ABSTRACT
Tuberculosis (TB) is one of the leading causes of death due to an infectious disease. The rise of multi-drug resistance (MDR) in Mycobacterium tuberculosis (Mtb), the causative agent of TB, presents a significant obstacle to TB control. While human studies report dysregulated immune responses during MDR TB, a clear understanding of the host-pathogen interactions of MDR Mtb is lacking. Here, we studied the immune responses induced by Mtb strains carrying two of the most common rifampicin drug-resistance (RDR)-conferring single-nucleotide polymorphisms (SNPs) in the RNA polymerase gene of Mtb, which accounts for nearly 90% of drug-resistance mutations found clinically in Mtb. During Mtb infection of primary human macrophages, we found that pro-inflammatory cytokine production was reduced upon infection with Mtb carrying the H445Y SNP but not the S450L SNP. Using a mouse model, we also characterized the host immune response in vivo following infection. Despite similar establishment of Mtb infection in the lung and dissemination to the peripheral organs, we show that infection with the RDR Mtb rpoB-H445Y strain, but not with the rpoB-S450L strain, resulted in a suppressed lung myeloid and lymphoid immune responses through type I IFN-dependent pathways, relative to wt Mtb. This suppressed host immunity had functional consequences in limiting control of RDR Mtb strains harboring rpoB-H445Y and led to worse pathology during chronic TB disease. Collectively, our results suggest that disease pathogenesis may be associated with specific RDR mutations in Mtb, which may differentially regulate immune responses.
IMPORTANCE
This study highlights the impact of specific rifampicin-resistance-conferring mutations on the host immune response to Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB). Clinical reports have previously suggested that multi-drug-resistant) TB patients exhibit altered peripheral immune responses as compared with their drug-sensitive TB counterparts. The murine model of infection with Mtb strains carrying drug-resistance-conferring mutations recapitulated these findings and allowed us to mechanistically interrogate the pathways responsible for driving the divergent immune responses. Our findings underscore the need for greater investigation into bacterial heterogeneity to better appreciate the diversity in host-pathogen interactions during TB disease.
KEYWORDS: Mycobacterium tuberculosis, type I interferons, inflammation, drug-resistant mutations, lung infection
INTRODUCTION
Tuberculosis (TB) is a major global health threat, with over 10 million people estimated to have developed active disease in 2021 (1). Emergence of Mycobacterium tuberculosis (Mtb) strains resistant to the frontline antibiotic, rifampicin, has significantly limited global efforts to control TB. In 2021 alone, nearly 600,000 TB cases were found to be rifampicin drug resistant (RDR), which requires expanded use of more toxic second-line antibiotics. A bottleneck to controlling RDR Mtb infection is our lack of clear understanding of the disease pathogenesis following DR Mtb infection. The immune responses and clinical outcomes in individuals infected with DR Mtb can differ when compared with TB patients infected with drug-susceptible (DS) Mtb. In particular, while there is evidence of impaired CD4+ T helper type 1 (Th1) responses and dysregulation of the Th1/T helper type 2 (Th2) balance in humans with multi-drug-resistant (MDR) TB (2 – 5), the underlying mechanisms that contribute to this impairment are not known.
The two most common DR mutations that confer resistance to rifampicin are H445Y and S450L in the RNA polymerase (rpoB) subunit of Mtb, which account for nearly 90% of drug-resistance mutations found clinically in Mtb (6 – 9). We recently showed that MDR Mtb harboring the mutation rpoB-H445Y, but not rpoB-S450L, differentially expressed Mtb surface lipids and induced macrophage reprogramming via a type I interferon (IFN) response in murine macrophages in vitro (10). In the current study, we investigated how drug-resistance-conferring mutations may impact the immune response following DR Mtb infection in vivo. We found that Mtb harboring the rpoB-H445Y SNP drove reduced pro-inflammatory cytokine production, namely IL-1, in infected human macrophages. Using a mouse model, we showed that Mtb infection with a strain carrying the H445Y SNP, but not the S450L SNP, resulted in impaired recruitment of myeloid and lymphoid cells in vivo. Importantly, we show that the suppressed immune response in the lung associated with infection with the rpoB-H445Y strain is dependent on type I IFN signaling mechanisms. Thus, our findings highlight how RDR mutations in Mtb may trigger type I IFNs to suppress immune responses and drive divergent immune responses and pathology when compared with DS Mtb. Our results contribute to our understanding of immune responses to major RDR Mtb infections and provide models and new avenues to mechanistically interrogate immunomodulation in DR and MDR TB patients.
RESULTS
The presence of drug-resistant rpoB-H445Y mutation drives differential pro-inflammatory cytokine production in Mtb-infected murine and human macrophages
We have previously reported that multi-drug-resistant clinical isolates, such as W_7642, harboring the specific drug-resistance-conferring mutation, H445Y, but not S450L in the RNA polymerase (rpoB) subunit of Mtb, modulate Mtb interactions with murine macrophages and drive differential cytokine production in macrophages (10), especially limiting interleukin-1 (IL-1) and increasing IFN production. We generated a collection of independently isolated drug-resistant mutants through plating on selective media containing rifampicin (Fig. S1). Comparative whole-genome sequence analysis between these Mtb mutants with the parental wt HN878 Mtb strain revealed various non-overlapping non-synonymous single-nucleotide polymorphisms (nsSNPs) (Fig. S1A). We observed that many of these nsSNPs, such as those in pks4 and pks12, are commonly occurring in independent isolates across the different rpoB mutants. When we infected bone marrow-derived macrophages (BMDMs) from C57Bl/6 (B6) mice, only the Mtb strains containing the rpoB-H445Y mutation, and a mutation in a probable transcriptional regulatory protein Rv0465c, significantly induced IFN production upon infection (Fig. S1B). Additionally, a comparative analysis of the MDR clinical isolate that we previously characterized (W_7642) (10) with three strains, HN878, HN13 (rpoB-S450L), and HN14 (rpoB-H445Y), revealed one shared nsSNP in rpoB that was present only in the rpoB-H445Y strain (Table 1). Despite the absence of isogenic strains due to technical limitations, these findings bolstered our interpretation of the role of a specific mutation in rpoB in altering host responses during Mtb infections.
TABLE 1.
Comparison of SNPs across spontaneous mutants a
| Pos | Wt | rpoB-S450L | rpoB-H445Y | AA_POS | Effect | Locus_tag | Gene | Product |
|---|---|---|---|---|---|---|---|---|
| 555619 | T | T | C | 230/474 | Missense_variant c.688A > G p.Met230Val | Rv0465c | Probably transcriptional regulatory protein | |
| 761139 | C | C | T | 445/1172 | Missense_variant c.1333C > T p.His445Tyr | Rv0667 | rpoB | DNA-directed RNA polymerase (beta chain) RpoB (transcriptase beta chain) (RNA polymerase beta subunit) |
| 761155 | C | T | C | 450/1172 | Missense_variant c.1349C > T p.Ser450Leu | Rv0667 | rpoB | DNA-directed RNA polymerase (beta chain) RpoB (transcriptase beta chain) (RNA polymerase beta subunit) |
| 972980 | C | T | C | 243/386 | Missense_variant c.727G > A p.Gly243Ser | Rv0874c | Conserved hypothetical protein | |
| 1155197 | A | A | G | 492/709 | Missense_variant c.1474A > G p.Ile492Val | Rv1030 | kdpB | Probable potassium-transporting P-type ATPase B chain KdpB |
| 1319222 | C | C | T | 1330/1582 | Missense_variant c.3989C > T p.Ala1330Val | Rv1181 | pks4 | Probably polyketide beta-ketoacyl synthase Pks4 |
| 2300546 | A | T | A | 2147/4151 | Missense_variant c.6441T > A p.His2147Gln | Rv2048c | pks12 | Polyketide synthase Pks12 |
| 3690948 | C | C | T | |||||
| 3804941 | G | G | A | 26/236 | Missense_variant c.77G > A p.Cys26Tyr | Rv3390 | lpqD | Probable conserved lipoprotein LpqD |
| 3846791 | A | G | A | |||||
| 3846851 | A | G | A | |||||
| 3846852 | G | C | G | |||||
| 3846853 | C | T | C | |||||
| 3846857 | A | G | A | |||||
| 3846860 | G | T | G | |||||
| 3846866 | A | C | A |
Based on the whole-genome sequencing, all unique non-synonymous SNPs, including the intergenic ones, for the rpoB-S450L and rpoB-H445Y strains were determined in reference to the wt parental Mtb strain HN878. The only SNP shared by any of these strains and the previously characterized W_7642 is boldfaced and is present in the rpoB-H445Y strain.
To extend the clinical relevance of our findings, we next studied the impact of either H445Y or S450L DR SNPs on cytokine responses following infection of human monocyte-derived macrophages (MDMs). We found that both DS wt Mtb (HN878) and the DR S450L mutant Mtb (HN13 rpoB-S450L) induced IL-1 production upon infection (Fig. 1A), while H445Y mutant Mtb (HN14 rpoB-H445Y) infection did not induce increased IL-1 production. These findings are in concert with our previously published findings, where the clinical W_7642 MDR Mtb strain also induced limited IL-1 production in murine macrophages upon infection (10). Additionally, infection with HN878 and rpoB-S450L Mtb also resulted in increased TNF production, while rpoB-H445Y infection did not (Fig. 1B). Infection with all three strains (HN878, rpoB-S450L, and rpoB-H445Y) resulted in increased production of other cytokines that were measured, such as the pro-inflammatory IL-6 and the anti-inflammatory IL-10 (Fig. 1C and D). Levels of IFN were below the level of detection. With our previously published findings (10), our data together show that the RDR Mtb carrying H445Y SNP and the S450L SNP elicit highly divergent host macrophage responses, with the rpoB-H445Y Mtb strain limiting the production of pro-inflammatory cytokines in infected human macrophages.
Fig 1.
Mycobacterium tuberculosis strains with different rpoB SNPs drive divergent cytokine responses in infected human monocyte-derived macrophages. CD14+ monocytes that were isolated from the peripheral blood of five different donors and differentiated into macrophages were infected with wt (HN878), rpoB-H445Y, or rpoB-S450L Mtb strains at an MOI of 1. After 6 days post infection (dpi), supernatants were collected. (A–D) Cytokine protein levels in the supernatants were measured by Lincoplex. Each data point shown represents the average of three technical replicates from an individual donor. All data sets were tested for normality using the Shapiro-Wilk and passed (P > 0.05). Significant differences are indicated with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) by ordinary one-way ANOVA with Tukey’s post-tests (A–D).
Infection with rpoB-H445Y Mtb results in limited lung immune responses in mice
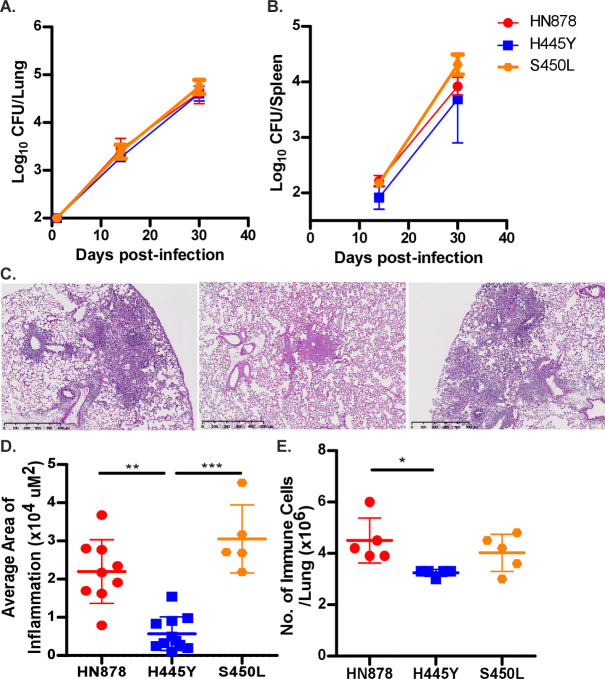
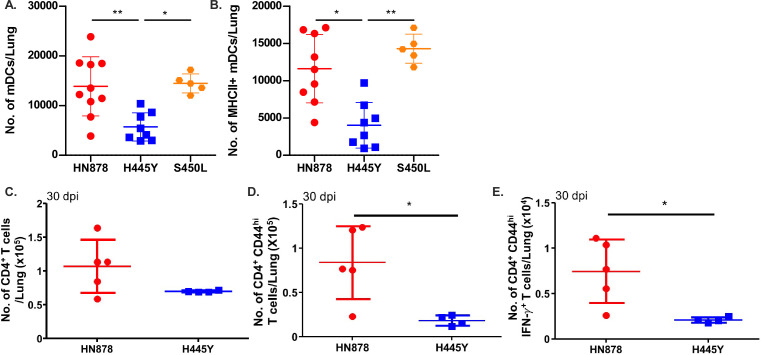
The impact of DR Mtb infection on cytokine responses likely impacts TB disease progression in vivo. Thus, C57BL/6J (B6) mice were infected with Mtb HN878, rpoB-H445Y, or rpoB-S450L (10 – 12). We found that infection with all Mtb strains established similar lung bacterial burden and disseminated effectively to the spleen at 14 and 30 dpi (Fig. 2A and B). Importantly, despite similar bacterial loads, rpoB-H445Y Mtb infection induced significantly lower pulmonary inflammation when compared to wt Mtb (HN878) and rpoB-S450L infections, with rpoB-H445Y Mtb infection inducing smaller-sized inflamed lesions (Fig. 2C and D). This coincided with reduced accumulation of lung immune cells after rpoB-H445Y Mtb infection, compared with wt HN878 and rpoB-S450L Mtb infections (Fig. 2E). Characterization of these immune populations revealed that the accumulation of lung myeloid dendritic cells (mDCs) was significantly reduced in rpoB-H445Y Mtb-infected mice when compared with either wt HN878 or rpoB-S450L Mtb-infected mice (Fig. 3A). Similar defects in the accumulation of lung neutrophils, recruited macrophages (RMs), and monocytes were also observed in rpoB-H445Y Mtb-infected mice when compared with Wt Mtb infection (Fig. S2A through C). Upregulation of Major Histocompatibility Complex (MHC) Class II expression on lung macrophages is associated with Mtb control (13). Fewer MHC Class II-expressing lung mDCs were also observed in rpoB-H445Y Mtb infection in contrast with either wt HN878 or rpoB-S450L Mtb infections (Fig. 3B). The numbers of other MHC Class II-expressing lung myeloid cells, RMs and monocytes, were also reduced in rpoB-H445Y Mtb-infected mouse lungs when compared to Wt Mtb infection (Fig. S2D and E). During Mtb infection, MHC Class II expression on mDCs is critical for antigen presentation to CD4+ T cells (13). Based on the fewer numbers of activated myeloid cells in rpoB-H445Y infection, we next examined T cell responses in the lungs of mice infected with Wt Mtb or rpoB-H445Y Mtb. While the total number of CD4+ T cells was similar in the lungs of mice infected with Wt or rpoB-H445Y Mtb (Fig. 3C), accumulation of activated CD4+CD44hi T cells was significantly reduced in the lungs of rpoB-H445Y Mtb-infected mice (Fig. 3D). Indeed, in mice infected with the rpoB-H445Y Mtb, there were significantly fewer Mtb-specific activated lung CD4+ CD44hi T cells, which produced IFN when compared with Wt Mtb infection (Fig. 3E). These results show that while wt and S450L SNP carrying Mtb drive similar activation of the immune response in vivo, Mtb harboring the H445Y SNP may be associated with a limited immune response in the lung in infection, characterized by reduced inflammation and decreased accumulation of activated myeloid and T cells.
Fig 2.
Mtb with different rpoB SNPs induce dissimilar pathology in infected mice. C57Bl/6 (B6) mice were infected with a low dose (~100 CFU) of Wt HN878, rpoB-H445Y, or rpoB-S450L Mtb by the aerosol route. Bacterial burden in (A) lung and (B) spleen was determined by plating at 14 and 30 dpi. (C) Formalin-fixed paraffin-embedded (FFPE) lung sections from mice infected for 30 days were hematoxylin and eosin (H&E) stained, and representative images for each group are shown: left (Wt HN878), middle (rpoB-H445Y), or right (rpoB-S450L Mtb). (D) The inflammatory area was measured. (E) Total numbers of lung immune cells in single-cell suspensions were determined. The data shown represent the means ± SD of 5–10 biological replicates per experiment. Significant differences are indicated with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001) by Kruskal-Wallis test with Dunn’s multiple comparisons tests (A–E). One of three independent experiments is shown.
Fig 3.
rpoB-H445Y Mtb infection induces limited myeloid dendritic cell accumulation and subsequent T cell responses. B6 mice were infected with a low dose (~100 CFU) of Wt, rpoB-H445Y, or rpoB-S450L Mtb by the aerosol route and sacrificed at 30 dpi for analysis. (A) Total number of mDCs and (B) MHC Class II+ mDCs were determined by flow cytometry. The numbers of (C) total, (D) CD44hi, and (E) IFN+ CD4+ T cells were determined by flow cytometry following ex vivo stimulation with purified Mtb antigens ESAT6 and Ag85B. The data shown represent the means ± SD of 4–10 biological replicates per experiment. Significant differences are indicated with asterisks (*P < 0.05; **P < 0.01) by Kruskal-Wallis test with Dunn’s multiple comparisons tests (A–E). One of three independent experiments is shown.
Infection with rpoB-H445Y Mtb resulted in higher lung IFN but lower IL-1 levels, relative to Wt Mtb infection (Fig. S3A and B). We also found reduced expression of chemokines in rpoB-H445Y Mtb-infected lungs, including keratinocytes-derived chemokine (KC), a neutrophil chemo-attractant, MIP1, a monocyte/macrophage chemo-attractant, and regulated upon activation, normal T cell expressed and presumably secreted (RANTES), a T cell and myeloid cell chemoattractant, in rpoB-H445Y Mtb infection (Fig. S3C through E). Assessment of markers of proliferation and cell death in the lung myeloid cell subsets at this time point showed no differences between Wt and rpoB-H445Y Mtb-infected mice (data not shown). Thus, these data collectively indicate that the reduction in chemokines may be driving reduced immune cell recruitment, which underlies the limited accumulation of immune cells in rpoB-H445Y Mtb infection.
Using the susceptible TB mouse model, C3HeB/FeJ (FeJ) mice, which exhibit necrotic TB granulomas similar to human TB (14), infection with rpoB-H445Y Mtb also resulted in reduced pulmonary inflammation and also induced lower IL-1 levels, relative to wt HN878 Mtb infection (Fig. S4A and B). Similar to our findings in B6 mice, rpoB-H445Y Mtb infection resulted in reduced accumulation of mDCs and fewer MHC Class II-expressing mDCs (Fig. S4C and D). When compared with Wt Mtb infection, FeJ mice infected with rpoB-H445Y Mtb also had fewer numbers of IFN-producing, activated lung CD4+CD44hi (Fig. S4E). These data suggest a likely association between the H445Y SNP and reduced lung myeloid and T cell recruitment and activation across resistant and susceptible mouse models of TB.
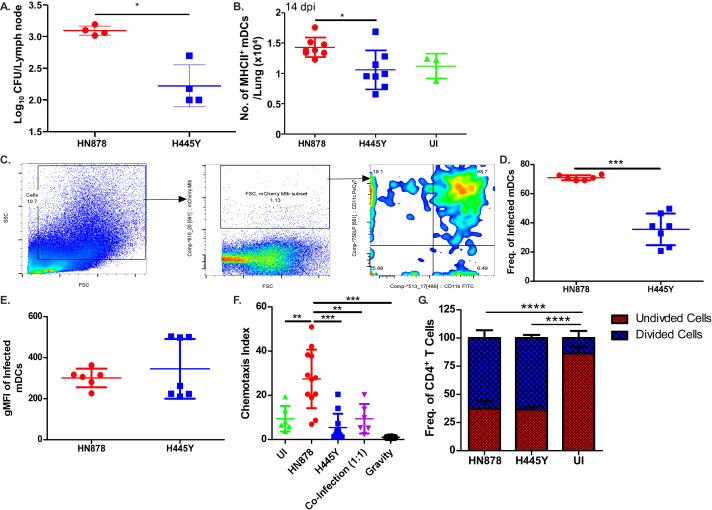
Reduced lung T cell responses in rpoB-H445Y Mtb infection are associated with decreased trafficking of mDCs to lymph nodes
After Mtb infection, T cell priming is initiated in the draining lymph nodes (DLNs), where Mtb antigens are presented by migrating mDCs (14, 15). To assess whether the impairment of T cell activation was linked to defects in DC-mediated Mtb transport to the DLNs, we enumerated Mtb burden in the DLNs early in infection at 14 dpi. We found significantly lower Mtb colony-forming unit (CFU) in the DLNs of mice infected with rpoB-H445Y Mtb compared to Wt HN878 Mtb (Fig. 4A). At this early time point, we also found decreased accumulation of MHC Class II-expressing mDCs in the lungs of rpoB-H445Y Mtb infection (Fig. 4B). These results suggest that the reduction in accumulation of activated T cells in the lung might be due to reduced activation of mDCS and trafficking of Mtb-infected lung mDCs to the DLNs. B6 mice were infected with Wt or rpoB-H445Y Mtb expressing mCherry reporter, and while there were no differences in the number of Mtb on a per cell basis in mDCs, there was decreased frequency of rpoB-H445Y Mtb-infected mDCs in contrast to Wt Mtb, which was mainly harbored within mDCs (12, 15) (Fig. 4D and E). To address if the rpoB-H445Y Mtb infection differentially impacted the migratory potential of mDCs toward chemokines (e.g., CCL-19), we used an in vitro chemotaxis assays and found that rpoB-H445Y Mtb-infected bone marrow-derived dendritic cells (BMDCs) did not migrate as effectively as Wt Mtb-infected BMDCs (Fig. 4F). In a co-infection model where BMDCs were infected with a 1:1 ratio of wt and rpoB-H445Y Mtb, the BMDCs recapitulated the limited migration observed in rpoB-H445Y Mtb infection. These results suggest that the immune suppression driven by rpoB-H445Y Mtb dominates and limits DC migration. We also sought to assess whether rpoB-H445Y Mtb infection of DCs impairs T cell activation in vitro by co-culturing naïve CD4+ T cells from Rag −/− ESAT61-20 TCR transgenic mice that recognize Mtb antigen with BMDCs were infected with either Wt Mtb or rpoB-H445Y Mtb. Interestingly, T cell proliferation was observed to similar levels independent of the Mtb strain (Fig. 4G). Together, our results suggest that the process of infected mDCs translocating Mtb to the DLNs for priming of T cells is impaired in mice infected with rpoB-H445Y Mtb infection.
Fig 4.
Impaired T cell responses are driven by defects in myeloid dendritic cell migration in rpoB-H445Y Mtb infection. B6 mice were aerosol infected with a low dose of either Wt or rpoB-H445Y Mtb and sacrificed at 14 dpi for analysis. (A) Bacterial burden in the lymph nodes was determined by plating. (B) Numbers of MHC Class II+ mDCs were determined by flow cytometry. Uninfected mice were also included (n = 3). B6 mice were aerosol infected with a low dose of either mCherry-expressing Wt or rpoB-H445Y Mtb. (C) Gating strategy for Mtb-infected cells in the lung is shown. (D) The frequencies of Mtb-infected lung cells that are mDCs (CD11c+ CD11b+) and (E) the geometric mean fluorescent intensity (MFI) of mCherry expression in infected mDCs were calculated. BMDCs were infected with either Wt or rpoB-H445Y Mtb or co-infected at a ratio of 1:1 Wt:rpoB-H445Y at a multiplicity of infection (MOI) of 1. (F) One day post infection, in vitro chemotaxis assays were carried out toward CCL-19, and a chemotaxis index was calculated. (G) One day post infection, BMDCs were co-cultured with isolated, enriched, and carboxyfluorescein succinimidyl ester (CFSE)-labeled CD4+ T cells from naïve ESAT-6 ab TCR mice at a ratio of 1:1. The data shown are the frequencies of divided and undivided cells, as represented by the expression of CFSE determined using flow cytometry. The data shown represent the means ± SD of four to seven biological replicates per experiment (A–E), technical replicates combined over two experiments (F), or technical replicates from a representative experiment (G). Significant differences are indicated with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001) by Kruskal-Wallis test with Dunn’s multiple comparisons tests (A–G).
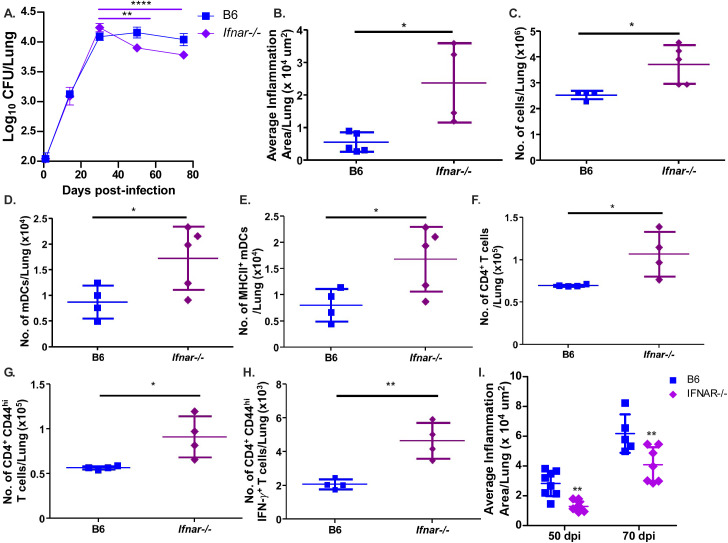
Type I IFN signaling suppresses myeloid and lymphoid cell recruitment and activation during rpoB-H455Y Mtb infection
From our cytokine data, we hypothesized that type I IFN production was playing a role in the induction of immune responses following infection. Thus, B6 mice and mice lacking the IFN-α/β receptor (Ifnar−/ −) were infected with rpoB-H445Y Mtb. While there were no differences in Mtb burden in the lungs between B6 and Ifnar−/− rpoB-H445Y Mtb-infected mice at 30 dpi (Fig. 5A), lung inflammation and immune cell accumulation were increased in rpoB-H445Y Mtb-infected Ifnar−/ − mice (Fig. 5B and C). Importantly, accumulation of mDCs and other myeloid subsets, including RMs and monocytes, was also increased in Ifnar−/− rpoB-H455Y Mtb-infected mice (Fig. 5D; Fig. S5A and B). Additionally, the number of activated mDCs that expressed MHC Class II was also increased in Ifnar−/− rpoB-H455Y Mtb-infected lungs when compared to their B6 counterparts (Fig. 5E). In contrast, when we infected B6 and Ifnar−/ − mice with wt HN878 Mtb, we found equivalent lung and splenic bacterial burden and no differences in inflammation and total immune cell accumulation between the lungs of these two groups of mice (Fig. S6A through D). The numbers of total and MHC Class II-expressing RMs and monocytes were not significantly different between B6 and Ifnar−/ − mice infected with wt HN878 Mtb (Fig. S5A through D). Next, we addressed whether improved mDC activation in Ifnar−/ − mice infected with rpoB-H455Y Mtb also improved activation of T cell responses. Indeed, we found that Ifnar−/− rpoB-H445Y Mtb-infected mice accumulated significantly higher numbers of total CD4+ T cells, as well as activated CD4+CD44hi T cells, when compared with B6 mice infected with rpoB-H445Y Mtb (Fig. 5F and G). The absence of type I IFN signaling also increased Mtb-specific IFN-producing CD4+ T cells in rpoB-H445Y Mtb infection (Fig. 5H). Together, these data demonstrate that H445Y and other associated SNPs in Mtb and its impact on bacterial determinants may suppress inflammation, immune cell recruitment, as well as myeloid cell activation and T cell responses through a type I IFN-dependent mechanism during Mtb infection. This suppression of the immune response modulated through type I IFN signaling appears to benefit rpoB-H445Y Mtb as Ifnar−/ − mice show a decrease in lung bacterial burden during the chronic stages of infection with this DR mutant (Fig. 5A). Coinciding with this decrease in bacterial loads, rpoB-H445Y Mtb-infected Ifnar−/ − mice also showed a decrease in the average size of inflammatory lesions as compared with their B6 counterparts (Fig. 5I). Thus, our results propose that the H445Y SNP may result in heightened type I IFN production in mice, which dampens the initial immune response and leads to increased bacterial abundance and worse pathology for the host.
Fig 5.
rpoB-H445Y Mtb infection suppresses lung immune responses through a type I IFN-dependent pathway. B6 and Ifnar −/− mice were aerosol infected with a low dose of rpoB-H445Y Mtb. Mice were sacrificed for subsequent analyses at the indicated time points. Bacterial burden in (A) lung was determined. (B) FFPE lung sections were stained with H&E, and lung inflammatory area was measured at 30 dpi. (C) Total numbers of lung immune cells in single cell suspensions were determined at 30 dpi. (D) Total numbers of (E) MHC Class II+ mDCs, as well as the numbers of (F) total, (G) CD44+, and IFN+ (H) CD4+ T cells, were determined by flow cytometry at 30 dpi. (I) FFPE lung sections were stained with H&E, and lung inflammatory area was measured at 50 and 70 dpi. The data shown represent the means ± SD of four to five biological replicates per experiment. Significant differences are indicated with asterisks (*P < 0.05; **P < 0.01; P < 0.0001) by Kruskal-Wallis test with Dunn’s multiple comparisons tests (A–H) or by two-way ANOVA with Bonferroni post test (I). One of two independent experiments is shown.
DISCUSSION
The rise of drug resistance has exacerbated efforts to combat and contain TB. Clinical reports have suggested that, in addition to being difficult to treat, MDR TB patients also exhibit dysregulated immune responses (2 – 5). Despite extensive studies that have described the effects of DR mutations in rpoB on rifampicin resistance and bacterial fitness (16), the impact of these SNPs on the host response has been unexplored and poorly understood. In our previous study, we showed that the presence of specific rifampicin drug-resistant mutations alters macrophage responses to Mtb infection in vitro through changes in bacterial cell wall lipid abundance and structure (10). In this work, we studied the association between drug-resistance-conferring mutations and in vivo host responses with the two most common mutations that account for nearly 90% of rifampicin-resistant MDR TB infections annually (6 – 9). While both Mtb strains, rpoB-H445Y and rpoB-S450L, are resistant to rifampicin, they are associated with vastly differing host responses during infection in the lung. Importantly, we found broader impacts on immunopathology of TB disease that are not attributable to bacterial burden. More specifically, we observed limits in mDC trafficking, which resulted in impairment of T cell generation and limited inflammation. This immunomodulation in the lung was achieved through a host type I IFN-dependent pathway, and removing this signaling not only lifted the suppression of the immune response but also led to improved control of Mtb during chronic infection. Together, our results demonstrate that infection with Mtb strains carrying drug-resistance-conferring and associated mutations results in divergent host immune responses and highlights that these responses have a regulatory role in immunopathogenesis in vivo.
Type I IFN signaling has a controversial role in Mtb infections. Individuals with TB disease and TB progressors show upregulation of a type I IFN transcriptional signature (17 – 20). In mice, type I IFN signaling has been shown to play a counter-regulatory role with IL-1 signaling and suggested to drive DS Mtb susceptibility by antagonizing IL-1 production and signaling (21 – 24). Conversely, other studies suggest that IFN-β may have a protective role through the induction of nitric oxide in infected macrophages (25, 26). Our study sheds light on a novel role for type I IFN signaling, specifically in some DR Mtb infections. We observed that rpoB-H445Y Mtb infection results in a heightened type I IFN production that suppressed the recruitment of activated immune cells to the site of infection and the downstream initiation of adaptive immune responses. This immune modulation appears to be important for Mtb pathogenesis, as lack of type I IFN signaling reversed innate and adaptive immune responses, leading to an enhanced bacterial control during chronic rpoB-H445Y Mtb infection. Therefore, induction of type I IFNs and suppression of host immune responses could serve as a mechanism for some DR Mtb strains to persist in the host. This finding stands in stark contrast to DS Mtb infection, where lack of type I IFN signaling had no discernable impact on inflammation or cell recruitment at the same time points.
In previous studies, the role of type I IFN signaling in DS Mtb infections has been discernible in hypersusceptible mouse strains (27, 28); hyper-IFN induction using poly IC or recombinant type I IFN co-treatment with Mtb infection (29, 30); or blockade of GM-CSF, a key mediator of Mtb immunity (31). In these studies, the observed role for type I IFNs was to drive the recruitment of Mtb-permissive cells and promote pathology, thus promoting TB disease. While type I IFN signaling during rpoB-H445Y Mtb infection also promoted Mtb pathogenesis, type I IFN signaling plays the opposite functions in that it suppressed immune responses and limited pathology. Also, our data suggest that heightened type I IFN signaling during rpoB-H445Y Mtb infection is not influencing the permissibility or frequency of apoptosis of the cells in which the DR Mtb is harbored. Additionally, type I IFN signaling did not impact T cell responses in DS Mtb infection of hypersusceptible mice (27), while during rpoB-H445Y Mtb infection, the restricted mDC accumulation results in reduced T cell responses through IFNAR-dependent pathways. It has been suggested that the hypervirulence of the DS Mtb clinical isolate, HN878, used in this study is tied to a limited induction of Th1 and associated with greater type I IFN induction, relative to other DS Mtb strains (30, 32). Here, we showed a link between heightened type I IFN induction and restricted Th1 immunity during infection with our rpoB-H445Y Mtb strain. Together, our findings suggest that, although type I IFN signaling might have a detrimental role in both Mtb infections, the detrimental effects observed in rpoB-H445Y DR Mtb infection are likely through distinct pathways.
There are a few potential explanations for the discrepancies between our findings and previously published work regarding the mechanism of action of type I IFN signaling between the DS and DR Mtb infections in the study. The nature of the H445Y mutation in rpoB does alter the transcriptional landscape and the production of various virulence factors in Mtb (16). We have previously shown that the presence of this specific H445Y SNP in rpoB is associated with changes to the bacterial cell wall lipid composition and abundance, which are sufficient to drive differential cytokine production in macrophages (10). Similarly, other important virulence factors that affect the impact of type I IFN signaling in cells infected with DS Mtb could be differentially transcribed in rpoB-H445Y DR Mtb. Banks and colleagues noted that DS Mtb inhibits type I IFN signaling in infected macrophages in a dose-dependent manner to promote intracellular survival (25). The addition of supraphysiological levels of IFN overcame Mtb suppression of type I IFN signaling and decreased bacterial loads. Various IFN-induced genes noted to be transcriptionally inhibited by DS Mtb, such as Ifit1, Ccl12, and Iigp1, were found to be transcriptionally upregulated in MDR Mtb-infected macrophages, relative to those infected with DS Mtb (10, 25). These findings suggest that DR and MDR Mtb with the H445Y SNP might not be inhibiting autocrine type I IFN signaling, potentially through decreased transcription of phosphatases known to interfere with various host signaling pathways (33). Thus, in the absence of these Mtb virulence factors, heightened type I IFN signaling could drive distinct outcomes between DS wt and rpoB-H445Y Mtb infections.
Other explanations can be found on the host side of this host-pathogen interaction. Desvignes and colleagues found a protective role for type I IFN signaling in the absence of type II IFN signaling by limiting the influx of permissive myeloid cells (34). Mice lacking both, type I and type II IFN, signaling pathways had exacerbated lung histopathology and succumbed to infection earlier. In the context of our study, we found fewer type II IFN-producing T cells during rpoB-H445Y Mtb infection, suggesting a lung environment with limited IFN. Therefore, the differences in the production and subsequent signaling of type II IFNs between DS and rpoB-H445Y Mtb could set the stage for different roles of type I IFN in Mtb infections. Another explanation for the different roles of type I IFN between the two Mtb infections is that the two strains of Mtb are interacting with different cell types. Our findings suggest that, while DS wt Mtb is mainly found in mDCs, rpoB-H445Y Mtb is not harbored to the same degree in this cell type. If rpoB-H445Y Mtb is found to be in different host cells than DS wt Mtb, then induction of type I IFN signaling in these other cell types could drive different cellular responses and not result in increased infiltration of permissive cells and loss of control of the infection.
Our study opens up several questions which need to be addressed to fully understand the pathogenesis of RDR and MDR TB. How does type I IFN signaling limit myeloid cell activation and recruitment as seen by us and others (34)? We observed a decrease in the levels of a few chemokines and cytokines in rpoB-H445Y Mtb infection of B6 mice and human MDMs, respectively, but it remains to be understood the pathways linking type I IFN signaling and inhibition of chemokine production and MHC Class II. By limiting the production and activity of potent pro-inflammatory molecules, type I IFNs could be curtailing the inflammatory cell recruitment in rpoB-H445Y Mtb infection. Similarly, how does rpoB-H445Y Mtb infection impair DC migratory ability? From the reduced accumulation of rpoB-H445Y Mtb in the DLNs early in infection and the decreased chemotactic sensitivity of infected BMDCs to CCL19, DCs infected with rpoB-H445Y Mtb likely have diminished expression of the cognate receptor, CCR7. Furthermore, a co-infection with an equal ratio of DS wt and rpoB-H445Y Mtb reveals that the suppressed chemotaxis response of rpoB-H445Y Mtb is dominant. Together, these data suggest that the rpoB-H445Y Mtb strain might suppress DC activity through a secreted virulence factor or heightened type I IFN signaling.
Although our study reveals intriguing insights into the intersection between drug resistance and host immunity, there are a few limitations of the work. One such limitation is the lack of matched mutant strains and the use of a complemented strain. While we generated the mutant RDR Mtb strains with the use of the wt Mtb strain, a few additional SNPs exist that separate the wt Mtb strain from the RDR Mtb strains beyond the RDR-conferring SNPs in rpoB. Despite our best efforts to genetically engineer the specific point mutation in rpoB in the wt Mtb strain or to generate a complemented Mtb strain with the wt sequence in the rpoB-H445Y Mtb strain, these tasks have proved to be technically difficult to execute. As such, it is possible that other SNPs outside of H445Y in rpoB could contribute to the phenotypes observed in the study. However, in our collection of independently generated rpoB mutant strains, we observed the repeated occurrence of these additional SNPs throughout the panel. This suggests that these mutations may be commonly co-occurring spontaneous mutations generated alongside rpoB mutations. Moreover, the presence of these additional SNPs did not phenotypically impact Mtb infection of BMDMs with strains not containing the rpoB-H445Y SNP. Yet, the question of causality with regard to the specific rpoB mutations and the observed phenotypes still remains open and needs to be confirmed in future studies. We also know little about how SNPs in rpoB lead to altered host-pathogen interactions. Previous studies have suggested that compensatory transcriptional changes might occur in Mtb strains as a result of the rpoB SNPs (27). Additionally, the location of these SNPs in the rpoB could also drive differences in transcription. However, the nature of these transcriptional changes and how they are linked to the phenotypes observed in the study remain to be determined.
In summary, our studies have mechanistically shown how the identities of the drug-resistance-conferring mutations are associated with differential host responses to infection and the relationship between drug resistance and TB disease pathogenesis. These altered host responses can be leveraged by the drug-resistant bacteria to promote its survival. The implications of these diverse immune responses underscore the importance of appreciating bacterial heterogeneity in studying host-pathogen interactions. While much of the focus of the field in studying host immunity to TB has centered around host factors and background, our study emphasizes the need to also approach the issue from the bacterial perspective. Fully delineating such immune mechanisms of antagonism by RDR Mtb infection and specific DR mutations in the future will allow us to develop targeted host-directed therapeutics and vaccines addressing the growing incidence of RDR and MDR TB.
MATERIALS AND METHODS
Mice
C57BL/6J (B6), C3HeB/FeJ, and Ifnar –/–mice on the B6 background were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). ESAT-6 TCR transgenic mouse line was established as previously described and maintained on the Rag1−/− background, with the αβ T cells recognizing IAb/ESAT61-20 (11, 35). All mice were maintained in the animal facility at Washington University in St. Louis and bred in-house. Experimental mice were age and sex matched and infected between the ages of 6 and 8 weeks. All mice were maintained and used in accordance with the approved Washington University in St. Louis Institutional Animal Care and Use Committee guidelines.
Generation of BMDMs and BMDCs
Bone marrow-derived macrophages and bone marrow-derived dendritic cells were generated as previously described (10, 12). Briefly, bone marrow cells were collected from the tibia and femur of B6 mice and cultured in complete Dulbecco’s modified Eagle’s medium (cDMEM) with 20 ng/mL of recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF). Cells were cultured at 37°C in 5% CO2 and supplemented with media on day 3. On day 7, non-adherent cells were collected as BMDCs and adherent cells were collected as BMDMs.
Generation of human monocyte-derived macrophages
Peripheral blood was obtained from the Mississippi Valley Regional Blood Center in LRS chambers through the Fehniger Laboratory. LRS chambers were flushed with PBS containing heparin (1 U/mL) and 2% HuAB (Sigma H3667-100ML, Lot #SLBZ1915) to collect blood. The diluted blood was then overlaid onto Ficoll (Lymphoprep, AXS-1114544, Axis Shield) and centrifuged to isolate peripheral blood mononuclear cells. CD14+ cells were enriched using CD14 Microbeads (130-050-201, Miltenyi Biotech) according to the manufacturer’s instructions. Cells were cultured in cRPMI1640 containing 750 IU/mL of human GM-CSF (Miltenyi Lot #5180329061) at 37°C in 5% CO2, with media changes every 2–3 days as needed for 7 days.
Mtb infections
Mtb strain HN878 was obtained from BEI resources (Manassas, VA, USA) under National Institutes of Health contract AI-75320. HN878-mCherry was generated by transforming the Mtb strain HN878 with a plasmid (pMSG432) encoding mCherry. Independent rifampicin-resistant Mtb HN878 colonies (biological replicates) were selected from rifampicin (2 µg/mL) containing 7H11 agar plates (36). The sequences of rpoB were confirmed by Sanger sequencing (Genewiz), and Mtb stocks were created for further experimentation. All Mtb strains were cultured in Proskauer Beck medium supplemented with 0.05% Tween 80 and frozen at −80°C while in mid-log phase. Colony forming units of the bacterial stocks were determined through serial dilutions on 7H11 agar plates. Mice were aerosol infected with low doses (~100 CFU) of indicated Mtb strains in sterile PBS using a Glass-col nebulizer (37). Mtb bacterial burden/organ was quantitated by plating serial dilutions of homogenized lung, spleen, or lymph node tissue or aspirated bone marrow on 7H11 agar plates (BD Biosciences, Franklin Lakes, NJ, USA). Plates were incubated for 2–3 weeks at 37°C, and the number of colonies was counted. In vitro infections were carried out at a multiplicity of infection (MOI) of 1 in respective antibiotic-free media for the indicated number of days.
Flow cytometry
Lungs were perfused with heparin in PBS, minced, digested with DNAse/collagenase, lysed for red blood cells, and pressed through a 70-µm filter to generate a single-cell suspension (37). For quantification of cytokine responses, single-cell suspensions were stimulated ex vivo with either Phorbol Myristic Acid, Ionomycin, and GolgiStop (BD Biosciences) for 5 h or purified Mtb antigens ESAT6 and Ag85B (10 µg/mL) overnight along with brefeldin and monensin (BD Biosciences) as previously described (38, 39). Cells were treated with Fc Block (CD16/CD32,2.4G2, Tonbo Biosciences) and stained with appropriate fluorochrome-labeled specific antibodies or isotype control antibodies. Intracellular cytokine staining was performed using the BD Cytofix/Cytoperm kit (BD Biosciences). Mouse antibodies used include CD11b (M1/70; Tonbo Biosciences), CD11c (HL3; BD Biosciences), Gr-1 (RB6–8C5, eBioscience), CD3 (500A2; BioLegend), CD4 (RM4–5; BD Biosciences), CD44 (IM7; Tonbo Biosciences), IFN (XMG1.2; Tonbo Biosciences) or isotype control IgG1 (A85-1; BD Biosciences), CD8 (53-6.7; BD Biosciences), MHC Class II (M5/114.15.2; Tonbo Biosciences), Ki-67 (16A8; BioLegend) or isotype control IgG2a, (RTK2758; BioLegend), 7AAD (BD Biosciences), and Annexin V (BD Biosciences).
Cells were processed with the Becton Dickinson (BD) Fortessa X-20 flow cytometer using FACS Diva software or the BD FACSJazz flow cytometer using FACS software (BD). Flow cytometry experiments were analyzed using FlowJo (Tree Star Inc.). As before (10, 12), monocytes were defined as CD11b+CD11c−Gr-1med cells, and recruited macrophages were defined as CD11b+CD11c−Gr-1low cells. mDCs were defined as CD11b+CD11c+ cells. T cells were defined as CD3+CD4+ or CD3+CD8+ cells. Total numbers of cells within each gate were back calculated based on cell counts/individual sample.
Histology
Lung lobes were perfused with 10% neutral buffered formalin and embedded in paraffin. Formalin-fixed paraffin-embedded (FFPE) lung sections were stained with hematoxylin and eosin (H&E), and inflammatory features were evaluated by light microscopy. Inflammatory lesions were outlined with the automated tool of the Zeiss Axioplan 2 microscope (Carl Zeiss), and the total inflammatory area in each lung lobe was measured.
Determination of proteins
Cytokine and chemokine production in the lung homogenates of Mtb-infected mice and the supernatants of Mtb-infected MDMs was analyzed using Milliplex Multiplex Assays (Millipore) according to the manufacturer’s protocol. IL-1 and IFN were measured using Quantikine ELISA kits (R&D Systems), following the manufacturer’s instructions.
Chemotaxis assay
BMDCs, generated using the above protocol, were infected, with their respective strain, at an MOI of 1 in antibiotic-free cDMEM. For co-infections, the amount of Mtb was titrated to the ratios specified (Wt Mtb:rpoB-H445Y Mtb). Chemotaxis was carried out as described previously (12, 40). Briefly, 1 dpi, BMDC migration in response to CCL-19 was determined through flow cytometry, using a standard number of fluorescent beads (Polysciences Inc.) to normalize the samples. The results are indicated as chemotaxis index, which is represented as the fold increase in the number of migrated cells in each sample, relative to spontaneous migration in the gravity control group.
T cell proliferation assay
BMDCs were similarly generated and infected for 1 day with the respective Mtb strain at an MOI of 1. T cell co-culture proliferation assay was carried out as previously described (11). Briefly, transgenic CD4+ T cells recognizing IAb/ESAT-61–20 were isolated from the spleens and lymph nodes of mice, enriched using CD4+ T cell MACS Microbeads (Miltenyi Biotech) and then stained with carboxyfluorescein succinimidyl ester (CFSE) for 15 min at 37°C. Infected or uninfected BMDCs and naïve, CFSE-labeled T cells were co-cultured with recombinant IL-2 (ThermoFisher) for 6 days. Cells were then stained and processed as described above.
Bacterial genomic DNA isolation and sequencing
In brief, Mtb cultures were expanded to mid-log phase and lysed through bead beating, and genomic DNA was extracted using phenol-chloroform-isoamyl alcohol. DNA was precipitated with isopropanol at −20°C overnight. DNA pellet was washed in 80% ethanol and dissolved in nuclease-free water. Genomic libraries were prepared using WGS KAPA Hyper-PCR free method and sequenced using the NovaSeq platform to target 15 Gb (GTAC MGI).
Whole-genome sequence data processing, variant calling, and chromosome visualization
Raw paired-end fastq reads were filtered and trimmed with Trim Galore v0.4.1 (Babraham Bioinformatics) and mapped to the H37Rv reference genome (NC_000962.3) using Burrows-Wheeler Aligner v0.7.12 (41). Samtools v1.5 (42) were used for removing duplicates and indexing with default settings. SNPs were then identified and filtered using Samtools v1.5 mpileup (42) and Bcftools v1.5 (42). SNPs with low quality (QUAL ≤100), low read consensus (≤75% reads supporting alternate allele), and high proximity to indels (within 15 bp) were filtered out. Additionally, polymorphisms in or within 50 base pairs of hypervariable PPE/PE gene families, repeat regions, and mobile elements were excluded. The resulting VCF files were used for SNP density visualization in R studio with package BioCircos (43) and Gviz (44).
Statistical analysis
All data sets were evaluated for normality using the Shapiro-Wilk test. Differences between the means of groups were analyzed using either the two-tailed Student’s t-test or the Mann Whitney U test, where applicable. Differences between the means of more than two groups were analyzed using one-way ANOVA or two-way ANOVA for time course studies, with Tukey’s post-tests for normally distributed distributions or Kruskal-Wallis test with Dunn’s multiple comparisons tests. All statistical analyses were done in GraphPad Prism 9. A P value <0.05 was considered significant. The data points across figures represent the mean (±SD or +SEM) of values as noted. * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; **** P ≤ 0.0001; ns, not significant (P > 0.05). All experiments were replicated for reproducibility.
ACKNOWLEDGMENTS
This work was supported by Washington University School of Medicine, NIH grant HL105427, AI111914-02, AI134236-02, and AI123780 to S.A.K.; NIH/NHLBI T32 HL007317-42 to S.B.; and NIH/NHLBI T32 HL007317-37 to N.C.H.
We thank Dr. David Sibley and Dr. Robyn S. Klein for generously providing mice; the Fehniger laboratory for distributing leukoreduction system (LRS) chambers; the NIH tetramer core facility for generously providing Ag85B and ESAT6 tetramers; and Mr. Andrew Beckendorf, Ms. Lan Lu, and Ms. Misty Veschak for technical support. We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomics services. The Center is partially supported by the NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR002345 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
S.A.K. designed the study, provided funding, and supervised the project. S.B., N.C.H., S.D., S.T., and M.A. performed experiments and compiled the results. S.B., N.C.H., M.H.L., B.M., M.D., and S.A.K. interpreted experiments, carried out data analysis, and/or provided reagents. S.A.K., S.B., and N.C.H. wrote the manuscript; all authors edited and approved the final version of the manuscript.
The authors have declared that no conflict of interest exists.
Footnotes
This article is a direct contribution from Shabaana A. Khader, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by M. Ramakrishna Vankayalapati, University of Texas Health Science Center at Tyler, and Chinnaswamy Jagannath, Houston Methodist Research Institute and Weill Cornell Medical School.
Contributor Information
Shabaana A. Khader, Email: khader@uchicago.edu.
Alan Sher, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA .
Joel Ernst, University of California, San Francisco, San Francisco, California, USA .
DATA AVAILABILITY
All data that support the findings of this study are available from the corresponding author upon request. DNA sequencing data have been submitted under BioProject ID PRJNA811702.
ETHICS APPROVAL
Protocols involving the use of animals were approved by IACUC at Washington University in St. Louis. All of the experiments were performed in accordance with the protocols.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.00946-23.
Mtb isolates containing rpoB-H445Y SNP drive altered type I interferon production during infection.
rpoB-H445Y Mtb infection limits myeloid cell recruitment and activation in mice.
rpoB-H445Y Mtb infection induces altered cytokine and chemokine production in mice.
rpoB-H445Y Mtb infection also results in a limited immune response in FeJ mice.
Ifnar -/- mice have significantly enhanced myeloid cell recruitment and activation in the lung only after infection with rpoB-H445Y Mtb.
Absence of type I IFN signaling does not impact wt HN878 Mtb infection.
Legends for all supplemental figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Published online 2022. Global Tuberculosis Report 2022. Available from: https://www.who.int/publications-detail-redirect/9789240061729. Accessed 3 January 2023
- 2. McDyer JF, Hackley MN, Walsh TE, Cook JL, Seder RA. 1997. Patients with multidrug-resistant tuberculosis with low CD4+ T cell counts have impaired Th1 responses. J Immunol 158:492–500. [PubMed] [Google Scholar]
- 3. Geffner L, Yokobori N, Basile J, Schierloh P, Balboa L, Romero MM, Ritacco V, Vescovo M, González Montaner P, Lopez B, Barrera L, Alemán M, Abatte E, Sasiain MC, de la Barrera S. 2009. Patients with multidrug-resistant tuberculosis display impaired Th1 responses and enhanced regulatory T-cell levels in response to an outbreak of multidrug-resistant Mycobacterium tuberculosis M and Ra strains. Infect Immun 77:5025–5034. doi: 10.1128/IAI.00224-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan Q, Xie WP, Min R, Dai GQ, Xu C-C, Pan HQ, Miao CD, Yang Z, Xu WG, Wang H. 2012. Characterization of Th1- and Th2-type immune response in human multidrug-resistant tuberculosis. Eur J Clin Microbiol Infect Dis 31:1233–1242. doi: 10.1007/s10096-011-1434-4 [DOI] [PubMed] [Google Scholar]
- 5. Geffner L, Basile JI, Yokobori N, Kviatcovsky D, Sabio y García C, Ritacco V, López B, Sasiain M del C, de la Barrera S. 2014. Mycobacterium tuberculosis multidrug resistant strain M induces an altered activation of cytotoxic CD8+ T cells. PLoS ONE 9:e97837. doi: 10.1371/journal.pone.0097837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prammananan T, Cheunoy W, Taechamahapun D, Yorsangsukkamol J, Phunpruch S, Phdarat P, Leechawengwong M, Chaiprasert A. 2008. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect 14:446–453. doi: 10.1111/j.1469-0691.2008.01951.x [DOI] [PubMed] [Google Scholar]
- 7. Thirumurugan R, Kathirvel M, Vallayyachari K, Surendar K, Samrot AV, Muthaiah M. 2015. Molecular analysis of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates by multiple allele specific polymerase chain reaction in Puducherry, South India. J Infect Public Health 8:619–625. doi: 10.1016/j.jiph.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 8. Chikaonda T, Ketseoglou I, Nguluwe N, Krysiak R, Thengolose I, Nyakwawa F, Rosenberg NE, Stanley C, Mpunga J, Hoffman IF, Papathanasopoulos MA, Hosseinipour M, Scott L, Stevens W. 2017. Molecular characterisation of rifampicin-resistant Mycobacterium tuberculosis strains from Malawi. Afr J Lab Med 6:463. doi: 10.4102/ajlm.v6i2.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kigozi E, Kasule GW, Musisi K, Lukoye D, Kyobe S, Katabazi FA, Wampande EM, Joloba ML, Kateete DP. 2018. Prevalence and patterns of rifampicin and isoniazid resistance conferring mutations in Mycobacterium tuberculosis isolates from Uganda. PLoS ONE 13:e0198091. doi: 10.1371/journal.pone.0198091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howard NC, Marin ND, Ahmed M, Rosa BA, Martin J, Bambouskova M, Sergushichev A, Loginicheva E, Kurepina N, Rangel-Moreno J, Chen L, Kreiswirth BN, Klein RS, Balada-Llasat J-M, Torrelles JB, Amarasinghe GK, Mitreva M, Artyomov MN, Hsu F-F, Mathema B, Khader SA. 2018. Mycobacterium tuberculosis carrying a rifampicin drug resistance mutation reprograms macrophage metabolism through cell wall lipid changes. Nat Microbiol 3:1327. doi: 10.1038/s41564-018-0281-9 [DOI] [PubMed] [Google Scholar]
- 11. Slight SR, Rangel-Moreno J, Gopal R, Lin Y, Fallert Junecko BA, Mehra S, Selman M, Becerril-Villanueva E, Baquera-Heredia J, Pavon L, Kaushal D, Reinhart TA, Randall TD, Khader SA. 2013. CXCR5+ T helper cells mediate protective immunity against tuberculosis. J Clin Invest 123:712–726. doi: 10.1172/JCI65728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunlap MD, Howard N, Das S, Scott N, Ahmed M, Prince O, Rangel-Moreno J, Rosa BA, Martin J, Kaushal D, Kaplan G, Mitreva M, Kim K-W, Randolph GJ, Khader SA. 2018. A novel role for C-C motif chemokine receptor 2 during infection with hypervirulent Mycobacterium tuberculosis. Mucosal Immunol 11:1727–1742. doi: 10.1038/s41385-018-0071-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srivastava S, Ernst JD. 2014. Cell-to-cell transfer of M. tuberculosis antigens optimizes CD4 T cell priming. Cell Host Microbe 15:741–752. doi: 10.1016/j.chom.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM. 2002. Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun 70:4501–4509. doi: 10.1128/IAI.70.8.4501-4509.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolf AJ, Desvignes L, Linas B, Banaiee N, Tamura T, Takatsu K, Ernst JD. 2008. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med 205:105–115. doi: 10.1084/jem.20071367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lahiri N, Shah RR, Layre E, Young D, Ford C, Murray MB, Fortune SM, Moody DB. 2016. Rifampin resistance mutations are associated with broad chemical remodeling of Mycobacterium tuberculosis . J Biol Chem 291:14248–14256. doi: 10.1074/jbc.M116.716704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466:973–977. doi: 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cliff JM, Lee J-S, Constantinou N, Cho J-E, Clark TG, Ronacher K, King EC, Lukey PT, Duncan K, Van Helden PD, Walzl G, Dockrell HM. 2013. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis 207:18–29. doi: 10.1093/infdis/jis499 [DOI] [PubMed] [Google Scholar]
- 19. Ottenhoff THM, Dass RH, Yang N, Zhang MM, Wong HEE, Sahiratmadja E, Khor CC, Alisjahbana B, van Crevel R, Marzuki S, Seielstad M, van de Vosse E, Hibberd ML. 2012. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS ONE 7:e45839. doi: 10.1371/journal.pone.0045839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA, ACS and GC6-74 cohort study groups . 2016. A prospective blood RNA signature for tuberculosis disease risk. Lancet 387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, Yuan X, Zhang G, Cai Y, Babu S, Catalfamo M, Salazar AM, Via LE, Barry CE 3rd, Sher A. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511:99–103. doi: 10.1038/nature13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, Feng CG. 2011. Mycobacterium tuberculosis triggers host type I interferon signaling to regulate IL-1Β production in human Macrophages. J Immunol 187:2540–2547. doi: 10.4049/jimmunol.1100926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, Oland S, Gordon S, Sher A. 2011. Innate and adaptive interferons suppress IL-1α and IL-1β production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection. Immunity 35:1023–1034. doi: 10.1016/j.immuni.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ji DX, Yamashiro LH, Chen KJ, Mukaida N, Kramnik I, Darwin KH, Vance RE. 2019. Type I interferon-driven susceptibility to Mycobacterium tuberculosis is mediated by interleukin-1 receptor antagonist IL-1Ra. Nat Microbiol 4:2128–2135. doi: 10.1038/s41564-019-0578-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banks DA, Ahlbrand SE, Hughitt VK, Shah S, Mayer-Barber KD, Vogel SN, El-Sayed NM, Briken V. 2019. Mycobacterium tuberculosis inhibits autocrine type I IFN signaling to increase intracellular survival. J Immunol 202:2348–2359. doi: 10.4049/jimmunol.1801303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Braverman J, Stanley SA. 2017. Nitric oxide modulates macrophage responses to Mycobacterium tuberculosis infection through activation of HIF-1α and repression of NF-ΚB. J Immunol 199:1805–1816. doi: 10.4049/jimmunol.1700515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dorhoi A, Yeremeev V, Nouailles G, Weiner J 3rd, Jörg S, Heinemann E, Oberbeck-Müller D, Knaul JK, Vogelzang A, Reece ST, Hahnke K, Mollenkopf H-J, Brinkmann V, Kaufmann SHE. 2014. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur J Immunol 44:2380–2393. doi: 10.1002/eji.201344219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang L, Jiang X, Pfau D, Ling Y, Nathan CF. 2021. Type I interferon signaling mediates Mycobacterium tuberculosis–induced macrophage death. J Exp Med 218. doi: 10.1084/jem.20200887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antonelli LRV, Gigliotti Rothfuchs A, Gonçalves R, Roffê E, Cheever AW, Bafica A, Salazar AM, Feng CG, Sher A. 2010. Intranasal poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J Clin Invest 120:1674–1682. doi: 10.1172/JCI40817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE 3rd, Freedman VH, Kaplan G. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc Natl Acad Sci U S A 98:5752–5757. doi: 10.1073/pnas.091096998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreira-Teixeira L, Stimpson PJ, Stavropoulos E, Hadebe S, Chakravarty P, Ioannou M, Aramburu IV, Herbert E, Priestnall SL, Suarez-Bonnet A, Sousa J, Fonseca KL, Wang Q, Vashakidze S, Rodríguez-Martínez P, Vilaplana C, Saraiva M, Papayannopoulos V, O’Garra A. 2020. Type I IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and netosis. Nat Commun 11:5566. doi: 10.1038/s41467-020-19412-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wiens KE, Ernst JD. 2016. The mechanism for type I interferon induction by Mycobacterium tuberculosis is bacterial strain-dependent. PLOS Pathog 12:e1005809. doi: 10.1371/journal.ppat.1005809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong D, Chao JD, Av-Gay Y. 2013. Mycobacterium tuberculosis-secreted phosphatases: from pathogenesis to targets for TB drug development. Trends Microbiol 21:100–109. doi: 10.1016/j.tim.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 34. Desvignes L, Wolf AJ, Ernst JD. 2012. Dynamic roles of type I and type II Ifns in early infection with Mycobacterium tuberculosis. J Immunol 188:6205–6215. doi: 10.4049/jimmunol.1200255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monin L, Griffiths KL, Slight S, Lin Y, Rangel-Moreno J, Khader SA. 2015. Immune requirements for protective Th17 recall responses to Mycobacterium tuberculosis challenge. Mucosal Immunol 8:1099–1109. doi: 10.1038/mi.2014.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8:369–377. doi: 10.1038/ni1449 [DOI] [PubMed] [Google Scholar]
- 38. Griffiths KL, Ahmed M, Das S, Gopal R, Horne W, Connell TD, Moynihan KD, Kolls JK, Irvine DJ, Artyomov MN, Rangel-Moreno J, Khader SA. 2016. Targeting dendritic cells to accelerate T-cell activation overcomes a bottleneck in tuberculosis vaccine efficacy. Nat Commun 7:13894. doi: 10.1038/ncomms13894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Das S, Marin ND, Esaulova E. n.d. Lung epithelial signaling mediates early vaccine-induced Cd4+ T cell activation and Mycobacterium Tuberculosis control. mBio:e01468–21. doi: 10.1128/mBio.01468-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khader SA, Partida-Sanchez S, Bell G, Jelley-Gibbs DM, Swain S, Pearl JE, Ghilardi N, Desauvage FJ, Lund FE, Cooper AM. 2006. Interleukin 12P40 is required for dendritic cell migration and T cell priming after Mycobacterium tuberculosis infection. J Exp Med 203:1805–1815. doi: 10.1084/jem.20052545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Heng, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and samtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cui Y, Chen X, Luo H, Fan Z, Luo J, He S, Yue H, Zhang P, Chen R. 2016. Biocircos js: an interactive circos javascript library for biological data visualization on web applications. Bioinformatics 32:1740–1742. doi: 10.1093/bioinformatics/btw041 [DOI] [PubMed] [Google Scholar]
- 44. Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and bioconductor. Methods Mol Biol 1418:335–351. doi: 10.1007/978-1-4939-3578-9_16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mtb isolates containing rpoB-H445Y SNP drive altered type I interferon production during infection.
rpoB-H445Y Mtb infection limits myeloid cell recruitment and activation in mice.
rpoB-H445Y Mtb infection induces altered cytokine and chemokine production in mice.
rpoB-H445Y Mtb infection also results in a limited immune response in FeJ mice.
Ifnar -/- mice have significantly enhanced myeloid cell recruitment and activation in the lung only after infection with rpoB-H445Y Mtb.
Absence of type I IFN signaling does not impact wt HN878 Mtb infection.
Legends for all supplemental figures.
Data Availability Statement
All data that support the findings of this study are available from the corresponding author upon request. DNA sequencing data have been submitted under BioProject ID PRJNA811702.