PROGRAMMED CELL DEATH IN EUKARYOTES
Programmed cell death (PCD) is the genetically controlled suicide of cells. The tight regulation of this program is essential to ensure that it is only activated in the required cells at the proper moment. Deregulation of apoptosis, the main form of PCD in animals, is associated with diseases such as cancer, autoimmune diseases, and neurodegenerative disorders (Shi, 2002). In plants, deregulation of PCD is often associated with pleiotropic developmental defects and lethality (Lorrain et al., 2003). PCD occurs in all eukaryotic kingdoms, both in unicellular and multicellular organisms, and is involved in many aspects of development as well as in responses to external stimuli. PCD has morphological similarities in animals and plants, and it regulates common processes such as morphogenesis of the embryo, organ abscission, formation of hollow tubular structures (vessels), and defense against pathogens (Woltering et al., 2002). In view of the ubiquitous occurrence of PCD throughout nature and the coincident morphological and functional features in eukaryotes, a question arises: Has PCD developed independently in the different kingdoms or has it evolved from a common ancestral cell death process and thus plants, fungi, and animals may share common regulatory mechanisms?
The most convincing evidence for a common origin of PCD is that some of the molecular components of the program are conserved in the different kingdoms. Proteins that activate or inhibit PCD in animals, such as the Bcl2 family of pro- and antiapoptotic proteins, or the baculovirus antiapoptotic proteins p35 and IAP, have a similar effect on PCD when expressed in plants or yeast (Saccharomyces cerevisiae; Lacomme and Santa Cruz, 1999; Mitsuhara et al., 1999; Sanchez et al., 2000; Lincoln et al., 2002; Danon et al., 2004). Moreover, homologs of genes that control apoptosis in animals, such as Defender against Apoptotic Death-1 and Bax-inhibitor-1, are encoded in plant genomes and serve the same function in PCD control (Matsumura et al., 2003; Danon et al., 2004). In contrast, there are no evident homologs in plant or fungal genomes of caspases, arguably the key regulators of PCD in animals. Caspases are a family of Cys proteases with specificity for Asp (hence the name) that function as molecular switches to activate the cell death program in metazoans as diverse as mammals, nematodes, or insects (Shi, 2002). PCD promoting signals induce the self-processing of inactive caspase zymogens. This irreversible activation triggers a proteolytic cascade that turns on enzymes involved in killing the cell. Paradoxically, although genome mining has not yielded candidate caspases, caspase inhibitors can block PCD in plants, and most instances of plant PCD are associated with the induction of caspase-like activities (del Pozo and Lam, 1998; Watanabe and Lam, 2004). Are these activities encoded by enzymes phylogenetically unrelated to caspases? Do they regulate PCD in a similar manner to animal caspases? We will address these questions in light of the recent identification of plant enzymes displaying caspase activity and regulating PCD.
PLANT CASPASES
Two groups of proteases have been proposed as candidates to encode caspase-like activities in plants: the vacuolar processing enzymes (VPEs) and the metacaspases (Woltering et al., 2002). They are related in sequence and in tertiary structure to animal caspases (Aravind and Koonin, 2002) and have been associated with PCD processes in plants (Hoeberichts et al., 2003; Rojo et al., 2003). Recent work has provided evidence that VPEs and metacaspases have caspase activity and function in regulating PCD in plants, suggesting that they are true caspase orthologs (Hatsugai et al., 2004; Rojo et al., 2004; Suarez et al., 2004). In addition, a Ser protease displaying caspase-like activity and possibly controlling PCD has been identified in oat (Avena sativa; Coffeen and Wolpert, 2004).
VACUOLAR PROCESSING ENZYMES
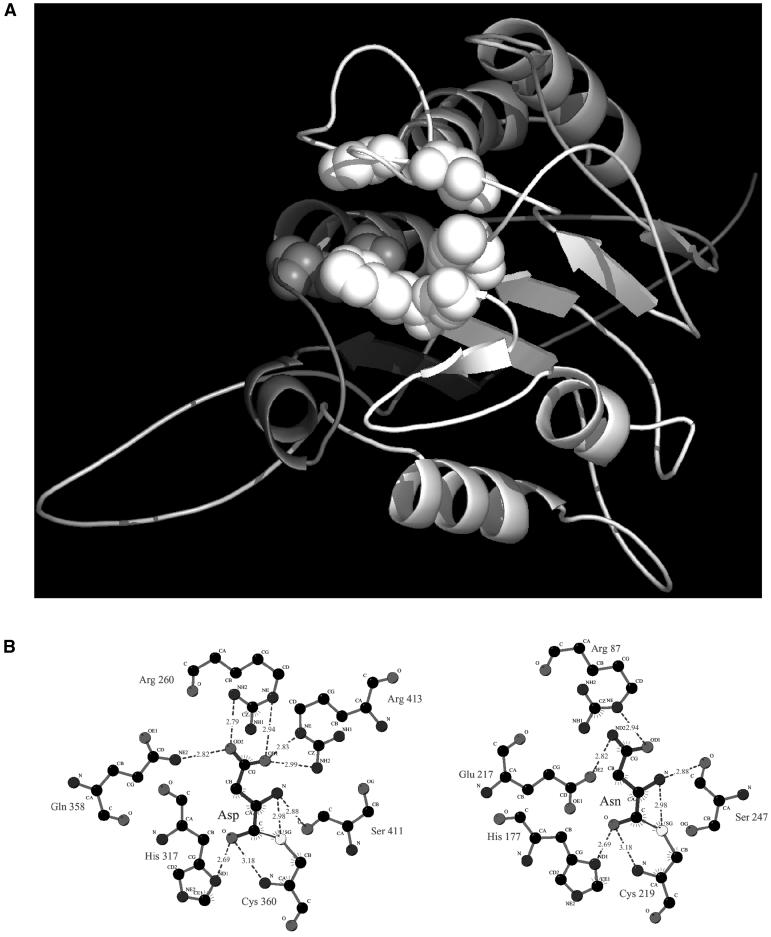
VPEs have been extensively studied for their role in the maturation of proteins in seed storage vacuoles, where they account for the bulk of processing activity (Shimada et al., 2003; Gruis et al., 2004). Recently, it was reported that members of this family are expressed in vegetative tissues where they are localized in protease precursor vesicles (PPVs) and in vacuoles (Kinoshita et al., 1999; Hayashi et al., 2001), organelles intimately associated with PCD in plants. In addition, the solved structures that are most similar to the folding predicted for VPEs are those of caspases, and modeling of the three-dimensional structure of Arabidopsis (Arabidopsis thaliana) VPEγ predicts a close alignment of its catalytic residues with those of caspase-8 (Fig. 1). Furthermore, VPEγ has been shown to regulate protein degradation during senescence, a type of PCD (Rojo et al., 2003). Taken together, these data suggest that VPEs may encode caspase activities associated with PCD progression in plants.
Figure 1.
The model VPEγ based on human caspase-8. A, The sequence of VPEγ was submitted to FFAS03 (Jaroszewski et al., 2000) and SUPERFAMILY (Gough et al., 2001) fold prediction servers. Both FFAS03 and SUPERFAMILY listed caspase fold as the most probable fold for the central region of VPEγ sequence (residues 50–330), and selected caspase-8 as the structure most similar to VPEγ. The alignment from FFAS03 was used for modeling using caspase-8 as a structural template. The model of the VPEγ molecule was built with the MODELER program. Residues aligned with binding site residues of caspase-8 are shown as spheres. B, The active site interactions in human caspase-8 (left) and predicted active site interactions in VPEγ (right). Replacement of Gln residue with Glu residue is connected with difference in specificity between caspases and VPEs (Asp and Asn residues in P1 position of the substrate, respectively).
Two recent reports have provided direct evidence that VPEs have caspase-like activity and regulate cell death in Nicotiana and Arabidopsis plants (Hatsugai et al., 2004; Rojo et al., 2004). Tobacco (Nicotiana tabacum) plants carrying the N resistance gene activate an acute PCD response when infected by tobacco mosaic virus (TMV), which is blocked by treatment with VPE-inhibitors or caspase1-inhibitors. A caspase activity induced during TMV infection is sensitive to VPE inhibitors, and, conversely, VPE activity in infected plants is inhibited by caspase1-inhibitors. In VPE-silenced Nicotiana benthamiana plants, the induction of caspase activity in response to TMV is suppressed, vacuolar collapse and PCD are blocked, and TMV proliferation increases (Hatsugai et al., 2004). These results suggest that the VPEs from Nicotiana may display caspase activity and initiate PCD during TMV infection by promoting vacuolar rupture. However, since many VPEs were silenced simultaneously, the activity and function of the individual N. benthamiana VPEs remains unknown. A study of the Arabidopsis VPEγ gene has provided direct in vivo evidence of its caspase-like activity and genetic evidence for the involvement of VPEγ in disease resistance and cell death progression (Rojo et al., 2004). VPEγ binds to caspase inhibitors that block self-maturation of this enzyme and all its downstream in vivo activities. An increase in caspase activity, concurrent with the initiation of a rapid PCD response, is observed in Arabidopsis plants infected with an incompatible strain of Pseudomonas syringae pv tomato DC3000 (Pst). The early induction of this caspase activity is compromised in vpeγ mutants, and vpeγ mutants are more susceptible to infection with the incompatible strain of Pst. The vpeγ mutants are also more susceptible to infection with turnip mosaic virus (TuMV), a pathogen that does not induce a classical PCD but rather affects the viability of infected cells. Thus, VPEs negatively regulate the growth of several biotrophic pathogens (TMV, Pst, TuMV), most likely by promoting cell death progression. What is their effect on the growth of necrotrophic pathogens that thrive on dead tissues? Interestingly, vpeγ plants are more susceptible to Botrytis cinerea, although this pathogen benefits from PCD in other conditions (Govrin and Levine, 2000; Dickman et al., 2001).
The picture that emerges from these studies is that VPEs are caspase orthologs that activate PCD in plants and influence the outcome of a wide range of interactions with pathogens.
METACASPASES
Iterative PSI-BLAST searches revealed the presence of a caspase-related family of proteases termed metacaspases in plant and fungal genomes (Uren et al., 2000). The yeast metacaspase YCA1 is required for the increased caspase-like activity and the subsequent PCD observed in H2O2-treated or in aging yeast cells. Moreover, YCA1 overexpression results in higher caspase activity and PCD in those conditions, suggesting that YCA1 encodes a caspase that activates PCD in yeast (Madeo et al., 2002). Plant genomes contain an extended complement of metacaspases, which have been classified as type I and type II based on their sequence and structural features (Fig. 2). Arabidopsis contains 3 type I (AtMCP1a–1c) and 6 type II (AtMCP2a–2f) metacaspases. AtMCP2d and AtMCP2f are Cys proteases that in vitro do not have caspase activity, instead cleaving substrates after P1 Arg/Lys residues (Vercammen et al., 2004). The in vitro or in vivo activity of the other metacaspases has not been tested. A type II metacaspase, mcII-Pa from Picea abies, is required for a burst of caspase activity that occurs during somatic embryogenesis in cells undergoing PCD (Suarez et al., 2004). The mcII-Pa transcript is expressed in embryonic tissues that are committed to death (suspensor and procambial strands), and its silencing by RNAi blocks the induction of caspase-like activity, inhibiting PCD and somatic embryogenesis (Suarez et al., 2004). These results suggest that mcII-Pa displays caspase-like activity and activates PCD, which is essential for embryo development (Bozhkov et al., 2004). A caveat of these studies in Picea is that the specificity of the RNAi for mcII-Pa was not analyzed, and the effects observed may be due to the silencing of related genes. Genetic analysis of an orthologous Arabidopsis gene could give further insights on the function of this gene. Unfortunately, Arabidopsis contains 4 genes highly similar to mcII-Pa, which may fulfill redundant functions and are located contiguously in chromosome 1 (AtMCP2a–2d), complicating the genetic analysis of their function. However, since all metacaspases have been identified in Arabidopsis, gene-specific silencing may be attempted and the specificity determined. The genetic analysis of the five remaining metacaspases in Arabidopsis should be more straightforward. Analysis of their expression may provide hints on their function (Table I). A tomato type II metacaspase (LeMCA1), which is most similar to the AtMCP2a to 2d gene cluster, is induced during infection with B. cinerea (Hoeberichts et al., 2003), suggesting that these genes may play a role in the PCD induced by this pathogen. We have performed in silico northerns of the Arabidopsis metacaspases in the GENEVESTIGATOR database (Zimmermann et al., 2004). The data retrieved shows that AtMCP2d is the most abundantly expressed metacaspase, and AtMCP1b expression is induced during compatible and incompatible interactions with Pst. These results corroborate previously reported data (Watanabe and Lam, 2004), indicating that the information obtained in the database for these probes is reliable. Other interesting in silico results have not been previously published (Table I). AtMCP1c is also induced in response to infection with compatible and incompatible strains of Pst at a higher-fold level than AtMCP1b. Interestingly, both AtMCP1b and AtMCP1c are induced when infected by a type III-secretion HrcC mutant of Pst and during a nonhost interaction with P. syringae pv phaseolicola, indicating that their expression is responsive to pathogen-associated molecular patterns (PAMPs) present in the bacterial surface rather than to virulent factors injected into the plant cell. Consistent with this, treatment with the flagellin peptide Flg22, a well-characterized bacterial PAMP (Zipfel et al., 2004), induces the expression of AtMCP1b and AtMCP1c. AtMCP2e, for which there was no previous evidence of expression (Vercammen et al., 2004; Watanabe and Lam, 2004), is also induced by Pseudomonas infection or by treatment with Flg22 and HrpZ. Moreover, infection with Phytophthora infestans or treatment with NPP1, a peptide PAMP from Phytophthora (Felbrich et al., 2002), induces the expression of AtMCP1b, AtMCP1c, and AtMCP2e. Thus, these three genes are coordinately induced by PAMPs from different pathogens and may be part of the innate immune response of plants, which in some cases includes the activation of cell death processes. AtMCP2f is induced in senescing organs of the flower and in senescing cell cultures, indicating that it may play a role in this type of PCD. Interestingly, 3 genes from the chromosome 1 cluster, AtMCP2a, AtMCP2c, and AtMCP2d, are expressed at higher levels in roots than in aerial organs, suggesting they may play a specialized role in these organs. Moreover, AtMCP2a is induced in roots under conditions (salt, drought, and genotoxic stress) that activate cell death. In summary, the expression data is consistent with a role of metacaspases in developmental or stress-induced PCD.
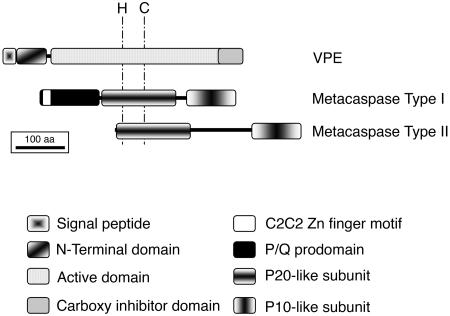
Figure 2.
Domain structure of plant VPEs and metacaspases. The position of the His-Cys catalytic dyad of these enzymes is shown. VPEs are synthesized as inactive zymogens with a signal peptide for insertion into the endomembrane system. The inhibitory C-terminal propeptide and the N-terminal propeptide of VPEs are sequentially removed, most likely in the acidic environment of the vacuole. Metacaspases are probably cytosolic enzymes that may be autoprocessed into 2 subunits, p20 and p10. In addition, type I metacaspases contain a prodomain rich in Pro and Gln residues, which contain a zinc finger motif similar to the ones present in LSD1, a negative regulator of PCD in Arabidopsis.
Table I.
Summary of Arabidopsis VPEs and metacaspases
The expression data from VPEs is from published works, whereas the data on metacaspases was obtained from the GENEVESTIGATOR database (Zimmermann et al., 2004) on December 5, 2004. Psp, P. syringae pv phaseolicola; Pi, P. infestans; HrpZ, harpin from P. syringae pv tomato DC3000; NPP1, cell wall elicitor protein from P. infestans.
| Gene | AGI-TIGR Locus | Signal Peptide | EST + cDNAs | T-DNA Mutants | Subcellular Location | Expression |
|---|---|---|---|---|---|---|
| VPEs | ||||||
| VPEα | At2g25940 | Amino acids 1–20 | 13 + 2 | Available | Secretory pathwaya,plasma membranea | Vegetative tissues. Induced in leaves by ethylene, salicylic acid (SA), wounding, and senescence. |
| VPEβ | At1g62710 | Amino acids 1–21 | 33 + 2 | Available | Secretory pathwaya, vacuolea | Seeds, flowers, root tips. |
| VPEγ | At4g32940 | Amino acids 1–25 | 129 + 5 | Available | PPV, vacuole | Vegetative tissues, induced by ethylene, SA, wounding, and senescence. Induced during infection with Pst avrRpm1, TuMV, B. cinerea. |
| VPEδ | At3g20210 | Amino acids 1–24 | 62 + 2 | Available | Secretory pathwaya,apoplasma | All stages in developing seeds, most abundant in cell division stage and germinating seeds. |
| Metacaspases | ||||||
| AtMCP1a | At5g64240 | 25 + 1 | Available | Mitochondriona | Ubiquitously expressed. Induced in senescing flowers. | |
| AtMCP1b | At1g02170 | 3 | Available | Chloroplasta | Induced in leaves by Pst, Pst avrRpm1, Psp, Pi, Flg22, HrpZ, and NPP1. | |
| AtMCP1c | At4g25110 | 4 | Available | Mitochondriona | Induced in leaves by Pst, Pst avrRpm1, Psp, Pi, Flg22, HrpZ, NPP1, SA, and AgNO3. | |
| AtMCP2a | At1g79310 | 1 | Available | Cytosola | Higher expression in roots than aerial organs. Induced in wounded roots or in roots under salt, osmotic, or genotoxic stress. | |
| AtMCP2b | At1g79330 | 2 | Available | Cytosola | Seed specific expression. | |
| AtMCP2c | At1g79320 | None | Available | Cytosola | Very low expression. Higher in roots than aerial organs | |
| AtMCP2d | At1g79340 | 49 + 2 | Available | Cytosola | Ubiquitously expressed. Higher expression in roots than aerial organs. | |
| AtMCP2e | At1g16420 | None | None | Cytosola | Induced in leaves by Pst, Pst avrRpm1, Psp, Pi, Flg22, HrpZ, and NPP1. Higher expression in roots than in aerial organs. | |
| AtMCP2f | At5g04200 | 9 + 2 | Available | Cytosola | Induced in senescing cell cultures and flowers. Induced in roots by osmotic stress. |
Localization predicted by TargetP.
SER PROTEASE WITH SPECIFICITY TOWARD ASPARTATE
Isolates of the necrotrophic fungus Cochliobolus victoriae that produce the toxin victorin activate a PCD response in susceptible oat cultivars, leading to disease progression (Navarre and Wolpert, 1999). Victorin-induced PCD is associated with two caspase-like activities that can be differentiated by their sensitivity to caspase inhibitors (Coffeen and Wolpert, 2004). One of these caspase-like enzymes is translocated to the apoplasm in response to victorin treatment or heat shock, correlating with the induction of PCD by these stimuli. This caspase activity was purified and partial sequence of the coeluting protein showed that it was homologous to subtilisin-like Ser proteases, thus its name saspase. Extensive biochemical characterization of the purified saspase showed that its substrate specificity is strict for Asp at the P1 position and thus it is distinct from all other known Ser proteases. Once the gene encoding the saspase is cloned, it will be possible to test its role in PCD activation.
PLANT AND ANIMAL CASPASES REGULATE PCD FROM DIFFERENT SUBCELLULAR COMPARTMENTS
Most animal caspases are soluble cytoplasmic proteins. Two exceptions are caspase-12, which is associated with the endoplasmic reticulum (ER) membrane from the cytosolic side and mediates ER-stress induced PCD (Nakagawa et al., 2000), and caspase-2, which is partially localized in the nucleus and mediates PCD triggered by DNA damage (Lassus et al., 2002). However, once activated, all animal caspases initiate a proteolytic cascade in the cytosol that results in the activation of numerous enzymes that degrade cellular components and kill the cell.
In plants, the enzymes displaying caspase activity may regulate PCD from several compartments, including the cytosol (Table I). Type II metacaspases do not contain signal peptides or transmembrane domains (Fig. 2), so they are predicted to be cytosolic and thus would function in the same location as animal caspases. Type I metacaspases are predicted to be localized in mitochondria and chloroplasts based on the presence of putative transit peptides at their N terminus of the proteins. However, the confidence level of these predictions is low and thus they may also be cytosolic enzymes. In contrast, VPEs and saspases probably function in other subcellular compartments. VPEs contain signal peptides for insertion into the endomembrane system (Fig. 2), and they have been localized to PPVs or vacuoles (Rojo et al., 2003). VPEs in N. benthamiana are required for the disruption of the tonoplast, a process that is common to most cases of PCD in plants and possibly constitutes an irreversible step in cell death commitment. Although the localization of N. benthamiana VPEs has not been reported, they are likely localized in vacuoles, and would activate PCD from the vacuolar lumen. In addition, VPEs may act after disruption of the tonoplast by processing cytosolic enzymes involved in PCD execution. This function would be similar to the role of animal cathepsins, which are lysosomal proteases that when released from damaged lysosomes activate caspase cascades in the cytosol, triggering apoptosis (Ferri and Kroemer, 2001). Subtilisin Ser proteases homologous to the saspase identified by Coffeen and Wolpert (2004) contain signal peptides and have been detected in vacuoles or in the extracellular space. The oat saspase is localized intracellularly in the absence of PCD-promoting stimuli, possibly retained in the ER or the Golgi apparatus. Secretion of the saspase to the apoplasm correlates with the initiation of PCD, suggesting that it activates cell death from the apoplasm.
REGULATION OF CASPASE ACTIVITY IN PLANTS
Animal caspases are expressed constitutively, but in a latent precursor form. The zymogens are processed and PCD is irreversibly initiated only in response to cell death promoting signals. Thus, in animals, the regulation of caspase activity is mainly posttranslational via proteolytic digestion of the zymogens.
This situation may be different in plants. For instance, the expression of VPEs is induced in conditions that activate PCD, such as TMV infection in tobacco (Hatsugai et al., 2004) or senescence and pathogen attack in Arabidopsis (Rojo et al., 2004), indicating that the regulation of their activity may be partly at the level of transcription. Similarly, metacaspases are induced in conditions that activate PCD (Table I). In addition, posttranslational regulation of VPEs by maturation of the zymogens may also occur. It has been postulated that VPEs are self-processed in the acidic environment of the vacuole. Therefore, VPEs may be maintained in a latent unprocessed form by keeping them in intermediate nonacidic compartments of the secretory pathway, such as PPVs (Rojo et al., 2003). In conditions inducing PCD, VPEs may be transferred to the vacuole where they are activated and promote vacuolar collapse. Whether this is achieved through a vacuolar proteolytic cascade similar to the cytosolic cascade activated by caspases of metazoans is not yet known. VPEγ has been shown to process a vacuolar protease and to regulate the degradation of vacuolar proteins, including a cystatin that may inhibit PCD progression (Rojo et al., 2004), suggesting that proteolytic events downstream of VPE activation may be important for PCD initiation. In contrast to VPEs and metacaspases, the oat saspase is expressed constitutively, most likely as an active enzyme (Coffeen and Wolpert, 2004). It has been proposed that this saspase is maintained inactive by its retention in the secretory pathway and is activated by secretion to the apoplasm where it may gain access to effector substrates that activate PCD. What are these effector proteins? P69, a pathogen-induced Ser protease from tomato, has been shown to cleave an extracellular Leu-rich repeat containing protein (LRP) during PCD induced by bacterial infection (Tornero et al., 1996). LRPs are in many cases signaling molecules, and some like the tomato Cf-2 are involved in disease resistance through the regulation of PCD. It is tempting to speculate that the oat saspase may cleave an LRP and activate a signaling cascade inducing PCD. In summary, several levels of regulation of caspase activity are functional in plants. The expression of caspase-like enzymes may be induced prior to PCD and their activity may be regulated by maturation of inactive zymogens or by subcellular relocalization.
A challenge for the future is to understand how developmental cues or external stimuli feed into these regulatory systems to activate plant caspases, and how they in turn promote cell death execution. As we have discussed, some of the plant caspases are functionally and evolutionarily related to animal caspases, suggesting that part of the information obtained from animal systems may be useful to further characterize PCD and caspase cascades in plants. However, fundamental differences in regulation and subcellular compartmentalization of plant and animal caspases have also been reported, indicating that many features of PCD will be plant specific.
Acknowledgments
We thank Dr. José J. Sánchez-Serrano and Dr. Raquel Martín for critically reading the manuscript.
This work was supported in part by the Spanish Ministerio de Educación y Ciencia (grant no. MBC2003–08039 to E.R.).
References
- Aravind L, Koonin E (2002) Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins 46: 355–367 [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S (2004) VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ 11: 175–182 [DOI] [PubMed] [Google Scholar]
- Coffeen WC, Wolpert TJ (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004) Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Dickman MB, Park YK, Oltersdorf T, Li W, Clemente T, French R (2001) Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc Natl Acad Sci USA 98: 6957–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nürnberger T (2002) NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32: 375–390 [DOI] [PubMed] [Google Scholar]
- Ferri KF, Kroemer G (2001) Organelle-specific initiation of cell death pathways. Nat Cell Biol 3: 255–263 [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919 [DOI] [PubMed] [Google Scholar]
- Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10: 751–757 [DOI] [PubMed] [Google Scholar]
- Gruis D, Schulze J, Jung R (2004) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, ten Have A, Woltering EJ (2003) A tomato metacaspase gene is upregulated during programmed cell death in Botrytis cinerea-infected leaves. Planta 217: 517–522 [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Godzik A (2000) Improving the quality of twilight-zone alignments. Protein Sci 9: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 19: 43–53 [DOI] [PubMed] [Google Scholar]
- Lacomme C, Santa Cruz S (1999) Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci USA 96: 7956–7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus P, Opitz-Araya X, Lazebnik Y (2002) Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297: 1352–1354 [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Richael C, Overduin B, Smith K, Bostock R, Gilchrist DG (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad-spectrum resistance to disease. Proc Natl Acad Sci USA 99: 15217–15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, et al (2002) A caspase-related protease regulates apoptosis in yeast. Mol Cell 9: 911–917 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L) cells. Plant J 33: 425–434 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Malik KA, Miura M, Ohashi Y (1999) Animal cell-death suppressors Bcl-x(L) and Ced-9 inhibit cell death in tobacco plants. Curr Biol 9: 775–778 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403: 98–103 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wolpert TJ (1999) Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell 11: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O, Lam E (1998) Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol 8: 1129–1132 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martin R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B, et al (2004) VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rojo E, Zouhar J, Carter C, Kovaleva V, Raikhel NV (2003) A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 7389–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, de Torres Zabala M, Grant M (2000) AtBI-1, a plant homologue of Bax inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J 21: 393–399 [DOI] [PubMed] [Google Scholar]
- Shi Y (2002) Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 9: 459–470 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV (2004) Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 14: 339–340 [DOI] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1996) Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA 93: 6332–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM (2000) Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 6: 961–967 [DOI] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279: 45329–45336 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2004) Recent advance in the study of caspase-like proteases and Bax inhibitor-1 in plants: their possible roles as regulator of programmed cell death. Mol Plant Pathol 5: 65–70 [DOI] [PubMed] [Google Scholar]
- Woltering EJ, van der Bent A, Hoeberichts FA (2002) Do plant caspases exist? Plant Physiol 130: 1764–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial resistance in Arabidopsis through flagellin perception. Nature 15: 764–767 [DOI] [PubMed] [Google Scholar]