difference in Na+ transport into or out of stelar cells surrounding the mature xylem. The genotypes could differ in export of Na+ to the xylem due to a difference in the number of transporters capable of loading Na+ into the xylem, in the selectivity of those transporters, or in other factors indirectly affecting Na+ export such as the membrane potential of xylem parenchyma cells or the control of a signaling pathway controlling shoot solute accumulation. Alternatively, the genotypes could export Na+ into the xylem at similar rates but differ in the rate of retrieval of Na+ back out of the xylem into stelar cells. This difference in xylem loading or retrieval was one of two major differences between the genotypes and is likely to be associated with one of the two major loci controlling leaf blade Na+. Positional cloning should indicate how this pathway of Na+ transport is differentially regulated between the two genotypes.
Sequestration of Na+ in Leaf Sheaths
The genotypes differed in the rate of Na+ delivery to leaves due to the large difference in the rate of xylem loading in the roots. This difference in leaf Na+ uptake was further amplified in the leaf blade by withdrawal of Na+ into the leaf sheath before it entered the blade. The genotypes appeared to have similar Na+ storage capacity in the sheath (although there was some evidence for greater storage capacity in salt tolerant line 149; Fig. 3). Thus, the genotypes could differ in the proportion of leaf Na+ allocated to the blade and sheath simply because the amount taken into Tamaroi leaves exceeds sheath storage capacity to a much greater extent than in line 149. However, it also appeared that the genotypes varied in the proportion of Na+ that the sheath could extract from the xylem sap before it reached the blade. Line 149 could extract up to 97% of the Na+ content of the xylem stream, versus a maximum of around 50% in Tamaroi (Fig. 3, C and F). A greater ability to sequester Na+ before it entered the blade would have the advantage that Na+ would accumulate more slowly in the leaf blade, prolonging leaf productivity. This trait of preferential sheath retention of Na+ was not observed in other varieties of durum or in a relatively salt tolerant bread wheat (Fig. 4), suggesting that it is associated with one of the loci involved in maintenance of low Na+ in line 149 leaf blades. This trait would interact with the low xylem loading trait. Sheath retention of Na+ in itself would delay the accumulation of Na+ in leaf blades but could not maintain low blade Na+ levels for very long (due to the small size of the sheath) if Na+ delivery to the shoot were rapid.
One interesting question regarding sequestration of Na+ in the leaf sheaths in line 149 is whether this mechanism is Na+-specific or reflects a general trait for solute or ion accumulation in leaf sheaths. Uptake of 86Rb+ (a K+ analog) into leaves of salt treated plants showed no differences between the genotypes (Fig. 8D). This suggests that sheath sequestration of Na+ could be Na+ specific. However, uptake of Rb+ was high relative to Na+ (when supplied in the ratio 2:25 Rb+:Na+) and may reflect mainly selective transport of K+ (Rb+) into the leaf blade and sheath with a minor contribution of nonselective pathways that sequester both Rb+ and Na+ into sheath cells. In Tamaroi, Rb+ and Na+ were partitioned between sheath and blade in similar proportions, whereas in line 49 Rb+ but not Na+ accumulated in the blade (Fig. 8, C and D). Selective uptake of K+ (Rb+) and not Na+ into the leaf blades in line 149 implies that the sheath acts as a selectivity filter, either by withdrawing specific cations such as Na+ from the xylem stream, or by absorbing most cations and then selectively rereleasing some such as K+ for onward transport to the blade.
Compartmentation of Na+ within the shoot has been proposed as a salt tolerance mechanism in barley on the basis of differences in net accumulation of Na+ in different leaves (Wolf et al., 1991). In the case of at least durum wheat, it appears that Na+ compartmentation can be achieved by withdrawal from the xylem stream into specific tissues (leaf sheath cells) and that this mechanism may be Na+ specific.
Na+ Recirculation
Leaf sheath sequestration of Na+ in line 149 would help to slow the rate of Na+ accumulation in the sensitive photosynthetic tissues of the leaf blade. However, this mechanism would cease to contribute to maintenance of low leaf blade Na+ once the sheath cells were filled. Leaf sheaths continued to accumulate 22Na+ at high rates even after several weeks of NaCl treatment (data not shown). This suggested that there may be some turnover of leaf Na+ that would help to maintain low leaf blade levels. Na+ has been proposed to recirculate via the phloem to either other shoot tissues or the roots for storage or for efflux (e.g. Munns et al., 1988; Lohaus et al., 2000). However, it is usually argued that phloem transport of Na+ is trivial relative to xylem transport rates because Na+ is toxic in the cytoplasmic environment of the phloem. We looked for evidence that Na+ was recirculated to the roots by loading plants with 22Na+ over 24 h and then measuring its disappearance from the shoot tissues. There was no evidence for retranslocation of 22Na+ from the shoot to the root except possibly in the salt sensitive Tamaroi (Fig. 9). However, the plants used were pretreated in NaCl for 7 d and it is possible that Na+ accumulation in leaf sheaths over this period was not sufficient to activate recirculation.
The Genetic Basis of Na+ Exclusion in Durum Wheats
We have identified two major physiological mechanisms controlling leaf blade Na+ in durum wheat. These traits are likely to be controlled at the gene level by the Nax1 and Nax2 loci. Work is under way to segregate the two loci to determine which locus is associated with which physiological mechanism of Na+ exclusion. The identification of the genes at these loci that control Na+ transport will provide major insights into the control of Na+ transport in cereals and will open up possibilities for modifying Na+ transport rates in other species and so increasing their salt tolerance. Analysis of salt sensitive mutants has implicated a small number of genes in root to shoot transport of Na+ in Arabidopsis (the SOS genes and sas loci including HKT1; for review, see Tester and Davenport, 2003). Whether homologs of these genes contribute to natural genotypic variation in salt tolerance in cereals remains to be seen. Several Na+-selective transporters (SOS1 and HKT1) have been identified in Arabidopsis, raising the possibility that Na+ has important physiological roles under nonsaline conditions. Discovery of the functions of Na+ transport pathways under nonsaline conditions is critical to understanding the consequences of manipulation of these pathways.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds were obtained from Dr. Ray Hare (New South Wales Department of Primary Industries, Tamworth, Australia). For nonradioactive experiments, plants were grown in a supported hydroponics system in half-strength nutrient solution (modified Hoagland and Arnon no. 2) in a controlled environment chamber as described in Rivelli et al. (2002). NaCl was added in 2 steps of 25 mm at 9 and 5 pm on day 0 with 2 mm CaCl2 to bring the total Ca2+ concentration to 4 mm. In experiments where the NaCl concentration was varied, CaCl2 was added to maintain a Na+:Ca2+ ratio below 15:1. For radioactive experiments, seeds were surface sterilized with 10% hypochlorite and imbibed in reverse osmosis-treated water overnight then allowed to germinate for 2 d on moist filter paper in petri dishes at 25°C before planting out. For uptake experiments, germinated seeds were transferred to lidless Eppendorf tubes with the bottom of the tube removed so that the roots protruded from the base. Tubes were placed in foam Eppendorf holders and floated on aerated hydroponic growth solution. Seedlings for the efflux experiment were grown hydroponically on mesh suspended over aerated growth solution. For the recirculation experiment, germinated seeds were placed in clear plastic 5-mL pipette tips suspended in holes in an opaque plastic lid over aerated growth solution. All seedlings were grown at 25°C with additional lighting (SON-T AGRO 400, Philips, Guildford, UK; light cycle 16 h/8 h light/dark) in a greenhouse in Cambridge in July and August 2003 and 2004. All seedlings were grown in half-strength nutrient solution as above. Five to 7 d before experiments, salt was added to a final concentration of 25 mm NaCl + 2 mm CaCl2. Solutions were changed every 5 d.
Radioactive Tracer Experiments
All radioactive solutions (except those for the 86Rb+ experiment) contained half-strength modified Hoagland with 25 mm NaCl + 2 mm CaCl2 solutions supplemented with various amounts of radioactive 22NaCl (Amersham, Buckinghamshire, UK). The relatively low NaCl concentration was used to exclude osmotic and toxic ion effects on growth and ion accumulation and to prevent ion toxicity. Genotypic differences in Na+ transport still occur at this concentration (Fig. 3). Experiments were conducted on a gently rotating shaker (to keep solutions aerated and reduce boundary layer effects) at 22°C to 25°C under a fluorescent light bank (120 μmol photons m−2 s−1 16 h/8 h light/dark). All samples were mixed with scintillation fluid (Optiphase HiSafe 3, Fisher Scientific, Loughborough, UK) and measured with a liquid scintillation counter (Tri-Carb 2000, Packard, Meriden, CT).
To measure 22Na+ uptake into whole plants, foam squares containing seedlings in Eppendorfs were transferred to fresh pretreatment solution (half-strength modified Hoagland with 25 mm NaCl + 2 mm CaCl2) and placed under a fluorescent light bank to adjust to the growth conditions used for the experiment. After a minimum of 1 h pretreatment, squares containing 2 plants were transferred to beakers containing 75 mL of radioactively labeled solution. After the uptake period, the squares were transferred to 2 successive ice-cold rinse solutions containing half-strength modified Hoagland with 25 mm NaCl + 10 mm CaCl2 for a total of 3 min. Roots were then excised 1 cm from the seed, blotted, and weighed. Shoots were excised 1 cm above the seed and either weighed whole or each leaf divided into sheath and blade portions before weighing. Uptake rates were calculated by fitting linear regression lines to the data by the least squares method using GraphPad Prism 3.0 (GraphPad Software, San Diego). For the 86Rb+ experiment, plants were pretreated in 2 successive solutions of modified Hoagland solution with 25 mm NaCl, 2 mm CaCl2, and 0 K+ (Na2HPO4 substituted for K2HPO4, RbNO3 substituted for KNO3 to give a Rb+ concentration of 3 mm) for 10 min total (to displace apoplastic K+ from roots) and then exposed to radioactively labeled 86Rb+ solution (of same composition as K+-free pretreatment solution). At the end of the 2 h uptake period, roots were rinsed in ice-cold K+-free pretreatment solution supplemented with 10 mm CaCl2 and plants processed as for 22Na+ uptake measurements.
To measure 22Na+ efflux from roots, whole seedlings were suspended with roots in radioactively labeled solution for 16 h. At the beginning of the efflux period, the roots were excised at the seed and rinsed briefly in deionized water, then placed in successive rinse solutions containing 25 mm NaCl and 2 mm CaCl2. The radioactivity in the rinse solutions and that remaining in the tissue were counted. Data were plotted as amount of 22Na+ remaining in the tissue and analyzed using a two compartment model of the root with bidirectional fluxes between each compartment (theoretically cytoplasm and vacuole). In addition, a linear component was subtracted from the first 1.33 min of data, which were considered to represent mainly apoplastic binding. The linear component was calculated by fitting a linear regression through the first three time points (20, 40, and 60 s). The remaining Na+ content in roots was fitted by the sum of two exponentials:
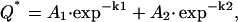 |
where Q* is root 22Na+ content, A1 and A2 are the initial contents (i.e. at t = 0) of the compartments assigned to cytoplasm and vacuole, respectively, and k1 and k2 are the half-times for exchange of the Na+ contents of the cytoplasm and vacuole, respectively (MacRobbie, 1981).
To measure 22Na+ recirculation, whole seedlings were suspended with roots in radioactively labeled solution for 24 h. The radioactive solution was then removed and the roots and container rinsed briefly with one volume of rinse solution (nonradioactive solution of the same composition as the labeled solution) and the container refilled with rinse solution. Plants were harvested at 24, 48, and 96 h after transfer to nonradioactive solution, and the solution was replaced at each harvest. Roots were rinsed and measured as for 22Na+ uptake experiments. Shoots were excised close to the seed and divided into basal and upper sheath (comprising the sheaths of all leaves) and individual leaf blades and weighed. Tissue 22Na+ content was converted to nominal Na+ content using the specific activity of the uptake solution. Shoot Na+ content was expressed per organ (e.g. blade of leaf 1) rather than per organ weight because the experiment was designed to test whether 22Na+ content of tissues reduced over time and so it was necessary to exclude the diluting effects of growth.
Supplementary Material
Acknowledgments
We thank Ray Hare for the wheat seeds, John Banfield for expert assistance with the radioactive experiments, and Alicia Munoz-Mayor for technical assistance.
This work was supported by the Royal Society (Dorothy Hodgkin fellowship to R.D.), by the Commonwealth Scientific and Industrial Research Organization (career development grant to R.A.J.), by the Nuffield Foundation (summer student bursary to A.Z.-P.), by the Biotechnology and Biological Sciences Research Council (research development fellowship to M.T.) and Australian Research Council (Federation fellowship to M.T.), and by the Grains Research and Development Corporation (to R.M.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057307.
References
- Davenport RJ, Reid RJ, Smith FA (1997) Sodium-calcium interactions in two wheat species differing in salinity tolerance. Physiol Plant 99: 323–327 [Google Scholar]
- Dubcovsky J, Santa Maria G, Epstein E, Luo M-C, Dvořák J (1996) Mapping of the K+/Na+ discrimination trait in wheat. Theor Appl Genet 2: 448–454 [DOI] [PubMed] [Google Scholar]
- Essah PA, Davenport R, Tester M (2003) Sodium influx and accumulation in Arabidopsis. Plant Physiol 133: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Yeo AR (1988) Ion relations of plants under drought and salinity. In D Baker, J Halls, eds, Solute Transport in Plant Cells and Tissues. Longman, Harlow, UK, pp 392–416
- Gorham J, Hardy C, Wyn Jones RG, Joppa LR, Law CN (1987) Chromosomal location of a K/Na discrimination character in the D genome of wheat. Theor Appl Genet 74: 584–588 [DOI] [PubMed] [Google Scholar]
- Gorham J, Wyn Jones RG, Bristol A (1990) Partial characterisation of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta 180: 590–597 [DOI] [PubMed] [Google Scholar]
- Husain S, Munns R, Condon AG (2003) Effect of sodium exclusion trait on chlorophyll retention and growth of durum wheat in saline soil. Aust J Agric Res 54: 589–597 [Google Scholar]
- James RA, Rivelli AR, Munns R, von Caemmerer S (2002) Factors affecting CO2 assimilation, leaf injury and growth in salt-stressed durum wheat. Funct Plant Biol 29: 1393–1403 [DOI] [PubMed] [Google Scholar]
- Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Lohaus G, Hussmann M, Pennewiss K, Schneider H, Zhu J-J, Sattelmacher B (2000) Solute balance of a maize (Zea mays L.) source leaf as affected by salt treatment with special emphasis on phloem retranslocation and ion leaching. J Exp Bot 51: 1721–1732 [DOI] [PubMed] [Google Scholar]
- MacRobbie EAC (1981) Ion fluxes in ‘isolated’ guard cells of Commelina communis L. J Exp Bot 32: 545–562 [Google Scholar]
- Munns R, Hare RA, James RA, Rebetzke GJ (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51: 69–74 [Google Scholar]
- Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253: 201–218 [Google Scholar]
- Munns R, Rebetzke GJ, Husain S, James RA, Hare RA (2003) Genetic control of sodium exclusion in durum wheat. Aust J Agric Res 54: 627–635 [Google Scholar]
- Munns R, Tonnet L, Shennan C, Gardner PA (1988) Effect of high external NaCl concentration on ion transport within the shoot of Lupinus albus. II. Ions in phloem sap. Plant Cell Environ 11: 291–300 [Google Scholar]
- Nevo E, Gorham J, Beiles A (1992) Variation for 22Na uptake in wild emmer wheat, Triticum dicoccoides in Israel: salt tolerance resources for wheat improvement. J Exp Bot 43: 511–518 [Google Scholar]
- Rivelli AR, James RA, Munns R, Condon AG (2002) Effect of salinity on water relations and growth of wheat genotypes with contrasting sodium uptake. Funct Plant Biol 29: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf O, Munns R, Tonnet ML, Jeschke WD (1991) The role of the stem in the partitioning of Na+ and K+ in salt-treated barley. J Exp Bot 42: 697–704 [Google Scholar]
- Xue Z-Y, Zhi D-Y, Xue G-P, Zhang H, Zhao Y-X, Xia G-M (2004) Enhanced salt tolerance of transgenic heat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167: 849–859 [Google Scholar]
- Yeo AR, Yeo ME, Flowers SA, Flowers TJ (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor Appl Genet 79: 377–384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.