ABSTRACT
Clostridioides difficile is a Gram-positive bacterium that can cause mild to severe diarrhea, inflammation, and colonic tissue damage in animal hosts. Symptoms of the disease can be attributed to the activity of toxin B (TcdB) secreted by C. difficile during infection. TcdB can engage multiple host cell surface receptors in vitro; however, little is known about where these receptors localize on colonic tissue and how these interactions promote disease. Here, we used immunofluorescence microscopy to first visualize TcdB interactions with two of the reported receptors, CSPG4 and Nectin-3, on cells in vitro. In cells expressing both receptors, we found that TcdB preferentially interacts with CSPG4. In moving to host colonic tissue, we observed novel localization of Nectin-3 within the brush border of epithelial cells and CSPG4 localization at epithelial cell junctions. The unexpected junctional CSPG4 signal led us to the hypothesis that the signal could represent a soluble form of the CSPG4 extracellular domain (ECD) being shed from fibroblasts in the underlying stromal layer of the tissue. To test, we set up a co-culture of epithelial cells and fibroblasts separated by transwell inserts. We observed CSPG4-ECD shed into the media of cultured fibroblasts and an accumulation in epithelial cells following co-culture. We also found that soluble CSPG4-ECD present in the conditioned media from fibroblasts can potentiate TcdB-mediated cytopathic responses in epithelial cells lacking CSPG4 expression. Based on these observations, we propose that Nectin-3 can facilitate the binding of TcdB at the epithelial surface and that a soluble form of CSPG4 derived from stromal cells can contribute to TcdB intoxication of epithelial cells in vivo.
Importance
Toxin B (TcdB) is a major virulence factor of Clostridioides difficile, a Gram-positive pathogen that is a leading cause of hospital-acquired diarrhea. While previous studies have established that TcdB can engage multiple cell surface receptors in vitro, little is known about how these interactions promote disease and where these receptors localize on colonic tissue. Here, we used immunofluorescence microscopy to visualize Nectin-3 and CSPG4 on tissue, revealing unexpected localization of both receptors on colonic epithelial cells. We show that Nectin-3, which was previously characterized as an adherens junction protein, is also localized to the brush border of colonocytes. Staining for CSPG4 revealed that it is present along epithelial cell junctions, suggesting that it is shed by fibroblasts along the crypt-surface axis. Collectively, our study provides new insights into how TcdB can gain access to the receptors Nectin-3 and CSPG4 to intoxicate colonic epithelial cells.
KEYWORDS: toxin-receptor interaction, Clostridium difficile, polarized epithelia
INTRODUCTION
Clostridioides difficile is a Gram-positive, spore-forming anaerobe and is a leading cause of hospital-acquired diarrhea in the United States (1, 2). Two major virulence factors of C. difficile are toxin A (TcdA) and toxin B (TcdB). These exotoxins damage the host epithelia and promote inflammation in the large intestine, causing symptoms ranging from diarrhea and inflammation in mild cases to pseudomembranous colitis, toxic megacolon, and death in severe cases. While TcdA contributes to symptoms of Clostridioides difficile infection (CDI), the activity of TcdB is necessary and sufficient for severe disease outcomes in various models of infection (3 – 5).
The intoxication of cells with TcdB begins with host receptor binding and endocytosis (6, 7). Acidification of the endosome facilitates the delivery of the toxin’s N-terminal glucosyltransferase domain (GTD) into the cytosol which can target and inactivate Rho family GTPases such as RhoA, Rac1, and Cdc42. The inactivation of these GTPases results in cytopathic responses such as cell rounding, disruption of tight junctions, and apoptotic cell death. In addition to an ability to bind glycans, multiple protein cell surface receptors for TcdB have been identified: chondroitin sulfate proteoglycan 4 (CSPG4), Frizzled (FZD) 1, FZD2, FZD7, Nectin-3, and tissue factor pathway inhibitor (TFPI) (8 – 13). The receptor specificity can vary depending on toxinotype as there is significant TcdB sequence variation among C. difficile strains. Among eight subtypes, TcdB1, TcdB2, and TcdB3 are the most potent when injected into mice, and the most prevalent among isolates with sequenced genomes (14). TcdB1 can be found in VPI10463, a commonly used laboratory strain, and has been shown to bind CSPG4, FZD1/2/7, and Nectin-3 with nanomolar affinities (15). The binding sites for CSPG4 and FZD1/2/7 are distinct and, in concept, can be engaged simultaneously, while the binding site for Nectin-3 remains unclear (16, 17). TcdB2 can be found in the epidemic ribotype 027 (RT027) strains and has been shown to bind CSPG4 and Nectin-3, but not FZD1/2/7 (15, 18, 19). TcdB3 can be found in ribotype 078 (RT078) strains and has been reported to bind TFPI (11, 12). While the knockout of each receptor can reduce TcdB-mediated cytopathic or cytotoxic effects in cell lines, the role of each receptor in the physiological context of infection remains unclear.
The Nectins are a family of four transmembrane proteins (Nectin-1, -2, -3, and -4) that are involved in the regulation of cell-cell adhesions (20). Nectin-3 (previously known as poliovirus receptor-like protein 3 or PVRL3) was identified as a receptor involved in TcdB-mediated cytotoxic responses in epithelial cell lines (10, 20). Nectin-3 binds TcdB1 and TcdB2 with affinities of 53 nM and 17 nM, respectively, but the interaction does not facilitate the cytopathic response, leading some to propose that Nectin-3 promotes cellular cytotoxic responses through cell surface signaling events (15, 21). While possible, Nectin-3 is known to undergo cellular entry and recycling, so the question of why Nectin-3 does not promote cytopathic responses in vitro is unclear (22).
Of the receptors that interact with TcdB, genetic deletion of CSPG4 confers the greatest protection from TcdB-mediated cell rounding (9). CSPG4, also known as neural-glial antigen 2 (NG2), is a ~300-kDa transmembrane protein involved in cell survival, migration, and angiogenesis (23, 24). The large extracellular domain (ECD) is post-translationally modified by chondroitin sulfate glycosaminoglycan chains and can serve as a scaffold for extracellular ligands to transmit signals into the cell (23). Residues within the ECD can be targeted by metalloproteinases, resulting in shed forms of soluble CSPG4-ECD (25, 26). TcdB binds to residues 410–551 of the ECD in an interaction that has now been defined with cryogenic electron microscopy (16). This same study showed that CSPG4 knockout mice infected with C. difficile experienced significantly reduced epithelial disruption and pathology. This result was somewhat puzzling as CSPG4 expression is proposed to be localized to various fibroblast populations of the intestines but not the epithelium (27).
Our initial goal in this study was to characterize TcdB interactions with Nectin-3 and CSPG4 on cells using confocal microscopy. As visualization of TcdB has been historically challenging, we screened various cell lines and identified 18Co cells, a human colonic fibroblast cell line, for robust TcdB binding and visualization. In these cells, TcdB preferentially co-localized with CSPG4. We next wanted to visualize the localization of Nectin-3 and CSPG4 in vivo, as the physiological roles of these proteins in the colon have not been fully established. In both human and mouse colonic tissues, we observed Nectin-3 at adherens junctions (as expected) but also on the apical surface as part of the brush border. We also observed co-localization of TcdB and Nectin-3 in intoxicated tissue, suggesting that Nectin-3 could have a role in capturing or concentrating TcdB at the apical surface of the epithelium. We observed CSPG4 localization in fibroblasts (as expected) but also in epithelial junctions, leading us to wonder whether this protein could represent CSPG4-ECD shed from the underlying stromal cells. We show that fibroblast-derived CSPG4-ECD can accumulate in epithelial cells and potentiate TcdB cytopathic effects. Together, these observations advance the mechanistic understanding of how specific TcdB-receptor interactions may contribute to the pathogenesis of C. difficile infection.
RESULTS
Visualization of TcdB with receptors on 18Co cells
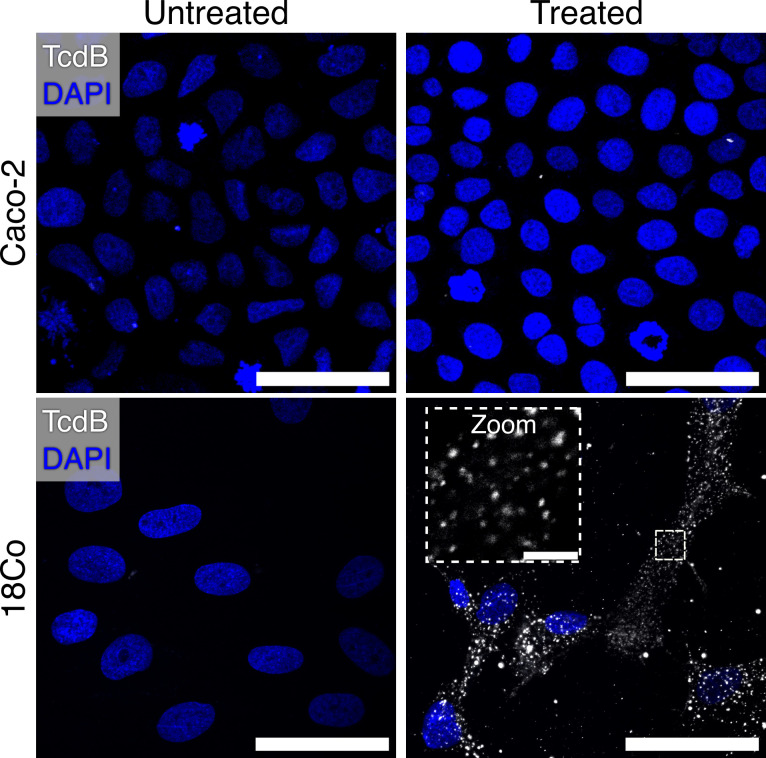
In our experience, the visualization of TcdB on cells has been challenging. For example, when Caco-2 cells are intoxicated with 10 nM Janelia Fluor 669-labeled TcdB1 (JF669-TcdB1) at 37°C for 30 minutes, no TcdB signal can be observed (Fig. 1). Knowing that cell lines can differ significantly in their cell surface protein profiles, we analyzed published transcriptomic data sets using the GEO RNA-seq Experiments Interactive Navigator to search for a cell line that expresses abundant levels of the reported TcdB receptors (28). We identified 18Co cells, derived from human colonic myofibroblasts, as cells that express all reported TcdB receptors (Fig. S1) (29 – 31). When imaged, we found that 18Co cells intoxicated with 10 nM JF669-TcdB1 contained abundant levels of TcdB signal (Fig. 1). While previous approaches used to visualize TcdB have required conditions with cold temperatures and long incubation periods, 18Co cells simply needed to be treated with labeled TcdB and incubated at 37°C (6).
Fig 1.
18Co cells can be used to visualize TcdB. Comparison between Caco-2 and 18Co cells intoxicated with 10 nM JF-669 TcdB1 for 30 minutes. Images are shown as max intensity projections while the zoomed panel is shown as a single Z-plane. Scale bars, 50 µm and 5 µm (zoom).
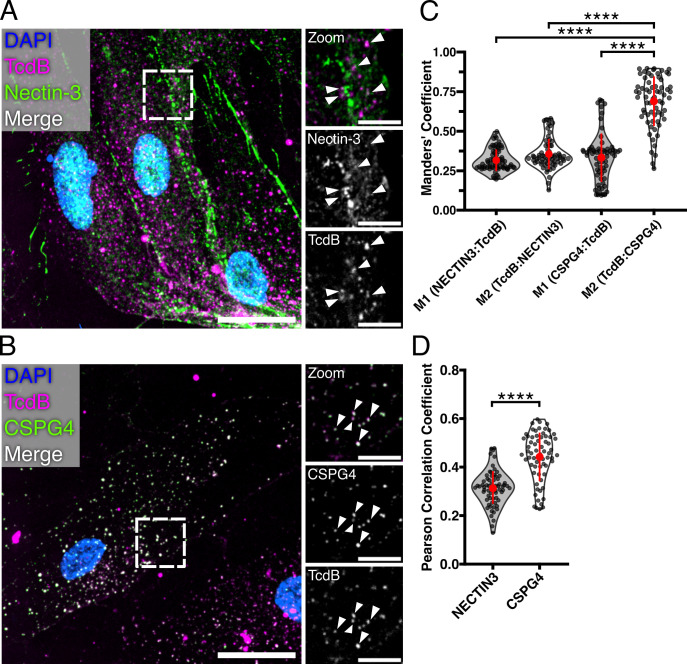
With the ability to now visualize TcdB on 18Co cells, we looked for TcdB co-localization with Nectin-3 and CSPG4 using immunofluorescence microscopy (Fig. 2). To quantify colocalization, we measured the overlap of receptors and TcdB signal using Manders’ coefficients (M1 = Receptor:TcdB and M2 = TcdB:Receptor) and the correlation of receptor signal with TcdB using the Pearson correlation coefficient (PCC). Analysis of confocal images of intoxicated 18Co cells immunostained for Nectin-3 revealed small levels of colocalization (average M1 = 0.31, M2 = 0.35) and positive correlation with TcdB (average PCC = 0.31) (Fig. 2A, C and D). Analysis of CSPG4 and TcdB revealed medium to large levels of colocalization (average M1 = 0.33, M2 = 0.68) and positive signal correlation (PCC = 0.44) (Fig. 2B, C and D). Thus, while Nectin-3 can colocalize with TcdB1 on 18Co cells, our analysis suggests that CSPG4 is the preferred interaction.
Fig 2.
TcdB primarily colocalizes with CSPG4 on 18Co cells. (A) Confocal image of 18Co cells immunostained for Nectin-3 and treated with JF669-TcdB1 for 30 minutes. The dashed box represents the location of zoomed panels. The main image is presented as a max intensity projection, while the zoomed panels are shown as a single Z-plane. Scale bars, 30 µm and 10 µm (zoom). Arrows indicate locations of colocalized signals. (B) Confocal image of 18Co cells treated with JF669-TcdB1 for 30 minutes and immunostained for CSPG4. The dashed box represents the location of zoomed panels. The main image is presented as a max intensity projection, while the zoomed panels are shown as single Z planes. Scale bars, 30 µm and 10 µm (zoom). Arrows indicate locations of colocalized signals. (C) Manders’ coefficient analyses of receptor signal overlap with TcdB (M1) or TcdB signal overlap with receptors (M2). Points represent individual Manders’ coefficients. The error bar in red represents the mean and standard deviation. ****P < 0.0001, Kruskal-Wallis ANOVA test and corrected for multiple comparisons using Dunn’s post hoc test. N = 5 (D) Pearson correlation coefficient analysis of TcdB colocalization with Nectin-3 or CSPG4. Points represent individual Pearson coefficients. The error bar in red represents the mean and standard deviation. ****P < 0.0001, Mann-Whitney U test. N = 5.
Nectin-3 localizes to the colonic epithelial brush border and apical cell junctions
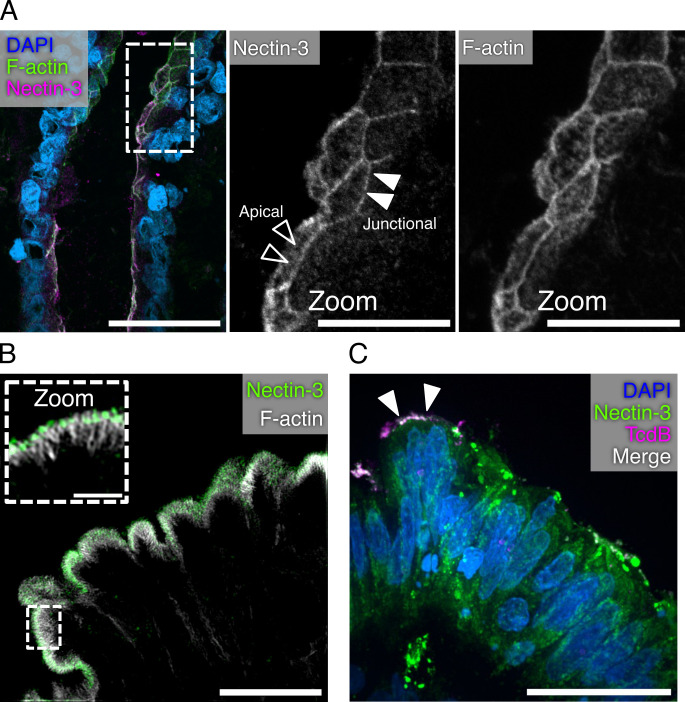
Nectin-3 belongs to a family of adhesion proteins that promote the formation of adherens junctions during the initial interactions between polarized cells (32). Since the 18Co cells do not form polarized junctions, we decided to turn to the epithelial cells of colonic tissue for imaging TcdB interactions with Nectin-3. Previous studies have established that Nectin-3 is expressed in vivo on mouse colonic epithelial cells with epifluorescence microscopy (33). We attempted to replicate these previous studies using human colonic tissue. As expected, we observed the Nectin-3 signal at the epithelial cell junctions (Fig. 3A). In addition, the Nectin-3 signal was present along the apical surface of the cell, potentially within the brush border (Fig. 3A, zoomed panels). The brush border is an array of microvilli supported internally by bundles of actin and organized externally by adhesion proteins (34). We acquired higher-resolution images of this brush border localized Nectin-3 signal using structured illumination microscopy (Fig. 3B, zoom). These images revealed that Nectin-3 is indeed present within the brush border on colonic epithelial cells. While the role of Nectin-3 within the microvilli is unresolved, Nectin-3 can also be observed within the brush border of polarized Caco-2 cells (Fig. S2). These findings suggest that Nectin-3, and perhaps other Nectin family proteins, has a role in brush border structure and function.
Fig 3.
NECTIN-3 localizes to the brush border of the colonic epithelium. (A) Confocal image of human colonic tissue stained for Nectin-3, F-actin, and DAPI. Dashed boxes represent the origin of the zoomed image. Black arrowheads indicate a signal localized to the apical surface of cells, while white arrowheads indicate areas of junction. Scale bars, 50 µm and 20 µm (zoom). (B) Structured illumination microscopy image of human colonic epithelium stained for Nectin-3 and F-actin. Dashed boxes represent the origin of zoomed image. The main image is presented as a max intensity projection, while the zoomed panels are shown as a single Z-plane. Scale bars, 10 µm and 2 µm (zoom). (C) Confocal image of a human colonic explant intoxicated with JF669-TcdB1 for 1 hour at 37°C and stained for Nectin-3. Scale bar, 30 µm.
With the unexpected finding that Nectin-3 localizes to the brush border of colonic epithelial cells, we wanted to determine whether TcdB could interact with Nectin-3 on colonic tissue. We intoxicated human colonic explants with JF669-TcdB1 and immunostained the tissue for Nectin-3 (Fig. 3C). While detection of JF669-TcdB1 was infrequently observed, colocalization between Nectin-3 and JF669-TcdB1 was observed along the apical surface of the colonic epithelium. These data suggest a role for Nectin-3 in binding to TcdB along the luminal surface of cells.
Colonic epithelial tissue is CSPG4(+)
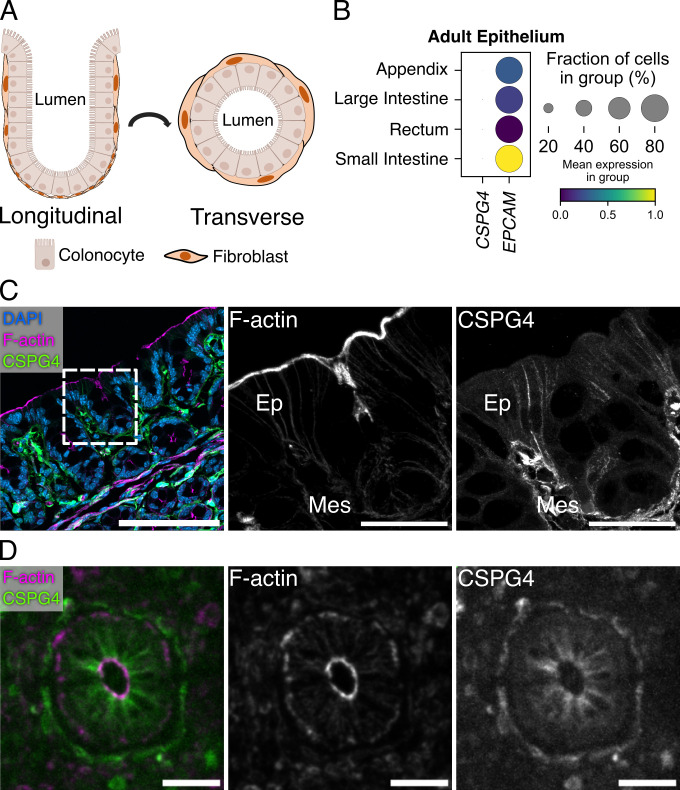
While single-cell sequencing studies have consistently shown CSPG4 expression among various fibroblast populations along the crypt-surface axis of the intestines (Fig. 4A), there is no evidence for CSPG4 expression in colonic epithelial cells (35, 36). We analyzed a published single-cell RNA-sequencing (scRNA-seq) data set from the Human Gut Cell Atlas derived from 77,341 EPCAM+ epithelial cells from the small intestine, large intestine, appendix, and rectum of healthy adults to determine whether CSPG4 expression could be found in any epithelial cell populations (37). Among these epithelial populations, we were unable to detect CSPG4 expression, confirming that CSPG4 expression is isolated to stromal populations in the colon (Fig. 4B).
Fig 4.
Mouse and human colonic epithelial cells stain positive for CSPG4. (A) Model of the colonic crypt-surface axis demonstrating the longitudinal and transverse views observed in panels C and D. (B) Analysis of single-cell RNA expression data from human adult intestinal epithelial cells (Gut Cell Atlas) reveals that CSPG4 is not expressed in epithelial cells of human intestines. (C) Confocal images of wild-type C57BL/6 mouse colon stained for CSPG4. Mouse epithelial cells and fibroblasts stained positive for CSPG4. Dashed boxes indicate the location of zoomed images. Scale bars, 50 µm and 30 µm (Zoom). (D) Confocal images of a cross-section of human ascending colon stained for CSPG4. Both fibroblasts and epithelial cells stained positive for CSPG4. Scale bars, 50 µm.
While imaging CSPG4 on mouse colonic tissue, we observed the predicted CSPG4 signal in fibroblasts that line the crypt surface axis (Fig. 4C, zoom). However, to our surprise, we also observed the CSPG4 signal along the junctions of epithelial cells. To verify this staining pattern, we stained colonic tissue from CSPG4−/− mice for CSPG4 (Fig. S3). These mice lacked CSPG4 signal on both fibroblasts and the junctions of colonic epithelial cells. We also stained human colonic tissue for CSPG4 and found that both fibroblasts and epithelial tissue stained positive for CSPG4 (Fig. 4D). These data led us to wonder whether fibroblasts along the crypt-surface axis could shed CSPG4 ECD that would then associate with the epithelium.
Caco-2 cells accumulate fibroblast--derived CSPG4-ECD
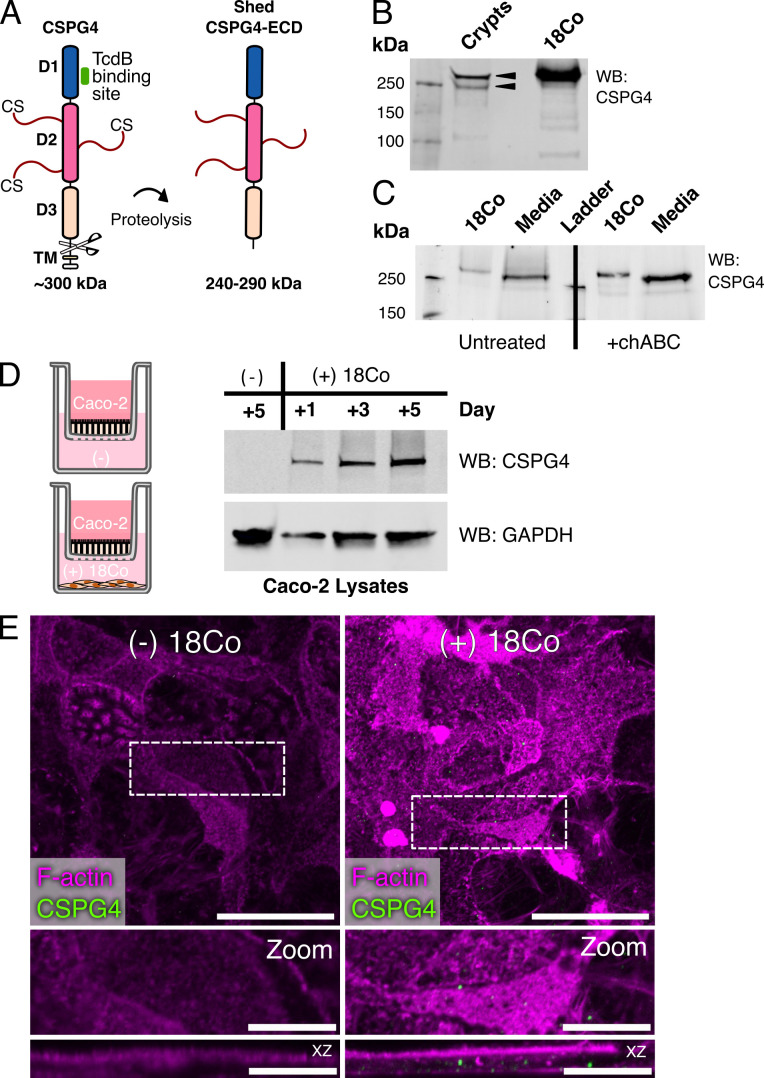
In humans, CSPG4 is a 2,322 amino acid integral membrane protein with a predicted protein mass of 250,537 Da, numerous glycosylation sites, and up to three glycosaminoglycan attachment sites. When viewed by SDS-PAGE, the CSGP4 core protein is often reported as ~300 kDa, with larger species visible as a smear (25, 38). In addition, the ECD of CSPG4 can be shed from the cell surface by various proteinases (Fig. 5A) (25, 26, 39 – 41). The MW range of the prevalent CSPG4-ECD fragments identified both in vitro and in vivo, is 240–290 kDa and can vary with cell type (24). We isolated and lysed crypts from mouse colons to determine if we could detect any cleaved forms of CSPG4 by western blot (Fig. 5B). Analysis of these lysates revealed two primary species, with bands above and below the 250 kDa protein standard. While we presume, but cannot confirm, that the upper band corresponds to the ~290 kDa ECD fragment, the presence of the smaller fragment running at ~240 kDa provides evidence that CSPG4 shedding is occurring in the mouse colon and that CSPG4 fragments associate with epithelial cells of the mouse colonic crypt.
Fig 5.
CSPG4-ECD from 18Co cells can enter Caco-2 cells. (A) A cartoon diagram of CSPG4. CSPG4 (~300 kDa) is a 2322 amino acid (aa) core glycoprotein-proteoglycan complex that consists of extracellular subdomains D1, D2, and D3, a transmembrane domain (TM), and an intracellular domain. The TcdB binding site is contained within the D1 subdomain. The D2 subdomain is predicted to contain multiple glycosylation and chondroitin sulfate (CS) sites. Residues between D3 subdomain and TM domain can be targeted by proteases to generate shed forms of the CSPG4 extracellular domain (ECD, 240–290 kDa) that can be released into the extracellular matrix or non-covalently associated with the cell. (B) A CSPG4 fragment corresponding to the putative MW of a full-length CSPG4-ECD is present in purified mouse colonic crypt lysates. Lysates from 18Co cells were used as a reference for the full-length CSPG4. Black arrowheads indicate the forms of CSPG4 present in the mouse crypt lysates. (C) 18Co cells were grown for 72 hours and the conditioned media and cells were individually treated with or without 0.1 units of chondroitinase ABC (chABC). Cell lysates (18Co) and conditioned media (media) were analyzed for CSPG4 by western blot. (D) Caco-2 cells were grown above 18Co cells in transwells, were collected and lysed on days 1, 3, and 5, and analyzed for CSPG4 by western blot. Caco-2 cells grown with 18Co cells contained CSPG4, while the Caco-2 cells grown without 18Co cells did not. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control for cell lysates. (E) Confocal images of 5 days post-confluency Caco-2 cells grown above 18Co cells in transwells and stained for CSPG4. Dashed boxes indicate the origin of zoomed panels and are shown as max projections in both xy and xz.
A possible source of shedding CSPG4 could be from stromal cells such as myofibroblasts found along the crypt-surface axis. As 18Co cells are a normal, non-transformed colonic myofibroblast cell line, we analyzed its conditioned media by western blot to determine whether these cells were capable of shedding CSPG4-ECD into the media (Fig. 5C). Compared to the band observed in the 18Co cell lysates, the conditioned media contained a smaller form that consistently ran just above the ~250 kDa protein standard. This was observed both in the absence and presence of chondroitinase ABC treatment, consistent with other reports showing that the chondroitin sulfate groups do not contribute to the size of the core complex observed by SDS-PAGE.
We next asked whether this 18Co-derived soluble CSPG4-ECD could be transferred to Caco-2 cells, a human colonic epithelial cell line through co-culture. Caco-2 cells were grown in cell inserts above 18Co cells for 5 days before collecting and lysing the Caco-2 cells. A control condition of Caco-2 cells grown for 5 days without 18Co cells was included. Over a 5-day period, we observed increasing intensities of CSPG4 in the Caco-2 lysates (Fig. 5D). Over the same 5-day period, we were unable to detect CSPG4 in the Caco-2 cells grown without 18Co cells. We then stained Caco-2 cells for CSGP4 after growing them for 5 days on cell inserts above 18Co cells. We observed CSPG4 signal both within and along the edges of Caco-2 cells (Fig. 5E). These data suggest that CSPG4-ECD derived from 18Co cells can enter Caco-2 cells.
CSPG4(+) conditioned media potentiate cell rounding
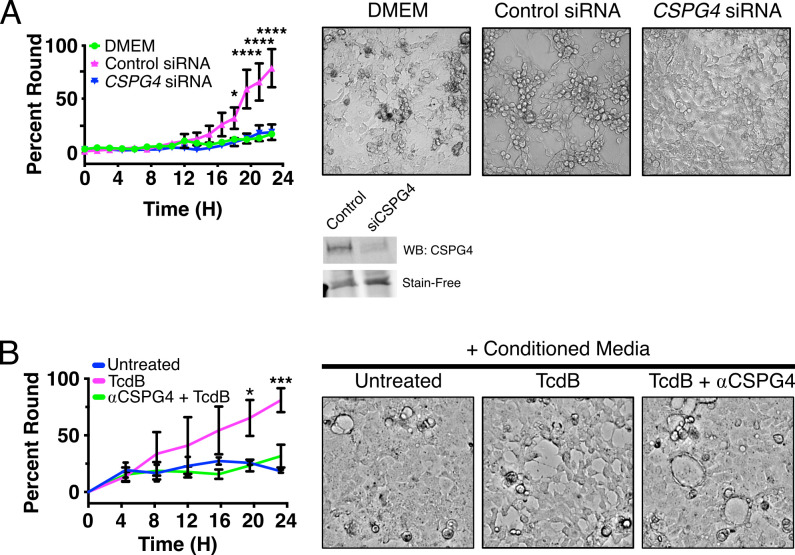
Given our observation that CSPG4-ECD can associate with Caco-2 cells, we next asked whether the 18Co-derived CSPG4-ECD would promote TcdB-induced cytopathic effects in Caco-2 cells. For these studies, we used TcdB2 to eliminate any FZD 1/2/7 responses. We intoxicated Caco-2 cells with 1 pM TcdB2 and 18Co conditioned media and found that these intoxicated cells rounded faster in the presence of conditioned media (Video S1). To determine whether this rounding was dependent on CSPG4-ECD, we collected conditioned media from 18Co cells treated with siRNA targeting CSPG4 or with a non-targeting control. Treatment of 18Co cells with siRNA targeting CSPG4 was effective at depleting CSPG4 in the media (Fig. 6A). We then intoxicated Caco-2 cells with 1 pM TcdB2 in 18Co conditioned media derived from these siRNA conditions and observed that this potentiation of cell rounding was CSPG4 dependent (Fig. 6A). Furthermore, we also confirmed that cell rounding is not promoted by 18Co conditioned media in the absence of TcdB (Fig. S4). As a complementary approach, we tested the effect of adding an anti-CSPG4 antibody to the conditioned media. Pre-incubation of conditioned media with an anti-CSPG4 antibody prior to intoxication of cells with 1 pM TcdB2 inhibited the potentiation of rounding (Fig. 6B). Together, these data suggest that soluble CSPG4-ECD can enhance the cytopathic effects of TcdB on epithelial cells, even in the absence of epithelial CSPG4 transcript expression.
Fig 6.
Fibroblast-derived conditioned media promotes Caco-2 cell rounding and is CSPG4 dependent. (A) Caco-2 cells were intoxicated with 1 pM TcdB2 in complete media (Dulbecco’s modified Eagle’s medium [DMEM]) or conditioned media from siRNA-treated 18Co cells targeting a non-targeting control (Control siRNA) or CSPG4. Rounding was quantified from images acquired over the course of 24 hours. Images to the right are representative images acquired at the end of the experiment. A representative western blot of CSPG4 in the 18Co conditioned media from the non-targeting control siRNA (Control) and CSPG4 siRNA (siCSPG4) conditions used in these experiments is shown below. A fluorescent image of the “stain-free” gel used in this western blot is shown to demonstrate equal loading of protein. Statistical analysis was performed using a two-tailed ANOVA with a Sidak post hoc test for multiple comparisons. N = 3 *P < 0.05, ****P < 0.0001. See also Fig. S4. (B) Conditioned media from 18Co cells were treated with antibodies targeting CSPG4 prior to the intoxication of Caco-2 cells with 1 pM TcdB2. Conditioned media lacking antibodies targeting CSPG4 rounded over a 24-hour period. Representative images from these assays are shown on the right that were acquired at the end of the experiment. Statistical analysis was performed using a two-tailed ANOVA with a Sidak post hoc test for multiple comparisons. Images to the right of the graph are representative images acquired at 24 hours. N = 3, *P < 0.05, ***P < 0.001.
DISCUSSION
TcdB is a key virulence factor in CDI. It is expressed by C. difficile in the lumen of the colon, and therefore the epithelial cells that form the barrier between the host and microbiota are expected to represent the first cells that the toxin will encounter. As intoxication requires receptor-mediated endocytosis, multiple studies have focused on the identification of epithelial cell surface proteins that can serve as TcdB receptors. The studies have largely been conducted in vitro using a combination of cell-based genetic screens and biochemical analyses and have led to the identification of multiple cell surface proteins that could be contributing to TcdB-induced symptoms (8 – 12). However, the understanding of whether and how each receptor may contribute to intoxication in vivo has been unclear for several reasons. First, the receptors that have been identified are not thought to be expressed on the apical surface of polarized cells (27, 33, 42, 43). FZD 1/2/7, Nectin-3, and TFPI have all been reported to have lateral or basolateral expression, and CSPG4 is not thought to be expressed at all in epithelial cells (27, 33, 42, 43). The proteins could still serve as receptors for TcdB that passes the barrier of the polarized epithelium, but the question of how the toxin makes initial contact with the epithelium has been unclear. Second, TcdB is a highly potent toxin capable of inducing cytopathic responses with EC50 values in the fM-pM range, depending on the cell type (44, 45). The protein receptors identified to date, each bind TcdB with nM affinities (8, 9, 11, 12, 15). While these affinities are tight by some standards, they are weak when considering the potency of TcdB. The potency of TcdB suggests the need for co-receptor interactions, but it is unclear which receptor pairs are co-expressed and co-localized in the same cells to be available for binding to TcdB. Lastly, the question of endocytosis has largely been examined indirectly, using the cytopathic rounding response as a proxy for cellular endocytosis. The direct visualization of TcdB requires high concentrations that are rapidly cytotoxic and likely not representative of what one would find in an early stage of CDI.
In this study, we characterized the localization of Nectin-3 and CSPG4 within the colon using immunofluorescence confocal microscopy. Our initial predictions that Nectin-3 would localize to the junctions of epithelial cells and CSPG4 would localize to fibroblasts below the crypt-surface axis were confirmed. However, we also found that Nectin-3 and CSPG4 localized to additional and unexpected locations on epithelial cells. In addition to the epithelial cell junctions (Fig. 3A), we found Nectin-3 localized at the epithelial brush border in microvilli (Fig. 3B). With CSPG4, we saw robust localization with fibroblasts along the crypt-surface axis but also at the junctions of epithelial cells (Fig. 4C). Since epithelial cells do not express CSPG4 transcripts (Fig. 4B), we reasoned that the CSPG4 signal represented soluble CSPG4-ECD shed by the underlying fibroblasts. Indeed, we showed that at least one proteolytic fragment of CSPG4 is present in lysates from murine colonic crypts (Fig. 5B), similar to an observation reported in another study showing that a shed form of CSPG4 is present in lysates from the small intestine of mice (27).
Since both receptors are capable of binding TcdB, we attempted to visualize fluorescently labeled TcdB with receptors on colonic tissue. While we could show evidence that TcdB colocalizes with Nectin-3 on colonic explants intoxicated with TcdB (Fig. 3C), the TcdB signal was infrequent, consistent with the challenge of trying to visualize a potent toxin in vivo. Therefore, we used cell culture models to ask whether TcdB is capable of colocalizing with these receptors. Historically, visualization of TcdB is challenging, with various cell lines showing little to no signal after intoxication (6). We show that 18Co cells provided strong signal for TcdB (Fig. 1) and that both Nectin-3 and CSPG4 are capable of colocalizing with TcdB with modest to high levels of correlation (Fig. 2D). Our measurement of colocalization using the Manders’ correlation coefficient showed that on average 68% TcdB signal overlapped with CSPG4 (Fig. 2C), revealing that CSPG4 is the preferred receptor on 18Co cells. The observation that TcdB had greater levels of colocalization with CSPG4 than Nectin-3 was surprising, as both receptors have similar reported affinities with TcdB. In considering the possible explanations for this difference, it could be that the receptors differ in their accessibility at the cell surface or that the affinity of TcdB for CSPG4 on the cell surface is tighter than what was observed with a recombinant fragment of the protein (8, 15). Alternatively, a higher affinity interaction between TcdB and CSPG4 could be mediated through co-receptor interactions with glycans on CSPG4 or with the various proteins CSPG4 interacts with along the cell surface (13, 24).
The observation that a shed form of CSPG4 exists within the colon led us to test if shed forms of CSPG4-ECD contribute to TcdB-mediated cytopathic effects. We found that 18Co cells shed abundant levels of CSPG4 into the media over time (Fig. 5C). We noted that Caco-2 cells, which lack CSPG4 expression, accumulate CSPG4 when supported by 18Co cells on transwells (Fig. 5D). Moreover, we found that 18Co conditioned media containing shed CSPG4 potentiated Caco2 cell rounding in response to concentrations of TcdB2 that typically do not induce a cytopathic response (Fig. 6A). The presence of CSPG4 in the conditioned media was required for this potentiation, as conditioned media derived from 18Co cells treated with siRNA targeting CSPG4 (Fig. 6A) or incubated with an antibody targeting CSPG4 (Fig. 6B) were unable to promote cell rounding. Together, these data suggest that shed forms of CSPG4-ECD can bind the cellular junctions of epithelial cells and could contribute to TcdB intoxication in vivo.
The observations of this study raise a new series of questions, however. First, while the co-localization of TcdB and Nectin-3 on the brush border provides conceptual evidence that Nectin-3 could represent a readily accessible binding partner for TcdB being made in the colon, we still do not know if Nectin-3 can mediate TcdB entry and intoxication in vivo. Further studies to map the Nectin-3-TcdB binding interaction are needed to dissect the functional importance of this interaction.
Second, while we showed CSPG4 localized at epithelial junctions in vivo and that shed CSPG4-ECD can potentiate TcdB-induced cytopathic responses on epithelial cells in vitro, we still do not know whether shed CSPG4-ECD potentiates TcdB intoxication of the epithelial layer in vivo. However, such a mechanism would be consistent with studies showing that mice infected with C. difficile experience significantly lower epithelial injury and overall disease if CSPG4 protein expression is depleted or genetically removed (16, 46).
Third, if the shed CSPG4 ectodomain is, in fact, acting as a TcdB receptor on epithelial cells, then what molecules are responsible for the binding and entry of TcdB-bound CSPG4-ECD? Given the vast number of known CSPG4 binding partners, which include growth factors as well as type V or VI collagens, perlecan, α-integrins, and galectin-3 (47 – 49), we were not able to answer this question in the scope of this study. However, it is also possible that the localization of CSPG4-ECD at the junctions provides TcdB with the necessary co-receptor pairing for Nectin-3, FZD1/2/7, and or TFPI-mediated events. Such pairings would not be plausible when considering cellular receptor expression profiles in isolation but could become relevant in the environment of colonic tissue. Further studies to examine these complex interactions in vivo are needed in the effort to identify the key intervention points for blocking TcdB-induced pathology in CDI.
MATERIALS AND METHODS
Cell lines and reagents
18Co (CCD-18Co, ATCC) and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), with high glucose, sodium pyruvate, L-glutamine (Gibco), and 10% fetal bovine serum (FBS, Corning). Caco-2 (C2BBe1, ATCC) cells were cultured with Eagle’s minimum essential medium (EMEM, ATCC) in 20% FBS. Cells were cultured in a humidified incubator at 37°C at 5% CO2.
Protein expression and purification
The vector for expressing TcdB2 (pBL598) was a gift from Dr. J Ballard (University of Oklahoma Health Sciences Center). Recombinant TcdB1 and TcdB2 were expressed and purified from Bacillus megaterium as previously described (Table 1) (6).
TABLE 1.
Bacterial strains and plasmids
| Plasmid or strain | Relevant genotype or features | Source, construction, or reference |
|---|---|---|
| Strains | ||
| B. megaterium | ||
| bm003 | TcdB1 expression strain containing pBL377 | (50) |
| bm074 | TcdB2 expression strain containing pBL598 | J. Ballard |
| Plasmids | ||
| pBL377 | TcdB1 expression vector | (50) |
| pBL598 | TcdB2 expression vector | J. Ballard |
| pLenti CMV GFP Puro (658-5) | Lentiviral expression vector encoding GFP under a CMV promoter | (51) |
| H2B-mCherry | Lentiviral expression vector encoding H2B fused to mCherry | (52) |
| psPAX2 | 2nd-Generation lentiviral packaging plasmid | Cat#12260,Addgene; RRID: Addgene_12260 |
| pMD2.G | VSV-G envelope expressing plasmid | Cat#12259, Addgene; RRID: Addgene_12259 |
Labeling TcdB
TcdB1 was purified as described above but with modifications. TcdB was isolated over an S200 column in 20 mM HEPES pH 8.3 in 50 mM NaCl. Fractions containing TcdB were pooled and concentrated to 20–25 µM using a 100-kDa MWCO filter (MilliporeSigma). A 20-molar excess of NHS-JF-669 (Tocris) Table 2was added to TcdB and incubated at room temperature in the dark with occasional pipetting for 2 hours. The labeling reaction was quenched with 50 mM Tris pH 8.0 and 50 mM NaCl for 10 minutes and applied through a PD-10 column (Cytiva) equilibrated with the same buffer to remove aggregates and unreacted dyes. Eluted samples were applied through an S200 column and collected in 20 mM HEPES pH 8 and 50 mM NaCl. Preparations of labeled TcdB resulted in ~1.5 dye labels per TcdB molecule.
TABLE 2.
Materials
| Reagent | Source | Identifier |
|---|---|---|
| Antibodies and cell stains | ||
| Rabbit monoclonal anti-CSPG4 | Abcam | Cat#ab255811; RRID: N/A |
| Mouse monoclonal anti-CSPG4 | Santa Cruz Biotechnology | Cat#sc-53389; RRID: AB_784821 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat#2118; RRID: AB_561053 |
| Rabbit anti-ZO-1 | Thermo Fisher Scientific | Cat#61–7300; RRID: AB_2533938 |
| Goat anti-Nectin-3 | Thermo Fisher Scientific | Cat#PA5-47441; RRID: AB_2577074 |
| Mouse anti-Nectin-3 | Santa Cruz Biotechnology | Cat#sc-271611; RRID: AB_10709881 |
| Goat anti-rabbit 488 | Thermo Fisher Scientific | Cat#A-11008; RRID: AB_143165 |
| Goat anti-rabbit 546 | Thermo Fisher Scientific | Cat# A-11035; RRID: AB_2534093 |
| Goat anti-mouse DyLight 680 | Cell Signaling Technology | Cat#5470; RRID: AB_10696895 |
| Goat anti-rabbit DyLight 800 | Cell Signaling Technology | Cat# 5151; RRID: AB_10697505 |
| Donkey anti-rabbit 488 | Thermo Fisher Scientific | Cat#A-21206; RRID: AB_2535792 |
| Donkey anti-goat 488 | Thermo Fisher Scientific | Cat#A-11055; RRID: AB_2534102 |
| Donkey anti-goat 555 | Thermo Fisher Scientific | Cat#A-21432; RRID: AB_2535853 |
| Alexa Fluor Plus 405 Phalloidin | Thermo Fisher Scientific | Cat#A30104 |
| Alexa Fluor 546 Phalloidin | Thermo Fisher Scientific | Cat#A22283 |
| Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | Cat#A22287 |
| Chemicals | ||
| NHS-Janelia Fluor 669 | Tocris | Cat#6420 |
| EZ-Link Sulfo-NHS-SS-Biotin | Thermo Fisher Scientific | Cat#21331 |
| Cell culture reagents | ||
| PDGF-AA | STEMCELL Technologies | Cat#78095 |
| Enzymes | ||
| Chondroitinase ABC from Proteus vulgaris | MilliporeSigma | Cat#C3667 |
Antibody staining
18Co cells were intoxicated with 10 nM JF669-TcdB1 for 30 minutes at 37°C. After intoxication, cells were washed once in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde in PBS for 15 minutes. Fixed samples were washed four times in PBS for 5 minutes of incubation per wash and blocked in 5% normal donkey serum (NDS, Jackson ImmunoResearch Labs) in PBS with 0.3% Triton X-100 for 1 hour at 37°C for samples to be stained for Nectin-3 or in 5% normal goat serum (NGS, Jackson ImmunoResearch Labs) in PBS with 0.3% Triton-X 100 for 1 hour at 37°C for samples to be stained for CSPG4. Coverslips were washed once in PBS prior to incubating overnight with primary antibodies (Table 2). Staining was performed using antibodies targeting Nectin-3 (Thermo Fisher Scientific, 1:40) prepared in 1% NDS in PBS with 0.3% TritonX-100 or CSPG4 (Abcam, 1:250) prepared in 1% NGS in PBS with 0.3% TritonX-100 overnight at 4°C. Coverslips were washed three times in PBS for at least 5 minutes per wash. Coverslips were then incubated in secondary antibody Alexa Fluor-488 donkey anti-goat (Thermo Fisher Scientific, 1:1,000) in 1% NDS in PBS with 0.3% TritonX-100 or Alexa Fluor-488 goat anti-rabbit (Thermo Fisher Scientific, 1:1,000) secondary antibody in 1% NGS in PBS with 0.3% Triton X-100 for 1 hour at room temperature. Next, coverslips were washed four times in PBS with at least 5 minutes per wash, stained with DAPI, mounted onto slides using ProLong Gold antifade (Thermo Fisher Scientific), and cured overnight in the dark. Images of 18Co cells are shown as max intensity projections unless otherwise noted in zoomed panels.
Optimal cutting temperature (OCT) compound from human and mouse colonic sections was removed by incubating sections in PBS for 5 minutes. For human tissue, slides were fixed in 4% paraformaldehyde for 10 minutes and washed three times in PBS with 5 minutes per wash. Human sections that were to be stained for Nectin-3 were blocked in 5% NDS in PBS with 0.3% Triton X-100 for 1 hour. Human and mouse tissue sections that were to be stained for CSPG4 were blocked in 5% NGS in PBS with 0.3% Triton X-100 for 1 hour at room temperature. Sections were briefly washed in PBS and then incubated overnight in primary antibodies targeting Nectin-3 (Thermo Fisher Scientific, 1:40) in 1% NDS in PBS with 0.3% Triton X-100 or in primary antibodies targeting CSPG4 (Abcam, 1:250) in 1% NGS in PBS with 0.3% Triton X-100 at 4°C. The following day, slides were washed three times in PBS for 5 minutes per wash and incubated for 1 hour at room temperature in secondary antibody Alexa Fluor-488 donkey anti-goat (Thermo Fisher Scientific, 1:200) and Alexa Fluor-546 phalloidin in 1% NDS in PBS with 0.3% Triton X-100 for slides stained for Nectin-3 or in secondary antibody Alexa Fluor-488 goat anti-rabbit (Thermo Fisher Scientific, 1:200) and Alexa Fluor-647 phalloidin (Thermo Fisher Scientific, 1:200) in 1% NGS in PBS with 0.3% Triton X-100 for slides stained for CSGP4. Finally, sections were briefly stained with DAPI and washed four times in PBS with 5 minutes per wash. Stained tissues were mounted in ProLong Gold antifade, covered with 1.5 coverglass, and cured overnight.
Caco-2 cells on coverslips were grown for 20 days before washing once in PBS and fixed in ice-cold 1:1 acetone:methanol at −20°C for 15 minutes. Fixed cells were washed four times in PBS with 3.5 minutes of incubation per wash and blocked in 5% NDS in PBS with 0.3% Triton X-100 for 1 hour at 37°C. Coverslips were washed once in PBS prior to incubating overnight with primary antibodies. Staining was performed using antibodies targeting Nectin-3 (Thermo Fisher Scientific, 1:40) and ZO-1 (Thermo Fisher Scientific, 1:50) prepared in 1% NDS in PBS with 0.3% Triton X-100 overnight at 4°C. Coverslips were washed three times in PBS for at least 5 minutes per wash. Coverslips were then incubated in Alexa Fluor-488 donkey anti-rabbit (Thermo Fisher Scientific, 1:1,000), Alexa Fluor-555 donkey anti-goat (Thermo Fisher Scientific, 1:1,000), and Alexa Fluor-647 phalloidin (Thermo Fisher Scientific, 1:200) in 1% NDS in PBS with 0.3% Triton X-100. Next, coverslips were washed four times in PBS with at least 5 minutes per wash and mounted onto slides using ProLong Gold antifade (Thermo Fisher Scientific) and cured overnight. Images are shown as max intensity projections and contrast enhanced using ImageJ (NIH). Orthogonal views were generated in ImageJ.
Caco-2 cells on transwell membranes that were fixed and permeabilized were blocked for 1 hour in 5% NGS in PBS with 0.3% Triton X-100 for 1 hour at room temperature and briefly washed in PBS, removing the supernatant by inverting the transwell insert. Blocked transwells were incubated in antibodies targeting CSPG4 (Abcam, 1:250) overnight in 1% bovine serum albumin fraction V (BSA, VWR) in PBS with 0.3% Triton X-100 at 4°C. The following day, the membranes were washed in PBS three times at 5 minutes per wash and incubated in secondary antibody Alexa Fluor-546 goat anti-rabbit (Thermo Fisher Scientific, 1:1,000) and Alexa Fluor Plus-405 phalloidin (Thermo Fisher Scientific, 1:200) in 1% BSA in PBS with 0.3% Triton X-100 for 1 hour at room temperature. Transwells were then washed four times in PBS, and the membrane was excised from the transwell insert using a scalpel. Membranes were placed on a slide and a 1.5 coverslip containing Prolong Gold antifade was placed onto each membrane. Mounted membranes were cured overnight in the dark. Images are shown as max intensity projections with en face views of the entire cell and were contrast enhanced using ImageJ (NIH). Cropped planes and X-Z images are shown as max projections along a single cell in the monolayer to present an intracellular CSPG4 signal within the cell.
Confocal and super-resolution microscopy
Confocal imaging of Caco-2 cells and 18Co cells intoxicated with only JF669-TcdB1 was conducted using a Zeiss LSM 710 META inverted microscope equipped with a Plan-Apochromat (Apo) 63×/1.4 NA objective lens and 405 and 633 nM excitation LASERs. Confocal imaging of Nectin-3 or CSPG4 immunostained human tissue and 18Co cells immunostained for receptors and intoxicated with JF669-TcdB1 were imaged using a Nikon Spinning Disk microscope equipped with a Plan-Apo 60×/1.4 NA objective lens and 405, 488, 561, and 647 nm excitation LASERs. Confocal imaging of CSPG4 immunostained mouse tissue, Caco-2 cells immunostained for Nectin-3, or Caco-2 cells grown on transwells and immunostained for CSPG4 was performed using a Zeiss LSM980 microscope with Airyscan 2 equipped with a Plan-Apo 63×/1.4 NA objective lens, an Airyscan detector, and 405, 445, 488, 514, 561, and 639 nm excitation LASERs. Super-resolution imaging of human colonic tissue stained for Nectin-3 was performed using a Nikon Structured Illumination microscope equipped with an Andor DU-897 EMCCD camera, a 100×/1.49 NA TIRF oil immersion objective, and 405, 488, 561, and 647 nm LASERs. The Nikon Elements software was used to reconstruct images. LASER intensities and camera gain were matched between samples for each experimental condition.
Analysis of colocalization of receptors and TcdB in 18co cells
Confocal images were analyzed in ImageJ (NIH) using the EZcolocalization plugin (53). Images generated from the same imaging experiments were thresholded with the same values prior to quantifying colocalization using EZcolocalization. No region of interest was used in this analysis. Manders’ and Pearson correlation coefficients generated from EZcolocalization were plotted in R (v4.2.0) using the R packages ggplot2 (v3.4.0), ggbeeswarm (v0.6.0), and ggprism (v1.0.4) (54 – 57).
siRNA knockdown of CSPG4
All siRNA stocks were purchased from Horizon Discovery and resuspended to 10 µm. To prepare the siRNA for transfection, 2 µL of the 10 µM stocks of non-targeting control siRNA (D-001210–2) or siRNA targeting CSPG4 (L-011632–01) was diluted into 18 µL of serum-free DMEM. Meanwhile, 12 µL of RNAiMax (Thermo Fisher Scientific) was added to 388 µL of serum free DMEM. The transfection reagent mixture was then mixed with the siRNA and incubated at room temperature for 15 minutes. This siRNA:RNAiMax solution was transferred into a six-well dish and a suspension of 170,000 18Co cells in 1.6 mL complete media was added. The molarity of siRNA in these conditions was 10 nM. After 24 hours, the media was replaced with fresh media and grown for additional 48 hours. Knockdowns were confirmed by western blot from cells lysed with TritonX-100 lysis buffer, consisting of 50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and HALT protease inhibitor (Thermo Fisher Scientific).
18co conditioned media purification
18Co cells were seeded into six-well dishes at 170,000 cells per well in 2 mL of complete media. The conditioned media from all conditions were collected at 72 hours, and the cellular debris was pelleted at 300 g for 10 minutes in a swinging-bucket centrifuge (58). The supernatant was filtered with a 0.45-µm filter and stored at −20°C for short-term storage or at −70°C for long-term storage.
Caco-2 CSPG4-ECD uptake assays
For experiments analyzed by western blotting, 70,000 Caco-2 cells were seeded on 12 mm polycarbonate cell inserts with 8.0 µm pores (MilliporeSigma), and 1,000,000 18Co cells were seeded into the bottom wells of the dish. The cell inserts were collected on days 1, 3, and 5 and lysed in 35 µL of Triton X-100 lysis buffer. Caco-2 cells grown for 5 days on the cell inserts were used as a negative control. The entire volume of cell lysates collected from these experiments was analyzed by western blot. For imaging experiments, 70,000 Caco-2 cells were seeded into transwells made with 12 mm polyester membranes with 0.4 µm pores (Corning) and grown above 100,000 18Co cells for 5 days. Caco-2 cells on transwells were washed twice in PBS and fixed for 20 minutes in 4% paraformaldehyde in PBS. Washes and removal of any reagents were performed by inverting the insert as mechanical forces could result in loss of the cell monolayer. After fixation, the cells were washed in PBS for 5 minutes per wash, permeabilized for 15 minutes using 0.3% Triton X-100 in PBS, and stained as described in the “Antibody staining” above.
For experiments analyzed by western blotting, 70,000 Caco-2 cells were seeded on 12 mm polycarbonate cell inserts with 8.0 µm pores (MilliporeSigma), and 1,000,000 18Co cells were seeded into the bottom wells of the dish. The cell inserts were collected on days 1, 3, and 5 and lysed in 35 µL of Triton X-100 lysis buffer. Caco-2 cells grown for 5 days on the cell inserts were used as a negative control. The entire volume of cell lysates collected from these experiments was analyzed by western blot. For imaging experiments, 70,000 Caco-2 cells were seeded into transwells made with 12 mm polyester membranes with 0.4 µm pores (Corning) and grown above 100,000 18Co cells for 5 days. Caco-2 cells on transwells were washed twice in PBS and fixed for 20 minutes in 4% paraformaldehyde in PBS. Washes and removal of any reagents were performed by inverting the insert as mechanical forces could result in loss of the cell monolayer. After fixation, the cells were washed in PBS for 5 minutes per wash, permeabilized for 15 minutes using 0.3% Triton X-100 in PBS, and stained as described in the “Antibody staining” aboveMouse crypt isolation
Crypt isolation was performed as previously described but with modifications (59). Mouse colonic tissue was prepared by first washing luminal contents with cold PBS to remove fecal pellets. The colon was then opened longitudinally and washed at least three times with cold PBS. Next, the samples were washed twice in cold 25 mM EDTA for approximately 3 minutes per wash. Colonic tissue was incubated in cold 25 mM EDTA on ice for 30 minutes with occasional inversions. After 30 minutes, the samples in 25 mM EDTA were shaken, and the supernatant was collected and kept on ice. This process was repeated two additional times. The collected supernatant was strained through a 70-µm mesh filter and pelleted at 600 g for 5 minutes. These crypt pellets were resuspended in PBS and visualized on a brightfield microscope at 10× magnification. Crypts were once again pelleted, and the supernatant was removed. Isolated crypts were then lysed in Triton X-100 lysis buffer and analyzed by western blot for CSPG4.
Chondroitinase ABC treatment of 18Co cells and conditioned media
18Co cells were seeded into six-well dishes at 170,000 cells per well in 2 mL of complete media. After 72 hours, the conditioned media was collected, and cells were washed 1 time in PBS. To remove chondroitin sulfates from 18Co cells, chondroitinase ABC (MilliporeSigma) prepared in 50 mM Tris pH 8.0 and 50 mM sodium acetate at a final concentration of 0.1 units/mL was applied to cells for 1 hour at 37°C. To remove chondroitin sulfates from shed CSPG4 in the conditioned media, 0.1 units/mL of chondroitinase ABC was added to 1 mL of conditioned media and incubated at 37°C for 1 hour. Cells were lysed in Triton X-100 lysis buffer, and conditioned media were pelleted and filtered as previously described.
Intoxication of Caco-2 for cell rounding
Caco-2 expressing GFP and H2B-mCherry were seeded into a 96-well plate at 25,000 cells per well in complete EMEM. After growing cells for 48 hours, media from wells were removed and replaced with 90 µL with complete DMEM, or, for experiments using 18Co conditioned media, with 90 µL of non-targeting siLuciferase control media or 90 µL siCSPG4 media. Finally, 10 µL of a stock solution of 10 pM TcdB2 was added to the cells. After intoxication, brightfield and fluorescence images were collected every 45 minutes on a Cytation 5 imager (BioTek) with an atmosphere of 5% CO2 and 37°C temperatures. A manuscript documenting the generation of Caco-2 expressing GFP and H2B-mCherry cell lines and the cell rounding analysis tools is in preparation. See supplemental methods for additional detail.
CSPG4 antibody-blocking experiments
Caco-2 cells expressing GFP and H2B-mCherry were plated into 96-well dishes at a density of 25,000 cells per well. After 48 hours, an antibody targeting CSPG4 (SantaCruz, 20 µg/mL) was added to the conditioned media and incubated at room temperature for 30 minutes. Untreated conditioned media or antibody-treated conditioned media was added to cells, and cells were intoxicated with 1 pM TcdB as previously described and imaged as previously described.
Western blotting
Cell lysate concentrations were determined with a BCA Protein Assay Kit (Thermo Fisher Scientific). Unless specified, 20 µg of protein from cell lysates was boiled in Laemmli buffer prior to loading on 4–20% Mini-PROTEAN TGX stain-free gels (BioRad) for SDS-PAGE. Stain-free gels were activated and imaged on a BioRad Gel Doc EZ or a BioRad ChemiDoc MP prior to transfer. Proteins were transferred onto polyvinylidene difluoride at 100 V for 70 minutes in Towbin buffer lacking methanol and membranes were blocked in 5% milk in PBS for 1 hour. Membranes were incubated overnight in primary antibodies against CSPG4 (Abcam, 1:2,000) or GAPDH (CST, 1:5,000) at 4°C in 5% milk in PBST when appropriate. Membranes were washed three times in PBST, with 5 minutes per wash, and incubated for 1 hour in secondary antibodies Dylight-680 goat anti-mouse (CST, 1:10,000), DyLight-800 goat anti-rabbit (CST, 1:10,000) in 5% Milk in PBST for 1 hour at room temperature. Membranes were washed three times in PBST and once in PBS prior to protein detection on a Licor Odyssey imager or BioRad ChemiDoc MP imager. Full images of uncropped westerns from immunofluorescent images or gels are available in Supplemental Materials (Fig. S5 and S6).
Tissue preparation
Human colonic tissue was obtained from the Cooperative Human Tissue Network from consenting and deidentified donors under Institutional Review Board-approved protocol 031078. To prepare human frozen colonic tissue samples, ascending colon tissue was first dissected into sections and washed with PBS supplemented with 1.2 mM CaCl2 and 1 mM MgCl2. If colonic sections were to be used for intoxication experiments, the dissected samples were then incubated in complete DMEM in the presence or absence of 10 nM TcdB1-JF669 at 37°C for 1 hour. Sections were then incubated in Tissue-Tek OCT compound and snap-frozen in a dry ice-cooled ethanol bath. Mouse colonic tissue from wild-type and CSPG4 knockout mice was prepared by flushing luminal contents with PBS supplemented with 1.2 mM CaCl2 and 1 mM MgCl2. Colons were cut along their length and fixed in 2% paraformaldehyde in PBS for 2 hours at room temperature with gentle agitation. Tissues were then washed three times in PBS and incubated overnight at 4°C in PBS supplemented with 30% sucrose and 1% sodium azide. The following day, samples were quickly dipped in the OCT compound, rolled along their length, submerged in OCT, and snap-frozen in a dry ice-cooled ethanol bath. All OCT-embedded samples were cut into 10 µM sections and mounted onto positively charged slides for immunofluorescence staining and confocal microscopy.
Single-cell RNA sequencing analysis
Pre-processed data sets of human colonic epithelial tissue were procured from the Space-Time Gut Cell Atlas (https://www.gutcellatlas.org/) (37). ScanPy (v1.9.1), NumPy (v1.22.4), Anndata (v0.8.0), Pandas (v1.1.2), and bbknn (v1.5.1) were used to analyze this published data set as previously described (60 – 64). Cells annotated as healthy adult tissue from the appendix, large intestine, rectum, and small intestines were subsetted from the original data set. Cell-type annotations were included by the original authors of the data set and methods were verified for consistency. Epithelial cells along the intestines were plotted for CSPG4 or EPCAM expression using the ScanPy dotplot function. Dot plot figures were generated with ScanPy using a standard_scale = var and vmax = 0.8.
ACKNOWLEDGMENTS
We gratefully acknowledge funding support from the Molecular Biophysics Training Grant T32GM008320 (to K.O.C.) and Vanderbilt University Medical Center. Research in the Lacy lab is supported by the NIH (AI095755) and the Department of Veterans Affairs (BX002943). The Translational Pathology Shared Resource is supported by NCI/NIH Cancer Center Support Grant P30CA068485. The Vanderbilt University Medical Center’s Digestive Disease Research Center is supported by NIH grant P30DK058404. Confocal microscopy was performed through the Vanderbilt Cell Imaging Shared Resource (supported by NIH Grants CA68485, DK20593, DK58404, DK59637, and EY08126). The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
We also thank Min Dong for providing CSPG4 knockout mouse tissue used in this study.
The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
This article is a direct contribution from Borden Lacy, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Jimmy Ballard, University of Oklahoma Health Sciences Center, and Roman Melnyk, University of Toronto, The Hospital for Sick Children.
Contributor Information
D. Borden Lacy, Email: borden.lacy@vanderbilt.edu.
Charles Darkoh, University of Texas Health Science Center, Houston, Texas, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01857-23.
Transcriptional analysis of 18Co cells indicates the expression of all TcdB receptors.
NECTIN-3 localizes to the brush border and cell junctions in Caco-2 cells.
Confirming the specificity of the anti-CSPG4 antibody staining.
18Co derived condition medium does not promote cell rounding.
Western blot images from Fig. 5.
Western blot and stain free gel images from Fig. 6.
Supplemental methods and figure legends.
18Co derived conditioned medium potentiates TcdB2 mediated cell rounding on Caco-2 cells. Caco-2 cells expressing GFP were intoxicated with 1 pM TcdB2 in the absence or presence of 18Co conditioned media and imaged every 45 minutes over 20 hours.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, Beldavs ZG, Kainer MA, Karlsson M, Gerding DN, McDonald LC, Emerging Infections Program Clostridioides difficile Infection Working Group . 2020. Trends in U.S burden of Clostridioides difficile infection and outcomes. N Engl J Med 382:1320–1330. doi: 10.1056/NEJMoa1910215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CDC . 2019. Antibiotic resistance threats in the United States, 2019. CDC; [Google Scholar]
- 3. Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397 [DOI] [PubMed] [Google Scholar]
- 5. Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551. doi: 10.1128/mBio.00551-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandrasekaran R, Kenworthy AK, Lacy DB. 2016. Clostridium difficile toxin A undergoes clathrin-independent, PACSIN2-dependent endocytosis. PLoS Pathog 12:e1006070. doi: 10.1371/journal.ppat.1006070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. 2010. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS One 5:e10673. doi: 10.1371/journal.pone.0010673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, He R, Li C, Guo S, Li S, Huang T, Perez-Cordon G, Feng H, Wei W. 2015. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25:157–168. doi: 10.1038/cr.2014.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, Zhang X, Stallcup WB, Miao J, He X, Hurdle JG, Breault DT, Brass AL, Dong M. 2016. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538:350–355. doi: 10.1038/nature19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. LaFrance ME, Farrow MA, Chandrasekaran R, Sheng J, Rubin DH, Lacy DB. 2015. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A 112:7073–7078. doi: 10.1073/pnas.1500791112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo J, Yang Q, Zhang X, Zhang Y, Wan L, Zhan X, Zhou Y, He L, Li D, Jin D, Zhen Y, Huang J, Li Y, Tao L. 2022. TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile. Cell 185:980–994. doi: 10.1016/j.cell.2022.02.010 [DOI] [PubMed] [Google Scholar]
- 12. Tian S, Xiong X, Zeng J, Wang S, Tremblay BM, Chen P, Chen B, Liu M, Chen P, Sheng K, Zeve D, Qi W, Breault DT, Rodríguez C, Gerhard R, Jin R, Doxey AC, Dong M. 2022. Identification of TFPI as a receptor reveals recombination-driven receptor switching in Clostridioides difficile toxin B variants. Nat Commun 13:6786. doi: 10.1038/s41467-022-33964-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartley-Tassell LE, Awad MM, Seib KL, Scarselli M, Savino S, Tiralongo J, Lyras D, Day CJ, Jennings MP. 2019. Lectin activity of the TcdA and TcdB toxins of Clostridium difficile. Infect Immun 87:e00676-18. doi: 10.1128/IAI.00676-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen E, Zhu K, Li D, Pan Z, Luo Y, Bian Q, He L, Song X, Zhen Y, Jin D, Tao L. 2020. ubtyping analysis reveals new variants and accelerated evolution of Clostridioides difficile toxin B. 1. Commun Biol 3:347. doi: 10.1038/s42003-020-1078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mileto SJ, Jardé T, Childress KO, Jensen JL, Rogers AP, Kerr G, Hutton ML, Sheedlo MJ, Bloch SC, Shupe JA, Horvay K, Flores T, Engel R, Wilkins S, McMurrick PJ, Lacy DB, Abud HE, Lyras D. 2020. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc Natl Acad Sci U S A 117:8064–8073. doi: 10.1073/pnas.1915255117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen P, Zeng J, Liu Z, Thaker H, Wang S, Tian S, Zhang J, Tao L, Gutierrez CB, Xing L, Gerhard R, Huang L, Dong M, Jin R. 2021. Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection. Nat Commun 12:3748. doi: 10.1038/s41467-021-23878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen P, Tao L, Wang T, Zhang J, He A, Lam KH, Liu Z, He X, Perry K, Dong M, Jin R. 2018. Structural basis for recognition of frizzled proteins by Clostridium difficile Toxin B. Science 360:664–669. doi: 10.1126/science.aar1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. López-Ureña D, Orozco-Aguilar J, Chaves-Madrigal Y, Ramírez-Mata A, Villalobos-Jimenez A, Ost S, Quesada-Gómez C, Rodríguez C, Papatheodorou P, Chaves-Olarte E. 2019. Toxin B variants from Clostridium difficile strains VPI 10463 and Nap1/027 share similar substrate profile and cellular intoxication kinetics but use different host cell entry factors. Toxins 11:348. doi: 10.3390/toxins11060348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan Z, Zhang Y, Luo J, Li D, Zhou Y, He L, Yang Q, Dong M, Tao L, . 2021. Functional analyses of epidemic Clostridioides difficile toxin B variants reveal their divergence in utilizing receptors and inducing pathology. PLoS Pathog 17:e1009197. doi: 10.1371/journal.ppat.1009197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takai Y, Nakanishi H. 2003. Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci 116:17–27. doi: 10.1242/jcs.00167 [DOI] [PubMed] [Google Scholar]
- 21. Schöttelndreier D, Seeger K, Grassl GA, Winny MR, Lindner R, Genth H. 2018. Expression and (lacking) internalization of the cell surface receptors of Clostridioides difficile toxin B. Front Microbiol 9:1483. doi: 10.3389/fmicb.2018.01483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams A, Wayne Vogl A. 2017. High resolution localization of Rab5, EEA1, and Nectin-3 to tubulobulbar complexes in the rat testis. Anat Rec 300:1160–1170. doi: 10.1002/ar.23563 [DOI] [PubMed] [Google Scholar]
- 23. Rolih V, Barutello G, Iussich S, De Maria R, Quaglino E, Buracco P, Cavallo F, Riccardo F. 2017. CSPG4: a prototype oncoantigen for translational immunotherapy studies. J Transl Med 15:151. doi: 10.1186/s12967-017-1250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamburini E, Dallatomasina A, Quartararo J, Cortelazzi B, Mangieri D, Lazzaretti M, Perris R. 2019. Structural deciphering of the NG2/CSPG4 proteoglycan multifunctionality. FASEB J 33:3112–3128. doi: 10.1096/fj.201801670R [DOI] [PubMed] [Google Scholar]
- 25. Nishiyama A, Lin XH, Stallcup WB. 1995. Generation of truncated forms of the NG2 proteoglycan by cell surface proteolysis. Mol Biol Cell 6:1819–1832. doi: 10.1091/mbc.6.12.1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nishihara T, Remacle AG, Angert M, Shubayev I, Shiryaev SA, Liu H, Dolkas J, Chernov AV, Strongin AY, Shubayev VI. 2015. Matrix metalloproteinase-14 both sheds cell surface neuronal glial antigen 2 (NG2) proteoglycan on macrophages and governs the response to peripheral nerve injury. J Biol Chem 290:3693–3707. doi: 10.1074/jbc.M114.603431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terada N, Ohno N, Murata S, Katoh R, Stallcup WB, Ohno S. 2006. Immunohistochemical study of NG2 chondroitin sulfate proteoglycan expression in the small and large intestines. Histochem Cell Biol 126:483–490. doi: 10.1007/s00418-006-0184-3 [DOI] [PubMed] [Google Scholar]
- 28. Mahi NA, Najafabadi MF, Pilarczyk M, Kouril M, Medvedovic M. 2019. GREIN: an interactive web platform for re-analyzing GEO RNA-seq data. Sci Rep 9:7580. doi: 10.1038/s41598-019-43935-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otte JM, Rosenberg IM, Podolsky DK. 2003. Intestinal myofibroblasts in innate immune responses of the intestine. Gastroenterology 124:1866–1878. doi: 10.1016/s0016-5085(03)00403-7 [DOI] [PubMed] [Google Scholar]
- 30. Hsia LT, Ashley N, Ouaret D, Wang LM, Wilding J, Bodmer WF. 2016. Myofibroblasts are distinguished from activated skin fibroblasts by the expression of AOC3 and other associated markers. Proc Natl Acad Sci U S A 113:E2162–71. doi: 10.1073/pnas.1603534113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simmons JG, Pucilowska JB, Keku TO, Lund PK. 2002. IGF-I and TGF-beta1 have distinct effects on phenotype and proliferation of intestinal fibroblasts. Am J Physiol Gastrointest Liver Physiol 283:G809–18. doi: 10.1152/ajpgi.00057.2002 [DOI] [PubMed] [Google Scholar]
- 32. Sato T, Fujita N, Yamada A, Ooshio T, Okamoto R, Irie K, Takai Y. 2006. Regulation of the assembly and adhesion activity of E-cadherin by Nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J Biol Chem 281:5288–5299. doi: 10.1074/jbc.M510070200 [DOI] [PubMed] [Google Scholar]
- 33. Tanaka-Okamoto M, Hori K, Ishizaki H, Itoh Y, Onishi S, Yonemura S, Takai Y, Miyoshi J. 2011. Involvement of afadin in barrier function and homeostasis of mouse intestinal epithelia. J Cell Sci 124:2231–2240. doi: 10.1242/jcs.081000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crawley SW, Mooseker MS, Tyska MJ. 2014. Shaping the intestinal brush border. J Cell Biol 207:441–451. doi: 10.1083/jcb.201407015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim JE, Fei L, Yin WC, Coquenlorge S, Rao-Bhatia A, Zhang X, Shi SSW, Lee JH, Hahn NA, Rizvi W, Kim KH, Sung HK, Hui C-C, Guo G, Kim TH. 2020. Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat Commun 11:334. doi: 10.1038/s41467-019-14058-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, Sharma E, Wills Q, Bowden R, Richter FC, Ahern D, Puri KD, Henault J, Gervais F, Koohy H, Simmons A. 2018. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175:372–386. doi: 10.1016/j.cell.2018.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elmentaite R, Kumasaka N, Roberts K, Fleming A, Dann E, King HW, Kleshchevnikov V, Dabrowska M, Pritchard S, Bolt L, Vieira SF, Mamanova L, Huang N, Perrone F, Goh Kai’En I, Lisgo SN, Katan M, Leonard S, Oliver TRW, Hook CE, Nayak K, Campos LS, Domínguez Conde C, Stephenson E, Engelbert J, Botting RA, Polanski K, van Dongen S, Patel M, Morgan MD, Marioni JC, Bayraktar OA, Meyer KB, He X, Barker RA, Uhlig HH, Mahbubani KT, Saeb-Parsy K, Zilbauer M, Clatworthy MR, Haniffa M, James KR, Teichmann SA. 2021. Cells of the human intestinal tract mapped across space and time. Nature 597:250–255. doi: 10.1038/s41586-021-03852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson BS, Imai K, Natali PG, Ferrone S. 1981. Distribution and molecular characterization of a cell-surface and a cytoplasmic antigen detectable in human melanoma cells with monoclonal antibodies. Int J Cancer 28:293–300. doi: 10.1002/ijc.2910280307 [DOI] [PubMed] [Google Scholar]
- 39. Iida J, Pei D, Kang T, Simpson MA, Herlyn M, Furcht LT, McCarthy JB. 2001. Melanoma chondroitin sulfate proteoglycan regulates matrix metalloproteinase-dependent human melanoma invasion into type I collagen. J Biol Chem 276:18786–18794. doi: 10.1074/jbc.M010053200 [DOI] [PubMed] [Google Scholar]
- 40. Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. 2007. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A). Biochem J 403:553–563. doi: 10.1042/BJ20061176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joo NE, Miao D, Bermúdez M, Stallcup WB, Kapila YL. 2014. Shedding of NG2 by MMP-13 attenuates anoikis. DNA Cell Biol 33:854–862. doi: 10.1089/dna.2014.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, de Punder K, Angers S, Peters PJ, Maurice MM, Clevers H. 2016. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530:340–343. doi: 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- 43. Sierko E, Wojtukiewicz MZ, Zimnoch L, Kisiel W. 2010. Expression of tissue factor pathway inhibitor (TFPI) in human breast and colon cancer tissue. Thromb Haemost 103:198–204. doi: 10.1160/TH09-06-0416 [DOI] [PubMed] [Google Scholar]
- 44. Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Lacy DB. 2016. Clostridium difficile toxins TcdA and TcdB cause colonic tissue damage by distinct mechanisms. Infect Immun 84:2871–2877. doi: 10.1128/IAI.00583-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lanis JM, Barua S, Ballard JD. 2010. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog 6:e1001061. doi: 10.1371/journal.ppat.1001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larabee JL, Doyle DA, Ahmed UKB, Shadid TM, Sharp RR, Jones KL, Kim YM, Li S, Ballard JD. 2023. Discovery of Hippo signaling as a regulator of CSPG4 expression and as a therapeutic target for Clostridioides difficile disease. PLoS Pathog 19:e1011272. doi: 10.1371/journal.ppat.1011272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fukushi J, Makagiansar IT, Stallcup WB. 2004. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and α3β1 integrin. Mol Biol Cell 15:3580–3590. doi: 10.1091/mbc.e04-03-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tillet E, Ruggiero F, Nishiyama A, Stallcup WB. 1997. The membrane-spanning proteoglycan NG2 binds to collagens V and VI through the central nonglobular domain of its core protein. J Biol Chem 272:10769–10776. doi: 10.1074/jbc.272.16.10769 [DOI] [PubMed] [Google Scholar]
- 49. Tang F, Lord MS, Stallcup WB, Whitelock JM. 2018. Cell surface chondroitin sulphate proteoglycan 4 (CSPG4) binds to the basement membrane heparan sulphate proteoglycan, perlecan, and is involved in cell adhesion. J Biochem (Tokyo) 163:399–412. doi: 10.1093/jb/mvy008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pruitt RN, Chumbler NM, Rutherford SA, Farrow MA, Friedman DB, Spiller B, Lacy DB. 2012. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem 287:8013–8020. doi: 10.1074/jbc.M111.298414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. 2009. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4:e6529. doi: 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nam H, Benezra R. 2009. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell 5:515–526. doi: 10.1016/j.stem.2009.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stauffer W, Sheng H, Lim HN. 2018. Ezcolocalization: an ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Sci Rep 8:15764. doi: 10.1038/s41598-018-33592-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dawson C. 2022. ggprism: a “ggplot2” extension inspired by “GraphPad Prism” (1.0.4)
- 55. Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K, Yutani H, Dunnington D, RStudio . 2022. ggplot2: create elegant data visualisations using the grammar of graphics (3.4.0)
- 56. Clarke E, Sherrill-Mix S. 2017. ggbeeswarm: categorical scatter (violin point) Plots (0.6.0)
- 57. R Core Team . 2022. R: A language and environment for statistical computing (4.2.0). R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 58. Nigro A, Finardi A, Ferraro MM, Manno DE, Quattrini A, Furlan R, Romano A. 2021. Selective loss of microvesicles is a major issue of the differential centrifugation isolation protocols. Sci Rep 11:3589. doi: 10.1038/s41598-021-83241-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Turksen K. 2019. Edited by Turksen K.. Isolation, propagation, and clonogenicity of intestinal stem cells BT - stem cell niche: methods and protocols, p 61–73. Springer, New York. doi: 10.1007/978-1-4939-9508-0 [DOI] [PubMed] [Google Scholar]
- 60. Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, Park J-E. 2020. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36:964–965. doi: 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Virshup I, Rybakov S, Theis FJ, Angerer P, Wolf FA. 2021. anndata: annotated data. bioRxiv. doi: 10.1101/2021.12.16.473007 [DOI]
- 62. Reback J, McKinney W, den Bossche JV, Augspurger T, Cloud P, Hawkins S, Klein A, Roeschke M, Tratner J, She C, Petersen T, Ayd W, Garcia M, Garcia J, Hayden A, Jancauskas V, Saxton D, McMaster A, Battiston P, Seabold S, Hoyer S, Dong DK, Overmeire W, Winkel M, jbrockmendel, gfyoung, Sinhrks, MomIsBestFriend, chris-b1, h-vetinari . 2020. pandas-dev/pandas: pandas 1.1.2. doi: 10.5281/ZENODO.4019559 [DOI]
- 63. McKinney W. 2010. Proceedings of the 9th python in science conference, p 56–61. In van der Walt S, Millman J (ed), Data structures for statistical computing in python. doi: 10.25080/Majora-92bf1922-00a [DOI] [Google Scholar]
- 64. Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, Del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE. 2020. Array programming with NumPy. Nature 585:357–362. doi: 10.1038/s41586-020-2649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptional analysis of 18Co cells indicates the expression of all TcdB receptors.
NECTIN-3 localizes to the brush border and cell junctions in Caco-2 cells.
Confirming the specificity of the anti-CSPG4 antibody staining.
18Co derived condition medium does not promote cell rounding.
Western blot images from Fig. 5.
Western blot and stain free gel images from Fig. 6.
Supplemental methods and figure legends.
18Co derived conditioned medium potentiates TcdB2 mediated cell rounding on Caco-2 cells. Caco-2 cells expressing GFP were intoxicated with 1 pM TcdB2 in the absence or presence of 18Co conditioned media and imaged every 45 minutes over 20 hours.