ABSTRACT
People with cystic fibrosis (pwCF) commonly test positive for the pathogenic fungus Aspergillus fumigatus, which is associated with a decline in lung function. Trikafta is a recently approved therapy for pwCF that improves quantity and function of the CFTR protein; however, it is not known how Trikafta affects microbial communities in the lung. Therefore, the aim of this study was to determine whether Trikafta directly affects A. fumigatus growth and biology. While Trikafta did not impact the viability of A. fumigatus conidia, treatment of A. fumigatus biofilms with Trikafta reduced overall biofilm biomass. This finding was associated with increased membrane permeability, decreased viability, and reduced metabolic activity following long-term treatment of biofilms. Trikafta-induced membrane permeability, and biomass reduction was partially blocked with the calcium channel inhibitor verapamil and fully blocked by the mammalian CFTR inhibitor GlyH-101. Trikafta-induced biomass reduction and metabolic activity was shown to be regulated by the high-osmolarity glycerol pathway gene sakA. Trikafta treatment also induced resistance to the cell wall stressor calcofluor white, susceptibility to the anti-fungal drug caspofungin, and decreased inflammatory responses from murine bone marrow cells. Collectively, these results reveal that Trikafta affects infection-relevant A. fumigatus biology and host-microbial interactions.
IMPORTANCE
PwCF commonly test positive for pathogenic fungi, and more than 90% of the cystic fibrosis patient population is approved for the modulator treatment, Trikafta. Therefore, it is critical to understand how fungal communities, specifically A. fumigatus, respond to Trikafta exposure. Therefore, we sought to determine whether Trikafta impacted the biology of A. fumigatus biofilms. Our data demonstrate that Trikafta reduces biomass in several laboratory strains as well as clinical strains isolated from the expectorated sputum of pwCF. Furthermore, Trikafta reduces fungal viability and the capacity of biofilms to recover following treatment. Of particular importance, Trikafta affects how A. fumigatus biofilms respond to cell wall stressors, suggesting that Trikafta modulates components of the cell wall. Since the cell wall directly affects how a host immune system will respond to and effectively neutralize pathogens, our work, demonstrating that Trikafta impacts the A. fumigatus cell wall, is potentially highly relevant to fungal-induced disease pathogenesis.
KEYWORDS: Aspergillus fumigatus, Trikafta, cystic fibrosis, anti-fungal agents, drug susceptibility
INTRODUCTION
The triple drug combination therapy Trikafta (elexacaftor/tezacaftor/ivacaftor) has been approved for cystic fibrosis (CF) patients harboring one or two copies of the dF508 cftr allele. The combination of two molecules involved in refining CFTR protein folding (elexacaftor/tezacaftor) and another molecule to potentiate CFTR ion gating function (ivacaftor) has improved lung function, sweat chloride levels, and overall quality of life in people with cystic fibrosis (pwCF) (1, 2). Recent data have also shown that elexacaftor can improve CFTR gating function in addition to folding (3). With the approval of Trikafta use in over 90% of pwCF, it is critical to understand the long-term impact of these combination treatments on the host and the associated microbial communities in the CF lung environment. Due to a thick buildup of sticky mucus, conventional host mechanisms that prevent microbial infection are perturbed in the CF lung and lead to chronic microbial colonization and infection. A wide variety of bacterial genera have been recovered from the CF lung, including Pseudomonas, Streptococcus, Prevotella, and many others. Additionally, diverse fungi (including Candida spp. and Aspergillus spp.) and viruses (influenza and respiratory syncytial viruses) have also been routinely identified [reviewed in reference (4)].
The role of fungi in CF lung disease progression remains ill-defined. Due to its frequent isolation from sputum samples of pwCF and its known role as a human fungal pathogen, the impact of A. fumigatus within the CF lung and subsequent pathogenesis are of particular importance. Invasive infections caused by A. fumigatus spp. typically occur in individuals with compromised immune systems, such as in patients undergoing chemotherapy or therapy with newer immune modulating biologics (5 – 9). After A. fumigatus conidia germinate in the lung environment and form hyphae, a biofilm that is resistant to anti-fungal therapy develops (10, 11) that contributes to morbidity and mortality (12 – 15). However, despite a functional immune system, A. fumigatus is detected in lung sputum samples of approximately 40%–50% of pwCF, and A. fumigatus presence in the lung has been found to be associated with a sharper decline in lung function (16 – 18) compared to pwCF who test negative for A. fumigatus. Ivacaftor treatment is correlated with reduced Aspergillus species in the lungs of pwCF (19, 20), and a more recent publication showed that Orkambi (ivacaftor/lumacaftor) reduced phagocyte-induced reactive oxygen species production induced by A. fumigatus (21). However, there have been no studies published directly addressing the potential impact of Trikafta on the biology of A. fumigatus itself and the potential consequences for the CF lung environment.
The overall goal of our study was to determine the impact of Trikafta on the biology of A. fumigatus and whether Trikafta treatment alters the host response to this important CF-associated fungus. We observed that Trikafta treatment has no minimum inhibitory concentration (MIC) against A. fumigatus conidia but does have a striking impact on biofilms from several laboratory and clinical isolates. Furthermore, our studies revealed that the effects of Trikafta on A. fumigatus biofilms are significantly exacerbated in combination with ionic stress. Collectively, we observed that Trikafta reduces the viability of A. fumigatus biofilms, increases fungal membrane permeability, alters susceptibility to anti-fungal cell wall agents, and reduces the inflammatory response of murine bone marrow cells to A. fumigatus.
RESULTS
Trikafta reduces growth of both laboratory and clinical A. fumigatus biofilms
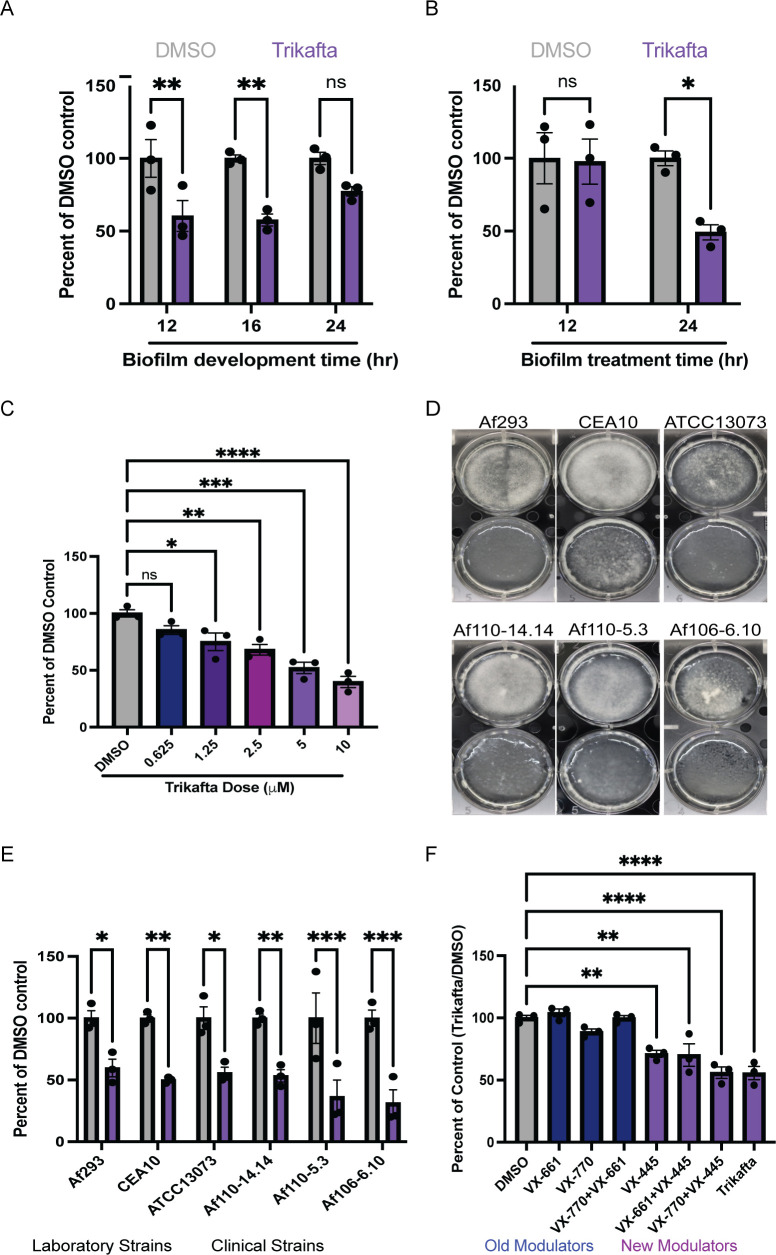
To determine whether Trikafta directly affects A. fumigatus growth and/or function, we first tested whether Trikafta treatment affected conidial growth. Conidia from the two most utilized laboratory reference strains, CEA10 and Af293, were grown in either standard CLSI RPMI-1640 with MOPS [3-(N-morpholino) propanesulfonic acid] buffer (22) or glucose minimal medium (GMM) with Trikafta doses ranging from 50.0 to 1.56 µM/individual modulator. The anti-fungal voriconazole was used as a positive control. While the MIC value of both strains to voriconazole was 0.25 µg/mL, consistent with previously published data, Trikafta did not affect fungal growth at any of the concentrations tested. We next tested whether Trikafta affected growth of A. fumigatus biofilms, which contain hyphae, at different levels of maturity. Initiating (12 hours), immature (16 hours), and mature (24 hours) CEA10 submerged biofilms were treated with either GMM plus dimethyl sulfoxide (DMSO) (vehicle control, 0.15%) or Trikafta (5 µM/molecule in every assay from this point forward) for an additional 24 hours before biomass was measured. Trikafta treatment of all stages of biofilm development reduced the total biomass, although only significantly in 12- and 16-hour biofilms (Fig. 1A). Since biomass reduction was most pronounced at 16 hours, 16-hour biofilms were treated with either GMM plus DMSO control or Trikafta for 12 and 24 hours. Surprisingly, biomass reduction occurred in the Trikafta-treated biofilms compared to DMSO-treated controls only after 24-hour treatment (Fig. 1B). We next tested a range of Trikafta doses on biomass reduction. We performed twofold serial dilutions of Trikafta (20.0–0.625 μM) on 16-hour biofilms and observed a smooth dose-response reduction in biofilm biomass, with 5 µM resulting in an approximate 50% reduction (Fig. 1C). To determine whether Trikafta effectively reduces biomass across A. fumigatus strains, we tested a selection of common laboratory strains (CEA10, Af293, and ATCC13073) as well as clinical isolates from pwCF respiratory samples (Af110-14.14, Af110-5.3, and Af106-6.10). After 16 hours of biofilm development, media were removed and biofilms were treated with GMM plus DMSO or Trikafta for an additional 24 hours. Biofilms were imaged and dry biomass was quantified (Fig. 1D and E). For all strains tested, Trikafta significantly reduced total fungal biomass between 50% and 70%. Strikingly, the Trikafta-treated biofilms macroscopically looked translucent and viscous, suggesting possible cell lysis (Fig. 1D).
Fig 1.
Trikafta reduces growth of both laboratory and clinical A. fumigatus biofilms. (A) CEA10 biofilms were grown in liquid glucose minimal medium (L-GMM) for 12, 16, or 24 hours. Media were removed and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours. Samples were collected, washed in water, and lyophilized. Dry biomass was measured, and all groups are represented as percentage of DMSO-treated average. (B) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 12 or 24 hours. Samples were processed and graphically represented as in panel A. (C) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO or the indicated dose of Trikafta (μM/molecule) were added for an additional 24 hours. Samples were processed and graphically represented as in panel A. (D) Biofilms of the indicated laboratory strains and clinical isolates were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours, and images were taken. (E) Samples were processed and graphically represented as in panel A. (F) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%), individual molecules, double combinations, or Trikafta (5 µM/molecule) were added for an additional 24 hours. Samples were processed and graphically represented as in panel A. For panels A, B, and E, a two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons was used, and for panels C and F, a one-way ANOVA with Dunnett’s multiple comparisons with all groups compared to DMSO was used. Data points are three biological replicates, which represent the average of technical replicates. ns, not significant.
Next we sought to define which molecule or combination of ivacaftor (VX-770), tezacaftor (VX-661), and elexacaftor (VX-445) caused the reduction in fungal biomass. CEA10 16-hour biofilms were treated with GMM plus DMSO, individual molecules, or different combinations of the individual molecules for an additional 24 hours before dry biomass was quantified (Fig. 1F). Single-molecule elexacaftor (VX-445) alone induced a significant reduction in total biomass (Fig. 1F). Interestingly, the elexacaftor/ivacaftor combination was as effective at reducing biomass as triple combination Trikafta, suggesting a potentially small but not significant effect for ivacaftor as well. Collectively, these data show that Trikafta significantly affects growth of established biofilms, primarily through the action of the dual corrector/potentiator, elexacaftor (VX-445).
Trikafta reduces viability and increases membrane permeability of A. fumigatus biofilms
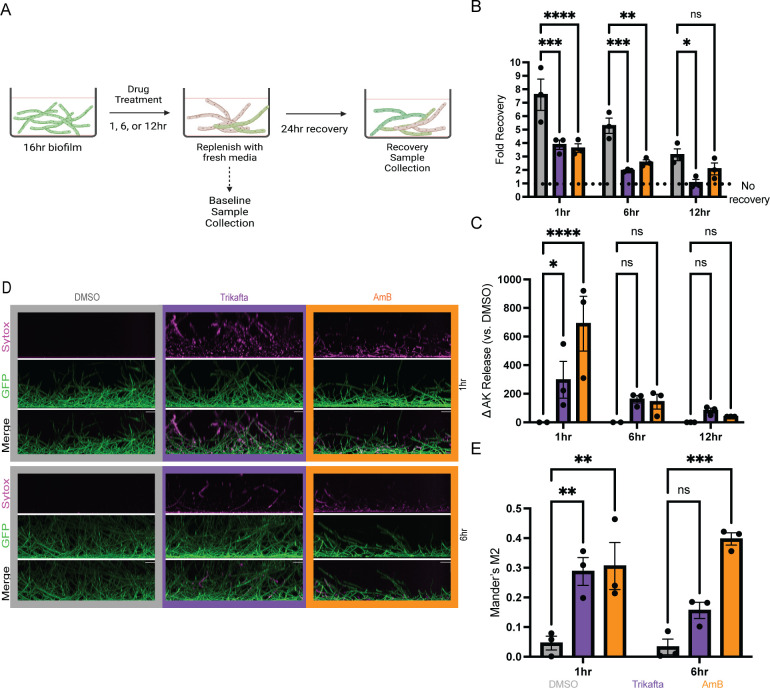
Given the macroscopic observations of Trikafta-treated biofilms and the substantial loss of biomass, we sought to determine if Trikafta treatment reduced viability of the biofilms. To test this hypothesis, we determined whether Trikafta-treated biofilms could recover growth in fresh media. CEA10 biofilms were grown for 16 hours and subsequently treated with GMM supplemented with either DMSO, Trikafta, or the anti-fungal drug amphotericin B (AmB, 1 µg/mL) as a positive control for an additional 1, 6, and 12 hours. At each time point, baseline samples were collected, and in parallel, a recovery sample was prepared where media were removed and replaced with fresh GMM for a further 24 hours of incubation before dry biomass weight was quantified (Fig. 2A). Biofilms treated with Trikafta and AmB for 1 and 6 hours had around 50% of the recovery of DMSO-treated biofilms. After 12-hour treatment with Trikafta, biofilms had no recovery compared to threefold recovery of DMSO-treated biofilms (Fig. 2B).
Fig 2.
Trikafta reduces viability and increases membrane permeability of A. fumigatus biofilms. (A) Experimental design for viability assay (B) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%), Trikafta (5 µM/molecule), or amphotericin B (AmB) (1 µg/mL) were added for an additional 1, 6, or 12 hours. Baseline samples were collected, and parallel biofilms had media removed and then replaced with fresh L-GMM for an additional 24 hours. Recovery biofilms were collected, washed in water, and lyophilized, and dry biomass was measured. Treatment groups were compared to their own controls, and data are represented as fold change from baseline control. (C) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%), Trikafta (5 µM/molecule), or AmB (1 µg/mL) was added for an additional 1, 6, or 12 hours. Supernatant was collected and adenylate kinase (AK) was measured. Data are shown as percent change over DMSO controls (DMSO set to 100%) and then subtracting 100 to get a delta in adenylate kinase release. (D) CEA10-gpdA:GFP biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%), Trikafta (5 µM/molecule), or AmB (1 µg/mL) were added for an additional 1 or 6 hours. Sytox Blue 1:1,000 was added for 5 minutes. Images were taken on Nikon spinning Disc confocal microscope ×20. (E) Images were quantified and represented as the fraction of GFP that overlaps with Sytox in each image (Mander’s M2). For (B, C, and E), a two-way ANOVA with Dunnett’s multiple comparisons was used, with both treatment groups compared to DMSO controls. Data points are three biological replicates, which represent the average of technical replicates.
Next, we determined whether Trikafta treatment affected overall membrane permeability of A. fumigatus biofilms. Supernatant from either DMSO or Trikafta-treated biofilms for 1, 6, or 12 hours was analyzed for adenylate kinase (AK) activity, a marker of membrane permeability and damage (23). Treatment with Trikafta or AmB for 1 hour significantly increased the change in AK release versus DMSO-treated controls approximately 300% and 600%, respectively. After 6- and 12-hour treatment with Trikafta and AmB, detectable levels of AK were observed to around 50%–100% change from DMSO controls, although to a much lesser extent than 1 hour treatment (Fig. 2C). To further validate this observation, the cell viability stain, Sytox Blue, was used to image damage to biofilms. The stain was used in conjunction with a fluorescently labeled A. fumigatus strain constitutively expressing green fluorescent protein (GFP) in the cytoplasm. The GFP fluorescence allowed for identifying the overlap in Sytox signal with hyphal cells in the biofilm. CEA10-gpdA:GFP conidia were grown into 16 hour biofilms and treated with GMM plus either DMSO, Trikafta, or Amphotericin B for an additional 1 and 6 hours prior to staining with Sytox Blue (12 hours treatment was too long for interpretable microscopy images). Biofilms treated with GMM plus DMSO showed no intracellular Sytox Blue staining while biofilms treated with AmB showed around 30–40% positive staining at both 1 and 6 hour treatments (Fig. 2D and E). Biofilms treated with Trikafta for 1 hour were also around 30% positive for Sytox Blue and around 15% positive after 6 hours of treatment (Fig. 2D and E). Collectively, these data demonstrate that Trikafta increases membrane permeability and reduces the viability of A. fumigatus biofilms.
Trikafta modulates ion channels in A. fumigatus biofilms
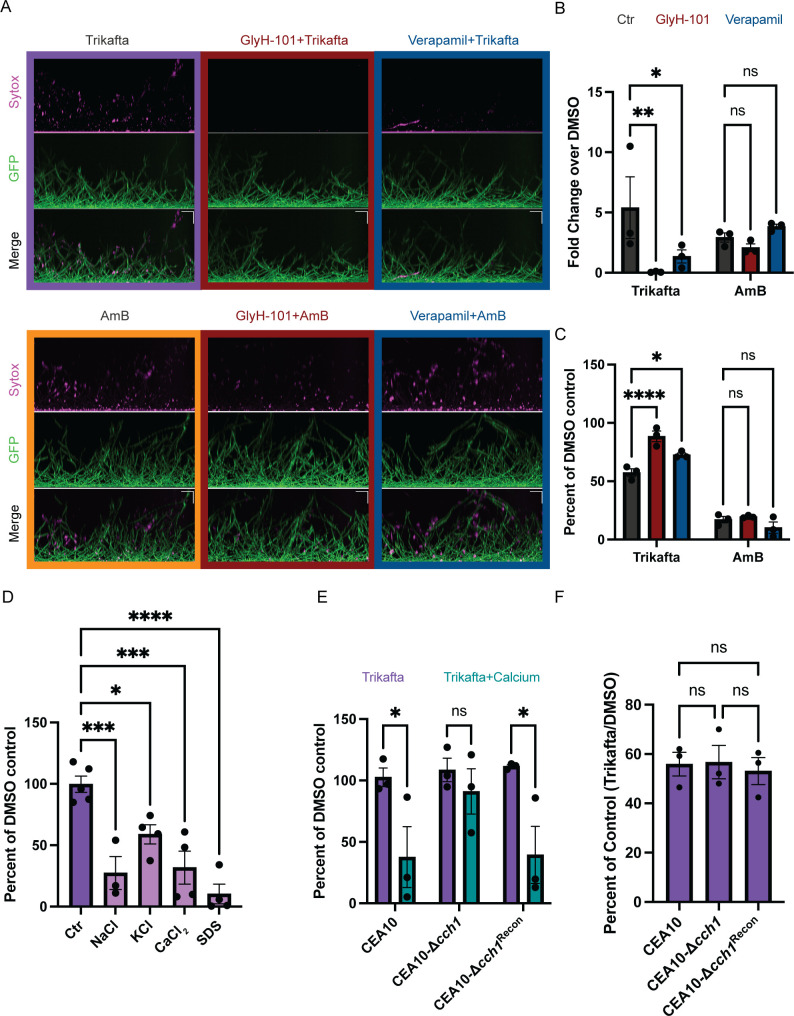
Trikafta modulates ion channel function in mammalian cells, so we next hypothesized that Trikafta-induced modulation of fungal ion channels causes membrane disruption and increased membrane permeability. We used a mammalian CFTR inhibitor GlyH-101 (24) and the calcium channel inhibitor verapamil to address this hypothesis. CEA10-gpdA:GFP conidia were grown into 16-hour biofilms and treated with either DMSO control, GlyH-101 (20 µM), or verapamil (1 mM) for 1 hour and then subsequently treated with Trikafta for an additional 1 hour prior to Sytox Blue membrane permeability staining. Pre-treatment of biofilms with GlyH-101 or verapamil prior to Trikafta treatment almost completely abolished Trikafta-induced Sytox Blue staining (Fig. 3A and B). To test if this was specific to Trikafta, we performed the same set of experiments with amphotericin B. Pre-treatment with GlyH-101 or verapamil had little to no effect on amphotericin B-induced Sytox staining (Fig. 3A and B). To determine whether ion channel modulation contributes to Trikafta-induced reduction in biomass, 16-hour biofilms were pre-treated with GlyH-101 or verapamil for 1 hour then treated with Trikafta or amphotericin B for 24 hours. GlyH-101 almost fully rescued the biomass reduction of Trikafta, while verapamil partially rescued it (Fig. 3C). GlyH-101 and verapamil had no effect on amphotericin B-induced biomass reduction (Fig. 3C). Therefore, the impact of ion channel inhibitors on Trikafta is likely specific to its mechanism of action, since the inhibitors did not affect AmB-induced membrane damage.
Fig 3.
Trikafta modulates ion channels in A. fumigatus biofilms. (A) CEA10-gpdA:GFP biofilms were grown in L-GMM for 16 hours. Media were removed and biofilms were treated with DMSO (control, 0.2%), verapamil (calcium channel inhibitor, 1 mM), or GlyH-101 (CFTR inhibitor, 20 µM) for 1 hour. Media with either DMSO (0.15%), Trikafta (final concentration 5 µM/molecule), or AmB (final concentration 1 µg/mL) was added for an additional 1 hour. Sytox Blue 1:1,000 was added for 5 minutes. Images were taken on Nikon spinning disc confocal microscope at ×20. (B) Images were quantified and represented as the fraction of GFP that overlaps with Sytox in each image (Mander’s M2), and data are represented as fold change over DMSO-treated control. (C) CEA10 biofilms were grown for 16 hours and media were removed. Fresh media with either DMSO (0.2%), GlyH-101 (20 µM), or verapamil (1 mM) was added for 1 hour. Subsequently, media with either DMSO (0.15%), AmB (final dose 1 µg/mL), or Trikafta (final dose 5 µM/molecule) was added for an additional 24 hours. Samples were processed and graphically represented as in Fig. 1A. (D) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added in either vehicle control (H20) or 0.5M NaCl, CaCl2, or KCl or 0.002% SDS for an additional hour. Metabolic activity was measured using XTT assay. Data are normalized as percentage of Trikafta/DMSO controls in each condition. (E) Wild-type (WT) CEA10, CEA10-Δcch1, and CEA10-cch1rec biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) was added in either control or 0.5-M CaCl2 for an additional hour, and an XTT assay was performed. Data are normalized as percentage of Trikafta/DMSO controls in either vehicle control (H2O) or CaCl2. (F) WT CEA10, CEA10-Δcch1, and CEA10-cch1rec biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours. Samples were processed and graphically represented as in Fig. 1A. For B, C, and E, a two-way ANOVA with Dunnett’s (B and C) or Sidak’s (E) multiple comparisons was performed and a one-way ANOVA (D and F) with Dunnett’s (D) or Tukey’s (F) multiple comparisons was performed. Data points are three biological replicates, which represent the average of technical replicates.
We next determined whether the effects of Trikafta were exacerbated in the presence of ionic stress. We chose to utilize a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT)-based metabolic assay to measure the metabolic activity of Trikafta-treated biofilms after 1-hour treatment because there was little to no effect of Trikafta on metabolic activity at that time point (Fig. 3D). Therefore, we treated 16-hour biofilms with either GMM plus DMSO control or Trikafta for 1 hour in either control, 0.5-M NaCl, 0.5-M CaCl2, or 0.5-M KCl. In parallel, we also included 0.002% SDS as a control for general membrane disruption. It is important to note that the low dose of SDS used did not cause any reduction in metabolic activity on its own in contrast to the different ions (data not shown). Strikingly, Trikafta treatment reduced metabolic activity by more than 70% in the presence of NaCl, CaCl2, and SDS. Trikafta also reduced metabolic activity by more than 40% in the presence of KCl (Fig. 3D). We next hypothesized that a strain harboring a null mutant in the calcium channel gene, cch1 (25), would be resistant to Trikafta-reduced metabolic activity in CaCl2 stress conditions. CEA10-Δcch1 biofilms treated with Trikafta in the presence of CaCl2 were resistant to reduced metabolic activity compared to wild-type (WT) CEA10 biofilms (Fig. 3E). However, the CEA10-Δcch1 biofilms treated with Trikafta for 24 hours had roughly equal susceptibility to Trikafta as WT controls, indicating that exogenous CaCl2 levels may impact Trikafta’s antifungal effects (Fig. 3E). Taken together, these data suggest that the antifungal effects of Trikafta are exacerbated in ionic stress conditions.
Trikafta transiently increases metabolic activity in A. fumigatus biofilms
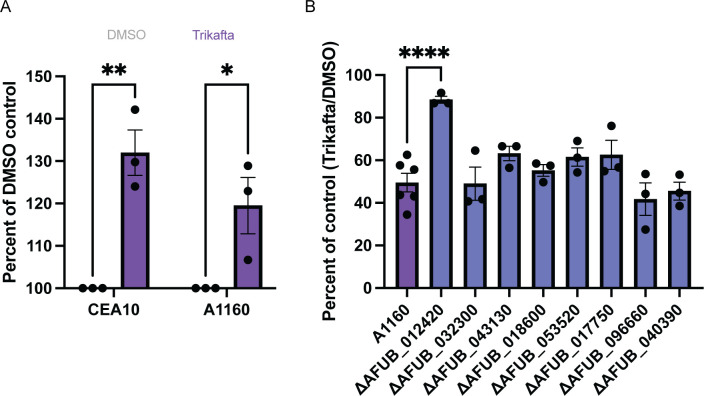
Since the CEA10-Δcch1 mutant biofilm was not resistant to Trikafta-induced biomass reduction, we further sought to determine how Trikafta reduced biofilm biomass after long-term exposure to the drug. To establish an assay for further exploring the metabolic changes in fungal biomass, we evaluated biofilms over time using the metabolic dye resazurin, as it allows for data collection over a range of time rather than a snapshot in time as with XTT-based metabolic assays. We treated 16-hour biofilms with either GMM plus DMSO or Trikafta for 30 minutes and then added resazurin for an additional 5 hours. Surprisingly, analysis of metabolic activity in Trikafta-treated biofilms compared to DMSO-treated controls showed a 30%–40% increase in metabolic activity in CEA10 biofilms (Fig. 4A). We hypothesized that this increased metabolic activity was likely due to increased mitochondrial activity.
Fig 4.
Trikafta acutely increases metabolic activity in A. fumigatus biofilms. (A) CEA10 or A1160 biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 30 min. The metabolic dye, resazurin, was added 10% vol for an additional 5 hours after Trikafta treatment and fluorescence 594 was captured. A two-way ANOVA with Sidak’s multiple comparisons was used. Data points are three biological replicates, which represent the average of technical replicates. (B) Biofilms of the indicated kinase library mutant strains were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours. Samples were processed and graphically represented as in Fig. 1A. Three to six technical replicates are represented.
To begin to delineate pathways by which Trikafta acts on A. fumigatus biofilm biomass reduction, we used this resazurin assay to perform a genetic screen on an A. fumigatus kinase null mutant library. Importantly, we observed a similar increase in metabolic activity in the WT A1160 background strain used to generate the null mutant library upon Trikafta treatment (Fig. 4A). A total of 108 putative kinase null mutants (Table 1) were grown into 16-hour biofilms, treated with either GMM plus DMSO or Trikafta for 30 minutes, and subsequently treated with resazurin for an additional 5 hours. For each run, the effect of Trikafta-induced metabolic activity in the parent strain (A1160) was calculated and used as the reference. A total of 18 mutant strains had altered metabolic activity in response to Trikafta compared to the control strain, based upon being either 20% below or 15% above the control strain (Table 2). Eight mutant strains (in bold) were selected to screen for biomass phenotypes, and it was found that a strain with a null mutation in ΔAFUB_012420 [high-osmolarity glycerol (HOG) pathway kinase, sakA] was the least susceptible to Trikafta-induced biomass reduction (Fig. 4B).
TABLE 1.
List of A. fumigatus kinase null mutant strains (A1160)
| Kinase null mutants | ||
|---|---|---|
| AFUB_045810 | AFUB_006320 | AFUB_096030 |
| AFUB_021710 | AFUB_052630 | AFUB_096080 |
| AFUB_053300 | AFUB_035220 | AFUB_099990 |
| AFUB_012420 | AFUB_019930 | AFUB_006190 |
| AFUB_032300 | AFUB_017750 | AFUB_089280 |
| AFUB_044560 | AFUB_011380 | AFUB_101210 |
| AFUB_030660 | AFUB_027890 | AFUB_044260 |
| AFUB_052450 | AFUB_056020 | AFUB_035990 |
| AFUB_043130 | AFUB_059390 | AFUB_096660 |
| AFUB_010510 | AFUB_059090 | AFUB_063270 |
| AFUB_018770 | AFUB_066150 | AFUB_020650 |
| AFUB_020560 | AFUB_071620 | AFUB_045550 |
| AFUB_029320 | AFUB_056110 | AFUB_060950 |
| AFUB_025560 | AFUB_082830 | AFUB_061120 |
| AFUB_038630 | AFUB_077790 | AFUB_040390 |
| AFUB_029240 | AFUB_075210 | AFUB_081220 |
| AFUB_016170 | AFUB_090090 | AFUB_076300 |
| AFUB_048440 | AFUB_078920 | AFUB_075230 |
| AFUB_006780 | AFUB_066030 | AFUB_074100 |
| AFUB_039620 | AFUB_095720 | AFUB_089460 |
| AFUB_045840 | AFUB_060320 | AFUB_093800 |
| AFUB_051750 | AFUB_087320 | AFUB_067080 |
| AFUB_029820 | AFUB_079830 | AFUB_017740 |
| AFUB_027640 | AFUB_078980 | AFUB_100220 |
| AFUB_038060 | AFUB_053960 | AFUB_036640 |
| AFUB_001600 | AFUB_070630 | AFUB_009070 |
| AFUB_053500 | AFUB_087120 | AFUB_096590 |
| AFUB_018600 | AFUB_075350 | AFUB_045710 |
| AFUB_010360 | AFUB_054020 | AFUB_030290 |
| AFUB_044400 | AFUB_081540 | AFUB_001940 |
| AFUB_027480 | AFUB_055480 | AFUB_095050 |
| AFUB_030570 | AFUB_056640 | AFUB_047210 |
| AFUB_053520 | AFUB_074550 | AFUB_063830 |
| AFUB_039100 | AFUB_071600 | AFUB_101530 |
| AFUB_007300 | AFUB_098230 | AFUB_071990 |
| AFUB_014350 | AFUB_099170 | AFUB_080040 |
TABLE 2.
Trikafta treatment alters metabolic activity in select kinase null mutants b
| AFUB no. | Putative function | % change from vehicle | % change from WT |
|---|---|---|---|
| AFUB_012420 | Mitogen-activated protein kinase | 176.5 | 139.9 |
| AFUB_053520 | Calcium/calmodulin-dependent protein kinase, putative | 156.6 | 118.6 |
| AFUB_017750 | Protein kinase, putative | 154.8 | 117.3 |
| AFUB_099170 | Protein kinase Yak1, putative | 129.1 | 112.3 |
| AFUB_018600 | Protein kinase, putative | 105.4 | 79.9 |
| AFUB_101530 | Sensor histidine kinase/response regulator, putative | 91.6 | 79.7 |
| AFUB_098230 | Inositol kinase kinase (UvsB), putative | 91.0 | 79.1 |
| AFUB_043130 | MAP kinase kinase Ste7 | 99.4 | 78.8 |
| AFUB_067080 | Putative uncharacterized protein | 90.1 | 78.4 |
| AFUB_055480 | Serine/threonine protein kinase, putative | 88.2 | 76.7 |
| AFUB_061120 | Protein kinase domain-containing protein | 87.9 | 76.5 |
| AFUB_045550 | Calcium/calmodulin-dependent protein kinase, putative | 86.6 | 75.4 |
| AFUB_020650 | Two-component osmosensing histidine kinase (Bos1) | 85.8 | 74.7 |
| AFUB_032300 | Protein kinase, putative | 93.2 | 73.9 |
| AFUB_071990 | N/A a | 84.6 | 73.6 |
| AFUB_063830 | Ubiquinone biosynthesis protein, putative | 84.0 | 73.0 |
| AFUB_096660 | Putative uncharacterized protein | 76.7 | 66.7 |
| AFUB_040390 | Protein kinase domain-containing protein | 72.6 | 63.2 |
N/A, not available.
The values in bold were selected for a secondary biomass screen.
Trikafta activates SakA and reduction of SakA activity increases resistance of A. fumigatus biofilms to Trikafta
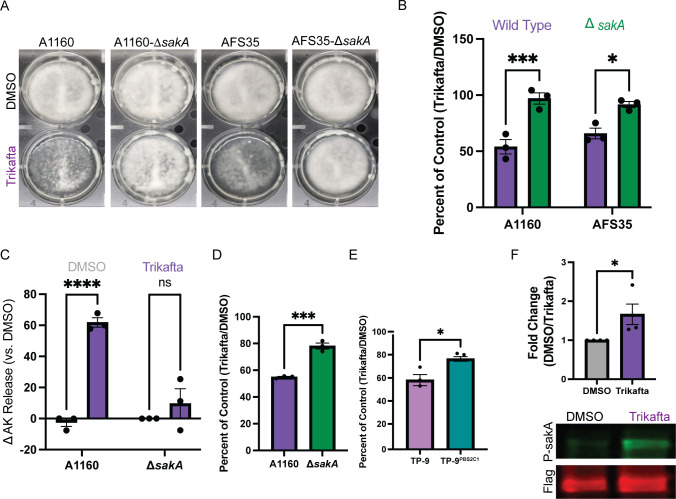
To confirm if loss of sakA alters biofilm biomass reduction to Trikafta, we next tested A1160-WT, A1160-ΔsakA, Afs35-WT, and Afs35-ΔsakA biofilms that were grown for 16 hours and treated with either DMSO or Trikafta for an additional 24 hours prior to imaging (Fig. 5A). Quantification of biomass showed that while both WT (A1160 and Afs35) biofilms had approximately 50%–60% reduction in biomass with Trikafta treatment compared to controls, both ∆sakA mutant biofilms had little to no reduction in biomass with Trikafta treatment (Fig. 5B). We also observed that A1160-ΔsakA biofilms were resistant to Trikafta-induced AK release after 12-hour treatment (Fig. 5C). To further determine the effects of Trikafta at this later time point, 16-hour A1160 and A1160-ΔsakA biofilms were treated with either DMSO or Trikafta for 12 hours, and metabolic activity was assessed by XTT reduction. Excitingly, Trikafta caused reduction of metabolic activity by 50% in WT A1160, while A1160-ΔsakA biofilms only had an approximately 20% reduction in metabolic activity (Fig. 5D). This further supports our hypothesis that the initial metabolic burst observed in response to Trikafta is followed by a marked reduction in metabolic activity and that Trikafta-induced changes in metabolic activity are regulated by sakA.
Fig 5.
Trikafta activates SakA, and reduction of SakA activity increases resistance of A. fumigatus biofilms to Trikafta. (A) WT (A1160 and Afs35) and ΔsakA conidia (in both WT strains) were grown for 16 hours into biofilms. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours and images were taken. (B) Samples were processed and graphically represented as in Fig. 1A. (C) WT A1160 and A1160-ΔsakA biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 12 hours. Supernatant was collected and adenylate kinase was quantified. Data are shown as percent change over DMSO controls (DMSO set to 100%) and then subtracting 100 to get a delta in AK release. (D) In parallel to panel C, metabolic activity of biofilms was measured by XTT. Data are represented as percentage of DMSO controls (Trikafta/DMSO). (E) TP9 and TP-9PBS2C1 biofilms were grown for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 24 hours. Samples were processed and graphically represented as in Fig. 1A. (F) TP-9PBS2C1: sakA Flag biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) were added for an additional 30 minutes. Biofilms were collected and protein extracted for Western blot analysis of either total SakA (Flag) or P-SakA. Samples were normalized to total protein and quantified using LI-COR. A two-way ANOVA with Sidak’s multiple comparisons (B and C) and Student’s t-test (D–F) were performed. Data points are three biological replicates, which represent the average of technical replicates.
Previous work from our laboratory observed that an A. fumigatus clinical isolate from a pwCF, designated TP-9, harbors mutations in the MAP kinase kinase pbs2 that regulates SakA pathway activity. This unique pbs2 allele arose from long-term growth in the CF lung environment and functionally leads to increased activation of SakA as indicated by phosphorylation (26). An allele swap with a pbs2 allele from a different A. fumigatus clade isolate from the same individual generated the strain TP-9PBS2C1 and resulted in reduced SakA activation compared to TP-9 (26). To further test the effects of SakA activity on the response to Trikafta, we next compared Trikafta-induced effects on these two strains. TP-9 and TP-9PBS2C1 biofilms were grown for 16 hours and treated with GMM plus either DMSO or Trikafta for an additional 24 hours. Quantification of biofilm biomass showed that while TP-9 biofilms had almost 45% reduction in biomass with Trikafta treatment compared to controls, TP-9PBS2C1 only had 20% reduction, further showing that a reduction in SakA activity conferred resistance to Trikafta-induced biomass reduction (Fig. 5E). Additionally, we wanted to confirm if Trikafta induced phosphorylation of SakA. Utilizing the TP-9PBS2C1:sakA-Flag strain (26), we treated 16-hour biofilms with GMM plus either DMSO or Trikafta for 30 minutes, collected the biomass, and subjected the protein to SDS-PAGE and subsequent Western blot analysis for total SakA (Flag) and P-SakA. Trikafta treatment increased P-SakA almost twofold in comparison to DMSO-treated controls, suggesting that Trikafta treatment activates SakA in A. fumigatus biofilms (Fig. 5F).
Trikafta modulates the biofilm cell wall and mediates inflammatory responses in host immune cells
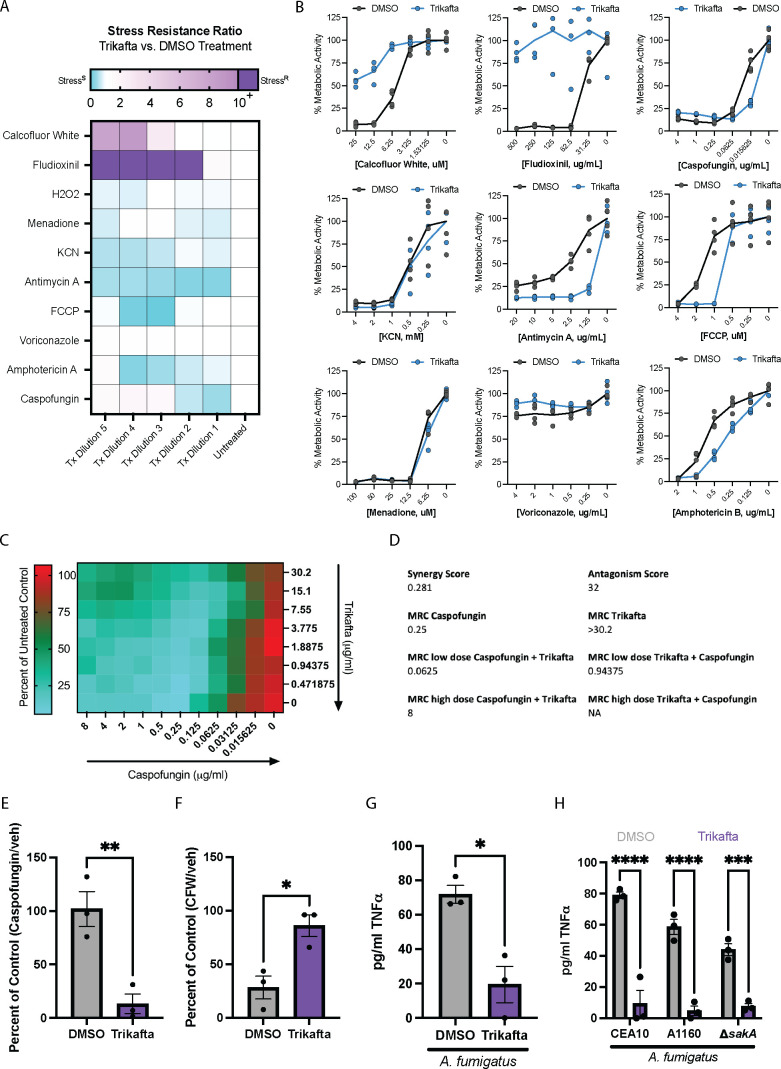
Since Trikafta treatment takes place in the complex CF lung environment, we next explored how Trikafta co-treatment with stress agents and conditions found in the CF lung environment affects A. fumigatus biofilms. We co-treated 16-hour CEA10 biofilms with either DMSO or Trikafta with increasing doses of anti-fungal drugs, cell wall stress agents, metabolic stress agents, and oxidative stress inducers and measured overall metabolic activity as a readout of fungal cell damage using the XTT assay (27) (Fig. 6A and B). Trikafta alone did not affect metabolic activity of biofilms compared to DMSO controls after 3 hours of treatment (data not shown). Voriconazole, an anti-fungal drug targeting ergosterol biosynthesis, only mildly damaged the biofilms by about 20%–30%, as previously published (12). Intriguingly, Trikafta modestly protected biofilms from voriconazole-induced damage at this time point. Trikafta co-treatment with amphotericin B, which damages the cell membrane through direct binding of ergosterol, increased fungal cell damage. The effects of the echinocandin β-(1,3)-glucan synthase inhibitor, caspofungin, were exacerbated by Trikafta at a lower dose (0.015 µg/mL). Trikafta also exacerbated the effects of mitochondrial inhibitors FCCP [carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone] and antimycin A. Unexpectedly, Trikafta strongly protected biofilms from calcofluor white (CFW)-induced damage (Fig. 6A and B).
Fig 6.
Trikafta modulates the biofilm cell wall and mediates inflammatory responses in host immune cells. (A) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed, and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) plus serial dilutions of the indicated stressors (CFW, Fludioxonil, caspofungin, KCN, antimycin A, FCCP, menadione, voriconazole, and AmB) were added for an additional 3 hours. Metabolic activity was measured by the reduction of the dye, XTT. Stress resistance ratio (Trikafta/DMSO controls) is represented by highly resistant (purple) through highly susceptible (blue) as a consequence of Trikafta co-treatment. (B) Graphs represent the percentage of stressor/control in the presence or absence of Trikafta. Data represent five technical replicates. (C) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with serial two-fold dilutions of Trikafta [30.2 µg/mL(20 µM/molecule)−0.471875 µg/mL (0.3125 µM/molecule)] in checkerboard combination with serial two-fold dilutions of caspofungin (8 µg/mL −0.015625 µg/mL). Heat map represents percent of untreated control values, with higher metabolic activity indicated by red and lower indicated by green. Data are averages of three biological replicates. (D) Synergy and antagonism scores for caspofungin and Trikafta based on values generated in C. (MRC = Metabolic reduction concentration). (E) CEA10 biofilms were grown in L-GMM for 16 hours. Media were removed and then fresh media with either DMSO (0.15%) or Trikafta (5 µM/molecule) plus 0.015625 µg/mL caspofungin or (F) 12.5 µM CFW for 3 hours. An XTT assay was performed to determine metabolic activity, and data are shown as percentage of stressor/control in either DMSO or Trikafta. (G) Bone marrow cells were isolated from C57Bl/6 mice harboring dF508 mutation in cftr and seeded 1 × 107/mL in co-culture with CEA10 conidia (1 × 1066/mL) for 16 hours in the presence of interferon gamma (IFN-γ) (10 ng/mL) with either DMSO (0.15%) or Trikafta (5 µM). 96 well plates were centrifuged and supernatants were analyzed for murine tumor necrosis factor alpha (TNF-α). (H) Bone marrow cells were isolated from WT C57Bl/6 mice and seeded 1 × 107 /mL in co-culture with CEA10, A1160, or A1160-ΔsakA conidia (1 × 106 /mL) for 16 hr in the presence of IFN-γ (10 ng/mL) with either DMSO (0.15%) or Trikafta (5 µM). Ninety-six-well plates were centrifuged and supernatants were analyzed for murine TNF-α. Student’s t-test was performed for panels E–G and a two-way ANOVA with Sidak’s multiple comparisons was performed for panel H. For panels C–H, data points are three biological replicates, which represent the average of technical replicates.
Since Trikafta had strong impacts on biofilm response to cell wall stressors CFW and caspofungin, we decided to further validate our screen results. We developed a modified checkerboard assay to test potential synergy/antagonism between Trikafta and caspofungin against A. fumigatus biofilms. We tested this by treating 16-hour biofilms with different dose combinations of caspofungin and Trikafta for 3 hours and measuring metabolic activity. We observed that at low doses of drug, Trikafta and caspofungin synergized with a score of 0.281 (Fig. 6C and D). Interestingly, we observed that at high doses of drug, Trikafta and caspofungin antagonized, with a score of 32 (Fig. 6C and D). To confirm if Trikafta and caspofungin synergize, we tested a low dose of Trikafta with a low dose of caspofungin on 16-hour biofilms and measured metabolic activity. We observed that Trikafta exacerbated a low dose of caspofungin-induced damage (Fig. 6E). We also observed that Trikafta protected biofilms from CFW-induced damage (Fig. 6F), demonstrating collectively that Trikafta alters biofilm responses to cell wall stressors.
Since Trikafta protected biofilms against CFW but exacerbated caspofungin effects, we hypothesized that Trikafta was likely increasing chitin and decreasing (1,3)-β-glucan content of the fungal cell wall. Furthermore, it has previously been shown that altering chitin content results in a compensatory effect that increases levels of (1,3)-β-glucan (28). Since mammalian cells recognize (1,3)-β-glucan, which triggers an inflammatory response, we hypothesized that Trikafta treatment would reduce the inflammatory response of host cells in co-culture with A. fumigatus. To test this hypothesis, CEA10 conidia were co-cultured with bone marrow (BM) cells from mice harboring the mouse dF508 mutation in both cftr alleles (29) for 16 hours. The supernatant was removed and assayed for tumor necrosis factor alpha (TNF-α) concentrations. Co-culture of murine bone marrow cells with A. fumigatus elicited an increase in TNF-α production; however, this effect was significantly reduced by Trikafta treatment (Fig. 6G). Finally, we tested whether Trikafta altered inflammatory cytokine production from WT BM cells. Intriguingly, Trikafta treatment also reduced TNF-α production from WT BM cells in co-culture with A. fumigatus compared to vehicle controls (Fig. 6H). Furthermore, A1160 and A1160-ΔsakA were equally susceptible to Trikafta-induced protection against the inflammatory response elicited in co-culture with murine BM cells, suggesting that the effects of Trikafta on the cell wall are upstream of SakA-mediated biofilm biomass reduction (Fig. 6H). These data collectively demonstrate that the effects of Trikafta alter the fungal cell wall, which subsequently change how host cells respond to A. fumigatus, independent of cftr mutations.
DISCUSSION
The objective of this study was to determine if the current state-of-the-art treatment for pwCF, Trikafta, affects the biology of a common CF associated fungus, A. fumigatus. To begin to address this question, standard microbroth dilution MIC assays were utilized, and no anti-fungal effect was observed. However, the MIC assay tests the effects of a drug or small molecule against conidia, so we tested whether Trikafta affected A. fumigatus biofilms, which are found in an established infection. Unexpectedly, we observed that long-term (24-hour) treatment of biofilms with Trikafta reduced A. fumigatus biomass in several laboratory and clinical strains (Fig. 1D and E). The reduction in biofilm biomass correlated with reduced cell viability and increased membrane permeability (Fig. 2). Interestingly, the effects of Trikafta on permeability and viability occurred acutely, whereas biofilm biomass decrease was not observed until between 12 and 24 hours post treatment (Fig. 1B).
Surprisingly, there are only three major classes of approved anti-fungal drug therapies for the treatment of aspergillosis: polyenes, echinocandins, and azoles (30). Polyenes and azoles target the fungal cell membrane, and echinocandins target the fungal cell wall. Our data strongly suggest that Trikafta reduces viability in A. fumigatus biofilms by increasing membrane permeability and subsequent metabolic dysfunction. Within 1 hour of treatment with Trikafta, biofilms showed a significant increase in Sytox Blue staining and AK release (Fig. 2) and exacerbation of SDS-induced cell damage (Fig. 3D). These effects occur in parallel to Trikafta’s impact on overall viability of the biofilm. AK, which is highly expressed in the cytosol, is released extracellularly after the fungal cell integrity is compromised. It is also secreted when A. fumigatus is not actively growing or germinating (23). AK release has been successfully leveraged to identify anti-fungal compounds in several fungal species, such as A. fumigatus (23), Candida albicans (31), and Cryptococcus neoformans (32). Although Trikafta does not reduce overall growth of biofilms to the same magnitude as AmB treatment (Fig. 3C), we observed a striking effect of Trikafta on biofilm membrane permeability, viability, and growth. Considering the drug-resistant nature of fungal biofilms, future studies examining how these observations are relevant in the context of in vivo-relevant stressors are warranted.
Our data strongly suggest that Trikafta modulates ion channels in A. fumigatus biofilms. Given that treatment with Trikafta took more than 12 hours to affect biofilm growth, we were surprised to observe membrane permeabilization and reduced viability within 1 hour of treatment (Fig. 2A through C). Since we observed an effect on the cell membrane, we hypothesized that Trikafta’s mode of action is potentially like its function in mammalian cells. Previous studies in the literature have shown that calcium-activated chloride channels can also act as dual scramblases, which alter membrane fluidity and function (33, 34). Verapamil has been successfully utilized to block calcium channels in Aspergillus fumigatus (35), and our data demonstrate that it inhibits Trikafta-induced membrane permeability and biomass reduction (Fig. 3A and B). Our data also support the hypothesis that Trikafta may target multiple ion channels due to its exacerbation in conditions with high NaCl, CaCl2, and KCl levels (Fig. 3C). Since our null mutant for cch1 is resistant to Trikafta damage in calcium stress, this lends support that there are multiple ion channel targets for A. fumigatus, particularly since the CEA10-Δcch1 strain is equally susceptible to Trikafta-induced biomass reduction as WT (Fig. 3E). We also searched for CFTR-like proteins using the human cftr sequence in the A. fumigatus genome and identified eight highly related sequences, with the top hit being AFUB_066250. This gene is a putative ABC multidrug transporter with orthologs shown to have roles in secondary metabolite biosynthetic processes. We generated a null mutant in this gene and did not observe differences in Trikafta-induced biomass reduction compared to WT and reconstituted controls (data not shown).
In addition to effects on membrane permeabilization and ion channel modulation, our data also suggest that Trikafta affects cell wall composition in fungal biofilms. Interestingly, when biofilms were exposed to Trikafta and lower concentrations of caspofungin, Trikafta increased biofilm susceptibility to caspofungin-induced damage at low doses of drug (Fig. 6A through E). Alternatively, Trikafta protected biofilms from high doses of caspofungin (Fig. 6A through D). When biofilms were exposed to Trikafta and the cell wall stressor, CFW, Trikafta protected the biofilms from CFW-induced damage (Fig. 6A through C). Since CFW directly binds and blocks chitin polymerization and higher doses of caspofungin induce chitin biosynthesis via the paradoxical effect (36), it is plausible that Trikafta increases chitin composition in the biofilm cell wall. The caspofungin paradoxical effect (CPE) occurs when (1,3)-β-glucan synthase is inhibited to such a degree that the cell compensates by increasing chitin content. CPE-induced chitin synthesis has been shown to be stimulated via the cell wall integrity pathway mitogen-activated protein kinase signaling cascade and the transcription factors RlmA and CrzA in A. fumigatus strain Af293 (37). However, this effect has been shown to be strain specific, as CrzA is not required for CPE in CEA10-derived strains (37). Furthermore, depletion of (1,3)-β-glucan synthases results in increased chitin in Candida albicans, which is dependent on calcium (38). Future studies examining the effects of calcium, CPE, and CWI pathway are critical in understanding how Trikafta affects cell wall composition.
Given our observations that Trikafta disrupts cell wall and cell membrane integrity, it is perhaps not surprising that we observed Trikafta-mediated phosphorylation of the A. fumigatus HOG pathway kinase SakA. However, two independent sakA null mutant strains were resistant to Trikafta-induced biomass reduction (Fig. 5A and B). Furthermore, sakA null mutant strains were not resistant to Trikafta effects on the inflammatory response (Fig. 6F), suggesting that the Trikafta/SakA phenotype is either downstream or independent of cell wall effects. One hypothesis we tested was that Trikafta induces an osmotic stress response in the biofilms; however, Trikafta did not increase glycerol accumulation, which is a classic response to osmotic stress (data not shown). In addition to the osmotic stress response, the HOG pathway has also been observed to be involved in general stress responses, such as those induced by oxidative stress. In Aspergillus nidulans, SakA has been shown to interact with other stress response proteins, such as SrkA, Mpkc, and AN6892 after treatment with hydrogen peroxide (39). This general stress response reduced mitochondrial function and caused cell cycle arrest. Furthermore, the SakA homolog Hog1 mutant has been shown to have increased respiration rates and altered mitochondrial membrane potential in Candida albicans (40). The widely used fungicide, fludioxonil, has been shown to cause fungal cell death by hyperactivating the Hog pathway (41, 42). In the context of Trikafta treatment of biofilms, increased ion channel activation could potentially cause SakA-dependent alterations in mitochondrial function and subsequent growth arrest, which was reduced in the biofilms lacking sakA (Fig. 5).
The fungal cell wall is critical in influencing the extent to which an immune response is generated against colonization or infection by fungal pathogens. The A. fumigatus cell wall contains galactomannan moieties as well as chitin and (1,3)-β-glucan, which all play a role in triggering an inflammatory response in the host [reviewed in reference (43)]. This is evidenced by early work showing that glucans can activate leukocytes, phagocytosis, and production of pro-inflammatory mediators (44). The C-type lectin receptor, Dectin-1, has been identified as the key host protein involved in recognition of fungal cell wall component (1,3)-β-glucan (45). Macrophages expressing high levels of Dectin-1 were shown to have an increased inflammatory response to A. fumigatus, which was completely blocked by neutralizing Dectin-1 antibodies (46). Furthermore, Dectin-1 can more efficiently recognize certain stages of A. fumigatus development that correspond with higher levels of β-glucans. Swollen conidia and germlings, which have the highest levels of β-glucan exposure, were shown to cause high levels of pro-inflammatory cytokine production (46). Our data demonstrate that Trikafta protects A. fumigatus biofilms from CFW (Fig. 6C) and causes increased susceptibility to caspofungin, which implies that there is less β-glucan content in the cell wall (Fig. 6D). This conclusion is supported by a decrease in TNF-α production in the supernatants of conidia and bone marrow cells in co-culture with Trikafta compared to vehicle controls (Fig. 6E and F). Since this phenotype is conserved in both WT and dF508 bone marrow cells, it is likely that this is attributed to the effect of Trikafta on the fungus itself, rather than host cells. It is also possible that this effect is due to non-CFTR-mediated effects (off-target) in the bone marrow cells. Further studies to determine how Trikafta affects the fungal cell wall and how this impacts host/pathogen interactions will be critical for determining ways to augment and/or leverage Trikafta’s antifungal activity in pwCF.
Our data suggest that the current CF treatment, Trikafta, causes biomass reduction, membrane permeabilization, and loss of viability in A. fumigatus biofilms. We observed that Trikafta alters A. fumigatus biofilm responses to cell wall stress, which correlated with reduced inflammatory cytokine production from bone marrow cells in co-culture with Trikafta-treated A. fumigatus. In our studies, we utilized drug concentrations that are potentially achievable in patients. U.S. Food and Drug Administration documentation showed Cmax levels were 9.2 µg/mL for elexacaftor (we used 5 μM = 2.99 µg/mL), 7.7 µg/mL for tezacaftor (we used 5 μM = 2.60 µg/mL), and 1.2 µg/mL for Ivacaftor (we used 5μM = 1.96 µg/mL) in serum (47). To understand the implications of these observations more fully, in vivo models of allergic and invasive aspergillosis are necessary to define the effect of Trikafta on A. fumigatus biofilms, and the subsequent host response, in the mammalian lung.
MATERIALS AND METHODS
Strains and growth conditions
All strains utilized in this study are listed in Table 3. All strains were initially grown in agar (1.5%) plates containing 1% GMM (1% glucose, 6-g/L NaNO3, 0.52-g/L KCl, 0.52-g/L MgSO4•7H2O, 1.52-g/L KH2PO4 monobasic, 2.2-mg/L ZnSO4•7H2O, 1.1-mg/L H3BO3, 0.5-mg/L MnCl2•4H2O, 0.5-mg/L FeSO4•7H2O, 0.16-mg/L CoCl2•5H2O, 0.16-mg/L CuSO4•5H2O, 0.11-mg/L (NH4)6Mo7O24•4H2O, and 5-mg/L Na4EDTA; pH 6.5). Conidia were collected for experiments after growth at 37°C and 5% CO2 for 72 hours with 0.01% Tween-80 and filtered through miracloth (Millipore Sigma) to exclude hyphae.
TABLE 3.
List of A. fumigatus strains used in the study
| Strain | Origin or background strain | Source or genetic modification |
|---|---|---|
| Laboratory “wild-type” strains | ||
| CEA10 (CBS144.89) | Patient with IPA a | CBS KNAW Fungal Biodiversity Centre |
| Af293 | Lung biopsy: neutropenic patient | David Denning Laboratory |
| ATCC13073 | Pulmonary lesion | American Type Culture Collection |
| Clinical isolates | ||
| Af110-14.14 | Sputum sample from pwCF | Dartmouth Hitchcock Medical Center |
| Af110-5.3 | Sputum sample from pwCF | Dartmouth Hitchcock Medical Center |
| Af106-6.10 | Sputum sample from pwCF | Dartmouth Hitchcock Medical Center |
| TP-9 | Sputum sample from pwCF | Dartmouth Hitchcock Medical Center |
| Mutants | ||
| A1160 | CEA10 | akuBKU80; pyrG (48) |
| Afs35 | D141 | akuAKU70−; ptrA (49) |
| CEA10-gpdA:GFP | CEA10 | gpdAp:GFP;hygB |
| CEA10-Δcch1 | CEA10 | cch1 null mutant; Hyg (AFUB_010540) |
| CEA10-cch1 rec | CEA10-Δcch1 | cch1 reconstituted mutant at atf4 save haven site; ptrA |
| A1160-ΔsakA | A1160 | sakA null mutant; Hyg (AFUB_012420 (50), |
| Afs35-ΔsakA | Afs35 | sakA null mutant; Hyg (51) |
| TP-9PBS2C1 | TP-9 | pbs2 allele swap; ptrA (26) |
| TP-9PBS2C1: sakA Flag | TP-9 | pbs2 allele swap; sakA:Flag; Hyg (26) |
IPA, invasive pulmonary aspergillosis.
Biofilm growth and drug treatments
For all experiments, conidia from the indicated strains were counted and seeded in liquid GMM at 1.0 × 105 conidia/mL for the indicated times in 6-well plates (for biomass assays) and 96-well plates for all other assays to develop immature biofilms. Media were then removed and replaced with GMM containing either DMSO or the indicated doses of ivacaftor (5 µM = 1.96 µg/mL), tezacaftor (5 µM = 2.60 µg/mL), elexacaftor (5 µM = 2.99 µg/mL), or the indicated combinations (Selleckchem). In ionic stress experiments, biofilms were grown in GMM, and at time of drug treatment, 500-mM ionic stressors were supplemented to the media. For drug co-treatments, all biofilms were grown as above. Biofilms were treated for 1 or 3 hours with the vehicle controls (DMSO or water), voriconazole (0.25- to 4.0-µg/mL DMSO, Sigma), amphotericin B (0.125- to 2.0-µg/mL DMSO, Cayman Chemicals), caspofungin (0.015625–4.0 µg/mL, Cayman Chemicals), calcofluor white (6.25–50.0 µg/mL, Sigma), GlyH-101 (20 µM, Selleckchem), verapamil (1 mM, Sigma), SDS (0.002%, Fisher), hydrogen peroxide (0.3125–5.0 mM, Sigma), FCCP (0.25–4.0 µM, Sigma), fludioxonil (31.25–500.0 µg/mL, Sigma), antimycin A (0.25–4.0 µg/mL, Sigma), and potassium cyanide (0.25–4.0 mM, Sigma).
Quantification of submerged biofilm biomass
After growth in six-well plates and treatment for the indicated times, excess supernatant was removed from the biofilms by tilting plates on the side and removing pooled liquid. Biomass was then carefully scraped and collected into preweighed tubes. Samples were vortexed in 1-mL ddH2O and then centrifuged at 15,000 rcf for 10 min to pellet mycelia and then repeated. Supernatants were removed, frozen, then lyophilized. Dry biomass was then quantified.
XTT/resazurin assay
After biofilm treatment with drugs or vehicle for the indicated times, media were removed. XTT solution (0.5-mg XTT/mL 1 × phosphate-buffered saline [PBS] with 25-µM menadione) (XTT sodium salt, VWR) was added at 150 µL per well and incubated at 37°C, 5% CO2, and 21% O2 until the positive vehicle control wells were reduced (1–2 hours). Next, 100 µL of the XTT solution supernatant was transferred to a 96-well plate, and optical density was measured at 450 nm. For resazurin experiments, a knockout library of putative A. fumigatus kinases was utilized (listed in Table 1) (50). Biofilms were grown to 16-hour maturity; media were removed; and fresh media with DMSO or Trikafta were added. After 30 minutes, media were supplemented with 10% resazurin and incubated for an additional 5 hours. Fluorescence at an excitation of 544 and an emission of 590 was used for quantification. To determine synergy/antagonism between Trikafta and caspofungin, a standard checkerboard assay (52) was modified for 16-hour biofilms. Trikafta working dose ranges used were 30.2 μg/mL (20 μM/molecule)–0.471875 μg/mL (0.3125 μM/molecule), and caspofungin working dose ranges were 8.0–0.015625 μg/mL in liquid GMM. Serial dilutions for Trikafta were inoculated in twofold dilutions vertically, and serial dilutions for caspofungin were inoculated in twofold dilutions horizontally. Media were removed from 16-hour biofilms, and media containing caspofungin/Trikafta dose combinations were transferred to biofilms for 3 hours. Media were then removed and XTT solution (described above) was added to biofilms for 1–2 hours, and the assay was performed as described above. The no-drug control well was set to 100%, and values of all other wells were calculated as a percentage of control. Fractional inhibitory concentration index was calculated based on the metabolic reduction concentration (MRC = less than 25% metabolic activity). Synergy/antagonism values were calculated as (MRC Drug A incombination with Drug B)/MRC Drug A + (MRC Drug B in combination with Drug A)/MRC Drug B. Any score less than 0.5 was considered synergy, and higher than 4 was antagonism.
Adenylate kinase assay
To quantify alkaline phosphatase in biofilm supernatants, samples were assayed as previously described (23). Briefly, supernatants were collected and kept at 4°C for no longer than 24 hours. AK detection reagent (100-µL Toxilight non-destructive cytotoxicity bioassay, Lonza) was added to each well of samples (20 µL) and incubated at room temperature for 5 minutes prior to luminescence measurements (1-hour, 5-minute intervals).
Sytox Blue stain of biofilms
To assess Sytox Blue staining in treated biofilms, biofilms were grown for 16 hours in filtered GMM; media were removed; and were treated as indicated. After treatment, biofilms were stained with Sytox Blue (Thermo Scientific) 1:1,000 for 5 minutes and then imaged using a Nikon spinning disc confocal microscope at ×20 magnification. Images were processed and analyzed using the Fiji image analysis software (53). Raw image stacks were resliced from top down without interpolation and maximum projected. The Fiji plugin JaCoP was used to analyze the co-localization of the cytosolic GFP signal with the Sytox signal (54). In the JaCoP plugin, the Sytox channel was set to channel 1, and GFP was set to channel 2. Channel thresholds were set manually to exclude the background signal. The Manders’ overlap was quantified between the two channels, and M2 was recorded as this represents the fraction of GFP-positive pixels that overlapped with Sytox-positive pixels as a readout for level of Sytox staining across conditions (55).
Phospho-SakA Western blot
To assess phosphorylation of SakA by Western blot, a protocol for submerged biofilms was adapted from stationary cultures (26). Briefly, submerged biofilms (TP9PBS2C1:sakA-Flag) were grown in six-well plates for 16 hours; media were removed; and indicated treatments in GMM were added for an additional 30 minutes. Biomass was scraped into 2-mL screw cap tubes, frozen, and lyophilized. They were then bead beaten in 1-mL protein extraction buffer, and total protein was quantified by Bradford kit. For Western blotting, 40-µg protein was used. Total protein was transferred from a 10% SDS-PAGE gel onto a nitrocellulose membrane using the Trans-Blot turbo transfer system (Bio-Rad). Total Flag-SakA was detected using the Flag M2 antibody 1:2,000 (Sigma), and phosphorylated SakA was detected using anti-P-p38 antibody 1:1,000 (Cell Signaling). To quantify fluorescence, imaging was performed using the LI-COR Odyssey CLX System according to manufacturer’s protocols. Total Flag and phospho-SakA were normalized to total protein using the REVERT total protein stain (LI-COR Biosciences).
Bone marrow co-culture
Bone marrow cells were collected from the tibia and femur of female C57Bl/6 WT and dF508 mice (10–12 weeks). The dF508 and littermate control breeders were obtained from Case Western Reserve University (Cystic Fibrosis Mouse Models Core). Bones were removed and BM cells were isolated by flushing bone marrow out using DMEM (Dulbecco's Modified Eagle's Medium)-based tissue culture media. Cells were pelleted; red blood cells were lysed; and cells were set up in co-culture with the indicated strains of conidia at an MOI (multiplicity of infection) of 1:10 supplemented with 20-ng/mL interferon gamma for 16 hours. Media were collected and assayed for total TNF-α concentration according to manufacturer’s instructions (R&D Systems).
Statistics
All experiments were performed with at least three biological replicates, and data points were averages of technical replicates for each individual experiment. For experiments comparing two groups, a standard Student t-test was performed. For experiments comparing two groups under multiple conditions, a two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons was used, or Dunnett’s multiple comparisons were used when comparing to one control. When comparing more than two groups, a one-way ANOVA with Tukey’s multiple comparisons was used. All statistical analyses were performed using GraphPad Prism version 9.4.1. For all graphs, ns (not significant), P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, ****, P ≤ 0.0001. All diagrams were generated using BioRender.com.
ACKNOWLEDGMENTS
This work was supported by the efforts of R.A.C. through funding by a Cystic Fibrosis Foundation research award (CRAMERGO19) and National Institutes of Health (NIH) NIAID R33-AI140878. K.A.M. was supported by NIH award T32-AI007519. Core facility support was provided by NIH grant P20-GM113132 to the Dartmouth BioMT COBRE. Additional support was provided by the Cystic Fibrosis Foundation Research Development Program (STANTO19R0) and NIH P30-DK117469 (Dartmouth Cystic Fibrosis Research Center).
The authors would like to thank Dr. Gustavo Goldman for AFS35 sakA mutant strains and the Dartmouth Translational Research Core led by Dr. Alix Ashare and Dr. Deborah Hogan for pwCF clinical samples.
Contributor Information
Robert A. Cramer, Email: robert.a.cramer.jr@dartmouth.edu.
J. Andrew Alspaugh, Duke University Hospital, Durham, North Carolina, USA .
DATA AVAILABILITY
All underlying data are available from the corresponding author upon request.
ETHICS APPROVAL
Murine studies for collection of bone marrow cells were approved by the Dartmouth Institutional Animal Care and Use Committee under protocol number 00002167.
REFERENCES
- 1. Heijerman HGM, McKone EF, Downey DG, Van Braeckel E, Rowe SM, Tullis E, Mall MA, Welter JJ, Ramsey BW, McKee CM, Marigowda G, Moskowitz SM, Waltz D, Sosnay PR, Simard C, Ahluwalia N, Xuan F, Zhang Y, Taylor-Cousar JL, McCoy KS, VX17-445-103 Trial Group . 2019. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet 394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polineni D, Ramsey BW, Taylor-Cousar JL, Tullis E, Vermeulen F, Marigowda G, McKee CM, Moskowitz SM, Nair N, Savage J, Simard C, Tian S, Waltz D, Xuan F, Rowe SM, Jain R, VX17-445-102 Study Group . 2019. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single phe508del allele. N Engl J Med 381:1809–1819. doi: 10.1056/NEJMoa1908639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laselva O, Bartlett C, Gunawardena TNA, Ouyang H, Eckford PDW, Moraes TJ, Bear CE, Gonska T. 2021. Rescue of multiple class II CFTR mutations by elexacaftor+ tezacaftor+ivacaftor mediated in part by the dual activities of elexacaftor as both corrector and potentiator. Eur Respir J 57:2002774. doi: 10.1183/13993003.02774-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filkins LM, O’Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanj A, Abdallah N, Soubani AO. 2018. The spectrum of pulmonary aspergillosis. Respir Med 141:121–131. doi: 10.1016/j.rmed.2018.06.029 [DOI] [PubMed] [Google Scholar]
- 6. Kousha M, Tadi R, Soubani AO. 2011. Pulmonary aspergillosis: a clinical review. Eur Respir Rev 20:156–174. doi: 10.1183/09059180.00001011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lass-Flörl C, Follett SA, Moody A, Denning DW. 2011. Detection of Aspergillus in lung and other tissue samples using the mycassay Aspergillus real-time PCR kit. Can J Microbiol 57:765–768. doi: 10.1139/w11-064 [DOI] [PubMed] [Google Scholar]
- 8. Messina JA, Wolfe CR, Hemmersbach-Miller M, Milano C, Todd JL, Reynolds J, Alexander BD, Schell WA, Cuomo CA, Perfect JR. 2018. Genomic characterization of recurrent mold infections in thoracic transplant recipients. Transpl Infect Dis 20:e12935. doi: 10.1111/tid.12935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marr KA, Carter RA, Crippa F, Wald A, Corey L. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin Infect Dis 34:909–917. doi: 10.1086/339202 [DOI] [PubMed] [Google Scholar]
- 10. Kowalski CH, Kerkaert JD, Liu K-W, Bond MC, Hartmann R, Nadell CD, Stajich JE, Cramer RA. 2019. Fungal biofilm morphology impacts hypoxia fitness and disease progression. Nat Microbiol 4:2430–2441. doi: 10.1038/s41564-019-0558-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loussert C, Schmitt C, Prevost M-C, Balloy V, Fadel E, Philippe B, Kauffmann-Lacroix C, Latgé JP, Beauvais A. 2010. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol 12:405–410. doi: 10.1111/j.1462-5822.2009.01409.x [DOI] [PubMed] [Google Scholar]
- 12. Kowalski CH, Morelli KA, Schultz D, Nadell CD, Cramer RA. 2020. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc Natl Acad Sci U S A 117:22473–22483. doi: 10.1073/pnas.2003700117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denning DW, Park S, Lass-Florl C, Fraczek MG, Kirwan M, Gore R, Smith J, Bueid A, Moore CB, Bowyer P, Perlin DS. 2011. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis 52:1123–1129. doi: 10.1093/cid/cir179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mowat E, Butcher J, Lang S, Williams C, Ramage G. 2007. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J Med Microbiol 56:1205–1212. doi: 10.1099/jmm.0.47247-0 [DOI] [PubMed] [Google Scholar]
- 15. Mowat E, Lang S, Williams C, McCulloch E, Jones B, Ramage G. 2008. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J Antimicrob Chemother 62:1281–1284. doi: 10.1093/jac/dkn402 [DOI] [PubMed] [Google Scholar]
- 16. O’Dea AL, Feng R, Glaser LJ, Kubrak C, Rubenstein RC, Dorgan DJ, Hadjiliadis D, Kawut SM, Hong G. 2023. The clinical association between Aspergillus fumigatus and respiratory outcomes in adolescents and adults with cystic fibrosis. Ann Am Thorac Soc 20:984–992. doi: 10.1513/AnnalsATS.202210-852OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noni M, Katelari A, Dimopoulos G, Doudounakis S-E, Tzoumaka-Bakoula C, Spoulou V. 2015. Aspergillus fumigatus chronic colonization and lung function decline in cystic fibrosis may have a two-way relationship. Eur J Clin Microbiol Infect Dis 34:2235–2241. doi: 10.1007/s10096-015-2474-y [DOI] [PubMed] [Google Scholar]
- 18. Saunders RV, Modha DE, Claydon A, Gaillard EA. 2016. Chronic Aspergillus fumigatus colonization of the pediatric cystic fibrosis airway is common and may be associated with a more rapid decline in lung function. Med Mycol 54:537–543. doi: 10.1093/mmy/myv119 [DOI] [PubMed] [Google Scholar]
- 19. Heltshe SL, Mayer-Hamblett N, Burns JL, Khan U, Baines A, Ramsey BW, Rowe SM, GOAL (the G551D Observation-AL) Investigators of the Cystic Fibrosis Foundation Therapeutics Development Network . 2015. Pseudomonas aeruginosa in cystic fibrosis patients with G551D-CFTR treated with ivacaftor. Clin Infect Dis 60:703–712. doi: 10.1093/cid/ciu944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frost FJ, Nazareth DS, Charman SC, Winstanley C, Walshaw MJ. 2019. Ivacaftor is associated with reduced lung infection by key cystic fibrosis pathogens. A cohort study using national registry data. Ann Am Thorac Soc 16:1375–1382. doi: 10.1513/AnnalsATS.201902-122OC [DOI] [PubMed] [Google Scholar]
- 21. Currie AJ, Main ET, Wilson HM, Armstrong-James D, Warris A. 2020. CFTR modulators dampen Aspergillus-induced reactive oxygen species production by cystic fibrosis phagocytes. Front Cell Infect Microbiol 10:372. doi: 10.3389/fcimb.2020.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. CLSI . 2017. Reference method for broth dilution antifungal susceptibility testing of Filamentous fungi. CLSI standard M38 [Google Scholar]
- 23. Beattie SR, Krysan DJ, Perlin DS. 2021. A unique dual-readout high-throughput screening assay to identify antifungal compounds with Aspergillus fumigatus. mSphere 6:e0053921. doi: 10.1128/mSphere.00539-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muanprasat C, Sonawane ND, Salinas D, Taddei A, Galietta LJV, Verkman AS. 2004. Discovery of glycine hydrazide pore-occluding CFTR inhibitors: mechanism, structure-activity analysis, and in vivo efficacy. J Gen Physiol 124:125–137. doi: 10.1085/jgp.200409059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Castro PA, Chiaratto J, Winkelströter LK, Bom VLP, Ramalho LNZ, Goldman MHS, Brown NA, Goldman GH. 2014. The involvement of the Mid1/Cch1/Yvc1 calcium channels in Aspergillus fumigatus virulence. PLoS One 9:e103957. doi: 10.1371/journal.pone.0103957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross BS, Lofgren LA, Ashare A, Stajich JE, Cramer RA. 2021. Aspergillus fumigatus in-host HOG pathway mutation for cystic fibrosis lung microenvironment persistence. mBio 12:e0215321. doi: 10.1128/mBio.02153-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowalski CH, Morelli KA, Schultz D, Nadell CD, Cramer RA. 2020. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc Natl Acad Sci U S A 117:22473–22483. doi: 10.1073/pnas.2003700117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verwer PEB, van Duijn ML, Tavakol M, Bakker-Woudenberg I, van de Sande WWJ. 2012. Reshuffling of Aspergillus fumigatus cell wall components chitin and β-glucan under the influence of caspofungin or nikkomycin Z alone or in combination. Antimicrob Agents Chemother 56:1595–1598. doi: 10.1128/AAC.05323-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeiher BG, Eichwald E, Zabner J, Smith JJ, Puga AP, McCray PB, Capecchi MR, Welsh MJ, Thomas KR. 1995. A mouse model for the delta F508 allele of cystic fibrosis. J Clin Invest 96:2051–2064. doi: 10.1172/JCI118253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perfect JR. 2017. The antifungal pipeline: a reality check. Nat Rev Drug Discov 16:603–616. doi: 10.1038/nrd.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krysan DJ, Didone L. 2008. A high-throughput screening assay for small molecules that disrupt yeast cell integrity. J Biomol Screen 13:657–664. doi: 10.1177/1087057108320713 [DOI] [PubMed] [Google Scholar]
- 32. Butts A, DiDone L, Koselny K, Baxter BK, Chabrier-Rosello Y, Wellington M, Krysan DJ. 2013. A repurposing approach identifies off-patent drugs with fungicidal cryptococcal activity, a common structural chemotype, and pharmacological properties relevant to the treatment of cryptococcosis. Eukaryot Cell 12:278–287. doi: 10.1128/EC.00314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malvezzi M, Chalat M, Janjusevic R, Picollo A, Terashima H, Menon AK, Accardi A. 2013. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun 4:2367. doi: 10.1038/ncomms3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Falzone ME, Rheinberger J, Lee B-C, Peyear T, Sasset L, Raczkowski AM, Eng ET, Di Lorenzo A, Andersen OS, Nimigean CM, Accardi A. 2019. Structural basis of ca2+dependent activation and lipid transport by a TMEM16 scramblase. Elife 8:e43229. doi: 10.7554/eLife.43229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Juvvadi PR, Muñoz A, Lamoth F, Soderblom EJ, Moseley MA, Read ND, Steinbach WJ. 2015. Calcium-mediated induction of paradoxical growth following caspofungin treatment is associated with calcineurin activation and phosphorylation in Aspergillus fumigatus. Antimicrob Agents Chemother 59:4946–4955. doi: 10.1128/AAC.00263-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fortwendel JR, Juvvadi PR, Perfect BZ, Rogg LE, Perfect JR, Steinbach WJ. 2010. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob Agents Chemother 54:1555–1563. doi: 10.1128/AAC.00854-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, de Assis LJ, Bertolini MC, Malavazi I, Goldman GH, Alspaugh JA. 2017. The Aspergillus fumigatus CrzA transcription factor activates chitin synthase gene expression during the caspofungin paradoxical effect. mBio 8:e00705-17. doi: 10.1128/mBio.00705-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han Q, Wang N, Pan C, Wang Y, Sang J. 2019. Elevation of cell wall chitin via ca2+-calcineurin-mediated PKC signaling pathway maintains the viability of candida albicans in the absence of β-1,6-glucan synthesis. Mol Microbiol 112:960–972. doi: 10.1111/mmi.14335 [DOI] [PubMed] [Google Scholar]
- 39. Jaimes-Arroyo R, Lara-Rojas F, Bayram Ö, Valerius O, Braus GH, Aguirre J. 2015. The SrkA kinase is part of the SakA mitogen-activated protein kinase interactome and regulates stress responses and development in Aspergillus nidulans. Eukaryot Cell 14:495–510. doi: 10.1128/EC.00277-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J. 2009. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen candida albicans. Microbiology (Reading) 155:413–423. doi: 10.1099/mic.0.023309-0 [DOI] [PubMed] [Google Scholar]
- 41. Lawry SM, Tebbets B, Kean I, Stewart D, Hetelle J, Klein BS. 2017. Fludioxonil induces Drk1, a fungal group III hybrid histidine kinase, to dephosphorylate its downstream target, Ypd1. Antimicrob Agents Chemother 61:e01414-16. doi: 10.1128/AAC.01414-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El-Mowafy M, Bahgat MM, Bilitewski U. 2013. Deletion of the HAMP domains from the histidine kinase caNik1P of candida albicans or treatment with fungicides activates the MAP kinase Hog1P in S. cerevisiae transformants. BMC Microbiol 13:209. doi: 10.1186/1471-2180-13-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beauvais A, Latgé JP. 2001. Membrane and cell wall targets in Aspergillus fumigatus. Drug Resist Updat 4:38–49. doi: 10.1054/drup.2001.0185 [DOI] [PubMed] [Google Scholar]
- 44. Williams DL. 1997. Overview of (1-->3)-beta-D-glucan immunobiology. Mediators Inflamm 6:247–250. doi: 10.1080/09629359791550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown GD, Herre J, Williams DL, Willment JA, Marshall ASJ, Gordon S. 2003. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med 197:1119–1124. doi: 10.1084/jem.20021890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, Kolls JK, Brown GD, Filler SG. 2005. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog 1:e42. doi: 10.1371/journal.ppat.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. FDA . 2021. In Highlights of prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/212273s008lbl.pdf.
- 48. da Silva Ferreira ME, Kress M, Savoldi M, Goldman MHS, Härtl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuB(Ku80) mutant deficient for Nonhomologous end joining is a powerful tool for analyzing Pathogenicity in Aspergillus Fumigatus. Eukaryot Cell 5:207–211. doi: 10.1128/EC.5.1.207-211.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagener J, Echtenacher B, Rohde M, Kotz A, Krappmann S, Heesemann J, Ebel F. 2008. The putative Alpha-1,2-Mannosyltransferase Afmnt1 of the opportunistic fungal pathogen Aspergillus Fumigatus is required for cell wall stability and full virulence. Eukaryot Cell 7:1661–1673. doi: 10.1128/EC.00221-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Rhijn N, Zhao C, Al-Furaji N, Storer I, Valero C, Gago S, Chown H, Baldin C, Fortune-Grant R, Shuraym HB, Ivanova L, Kniemeyer O, Krüger T, Bignell E, Goldman G, Amich J, Delneri D, Bowyer P, Brakhage A, Haas H, Bromley M. 2023. Functional analysis of the Aspergillus fumigatus kinome reveals a DYRK kinase involved in septal plugging is a novel antifungal drug target. Research square:rs.3.rs-2960526. doi: 10.21203/rs.3.rs-2960526/v1 [DOI]
- 51. Bruder Nascimento A de O, Dos Reis TF, de Castro PA, Hori JI, Bom VLP, de Assis LJ, Ramalho LNZ, Rocha MC, Malavazi I, Brown NA, Valiante V, Brakhage AA, Hagiwara D, Goldman GH. 2016. Mitogen activated protein Kinases Saka(Hog1) and Mpkc collaborate for Aspergillus fumigatus virulence. Mol Microbiol 100:841–859. doi: 10.1111/mmi.13354 [DOI] [PubMed] [Google Scholar]
- 52. 2023. M38Ed3 Filamentous Fungi Antifungal Susceptibility Test. Available from: https://clsi.org/standards/products/microbiology/documents/m38
- 53. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bolte S, Cordelières FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 55. Manders EMM, Verbeek FJ, Aten JA. 1993. Measurement of co-localization of objects in dual-colour confocal images. J Microsc 169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All underlying data are available from the corresponding author upon request.