Abstract
Isoflavonoids are ecophysiologically active secondary metabolites of the Leguminosae and known for health-promoting phytoestrogenic functions. Isoflavones are synthesized by 1,2-elimination of water from 2-hydroxyisoflavanones, the first intermediate with the isoflavonoid skeleton, but details of this dehydration have been unclear. We screened the extracts of repeatedly fractionated Escherichia coli expressing a Glycyrrhiza echinata cDNA library for the activity to convert a radiolabeled precursor into formononetin (7-hydroxy-4′-methoxyisoflavone), and a clone of 2-hydroxyisoflavanone dehydratase (HID) was isolated. Another HID cDNA was cloned from soybean (Glycine max), based on the sequence information in its expressed sequence tag library. Kinetic studies revealed that G. echinata HID is specific to 2,7-dihydroxy-4′-methoxyisoflavanone, while soybean HID has broader specificity to both 4′-hydroxylated and 4′-methoxylated 2-hydroxyisoflavanones, reflecting the structures of isoflavones contained in each plant species. Strikingly, HID proteins were members of a large carboxylesterase family, of which plant proteins form a monophyletic group and some are assigned defensive functions with no intrinsic catalytic activities identified. Site-directed mutagenesis with soybean HID protein suggested that the characteristic oxyanion hole and catalytic triad are essential for the dehydratase as well as the faint esterase activities. The findings, to our knowledge, represent a new example of recruitment of enzymes of primary metabolism during the molecular evolution of plant secondary metabolism.
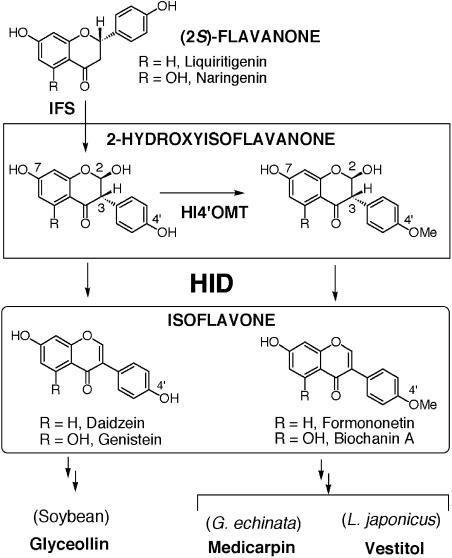
Isoflavonoids are the characteristic metabolites of the Leguminosae, the third largest family of the higher plants. Isoflavonoids play significant roles in the adaptation of the producer plants to their biological environments as, for example, defense substances (phytoalexins) and a host signal to symbiotic nitrogen-fixing rhizobial bacteria (Dewick, 1993; Dixon, 1999; Aoki et al., 2000). Isoflavones are early products of the isoflavonoid pathway. Free and glycoconjugated daidzein (7,4′-dihydroxyisoflavone) and genistein (5,7,4′-trihydroxyisoflavone) supplied by soybeans (Glycine max) in the diet are health-promoting and disease-preventing phytoestrogens (Dixon, 1999). Several legume-specific enzymes are essential for isoflavone biosynthesis. The isoflavonoid skeleton is constructed from a class of general flavonoid intermediates, (2S)-flavanones, by a member of the CYP93C subfamily of cytochrome P450 (P450), 2-hydroxyisoflavanone synthase (IFS; Fig. 1; Akashi et al., 1999; Steele et al., 1999). The IFS product, 2,5,7,4′-tetrahydroxyisoflavanone or 2,7,4′-trihydroxyisoflavanone, is then dehydrated to produce a double bond between C-2 and C-3, yielding genistein or daidzein. The soybean phytoalexins, glyceollins, are derived from daidzein. 4′-Methoxylated isoflavonoids, such as medicarpin and vestitol, constitute the major group of legume phytoalexins (Dixon, 1999). In their biosynthesis, the reaction of 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase (HI4′OMT) occurs, and subsequent dehydration yields formononetin (7-hydroxy-4′-methoxyisoflavone), the central biosynthetic intermediate (Akashi et al., 2000, 2003). 5-Deoxylated (iso) flavonoid structure represented by daidzein and formononetin is another feature of leguminous flavonoids, and chalcone polyketide reductase (Welle et al., 1991; Akashi et al., 1997) and type II chalcone isomerase (Shimada et al., 2003) are responsible for their biosynthesis. The dehydration yielding isoflavones has been assumed to be catalyzed by a soluble enzyme, but it is also possible that unstable 2-hydroxyisoflavanones spontaneously lose water to produce isoflavones in plant cells (Kochs and Grisebach, 1986; Hashim et al., 1990).
Figure 1.
Isoflavonoid biosynthesis in leguminous plants. 4′-Hydroxylated isoflavones are biosynthesized from (2S)-flavanones by the reaction catalyzed by IFS and subsequent intramolecular 1,2-dehydration of 2-hydroxyisoflavanones. 4′-Methoxylated isoflavones are produced from the IFS reaction products in 2 steps: 4′-O-methylation catalyzed by HI4′OMT and subsequent dehydration. In this study, HIDs of distinct substrate specificity toward 4′-hydroxylated and 4′-methoxylated 2-hydroxyisoflavanones were characterized.
IFS cDNAs were characterized from a medicinal legume, Glycyrrhiza echinata (Akashi et al., 1999), soybean (Steele et al., 1999; Jung et al., 2000), and a model legume, Lotus japonicus (Shimada et al., 2000). The in vitro assay with recombinant IFS in yeast (Saccharomyces cerevisiae) microsomes produced a substantial quantity of isoflavone in addition to the original product, 2-hydroxyisoflavanone, via spontaneous dehydration (Akashi et al., 1999; Jung et al., 2000; Shimada et al., 2000). Also, the enzyme expressed in insect cells was reported to yield only isoflavone due to the presence of nonspecific dehydratases (Steele et al., 1999). On the other hand, a biochemical study of the 2-hydroxyisoflavanone dehydratase (HID) of Pueraria lobata has been reported (Hakamatsuka et al., 1998). The substrate, 2,7,4′-trihydroxyisoflavanone, was prepared using microsomes of elicited P. lobata cells, and the production of daidzein was measured during the enzyme purification steps. The P. lobata HID protein was purified to apparent homogeneity, but, unfortunately, no information on the amino acid sequence was provided. More recently, we observed that a cell-free extract of G. echinata converted 2,7-dihydroxy-4′-methoxyisoflavanone into formononetin, but did not convert 2,7,4′-trihydroxyisoflavanone into daidzein (Akashi et al., 2000). This result indicated that G. echinata HID has a clear substrate specificity and distinguishes differently substituted 2-hydroxyisoflavanones.
The combination of in vitro reactions with microsomes of yeast expressing IFS (Akashi et al., 1999) as well as purified recombinant HI4′OMT protein (Akashi et al., 2003) allowed us to prepare a sufficient quantity of 2-hydroxyisoflavanone and its radiolabeled tracer. In this study, we examined HID activities in G. echinata and soybean for the presence of species-specific enzymes. To isolate the cDNA encoding HID, a cDNA library of elicited G. echinata cell cultures expressed in Escherichia coli was screened, assaying the activity in fractionated bacterial extracts. The G. echinata HID sequence was further used for the isolation of soybean HID cDNA. Surprisingly, the amino acid sequences of both HIDs possess the motifs characteristic of carboxylesterases, to which dehydratase activities have never been attributed. Analogous proteins, including human carboxylesterases, are widely distributed in living organisms, and plant carboxylesterase-like proteins form a monophyletic group. Furthermore, some plant proteins of this family are known for defensive functions with no intrinsic catalytic activities identified. Possibly, some of the dehydration in natural product biosynthesis is mediated by this group of enzymes. The implication of this finding to the molecular evolution of plant secondary metabolism is discussed.
RESULTS
HID Activities in G. echinata and Soybean
G. echinata cells constitutively accumulate formononetin and rapidly produce medicarpin on elicitor treatment (Fig. 1; Nakamura et al., 1999). Both products are 4′-methoxyisoflavonoids. Soybean mainly produces glycoconjugates of 4′-hydroxyisoflavones, such as daidzein, genistein, and glycitein (7,4′-dihydroxy-6-methoxyisoflavone), but minor quantities of formononetin and its derivatives also accumulate in soybean seeds (Dewick, 1993).
The crude enzyme preparation of elicited G. echinata cells efficiently converted 2,7-dihydroxy-4′-methoxyisoflavanone to formononetin (Table I). Only very low activity for the production of daidzein from 2,7,4′-trihydroxyisoflavanone and genistein from 2,5,7,4′-tetrahydroxyisoflavanone (0.7% and 3% of the formononetin production, respectively) with the same extract was detected. In contrast, the enzyme preparation of soybean seedlings was highly active to 2,5,7,4′-tetrahydroxyisoflavanone and yielded genistein, while it also showed activity toward 2,7,4′-trihydroxyisoflavanone and 2,7-dihydroxy-4′-methoxyisoflavanone, producing daidzein and formononetin (34% and 18% of genistein production, respectively; Table I). The production of isoflavones by spontaneous dehydration of 2-hydroxyisoflavanones was negligible during 5-min incubation at the substrate concentration of 100 μm in the neutral buffer, pH 7.5. Thus, the enzyme-dependent dehydration of 2-hydroxyisoflavanones to form isoflavones was clearly demonstrated, and the substrate specificities of HIDs of G. echinata and soybean were shown to be different.
Table I.
Specific HID and carboxylesterase activity of the plant cell-free extracts and recombinant proteins
| Substrate
|
Product
|
Specific Activityab
|
||||
|---|---|---|---|---|---|---|
|
G. echinata
|
Soybean
|
Porcine Liver Carboxylesterase
|
||||
| Cell-Free Extract c | HIDMd | Cell-Free Extractc | HIDHd | |||
| pkatal mg−1 | nkatal mg−1 | pkatal mg−1 | nkatal mg−1 | nkatal mg−1 | ||
| 2,7-Dihydroxy-4′-methoxyisoflavanonee | Formononetin | 120 ± 8 | 153 ± 12 | 100 ± 10 | 18 ± 4.2 | NDf |
| 2,7,4′-Trihydroxyisoflavanonee | Daidzein | 0.82 ± 0.04 | 0.9 ± 0.3 | 190 ± 19 | 44 ± 8 | NDf |
| 2,5,7,4′-Tetrahydroxyisoflavanonee | Genistein | 3.5 ± 1.1 | 0.9 ± 0.3 | 550 ± 24 | 110 ± 10 | NDf |
| p-Nitrophenyl butyrate | p-Nitrophenol | −g | 0.32 ± 0.07h | −g | 0.73 ± 0.10h | 425 ± 7h |
Mean ± sd from three independent experiments.
Specific activity was calculated from the production rates of isoflavones and p-nitrophenol.
Ammonium sulfate (30% to 80% saturation) precipitates of the cell-free extracts from elicited G. echinata cultures (0.2% yeast extract, 24 h) and 7-d-old soybean seedlings were used for the assays.
Purified recombinant proteins were used for the assays.
Specific activity was determined using 100 μm substrate.
Not detected.
Not examined.
No hydrolyzing activity was detected in assays with boiled (100°C, 10 min) proteins and p-nitrophenyl butyrate.
Cloning of G. echinata HID cDNA by Screening of the Fractionated Expression Library
A specific and highly sensitive assay of dehydratase activity yielding formononetin was achieved using [methyl-14C]-2,7-dihydroxy-4′-methoxyisoflavanone that was prepared from 2,7,4′-trihydroxyisoflavanone, the product of the in vitro IFS reaction (Akashi et al., 1999), and [methyl-14C]-S-adenosyl-l-Met with the recombinant HI4′OMT protein (Akashi et al., 2003). The first screening of the cDNA expression library of G. echinata (Akashi et al., 2003) was carried out with five cDNA pools (about 30,000 transformants/pool). The crude extracts of the pools were reacted with [methyl-14C]-2,7-dihydroxy-4′-methoxyisoflavanone, and reaction mixtures were analyzed by thin-layer chromatography-autoradiography. Two pools were found to produce [14C]formononetin, and one of the arbitrarily selected positive pools was subdivided into 10 pools of smaller size (about 3,000 clones/pool). Expression of enzyme proteins and the assay were performed again, and the one resultant positive pool was subjected to the next round of subdivision and assay. One positive pool among the 10 pools was repeatedly identified in the successive third (about 300 clones/pool) and fourth (30 clones/pool) screenings. A single E. coli clone showing 2,7-dihydroxy-4′-methoxyisoflavanone dehydratase activity was then isolated from the fourth pool.
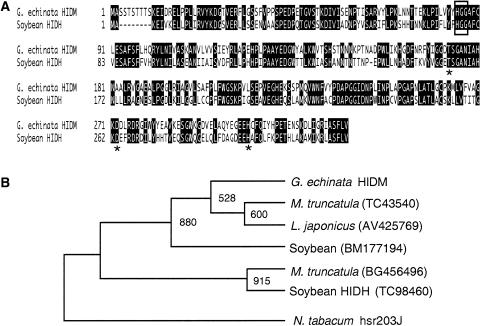
The cDNA, designated HIDM (HID methoxy type; accession no. AB154414), had 1,178-bp nucleotides and encoded 328 amino acids (Fig. 2A). A conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) suggested that HIDM is structurally related to the α/β-hydrolase fold family protein (Ollis et al., 1992) and has a conserved sequence motif recorded for carboxylesterase (about 40–180 amino acids from the N terminus). A conserved sequence (His-85-Gly-86-Gly-87) constituting an oxyanion hole within the carboxylesterase motif (Satoh and Hosokawa, 1995), and a triad of amino acid residues (Thr-173, Asp-272, and His-304) that acts as the catalyst for ester hydrolysis outside the carboxylesterase motif, were also present, although a more general Ser residue in the catalytic triad of lipases and esterases (Nardini and Dijkstra, 1999; Manco et al., 2000) is substituted to Thr.
Figure 2.
Amino acid sequence of G. echinata HIDM aligned with soybean HIDH (A) and the phylogenetic relationship of G. echinata HIDM and homologous proteins found in L. japonicus, M. truncatula, and soybean EST databases (B). A, The amino acid residues of which two sequences are identical are in reverse type. Gaps (-) are inserted to optimize alignment. Putative residues that constitute the catalytic triad (Thr, Asp, and His) are indicated with asterisks (*), and the oxyanion hole (His-Gly-Gly) is boxed. B, A total of seven amino acid sequences were analyzed using the ClustalW program (Thompson et al., 1994), and the tree was displayed by TreeView software (Page, 1996). Numbers indicate bootstrap value from 1,000 replicates. Nicotiana tabacum hsr203j was defined as the out group.
Cloning of a Soybean HID cDNA
Searches of expressed sequence tag (EST) databases of soybean, Medicago truncatula, and L. japonicus, leguminous plants known to produce isoflavones, revealed the presence of cDNAs homologous to G. echinata HIDM (>50% amino acid identity) in these plants. All these sequences were, however, only annotated as putative proteins. Phylogenetic analysis (Fig. 2B) indicated that soybean BM177194, L. japonicus AV425769, and M. truncatula TC43540 proteins form the same branch with G. echinata HIDM (>80% identity at the amino acid level). The soybean TC98460 protein had >60% identity with the 4 proteins and formed the adjacent branch with M. truncatula BG456496. The TC98460 sequence had the predicted initiation and stop codons. The cDNA of the coding region was cloned from soybean seedlings by reverse transcription (RT)-PCR and named HIDH (HID hydroxy type; accession no. AB154415): It showed 61% identity with HIDM at the amino acid level, and the characteristic carboxylesterase motifs were also conserved. On the other hand, the sequence of BM177194 contained only 5′-end 437 bps, and neither the fragment nor the full cDNA were amplified by RT-PCR and 3′-RACE, respectively, from the same soybean seedlings. Thus, the gene corresponding to BM177194 may not be expressed in the seedlings used in this study.
Biochemical Characterization of HIDs of G. echinata and Soybean
HIDM and HIDH proteins were expressed in E. coli, and purified recombinant proteins with six His residues at the N terminus were assayed for substrate specificities using HPLC for the product analysis. The chemical structures of the isoflavones were confirmed by comparison of the retention time values with those of authentic samples and electron ionization mass spectrometry (data not shown). Specific activities at the substrate concentration of 100 μm are listed in Table I. A high activity of formononetin production from 2,7-dihydroxy-4′-methoxyisoflavanone by G. echinata HID (HIDM protein) was observed. HIDM protein yielded only very small amounts of daidzein and genistein from respective substrates: The specific activity toward both 2,7,4′-trihydroxyisoflavanone and 2,5,7,4′-tetrahydroxyisoflavanone was 1/170th of that toward 2,7-dihydroxy-4′-methoxyisoflavanone. Thus, the substrate preference of HIDM protein coincided with that of the G. echinata cell-free extract. On the other hand, recombinant HID of soybean (HIDH protein) had the highest activity toward 2,5,7,4′-tetrahydroxyisoflavanone (relative activity, 100%) and also showed significant activity toward 2,7,4′-trihydroxyisoflavanone (40%) and 2,7-dihydroxy-4′-methoxyisoflavanone (16%). Thus, again, the substrate preference of HIDH protein coincided with that of soybean extract.
Recombinant G. echinata HIDM and soybean HIDH proteins hydrolyzed p-nitrophenyl butyrate in the standard assay of carboxylesterases (Heymann and Mentlein, 1981), but the magnitude of the activity was only 0.2% (HIDM) or 0.7% (HIDH) of the respective activity to the most preferred substrate (Table I). On the other hand, commercially available porcine liver carboxylesterase did not act on 2-hydroxyisoflavanones at all (Table I).
To further characterize the enzyme properties, steady-state kinetic parameters for the recombinant HIDs were determined (Table II). HIDM and HIDH proteins showed comparable Km values to the three substrates, with the HIDH Kms displaying about one-half the levels of the HIDM Kms. Interestingly, the relative ratios of Km values for the three substrates displayed by HIDM and HIDH proteins were roughly the same (about 1:4:6 for both Kms toward 2,7-dihydroxy-4′-methoxyisoflavanone, 2,7,4′-trihydroxyisoflavanone, and 2,5,7,4′-tetrahydroxyisoflavanone, respectively), indicating that the affinities of the two enzymes to these substrates are conserved. However, these proteins exhibited quite different turnover numbers (kcat values) for differently substituted substrates. Thus, G. echinata HIDM protein had a high kcat for 2,7-dihydroxy-4′-methoxyisoflavanone, but only 1% to 2% kcat levels for 4′-hydroxylated substrates. In contrast, soybean HIDH protein displayed the highest kcat for 2,5,7,4′-tetrahydroxyisoflavanone and about 30% and 10% kcat levels for 2,7,4′-trihydroxyisoflavanone and 2,7-dihydroxy-4′-methoxyisoflavanone, respectively. These differences in kcat values mainly caused different specificity constants (kcat/Km ratios) of the two enzymes to the three substrates. Consequently, HIDM protein possessed a narrow substrate specificity, 2,7-dihydroxy-4′-methoxyisoflavanone being the most favored substrate, while HIDH protein exhibited broader substrate specificity toward both 4′-hydroxy- and 4′-methoxy-2-hydroxyisoflavanones.
Table II.
Kinetic properties of recombinant HIDH and HIDM
The values are the averages from two independent experiments (maximum deviation was about 10%).
| Substrate
|
Product
|
G. echinata HIDM
|
Soybean HIDH
|
||||
|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | ||
| μm | s−1 | s−1mm−1 | μm | s−1 | s−1 mm−1 | ||
| 2,7-Dihydroxy-4′-methoxyisoflavanone | Formononetin | 58 | 9.8 | 170 | 29 | 1.6 | 46 |
| 2,7,4′-Trihydroxyisoflavanone | Daidzein | 210 | 0.12 | 0.56 | 114 | 5.3 | 47 |
| 2,5,7,4′-Tetrahydroxyisoflavanone | Genistein | 304 | 0.19 | 0.62 | 170 | 18.1 | 106 |
The spontaneous dehydration of 2,7,4′-trihydroxyisoflavanone yielding daidzein in the neutral buffer, pH 7.5 at 30°C was shown to be a first-order reaction. The velocity is dependent on the substrate concentration, and the calculated rate constant was 3.1 × 10−5 s−1, a much smaller value than the kcat (5.3 s−1) displayed by the HIDH protein toward the same substrate. The half-life period of 2,7,4′-trihydroxyisoflavanone under this condition was 6.3 h.
Site-Directed Mutagenesis of Soybean HID
During ester hydrolysis by carboxylesterases, the amino acid residues of an oxyanion hole stabilize the negatively charged intermediate, and the nucleophilic hydroxyl of the Ser residue in the catalytic triad acts as the catalyst. To examine whether these residues also play catalytic roles in the HID reaction, mutations were introduced into the catalytic triad (Thr-164, Asp-263, and His-295) and oxyanion hole (Gly-78 and Gly-79) of soybean HIDH protein, and the purified recombinant proteins with six His residues were assayed for the activity. The mutants (D263N and H295A), in which a hydrogen bond between Asp-263 and His-295 within the triad was destroyed completely, lost both HID and carboxylesterase activity (Table III). When the Thr-164 residue of the triad was replaced by Ser (a more general amino acid of the triad in hydrolases) and Ala (without hydroxyl), both HID and carboxylesterase activity were reduced to 3% to 4% and 1% of the wild type, respectively. Mutation in the oxyanion hole (G78A and G79A) also reduced both activities to about 10% of the wild type. Because the relative ratios of the reduction of HID and carboxylesterase activity with mutant proteins were almost identical (Table III), it was strongly suggested that these amino acid residues function in a similar manner during the catalytic process of both dehydration and ester hydrolysis.
Table III.
Specific HID and carboxylesterase activity of recombinant soybean HIDH protein and site-directed mutants
| 2,7,4′-Trihydroxyisoflavanone Dehydratase Activity | p-Nitrophenyl Butyrate Hydrolyzing Activity | |
|---|---|---|
| %a | %c | |
| Soybean HIDH | 100b | 100d |
| G78A | 12 | 14 |
| G79A | 14 | 12 |
| T164S | 4 | 3 |
| T164A | 1 | 1 |
| D263N | 0 | 0 |
| H295A | 0 | 0 |
Average of two independent experiments (maximum deviation was about 10%). Specific activity was determined using 100 μm phenolic substrate.
Specific activity is 43.6 nkatal mg−1 protein.
Average of two independent experiments (maximum deviation was about 40%).
Specific activity is 730 pkatal mg−1 protein.
Phylogenetic Analysis of HIDs and the Carboxylesterase Gene Family
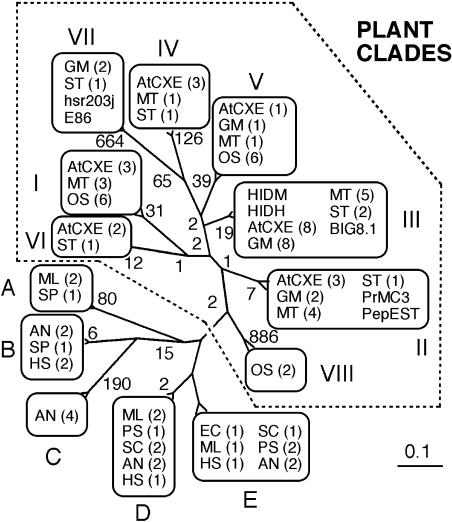
The protein-protein BLAST search revealed that HID proteins had 30% to 40% identity with Nicotiana tabacum hsr203j (accession no. CAA54393; Pontier et al., 1994), Pisum sativum E86 (accession no. BAA85654; Ichinose et al., 2001), and some Arabidopsis (Arabidopsis thaliana) putative proteins. The genes of all these proteins are categorized into the plant carboxylesterase gene family, which was recently reported to consist of 7 clades by phylogenetic analysis of 20 Arabidopsis genes (representing 6 clades) and other plant carboxylesterase-like genes (Marshall et al., 2003). We extended the analysis of HID-like carboxylesterase proteins into those from evolutionarily more divergent organisms, including microorganisms and human (Homo sapiens). The phylogenetic tree comprising all the genes examined (102 sequences) is complicated (see Supplemental Figure 1), and a simplified scheme is shown in Figure 3. The whole carboxylesterase family contained monophyletic plant clades and five other clades (tentatively named clades A–E) to which genes of microbial and human origins belong. Except for clade C, consisting of Aspergillus nidulans genes, nonplant clades contain genes from evolutionarily separated species: For example, clade D includes genes of eubacteria (Mesorhizobium loti and Pseudomonas syringae), fungi (A. nidulans and Schizosaccharomyces pombe), and human. The phylogenetic relationships among plant carboxylesterases are essentially in agreement with the reported classification (Marshall et al., 2003), and eight clades are identified. The genes of Arabidopsis and rice (Oryza sativa), two species for which genome information is available, as well as those in ESTs of leguminous plants (M. truncatula and soybean), are scattered into diverse clades. HIDs are located in a small group of proteins from leguminous plants in clade III, which also contains Arabidopsis, rice, and potato (Solanum tuberosum) genes.
Figure 3.
The phylogenetic relationship of G. echinata HIDM and the carboxylesterase family in various organisms. The complete phylogenetic tree comprising all the proteins examined (102 sequences) is shown in Supplemental Figure 1. The nomenclature of proteins in the figure is either after the names used in the original reports or abbreviated names of the species producing the proteins, followed by the numbers of distinct sequences in parentheses: AtCXE, Arabidopsis; AN, Aspergillus nidulans; EC, Escherichia coli; GM, Glycine max; HS, Homo sapiens; ML, Mesorhizobium loti; MT, Medicago truncatula; OS, Oryza sativa; PS, Pseudomonas syringae; SC, Streptomyces coelicolor; SP, Schizosaccharomyces pombe; ST, Solanum tuberosum; BIG8.1, Vitis vinifera BIG8.1; E86, Pisum sativum E86; hsr203j, Nicotiana tabacum hsr203j; PepEST, Capsicum annuum PepEST; PrMC3, Pinus radiata PrMC3. Plant clades are enclosed by a dotted line.
DISCUSSION
In this study, two cDNAs encoding HIDM of G. echinata and HIDH of soybean were cloned. HIDM and HIDH display different substrate specificities to 2-hydroxyisoflavanones. The substrate preferences reflect the structures of isoflavonoids contained in each plant species. The spontaneous dehydration of the substrates was slow compared to the enzyme-catalyzed reaction, suggesting that dehydration of 2-hydroxyisoflavanones to form isoflavones in plant cells is primarily enzyme dependent. However, a prolonged incubation of the substrates yields isoflavones, and the calculated ratio (23%) of spontaneous dehydration of 2,7,4′-trihydroxyisoflavanone in 2 h from the rate constant (pH 7.5, 30°C) can explain the previous observation of 38% daidzein and 62% 2,7,4′-trihydroxyisoflavanone among the isoflavonoids produced from 7,4′-dihydroxyflavanone by IFS with recombinant yeast microsomes (Akashi et al., 1999). Nonenzymatic slow production of isoflavones from 2-hydroxyisoflavanones may be an alternative process in plant organs where rapid isoflavone production is not necessary.
The specific activity of the HID reaction by recombinant G. echinata HIDM protein is about 1,200 times higher than that of the crude extract of G. echinata, and both the recombinant protein and the crude extract displayed a very high preference for 2,7-dihydroxy-4′-methoxyisoflavanone. Therefore, the activity in the crude extract should be attributed to the HIDM protein. Also, a preliminary RT-PCR analysis with G. echinata cells revealed a constitutive as well as an increased accumulation of HIDM mRNA upon yeast extract treatment in a coordinated manner with IFS and HI4′OMT mRNAs (Supplemental Fig. 2), coinciding with the constitutive and elicitor-induced production of the isoflavonoids (Nakamura et al., 1999). The HIDM-type enzyme should also be involved in the biosynthesis of biochanin A (5,7-dihydroxy-4′-methoxyisoflavone) in legumes such as chickpea (Barz and Welle, 1992), because, in a preliminary experiment, HIDM did actually dehydrate 2,5,7-trihydroxy-4′-methoxyisoflavanone into biochanin A (data not shown). On the other hand, both the soybean extract and recombinant HIDH protein showed high substrate preferences for 2,5,7,4′-tetrahydroxyisoflavanone and also displayed significant activity toward 2,7,4′-trihydroxyisoflavanone and 2,7-dihydroxy-4′-methoxyisoflavanone. Thus, the HID activity of the crude extract of soybean could be attributed to HIDH protein. However, soybean EST libraries exhibited another HID candidate giving rise to the fragment clone BM177194, although in this study, its full sequence could not be obtained. Because this putative protein seems to be more similar to HIDM than to HIDH, its substrate specificity and expression in soybean will be quite interesting. Recent comprehensive studies on the expression of two soybean IFS genes have indicated distinct physiological functions of paralogous genes in the tissue-specific and pathogen- or rhizobia-responsive production of isoflavones (Dhaubhadel et al., 2003; Subramanian et al., 2004). The biochemical characteristics and physiological functions of similar enzymes encoded by multiple genes in secondary metabolism are challenging subjects to be examined in the future.
The characteristics of the purified P. lobata HID (Hakamatsuka et al., 1998) agree with the properties of soybean HIDH, except for the unusual Km value of P. lobata HID for 2,7,4′-trihydroxyisoflavanone (about 64-fold higher than that of soybean HIDH). Thus, the molecular mass (38 kD) of P. lobata protein is similar to the calculated average Mr (35,137) of soybean HIDH protein, and the specific activity of P. lobata HID toward 2,7,4′-trihydroxyisoflavanone (56.8 nkatal mg−1) is roughly identical to the activity of recombinant soybean HIDH protein (Vmax value, 143 nkatal mg−1). Characteristically, inhibition experiments indicated that His residues in P. lobata HID are important for the activity, and this is consistent with our finding that His is the central residue for the HID catalysis. Therefore, HID of P. lobata is likely to be a protein of the carboxylesterase family.
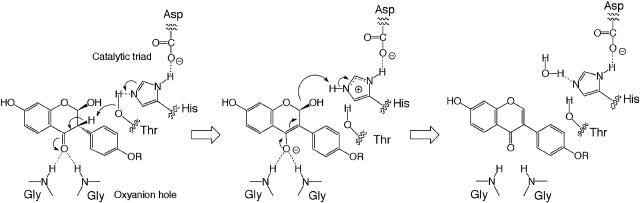
1,2-Dehydratases are grouped into the hydro-lyases (EC 4.2.1), which contain several types of proteins, but no hydrolase-like proteins are found among them. HID reaction is the first example of 1,2-dehydration by proteins classified into the carboxylesterases of the hydrolase family. Both mammalian carboxylesterases and plant carboxylesterase-like proteins have the α/β-hydrolase fold common to Ser hydrolases that carries an oxyanion hole and the catalytic triad. In fact, site-directed mutagenesis of HIDH protein indicated that amino acid residues comprising the oxyanion hole and the catalytic triad are important for both HID and ester hydrolysis activities (Table III). Considering the established catalytic processes of carboxylesterase (Blow et al., 1969; Kraut, 1977) and microbial scytalone dehydratase, which is known to use a catalytic His residue for the dehydration of the substrate resembling 2-hydroxyisoflavanone (Lundqvist et al., 1994; Zheng and Bruice, 1998), a possible scheme for the HID reaction can be drawn, as in Figure 4. In this scheme, the nucleophilic oxygen of Thr in the catalytic triad acts as a base to abstract hydrogen from C-3 of the substrate, and the C-4 carbonyl is enolized to yield a negatively charged intermediate stabilized by the oxyanion hole. Subsequent abstraction of the hydroxyl from C-2 of the intermediate by the positively charged His residue yields an isoflavone. The initial hydrogen abstraction at C-3 is not necessarily accomplished by the Thr: The His can substitute for it. In the HID reaction shown in Figure 4, the stereochemistry of the 1,2-elimination of water is syn-oriented. This is in agreement with the predicted structure of the IFS reaction product, i.e. (2R, 3S)-2-hydroxyisoflavanone (Sawada et al., 2002), and the syn-elimination of water from this enantiomer. Further studies are, however, indispensable to establish the mechanism of the reaction.
Figure 4.
Putative reaction mechanism for HID. The amino acid residues of the catalytic triad and the oxyanion hole cooperatively eliminate water from a 2-hydroxyisoflavanone to produce an isoflavone. His, instead of Thr, of the catalytic triad could perform the initial proton abstraction (see “Discussion”).
The phylogenetic tree of the carboxylesterase family (Fig. 3; Supplemental Fig. 1) implies an interesting evolutionary process of this group of proteins. Several genes in the same clades are distributed to broad organisms of eubacteria, fungi, and human, and thus thought to be very old: primitive in evolution, in other words. However, whereas human carboxylesterases are known to catalyze the hydrolysis of a variety of ester-containing xenobiotics, fatty acids, and steroids (Satoh and Hosokawa, 1995), no clear intrinsic biochemical functions of carboxylesterases in microbes and plants have been identified. A very striking feature is that plant carboxylesterases are monophyletic, clearly distinguished from those of other higher and lower organisms. This scheme suggests that some specific function(s) that arose in an ancestral protein of the plant carboxylesterase family was significant, and the proteins were conserved and diverged during plant evolution. Regarding their physiological roles, it is noteworthy that P. sativum E86 (Ichinose et al., 2001), N. tabacum hsr203j (Pontier et al., 1994; Kiba et al., 2003; Takahashi et al., 2004), and grapevine (Vitis vinifera) BIG8.1 (accession no. AAN77692; Bézier et al., 2002) are induced in response to pathogenesis, and the L. japonicus hsr203J homolog (gene identity MDP051d06_f) is induced during the early stage of infection by symbiotic Mesorhizobium loti (Kouchi et al., 2004). Also, homologous sequences are also found in the EST database of Pinus radiata male cones (Walden et al., 1999) and a root hair-enriched M. truncatula cDNA library (Covitz et al., 1998). HIDs are the only proteins of the plant carboxylesterase family whose catalytic function is now clearly demonstrated, and, surprisingly, it is not ester hydrolysis. HIDs must have emerged within the clade III carboxylesterases in leguminous plants to dehydrate 2-hydroxyisoflavanones yielding isoflavones, the ecophysiologically significant substances such as biosynthetic intermediates of antimicrobial phytoalexins and chemical signals to nitrogen-fixing bacterial symbionts. The roles of other carboxylesterases in metabolic processes related to plant defense responses and/or organ-specific production of phytochemicals must be fascinating subjects for future studies.
Following the cloning of IFS (Akashi et al., 1999) and HI4′OMT (Akashi et al., 2003), together with characterization of chalcone polyketide reductase (Welle et al., 1991; Akashi et al., 1997) and type II chalcone isomerase (Shimada et al., 2003) producing 5-deoxyflavonoids, this work completes identification of enzymes of the legume-specific 5-deoxy- and either 4′-methoxy- or 4′-hydroxy-isoflavone pathways branching from general flavonoid metabolism. Among these, IFS should have evolved from the ancestral P450, which is also the ancestor of flavone synthase II, a P450 of the general flavonoid pathway, through critical events like the introduction of limited numbers of key amino acid residues for the unique aryl migration (Sawada et al., 2002; Y. Sawada and S. Ayabe, unpublished data). Also, type II chalcone isomerase must have emerged in the Leguminosae genome from the general type I chalcone isomerase through local gene duplication and subsequent mutations, resulting in the expansion of substrate specificity to 6′-deoxychalcone (Shimada et al., 2003). Extensive gene clusters of chalcone isomerases and other legume-specific (e.g. chalcone polyketide reductase and isoflavone reductase), as well as general enzymes (e.g. dihydroflavonol-4-reductase), in a model legume L. japonicus (Shimada et al., 2003; N. Shimada, T. Aoki, S. Sato, Y. Nakamura, S. Tabata, and S. Ayabe, unpublished data) further support the idea that gene duplication and mutation to gain new catalytic functions caused the diversity of secondary metabolism (Pichersky and Gang, 2000). During these processes, enzymes of primary metabolism also may have been recruited to secondary metabolism, as evidenced, for example, by the fact that chalcone polyketide reductase of the legume 5-deoxyflavonoid pathway and analogous codeinone reductase of morphinan alkaloid biosynthesis in Papaveraceae (Unterlinner et al., 1999) are members of the superfamily of aldo-keto reductases participating mainly in sugar metabolism. The findings in this study add another example of the recruitment: from hydrolases to hydro-lyases of secondary metabolism. In this respect, it is interesting to note that Ser carboxypeptidase-like proteins catalyze the transacylation in plant natural product biosynthesis (Milkowski and Strack, 2004; Stout and Chapple, 2004). The proteins also belong to the class of α/β hydrolases, and the residues composing the catalytic triad are assumed to be important for the acyl-transfer reaction. Thus, the vast array of plant proteins classified as α/β hydrolases can be interesting objectives to experimentally examine the recruitment mechanism during the molecular evolution of secondary metabolism. As the initial step to such a direction of research, we are planning to identify the biochemical basis for HID proteins to distinguish hydrolysis and dehydration through protein-engineering approaches.
MATERIALS AND METHODS
Chemical Materials
Daidzein and genistein were obtained from Extrasynthése (Genay, France), and p-nitrophenyl butyrate from Sigma (St. Louis). Formononetin was from our laboratory stock. 2,7,4′-Trihydroxyisoflavanone (Ayabe et al., 2002), 2,7-dihydroxy-4′-methoxyisoflavanone (Akashi et al., 2003), and 2,5,7,4′-tetrahydroxyisoflavanone (Akashi et al., 1999) were prepared as described.
Preparation of Plant Cell-Free Extracts
Glycyrrhiza echinata cells (Ak-1 line; Akashi et al., 1999) were elicited with 0.2% (w/v) yeast extract (Invitrogen, Carlsbad, CA) for 24 h (Nakamura et al., 1999). Soybean (Glycine max L. Merr. cv Mikawashima; Tohoku, Tochigi, Japan) seeds were sown on filter paper in an Erlenmeyer flask, and seedlings were grown under 12-h-light/12-h-dark cycles at room temperature for 1 week. Filtered homogenates of these plant materials (10 g) in 10 mL of 100 mm potassium phosphate buffer, pH 7.5, containing 10% Suc and 14 mm 2-mercaptoethanol were centrifuged at 10,000g for 10 min. The ammonium sulfate fraction (30%–80% saturation) of the 10,000g supernatant, which had been treated with 2.5 g Dowex 1-X2 (20 min), was suspended in the same buffer for the initial extraction and, after desalting by passing through a Sephadex G-25 column, used as the enzyme (approximately 600 μg protein mL−1).
Enzyme Assays
The buffer used was 100 mm potassium phosphate, pH 7.5, supplemented with 10% Suc and 14 mm 2-mercaptoethanol. For the HID assay, 5 nmol of each the 2-hydroxyisoflavanone sample in 2 μL 2-methoxyethanol was incubated with the enzyme preparation in a total volume of 50 μL at 30°C for 5 min. The ethyl acetate extract of the reaction mixture was analyzed by HPLC. HPLC conditions for daidzein and formononetin were the same as described (Akashi et al., 2003). Genistein was analyzed using 50% methanol in water as the HPLC solvent. Specific activities were determined for cell-free extracts of G. echinata and soybean (approximately 10–100 μg protein) and also for purified recombinant HIDM and HIDH proteins (approximately 20–800 ng) from the peak areas of the isoflavones calibrated with those of standard samples. To determine the kinetic parameters, 2,7-dihydroxy-4′-methoxyisoflavanone (5, 20, 50, 100, and 200 μm), 2,7,4′-trihydroxyisoflavanone (5, 20, 50, 100, 200, and 400 μm), or 2,5,7,4′-tetrahydroxyisoflavanone (10, 20, 50, 100, and 200 μm) was incubated with the recombinant protein (HIDH [50 ng] or HIDM [21 ng for 2,7-dihydroxy-4′-methoxyisoflavanone dehydratase activity and 820 ng for 2,7,4′-trihydroxyisoflavanone and 2,5,7,4′-tetrahydroxyisoflavanone dehydratase activities]) in a total volume of 50 μL at 30°C for 5 min. Km and kcat values were calculated using a Lineweaver-Burk plot. In all the assays, enzymatically produced isoflavones were determined after the correction of preexisting and spontaneously formed isoflavones in the samples with the controls using the same concentrations of substrates in the buffer. To measure the rate of spontaneous dehydration, 2,7,4′-trihydroxyisoflavanone (0.1, 1, 10, 50, and 100 μm) was incubated in the buffer in a total volume of 50 μL at 30°C for 0.5, 1, 2, 4, 8, 12, and 20 h. Daidzein production was determined by HPLC after the correction with the daidzein concentration at time 0. The velocity of dehydration was calibrated from the first 30-min incubations when the daidzein formation was linear with all the substrate concentrations tested. The initial velocities were proportional to the substrate concentrations, and the rate constant of dehydration was calculated from these values. The carboxylesterase activity of recombinant HID proteins was determined by the rate of p-nitrophenol production from p-nitrophenyl butyrate (Heymann and Mentlein, 1981). Porcine liver carboxylesterase (Sigma) was used as a positive control.
Functional Expression Fractionation Screening of G. echinata HIDM
The cDNA expression library (Akashi et al., 2003) constructed from yeast extract-treated G. echinata cells was the starting material. Five independent pools of cDNA fractions, each containing about 30,000 Escherichia coli clones, were prepared from the mother plate. Expression of protein and preparation of the crude extract of E. coli were performed as previously described (Akashi et al., 2003). 2,7,4′-Trihydroxyisoflavanone (0.4 nmol) dissolved in 2 μL 2-methoxyethanol was preincubated with recombinant G. echinata HI4′OMT (50 ng; Akashi et al., 2003) in the presence of 0.4 nmol [methyl-14C]-S-adenosyl-l-Met (2.26 GBq mmol−1; Amersham Biosciences, Buckinghamshire, UK) in a total volume of 50 μL at 30°C for 3 min. The crude extract (100 μL) of the E. coli pool was added to the mixture and incubated at 30°C for 10 min. The reaction was terminated by the addition of ethyl acetate, then the ethyl acetate extract of the mixture was subjected to silica gel thin-layer chromatography (LK6DF; Whatman, Maidstone, UK); solvent, chloroform:acetone:25% aqueous ammonia solution (70: 29: 1, v/v), 2,7-dihydroxy-4′-methoxyisoflavanone (RF 0.15), and formononetin (RF 0.30); and analyzed by an image analyzer (Typhoon 8600; Amersham Biosciences). A positive pool that produced [14C]formononetin was subdivided into 10 pools of smaller size (about 3,000 clones/pool) for the next round of screening. The fractionation of positive pools and assay was repeated three times. The nucleotide sequence of the isolated cDNA (HIDM) was determined.
EST Database Search and Cloning of the Soybean HIDH
The EST databases of soybean (http://www.tigr.org/tdb/tgi/gmgi), Medicago truncatula (http://www.tigr.org/tdb/tgi/mtgi), and Lotus japonicus (http://www.kazusa.or.jp/en/plant/lotus/EST; Asamizu et al., 2000) were searched for cDNAs homologous to HIDM. Two specific primers containing NdeI or BamHI sites (underlined) were designed from the HIDM-like sequence of soybean (accession no. TC98460): TC98-F, 5′-GTCATATGGCGAAGGAGATAGTGAA-3′ and TC98-R, 5′-AGGGATCCATCAAACCAGAAAAGA-3′. Total RNA was isolated from soybean seedlings using RNeasy Plant Mini kit (Qiagen, Hilden, Germany). cDNAs were synthesized by Ready-To-Go T-Primed First Strand kit (Amersham Biosciences). cDNA (HIDH) obtained by the RT-PCR with the primers and soybean cDNAs as the template was cloned into pT7Blue T-vector (Novagen, Madison, WI).
Expression and Purification of Recombinant Proteins
Two primers containing NdeI or BamHI sites (underlined) were designed from G. echinata HIDM: HID-F, 5′-GTCATATGGCTTCTTCAACCTCAAC-3′; HID-R, 5′-CTGGATCCTCAAACAAGGAAGGAAG-3′. The coding region of HIDM was amplified by PCR with KOD polymerase (Toyobo, Tokyo) using HIDM cDNA as template. The NdeI-BamHI fragments of the PCR product from G. echinata HIDM and of the soybean HIDH were subcloned into corresponding sites of pET28a (Novagen) to produce pET28a-HIDM and pET28a-HIDH. The vectors were transformed into E. coli BL21(DE3), and recombinant proteins were expressed and purified as described (Akashi et al., 2003).
Site-Directed Mutagenesis
Mutagenic primers (mutated sites in each primer are underlined) were designed from soybean HIDH: T164S primer, 5′-GTAGGAGGTGAATCCAGCGGTGCTAAC-3′; T164A primer, 5′-GTAGGAGGTGAAGCCAGCGGTGCTAAC-3′; D263N primer, 5′-CACTGGCAAAAACGAGTTCAGAGAC-3′; H295A primer, 5′-GGCGATGAGGAGGCTGCTTTCCAGCTC-3′; G78A primer, 5′-CTCTACTTCCACGCTGGCGCCTTTTG-3′; and G79A primer, 5′-CTCTACTTCCACGGTGCCGCCTTTTG-3′. Point mutations were introduced using pET-28a+HIDH and Transformer Site-Directed Mutagenesis kit (CLONTECH Laboratories, Palo Alto, CA). Transformation of the vectors into E. coli BL21(DE3) expression and protein purification were performed as described above.
Phylogenetic Analysis
The DNA sequences used for phylogenetic analysis were selected using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST). Those were: (1) DNAs encoding proteins similar to HIDM (having E-values smaller than 1 × 10−5 at amino acid level) in the National Center for Biotechnology Information genome database (http://www.ncbi.nih.gov/Genomes) of Mesorhizobium loti (5 sequences), E. coli K12 (1 sequence), Pseudomonas syringae pv. tomato str. DC3000 (3 sequences), Streptomyces coelicolor (3 sequences), Aspergillus nidulans FGSC A4 (10 sequences), Schizosaccharomyces pombe (2 sequences), human (Homo sapiens; 4 sequences), rice (Oryza sativa; 14 sequences), and Arabidopsis (Arabidopsis thaliana; 20 sequences); (2) cDNAs encoding >200 N-terminal amino acids having E-values smaller than 1 × 10−5 compared to HIDM from the EST libraries in The Institute for Genomic Research Gene Indices (http://www.tigr.org/tdb/tgi/plant.shtml) of soybean (14 sequences), M. truncatula (14 sequences), and potato (Solanum tuberosum; 6 sequences); and (3) cDNAs reported to encode carboxylesterase-like protein of higher plants, i.e. Pinus radiata PrMC3 (accession no. AF110333; Walden et al., 1999), Pisum sativum E86 (Ichinose et al., 2001), Nicotiana tabacum hsr203j (Pontier et al., 1994), Capsicum annuum PepEST (accession no. AAF77578; Kim et al., 2001), grapevine (Vitis vinifera) BIG8.1 (Bézier et al., 2002), and G. echinata HIDM. A total of 102 amino acid sequences were analyzed using the ClustalW program (Thompson et al., 1994) of the DNA Data Bank of Japan (Shizuoka, Japan), and a neighbor-joining tree was produced from the results of 1,000 bootstrap replicates. The tree was displayed by TreeView (Page, 1996) or NJPlot software (Perrière and Gouy, 1996).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AB154414 and AB154415.
Supplementary Material
Acknowledgments
The authors thank Dr. Yuji Sawada and Takuro Yamashita (Nihon University, Japan) for technical assistance and useful discussions.
This work was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research [C] nos. 15510183 and 14540603), the Ministry of Education, Culture, Sports, Science and Technology of Japan (21st Century Center of Excellence Program), and New Energy and Industrial Technology Development Organization (the Green Biotechnology Program).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056747.
References
- Akashi T, Aoki T, Ayabe S (1999) Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 121: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Saito N, Hirota H, Ayabe S (1997) Anthocyanin-producing dandelion callus as a chalcone synthase source in recombinant polyketide reductase assay. Phytochemistry 46: 283–287 [DOI] [PubMed] [Google Scholar]
- Akashi T, Sawada Y, Aoki T, Ayabe S (2000) New scheme of the biosynthesis of formononetin involving 2,7,4′-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci Biotechnol Biochem 64: 2276–2279 [DOI] [PubMed] [Google Scholar]
- Akashi T, Sawada Y, Shimada N, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44: 103–112 [DOI] [PubMed] [Google Scholar]
- Aoki T, Akashi T, Ayabe S (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J Plant Res 113: 475–488 [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) Generation of 7137 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res 7: 127–130 [DOI] [PubMed] [Google Scholar]
- Ayabe S, Akashi T, Aoki T (2002) Cloning of cDNAs encoding P450s in the flavonoid/isoflavonoid pathway from elicited leguminous cell cultures. Methods Enzymol 357: 360–369 [DOI] [PubMed] [Google Scholar]
- Barz W, Welle R (1992) Biosynthesis and metabolism of isoflavones and pterocarpan phytoalexins in chickpea, soybean and phytopathogenic fungi. In HA Stafford, RK Ibrahim, eds, Recent Advances in Phytochemistry, Vol. 26. Phenolic Metabolism in Plants. Plenum Press, NY, pp 139–164
- Bézier A, Lambert B, Baillieul F (2002) Cloning of a grapevine Botrytis-responsive gene that has homology to the tobacco hypersensitivity-related hsr203J. J Exp Bot 53: 2279–2280 [DOI] [PubMed] [Google Scholar]
- Blow DM, Birktoft JJ, Hartley BS (1969) Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221: 337–340 [DOI] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR (1998) Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol 117: 1325–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick PM (1993) Isoflavonoids. In JB Harborne, ed, The Flavonoids. Advances in Research since 1986. Chapman and Hall, London, pp 117–238
- Dhaubhadel S, McGarvey BD, Williams R, Gijzen M (2003) Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol Biol 53: 733–743 [DOI] [PubMed] [Google Scholar]
- Dixon RA (1999) Isoflavonoids: biochemistry, molecular biology, and biological functions. In U Sankawa, ed, Comprehensive Natural Products Chemistry, Vol 1. Polyketides and Other Secondary Metabolites Including Fatty Acids and Their Derivatives. Elsevier, Amsterdam, pp 773–823
- Hakamatsuka T, Mori K, Ishida S, Ebizuka Y, Sankawa U (1998) Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata. Phytochemistry 49: 497–505 [Google Scholar]
- Hashim MF, Hakamatsuka T, Ebizuka Y, Sankawa U (1990) Reaction mechanism of oxidative rearrangement of flavanone in isoflavone biosynthesis. FEBS Lett 271: 219–222 [DOI] [PubMed] [Google Scholar]
- Heymann E, Mentlein R (1981) Carboxylesterases-amidases. Methods Enzymol 77: 333–344 [DOI] [PubMed] [Google Scholar]
- Ichinose Y, Hisayasu Y, Sanematsu S, Ishiga Y, Seki H, Toyod K, Shiraishi T, Yamada T (2001) Molecular cloning and functional analysis of pea cDNA E86 encoding homologous protein to hypersensitivity-related hsr203J. Plant Sci 160: 997–1006 [DOI] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau SM, O'Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18: 208–212 [DOI] [PubMed] [Google Scholar]
- Kiba A, Tomiyama H, Takahashi H, Hamada H, Ohnishi K, Okuno T, Hikichi Y (2003) Induction of resistance and expression of defense-related genes in tobacco leaves infiltrated with Ralstonia solanacearum. Plant Cell Physiol 44: 287–295 [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee HH, Ko MK, Song CE, Bae CY, Lee YH, Oh BJ (2001) Inhibition of fungal appressorium formation by pepper (Capsicum annuum) esterase. Mol Plant Microbe Interact 14: 80–85 [DOI] [PubMed] [Google Scholar]
- Kochs G, Grisebach H (1986) Enzymic synthesis of isoflavones. Eur J Biochem 155: 311–318 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu G-J, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11: 263–274 [DOI] [PubMed] [Google Scholar]
- Kraut J (1977) Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem 46: 331–358 [DOI] [PubMed] [Google Scholar]
- Lundqvist T, Rice J, Hodge CN, Basarab GS, Pierce J, Lindqvist Y (1994) Crystal structure of scytalone dehydratase: a disease determinant of the rice pathogen, Magnaporthe grisea. Structure 2: 937–944 [DOI] [PubMed] [Google Scholar]
- Manco G, Camardella L, Febbraio F, Adamo G, Carratore V, Rossi M (2000) Homology modeling and identification of serine 160 as nucleophile of the active site in a thermostable carboxylesterase from the archaeon Archaeoglobus fulgidus. Protein Eng 13: 197–200 [DOI] [PubMed] [Google Scholar]
- Marshall SD, Putterill JJ, Plummer KM, Newcomb RD (2003) The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol 57: 487–500 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Strack D (2004) Serine carboxypeptidase-like acyltransferases. Phytochemistry 65: 517–524 [DOI] [PubMed] [Google Scholar]
- Nakamura K, Akashi T, Aoki T, Kawaguchi K, Ayabe S (1999) Induction of isoflavonoid and retrochalcone branches of the flavonoid pathway in cultured Glycyrrhiza echinata cells treated with yeast extract. Biosci Biotechnol Biochem 63: 1618–1620 [DOI] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW (1999) α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9: 732–737 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al (1992) The α/β hydrolase fold. Protein Eng 5: 197–211 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Pontier D, Godiard L, Marco Y, Roby D (1994) hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Satoh T, Hosokawa M (1995) Molecular aspects of carboxylesterase isoforms in comparison with other esterases. Toxicol Lett 82/83: 439–445 [DOI] [PubMed] [Google Scholar]
- Sawada Y, Kinoshita K, Akashi T, Aoki T, Ayabe S (2002) Key amino acid residues required for aryl migration catalyzed by the cytochrome P450 2-hydroxyisoflavanone synthase. Plant J 31: 555–564 [DOI] [PubMed] [Google Scholar]
- Shimada N, Akashi T, Aoki T, Ayabe S (2000) Induction of isoflavonoid pathway in the model legume Lotus japonicus: molecular characterization of enzymes involved in phytoalexin biosynthesis. Plant Sci 160: 37–47 [DOI] [PubMed] [Google Scholar]
- Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol 131: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367: 146–150 [DOI] [PubMed] [Google Scholar]
- Stout J, Chapple C (2004) The phenylpropanoid pathway in Arabidopsis: lessons learned from mutants in sinapate ester biosynthesis. In JT Romeo, ed, Recent Advances in Phytochemistry, Vol. 38. Secondary Metabolism in Model Systems. Elsevier, Amsterdam, pp 39–67
- Subramanian S, Hu X, Lu G, Odelland JT, Yu O (2004) The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol Biol 54: 623–639 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Uehara Y, Berberich T, Ito A, Saitoh H, Miyazaki A, Terauchi R, Kusano T (2004) A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J 40: 586–595 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterlinner B, Lenz R, Kutchan TM (1999) Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy Papaver somniferum. Plant J 18: 465–475 [DOI] [PubMed] [Google Scholar]
- Walden AR, Walter C, Gardner RC (1999) Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol 121: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle R, Schröder G, Schiltz E, Grisebach H, Schröder J (1991) Induced plant responses to pathogen attack. Analysis and heterologous expression of the key enzyme in the biosynthesis of phytoalexins in soybean (Glycine max L. Merr. cv. Harosoy 63). Eur J Biochem 196: 423–430 [DOI] [PubMed] [Google Scholar]
- Zheng YJ, Bruice TC (1998) Role of a critical water in scytalone dehydratase-catalyzed reaction. Proc Natl Acad Sci USA 95: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.