Abstract
Although nitric oxide (NO) has grown into a key signaling molecule in plants during the last few years, less is known about how NO regulates different events in plants. Analyses of NO-dependent processes in animal systems have demonstrated protein S-nitrosylation of cysteine (Cys) residues to be one of the dominant regulation mechanisms for many animal proteins. For plants, the principle of S-nitrosylation remained to be elucidated. We generated S-nitrosothiols by treating extracts from Arabidopsis (Arabidopsis thaliana) cell suspension cultures with the NO-donor S-nitrosoglutathione. Furthermore, Arabidopsis plants were treated with gaseous NO to analyze whether S-nitrosylation can occur in the specific redox environment of a plant cell in vivo. S-Nitrosylated proteins were detected by a biotin switch method, converting S-nitrosylated Cys to biotinylated Cys. Biotin-labeled proteins were purified and analyzed using nano liquid chromatography in combination with mass spectrometry. We identified 63 proteins from cell cultures and 52 proteins from leaves that represent candidates for S-nitrosylation, including stress-related, redox-related, signaling/regulating, cytoskeleton, and metabolic proteins. Strikingly, many of these proteins have been identified previously as targets of S-nitrosylation in animals. At the enzymatic level, a case study demonstrated NO-dependent reversible inhibition of plant glyceraldehyde-3-phosphate dehydrogenase, suggesting that this enzyme could be affected by S-nitrosylation. The results of this work are the starting point for further investigation to get insight into signaling pathways and other cellular processes regulated by protein S-nitrosylation in plants.
Overwhelming evidence suggests that nitric oxide (NO) is an integral part of normal physiological processes in animals (Nathan, 1995; Bogdan, 2001). By the late 1990s, NO was identified as an important messenger in plant defense signaling against microbial pathogens (Delledonne et al., 1998; Durner et al., 1998); it subsequently was shown to be a crucial player in the regulation of normal plant physiological processes including stomatal closure, growth, and development (Neill et al., 2002; Wendehenne et al., 2004), and recently, a hormone-activated NO producing enzyme was identified in Arabidopsis (Arabidopsis thaliana; Guo et al., 2003; Zeidler et al., 2004).
As a readily diffusible free radical, NO reacts with a variety of intracellular and extracellular targets and can act as activator or inhibitor of enzymes, ion channels, or transcription factors and in this way regulates specific processes during plant development and abiotic or biotic stress situations. The alteration of protein function/activity can be achieved by reaction of NO with sulfhydryl groups and transition metals (Stamler, 1994), and the resulting products, S-nitrosothiols and metal nitrosyls, respectively, have intrinsic reactivities that enable local action.
The majority of all NO-affected proteins seem to be regulated by S-nitrosylation of a single critical Cys residue, which occurs by oxygen-dependent chemical reactions or by the transfer of NO from a nitrosothiol to a protein sulfhydryl group (transnitrosylation). Because of their reactivity with intracellular reducing agents, e.g. ascorbic acid or glutathione (GSH), and with reduced metal ions, especially Cu+, nitrosothiols are exceptionally labile. This lability results in tissue half-lives of seconds to a few minutes and therefore provides a very sensitive mechanism for regulating cellular processes. S-Nitrosylation is now regarded as posttranslational modification similar to phosphorylation.
In mammalians, especially S-nitrosoalbumin, S-nitrosohaemoglobin, and S-nitrosoglutathione (GSNO) are discussed as in vivo NO reservoirs and NO donors (Stamler et al., 1992a; Jia et al., 1996; Kluge et al., 1997; Tsikas et al., 1999). In plants, a strong GSNO reductase activity was demonstrated recently for GSH-dependent formaldehyde dehydrogenase (Diaz et al., 2003), an enzyme previously identified as formaldehyde detoxifying protein in maize (Zea mays; Fliegmann and Sandermann, 1997; Wippermann et al., 1999) indicating that GSNO and the formation of other nitrosothiols might play an important role in NO signaling in plants as well (Durner et al., 1999; Diaz et al., 2003). On the other side, it is also speculated whether metal nitrosyls are the dominant compounds for NO storage and NO transport in plants (Garcia-Mata and Lamattina, 2003).
Until now, little has been known about the dimension let alone the physiological function of S-nitrosylation in plants, and no endogenous S-nitrosylated plant protein has been described. To identify possible targets of S-nitrosylation, SwissProt database was searched for the consensus motif of S-nitrosylation sensitive Cys residues (Stamler et al., 1997). This search revealed 103 matches in 99 sequences from the deduced Arabidopsis proteome (Huber and Hardin, 2004). Except for this bioinformatics strategy, no approaches were undertaken to identify S-nitrosylated plant proteins.
The aim of this study was to identify possible candidates for S-nitrosylation in Arabidopsis cell suspension cultures and leaves to get insight into the regulatory function of NO on protein level in plants. Recently, Jaffrey et al. (2001) developed a highly specific biotin switch method for detection and purification of S-nitrosylated proteins in animals (Jaffrey et al., 2001; Kuncewicz et al., 2003; Foster and Stamler, 2004; Martinez-Ruiz and Lamas, 2004). A proteomics approach using this method in combination with nano liquid chromatography and tandem mass spectrometry (nanoLC/MS/MS) allowed us to identify 63 proteins from GSNO-treated cell culture extracts and 52 proteins from NO-treated Arabidopsis leaves, which represent targets for S-nitrosylation in plants. These proteins include stress-related proteins, signaling/regulating proteins, redox-related proteins, and cytoskeleton proteins as well as metabolic enzymes. Strikingly, about 60% of the identified proteins were already described in the animal system in context with S-nitrosylation or S-glutathionylation, underlining the specificity of the method and indicating that NO-regulated processes in plants and animals have common features.
RESULTS
Generation of S-Nitrosothiols
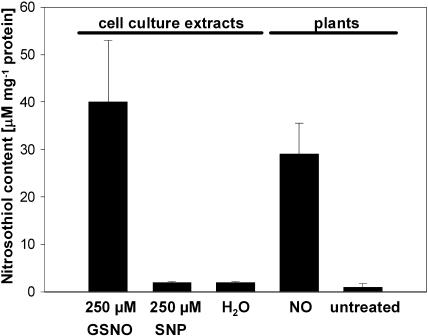
To generate S-nitrosylated Arabidopsis proteins, we treated extracts from cell suspension cultures with the NO-donors GSNO and sodium nitroprusside (SNP). Furthermore, we exposed Arabidopsis plants to NO gas to investigate nitrosothiol formation in intact plants. Nitrosothiol contents were determined using the colorimetric assay developed by Saville (1958). Treatment of cell culture extracts with 250 μm GSNO resulted in a nitrosothiol content of about 40 μm/mg protein (Fig. 1). Although SNP is described to also have a strong S-nitrosylating potential (Stamler et al., 1992b), we could detect only very low amounts of nitrosothiols (2 μm/mg protein) in the SNP-treated extracts (Fig. 1). The nitrosothiol content in NO-treated plants reached values up to 30 μm/mg protein. In untreated cell culture extracts as well as in untreated plants, only traces of nitrosothiols (1–2 μm/mg protein) could be measured (Fig. 1).
Figure 1.
Nitrosothiol content in Arabidopsis cell suspension culture extracts after treatment with NO donors and in leaves of NO-treated Arabidopsis plants. After treatment of cell culture extracts with either 250 μm GSNO, 250 μm SNP, or water, remaining NO donors were removed by chromatography on a Sephadex G-25M column. Nitrosothiol contents were determined according to Saville (1958). Additionally, nitrosothiol content from leaves of Arabidopsis plants treated with NO gas and from leaves of untreated plants was measured. Values represent mean of at least two independent determinations.
Detection of S-Nitrosylated Proteins
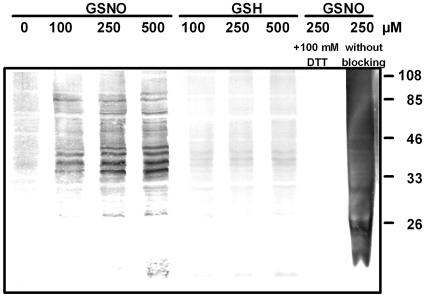
To detect S-nitrosylated proteins, we chose the biotin switch method that is based on the labeling of S-nitrosylated proteins with a biotin moiety specifically on S-nitrosylated Cys (Jaffrey et al., 2001). Biotinylated proteins can then be made visible by immunoblotting using anti-biotin antibody. After treatment of cell culture extracts with up to 500 μm GSNO, a group of about 12 protein bands was detected representing S-nitrosylated proteins (Fig. 2). In the samples treated with increasing concentrations of GSH, however, only some weak protein bands could be seen, confirming that the detected band in the GSNO samples are attributed to NO (Fig. 2). Furthermore, in GSNO-treated samples that underwent the biotin switch procedure following treatment with 100 mm dithiothreitol (DTT), no protein bands could be detected (Fig. 2). This demonstrates that the anti-biotin antibody shows no unspecific cross reaction with unlabeled proteins and that no in vivo biotinylated proteins were detected. Analyses of GSNO-treated extracts that underwent the biotin switch method without a blocking step resulted in high grade of unspecific biotinylation (Fig. 2). In extracts treated with 250 μm oxidized glutathione, 20 mm DTT, or water, no protein bands could be detected with the anti-biotin antibody (data not shown). Taken together, these results underline the specificity of the biotin switch method for detection of S-nitrosylated proteins.
Figure 2.
Detection of S-nitrosylated proteins of Arabidopsis cell culture extracts. Extracts containing 100 μg protein were treated with different concentrations of GSNO or GSH and labeled with biotin using the biotin switch method. Additionally, proteins were S-nitrosylated with 250 μm GSNO and reduced with 100 mm DTT after biotinylation. The sample on the right side was treated with 250 μm GSNO and underwent biotin switch method without MMTS treatment (blocking step). Proteins were separated by SDS-PAGE and blotted onto polyvinylidene difluoride-membrane. Detection of biotinylated proteins was achieved using anti-biotin antibody. The relative masses of protein standards are shown on the right.
Identification of Candidates for Protein S-Nitrosylation
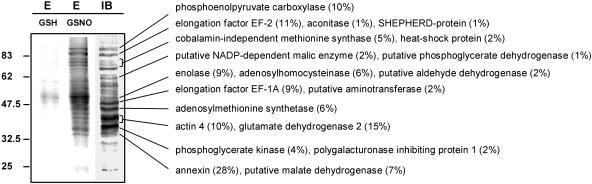
Arabidopsis cell cultures were treated with 250 μm GSNO or GSH and S-nitrosylated proteins were subjected to the biotin switch method. Biotin-labeled proteins were purified by affinity chromatography on a neutravidin matrix and separated by SDS-PAGE (Fig. 3). In the eluate derived from GSNO treatment, 16 prominent protein bands could be detected, whereas only a few protein bands were visible in the GSH-derived eluate. Twelve protein bands that could be assigned to predominant bands of the immunoblot of GSNO-treated cell culture extracts were digested with trypsin and the resulting peptides were subjected to nanoLC/MS/MS (Fig. 3).
Figure 3.
S-Nitrosylated proteins of Arabidopsis cell cultures. A total of 10 mg of cell culture proteins were treated with 250 μm GSH or GSNO, subjected to the biotin switch method, and biotinylated proteins were purified by affinity-chromatography using neutravidin-agarose. Eluates (E) were separated by SDS-PAGE and visualized by Coomassie Blue staining. Protein bands corresponding to predominant bands of the immunoblot analysis (IB) were identified by nanoLC/MS/MS. The percentage of protein covered by the matched peptides is given in brackets. The relative masses of protein standards are shown on the left.
To also identify low abundant candidates for protein S-nitrosylation, proteins of GSNO- and GSH-treated extracts were labeled with biotin, affinity purified as described above, and the eluates were subjected to nanoLC/MS/MS analysis. In the samples treated with the NO donor, 57 proteins could be identified comprising members of different functional families including stress-related proteins, signaling/regulating proteins, redox-related proteins, cytoskeleton proteins, and metabolic enzymes (Table I; supplemental material). More than 60% of the identified proteins are already described in the context of S-nitrosylation, S-glutathionylation, or redox-regulated processes (for references, see Table I and supplemental material), confirming that the identified proteins represent promising candidates for S-nitrosylation in plants. In the GSH-treated samples, 30 proteins could be identified (Table I; supplemental material). Twenty-seven of these are also identified in the GSNO-treated samples, and 18 of them are identified as targets for S-nitrosylation or S-glutathionylation in the animal system or have been reported to be involved in redox-regulated processes (for references, see Table I and supplemental material).
Table I.
Selected candidates of identified S-nitrosylated proteins from Arabidopsis cell cultures and leaves
Cell culture extracts treated with GSNO or GSH and leaf extracts of NO-treated or -untreated Arabidopsis plants were subjected to the biotin switch method and analyzed by nanoLC/MS/MS after trypic digestion. The MASCOT search engine was used to parse MS data to identify proteins from primary sequence databases. The best-matching peptide identifying the protein is given. If there were further peptides found, the number of the peptides is given. Hints confirming that the identified protein is a candidate for S-nitrosylation are given in the right column. The results of two separate experiments of each treatment were summarized in the table. Acc. No., Accession number.
| Protein | Acc. No. | Molecular Mass | Identified Peptides (Score) GSNO/NO | Identified Peptides (Score) GSH/Untreated | Hints to S-Nitrosylation |
|---|---|---|---|---|---|
| D | |||||
| Stress-Related Proteins | |||||
| GST, putative | NP_565178 | 25,634 | VTEFVSELR (79) | NPILPSDPYLR (38) | Sies et al. (1998); Ji et al. (2002) |
| Hsp 90, putative | AAL49788 | 80,036 | ADLVNNLGTIAR (50) + 1 | Jaffrey et al. (2001); Fratelli et al. (2002, 2003); Lind et al. (2002); Shenton and Grant (2003) | |
| Cu/Zn-superoxide dismutase | NP_172360 | 15,088 | AVVVHADPDDLGK (55) + 1 | Lawler and Song (2002) | |
| Redox-Related Proteins | |||||
| Glutathione peroxidase, putative | NP_192897 | 25,568 | FAPTTSPLSIEK (65) + 1 | FAPTTSPLSIEK (51) | Koh et al. (2001) |
| Peroxiredoxin-related | AAF66133 | 21,230 | AVNVEEAPSDFK (37) | Motohashi et al. (2001); Fratelli et al. (2002, 2003); Lind et al. (2002) | |
| Type 2 peroxiredoxin-related | NP_176773 | 17,417 | APIAVGDVVPDGTISFFDENDQLQTASVHSLAAGK (51) | ||
| Glutaredoxin, putative | NP_198853 | 11,749 | LVPLLTEAGAIAGK (59) | Klatt et al. (2000); Song and Lee (2003) | |
| Signaling/Regulating Proteins | |||||
| Elongation factor eEF-1α-chain | S08534 | 49,457 | VETGMIKPGMVVTFAPTGLTTEVK (61) + 4 | IGGIGTVPVGR (58) +2 | Fratelli et al. (2002); Shenton and Grant (2003) |
| Elongation factor EF-2 | A96602 | 94,185 | AYLPVVESFGFSSQLR (80) + 4 | LWGENFFDPATR (43) +2 | |
| Elongation factor 1B α-subunit | NP_568375 | 24,186 | TYISGDQLSVDDVK (67) + 1 | ||
| Elongation factor 1B γ, putative | AAK59587 | 46,371 | VPVLETPEGPIFESNAIAR (40) + 1 | ||
| Initiation factor eIF-4A1 | CAC43286 | 41,823 | GLDVIQQAQSGTGK (41) | GLDVIQQAQSGTGK (38) | Lind et al. (2002); Shenton and Grant (2003) |
| Initiation factor5A-4-related | NP_173985 | 17,129 | TYPQSAGNIR (58) | ||
| Cytoskeleton Proteins | |||||
| Tubulin α 6 chain | JQ1597 | 49,506 | AVFVDLEPTVIDEVR 84) + 1 | DVNAAVGTIK (34) +1 | Jaffrey et al. (2001) |
| Tubulin β 4 chain | S68122 | 49,876 | AVLMDLEPGTMDSLR (82) + 2 | GHYTEGAELIDSVLDVVR (71) | Jaffrey et al. (2001); Lind et al. (2002) |
| Actin 2/7 | NP_196543 | 41,709 | NYELPDGQVITIGAER (114) + 3 | AGFAGDDAPR (53) +1 | Dalle-Donne et al. (2000, 2003); Jaffrey et al. (2001); Fratelli et al. (2002, 2003); Lind et al. (2002) |
| Actin depolymerizing factor 3-like | NP_851227 | 15,912 | YAIFDFDFVSSEGVPR (54) | ||
| Annexin | CAA67608 | 35,757 | HYNDEDVIR (48) + 2 | DALLANEATK (48) +2 | Liu et al. (2002); Kuncewicz et al. (2003) |
| Metabolic Enzymes | |||||
| Fru 1,6-biphosphate aldolase, putative | NP_190861 | 38,516 | VSPEVIAEHTVR (85) + 4 | LASINVENVETNR (64) +3 | Fratelli et al. (2002); Lind et al. (2002); Ito et al. (2003); Shenton and Grant (2003) |
| Triosephosphate isomerase | T50646 | 27,138 | VASPAQAQEVHDELR (109) + 3 | VASPAQAQEVHDELR (68) +3 | Fratelli et al. (2002); Ito et al. (2003); Shenton and Grant (2003) |
| GAPDH C-subunit | NP_187062 | 36,891 | GILGYTEDDVVSTDFVGDNR (92) + 4 | AASFNIIPSSTGAAK (46) +3 | Mohr et al. (1996, 1999); Klatt et al. (2000); Motohashi et al. (2001); Kuncewicz et al. (2003); Shenton and Grant (2003) |
| 2-Phosphoglycerate hydrolase (enolase) | NP_181192 | 47,689 | VTAAVPSGASTGIYEALELR (86) + 3 | Fratelli et al. (2002); Lind et al. (2002); Shenton and Grant (2003) | |
| Phosphoglycerate kinase | NP_178073 | 42,105 | GVTTIIGGGDSVAAVEK (62) + 2 | Fratelli et al. (2002) | |
| Aconitase | Q8L784 | 108,133 | INPLVPVDLVIDHSVQVDVAR (43) | Navarre et al. (2000) | |
| SAM synthetase, putative | AAO11581 | 42,769 | KPEEVGAGDQGHMFGYATDETPELMPLTHVLATK (70) + 5 | FVIGGPHGDAGLTGR (76) +4 | Ruiz et al. (1998); Perez-Mato et al. (1999) |
| Adenosylhomocysteinase | CAB09795 | 51,447 | LVGVSEETTTGVK (48) | ||
| Met synthase | NP_197294 | 84,304 | IPSSEEIADR (82) + 1 | FALESFWDGK (53) +1 | Yamazaki et al. (2004) |
| Cys synthase | CAA58893 | 33,842 | IDGFVSGIGTGGTITGAGK (59) | LFVAIFPSFGER (35) | |
| ATP synthase CF1 α-chain | NP_051044 | 55,294 | LIESPAPGIISR (49) + 2 | Eaton et al. (2003) | |
| ATP synthase CF1 β-chain | NP_051066 | 53,900 | YKELQDIIAILGLDELSEEDR (96) + 14 | GIYPAVDPLDSTSTMLQPR (108) +6 | |
| Proteins Involved in Photosynthetic Processes | |||||
| GAPDH precursor, chloroplast | JQ1285 | 42,481 | VVDLADIVANNWK (74) + 1 | Mohr et al. (1996, 1999); Klatt et al. (2000); Motohashi et al. (2001); Kuncewicz et al. (2003); Shenton and Grant (2003) | |
| Rubisco large chain | NP_051067 | 52,922 | ESTLGFVDLLR (90) + 6 | LSGGDHIHAGTVVGK (56) +2 | Marcus et al. (2003) |
| Rubisco small chain 1a precursor | RKMUA1 | 20,189 | IIGFDNTR (45) + 1 | IIGFDNTR (58) +1 | Motohashi et al. (2001) |
| Rubisco activase, large subunit | AAA20202 | 51,948 | VPLILGIWGGK (34) | GLAYDTSDDQQDITR (81) +3 | Zhang and Portis (1999); Zhang et al. (2002) |
| PSII P680 47-kD protein | NP_051084 | 56,001 | MPTFFETFPVVLVDGDGIVR (85) | ||
| PSII D2 protein,fragment | AAO13251 | 35,220 | NILLNEGIR (31) | Carlberg et al. (1999) | |
| PSII oxygen-evolving complex 33 | NP_190651 | 34,998 | NTAASVGEITLK (53) + 4 | ||
| Rieske Fe-S protein | CAC03598 | 24,334 | VLFVPWVETDFR (63) |
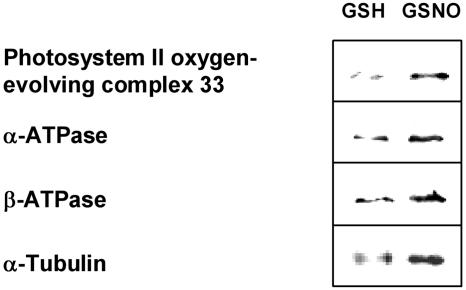
To get insight into NO-dependent protein regulation in plants, we also analyzed plants exposed to NO gas. Leaf extracts of NO-treated and untreated plants were subjected to the biotin switch method and affinity purified proteins were analyzed by nanoLC/MS/MS after tryptic digestion. The identified candidates for protein S-nitrosylation were divided into metabolic enzymes, proteins involved in photosynthetic processes, redox-related proteins, signaling/regulating proteins, stress-related proteins, and others (Table I; supplemental material). A total of 41 proteins have been identified in NO-treated extracts, and 19 of them have been described in context of redox-related processes, S-glutathionylation, or S-nitrosylation in plants or animals (for references, see Table I and supplemental material). In extracts of untreated plants, 25 proteins could be identified, and more than one-half of them (14) were found in the NO-treated samples. Western-blot analysis using antibodies raised against several of the identified proteins provide further evidence that these proteins are indeed retained by the biotin switch method (Fig. 4).
Figure 4.
Immunoblot analysis of in vitro S-nitrosylated proteins. Leaf extracts were treated with 250 μm GSNO or GSH and analyzed with the biotin switch method. After biotinylation, proteins were purified with neutravidin-agarose, separated by SDS-PAGE, and immunoblotted with anti-PSII oxygen-evolving complex 33, anti-α-ATPase, and anti-β-ATPase antibodies. Additionally, cell culture extracts were treated and prepared in the same way and analyzed with anti-α-tubulin antibody.
In GSH-treated cell culture extracts and in untreated leaves, many proteins were identified that are also present in the NO-treated samples. Especially in the case of the untreated leaves of Arabidopsis plants, we think that these proteins represent in vivo S-nitrosylated proteins. Cell cultures as well as plants are known to show constitutive NO production due to the activity of NO producing enzymes and due to an enzyme-independent process in the apoplast of plant tissues (Guo et al., 2003; Bethke et al., 2004). However, despite constitutive basal S-nitrosylation, the detection of S-nitrosylated proteins in GSH-treated extract seemed to be contradictory because of the reducing activity of GSH. The reducing power of GSH is probably not strong enough for an effective reduction of all S-nitrosylated Cys residues. Furthermore, some S-nitrosylated proteins can only be reduced by the action of reducing enzymes like thioredoxin or glutaredoxin (Stamler et al., 2001). Taken together with the high specificity of the biotin switch method, we concluded that most of the identified proteins from the GSH-treated cell culture extracts and from the untreated leaves represent in vivo S-nitrosylated proteins, although we cannot exclude unspecific interactions of proteins with the biotinylation agent biotin-HPDP.
Inhibition of Glyceraldehyde-3-Phosphate Dehydrogenase Activity by NO
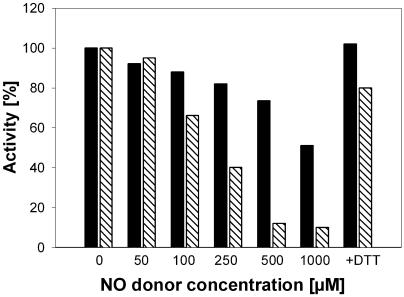
Although all of the proteins identified as candidates for S-nitrosylation contain at least one Cys residue, the effects of S-nitrosylation on enzyme activity or protein structure have to be elucidated. We have chosen glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the model enzyme since an easy and fast activity assay for this enzyme is already established (Mohr et al., 1996). Crude extracts of Arabidopsis cell cultures were incubated with different concentrations of GSH or the NO donors GSNO and SNP (Fig. 5). Whereas 1,000 μm GSNO reduced the GAPDH activity up to 90%, the same concentration of SNP reduced the GAPDH activity up to only 50%. Addition of 10 mm DTT to the inhibited enzymes completely restored the activity, confirming that the effect of the NO donors was due to S-nitrosylation of one or more critical Cys residues. The treatment of crude cell culture extracts with up to 1,000 μm GSH did not affect GAPDH activity (data not shown).
Figure 5.
Effect of GSNO and SNP on GAPDH activity. Crude extracts of Arabidopsis cell cultures were treated with different concentrations of GSNO (diagonal lines) or SNP (black) and enzyme activity was determined according to Mohr et al. (1996). The GAPDH activity in untreated control extract was set at 100%. For restoring GAPDH-activity, 10 mm DTT was added to extracts with inhibited enzymes. For each concentration, measurements were done at least in triplicates.
DISCUSSION
In mammals, protein S-nitrosylation is an accepted and intensively studied posttranslational modification and until now more than 50 S-nitrosylated proteins could be identified using the biotin switch method (Jaffrey et al., 2001; Kuncewicz et al., 2003; Foster and Stamler, 2004). In plants, however, the knowledge about the posttranslational modification of Cys residues by S-nitrosylation is rather limited and restricted to results obtained by protein database search for a putative S-nitrosylation motif (Huber and Hardin, 2004). To identify plant proteins sensitive to this posttranslational process, we treated extracts of Arabidopsis cell cultures with the NO donor GSNO, resulting in an effective formation of nitrosothiols (Fig. 1) and S-nitrosylated proteins (Fig. 2).
The proteomic analyses by nanoLC/MS/MS resulted in the identification of 67 proteins belonging to stress-related proteins, signaling/regulating proteins, redox-related proteins, cytoskeleton proteins, metabolic proteins, and others. Within the stress-related proteins, several proteins were identified that are already described to undergo S-nitrosylation in the animal systems. An interesting target in Arabidopsis is glutathione S-transferase (GST). In rats, microsomal and cytosolic GSTs showed differential activation and inhibition after treatment with GSNO, respectively (Ji et al., 2002). These results together with previous findings of increased microsomal GST activity after exposure to oxidants such as hydrogen peroxide and superoxide (Aniya and Anders, 1989) have led to the suggestion that this GST isoform may play a protective role under conditions of oxidative and nitrosative stress (Ji et al., 2002).
Another cluster of S-nitrosylated proteins in Arabidopsis includes several signaling and regulating factors. The identification of different elongation (eEF-1 and eEF-2) and initiation (4A-1 and 5A-4-related) factors as targets for S-nitrosylation suggest the assumption that NO encroaches in protein synthesis. Shenton and Grant (2003) identified elongation factors EF-1α and EF-1β and initiation factor Nip 1 of yeast as targets for S-glutathionylation and demonstrated that protein synthesis is rapidly and reversibly inhibited by H2O2 treatment.
Other candidates for S-nitrosylation in Arabidopsis are cytoskeleton proteins such as tubulin α, tubulin β, actin-depolymerizing factor, and actin. Both tubulin variants and actin have previously been described to undergo S-nitrosylation in neuronal cells of mammals (Jaffrey et al., 2001). The dynamic nature of the cytoskeleton filaments allows cells a rapid response to intracellular and extracellular signals by changing shape and translocating intracellular organelles or vesicles. Conformational changes of cytoskeleton components due to S-nitrosylation might be involved in directing vesicles loaded with toxic metabolites to the infection site and deflating the contents into the extracellular space (Collins et al., 2003).
The proteomic analysis of Arabidopsis also revealed proteins related to enzymes of the antioxidant system. In plants, NO dramatically affects redox balance and genes involved in redox control (Wendehenne et al., 2004). Glutaredoxin, peroxiredoxin, and glutathione peroxidase are together with GSH and thioredoxin important components of the cellular redox-status controlling system. Thioredioxin and glutaredoxin function as redox-regulators of target enzymes and transcription factors and serve as hydrogen donor to peroxiredoxin and dehydroascorbate (Sha et al., 1997; Rouhier et al., 2001). Both proteins harbor two highly conserved Cys residues within their active site and represent possible targets for S-nitrosylation.
Additionally, several metabolic enzymes of Arabidopsis were identified as potential candidates for S-nitrosylation. Five enzymes of the glycolysis are sensitive to S-nitrosylation, however, only GAPDH with a Cys residue in the active center is shown to be inhibited by NO (Padgett and Whorton, 1995; Mohr et al., 1996; Fig. 5). Fru 1,6-biphosphate aldolase, triosephosphate isomerase, and 2-phosphoglycerate hydrolase undergo S-glutathionylation during oxidative stress (H2O2 treatment) without dramatic effect on the enzyme activity (Shenton and Grant, 2003). However, recombinant Arabidopsis triosephosphate isomerase is reversibly inactivated by oxidized glutathione (Ito et al., 2003). The functional aspect of inhibition of GAPDH activity could be the diversion of Glc equivalents into the pentose phosphate cycle and production of NADPH for controlling the redox status of the cell.
The second important group of metabolic enzymes undergoing S-nitrosylation includes enzymes involved in sulfur metabolism, such as Cys synthase, S-adenosylhomocysteinase, vitamin B12-independent Met synthase, and S-adenosylmethione (SAM) synthetase. In plants, the latter three proteins are part of the methylMet cycle that provides activated methyl groups in the form of SAM for methylation of many different cell components and ingredients such as DNA, lignin, and flavonoids. Inhibition of SAM synthetase by NO is already described for the rat enzyme (Ruiz et al., 1998; Perez-Mato et al., 1999). In plants, SAM is a precursor for ethylene biosynthesis. NO regulates ethylene production in plants (Leshem and Haramaty, 1996), and S-nitrosylation of SAM synthetase or other enzymes of the methylMet cycle might mediate the crosstalk between ethylene and NO signaling.
To analyze S-nitrosylation processes in photosynthetic active tissue, we exposed Arabidopsis leaves to NO gas. Many chloroplast proteins identified as targets for S-nitrosylation are regulated in a redox-dependent manner including Gln synthase, NADPH-dependent GAPDH, Rubisco, and Rubisco activase (Ruelland and Miginiac-Maslow, 1999; Motohashi et al., 2001; Zhang et al., 2002; Marcus et al., 2003). The latter one plays a pivotal role in regulating the activity of the Calvin-Benson cycle and is regulated by thioredoxin-f (Zhang et al., 2002), a disulfide oxidoreductase, which controls the activity of many enzymes by reducing disulfide bridges and protecting single Cys residues from oxidation (Ruelland and Miginiac-Maslow, 1999; Motohashi et al., 2001; Sparla et al., 2002). In the large subunit of Rubisco, a Cys residue is adjacent to the active site and has been suggested to play a role in Rubisco activity and degradation (Marcus et al., 2003). Oxidizing conditions, both in vitro and in vivo, inhibited Rubisco activity and stimulated its degradation that in several instances was prevented by thiol reducing agents (Mehta et al., 1992; Desimone et al., 1996). In addition to these key enzymes of the Calvin-Benson cycle, proteins of PSII seemed especially to be targets for NO. Phosphorylation of PSII reaction center proteins D1 and D2, as well as other subunits of PSII, was found to be stimulated by moderately thiol-reducing conditions and kept at a high level also under highly reducing conditions (Carlberg et al., 1999). Furthermore, reversible inhibition of photophosphorylation in chloroplasts by NO was demonstrated (Takahashi and Yamasaki, 2002), suggesting that proteins of the energy transduction system in chloroplast thylakoids could be affected by S-nitrosylation.
In sum, the identification of plant proteins being potential targets for S-nitrosylation in vivo is a promising starting point to get insight in physiological as well as regulatory functions of NO in plants. The effect of S-nitrosylation on the identified plant proteins, if enzyme activities are inhibited or enhanced due to S-nitrosylation or if a structural alteration followed by change of the protein function is the result of the modification, has to be analyzed. Additionally, these results will probably give hints to the regulation of crosstalk between more plant specific NO-, salicylic acid-, and jasmonic acid/ethylene-dependent signaling pathways.
MATERIALS AND METHODS
Chemicals
The NO donors GSNO and SNP were purchased from Alexis (Grünberg, Germany) and Fluka (Neu-Ulm, Germany), respectively. GSH, methyl methanethionsulfonate (MMTS), neocuproine, and anti-biotin mouse monoclonal antibody were from Sigma (Taufkirchen, Germany). N-[6-(biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP) and neutravidin-agarose were purchased from Perbio (Bonn).
Standard chemicals of analytical grade were from Sigma (Taufkirchen, Germany), Roth (Munich), and Boehringer (Mannheim, Germany).
Plant Material
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia) were cultivated in a growth chamber for 6 weeks at 23°C during the day and 18°C at night (14 h). The experimental setups to study the effect of NO on whole plants consisted of controlled-environment cabinets as well as complete instrumentation to adjust and control gaseous NO through an electrochemical sensor. Arabidopsis plants were treated with NO concentrations of 1,250 μL/L for 10 min under light (Huang et al., 2004). At this NO concentration, the plants did not show any symptoms. Twenty minutes after the treatment, leaves were harvested and stored at −20°C. Cell suspension cultures of Arabidopsis (ecotype Columbia) were grown in dark at 26°C (126 rpm) in PS medium as described (Huang et al., 2002).
Determination of Nitrosothiol Content
Nitrosothiol content was determined according to the procedure of Saville (1958). Briefly, 180 μL of each sample were incubated with 30 μL of 0.5% ammonium sulfamate in water for 2 min. A total of 300 μL of 2.7% sulfanilamide and 0.25% HgCl2 in 0.4 n HCl were added, followed by 240 μL of 0.1% N-(1-naphthyl) ethylenediamine in water. The reference was incubated without HgCl2. The concentration of formed azo compound was determined after 20 min by measuring the absorption at 540 nm. The nitrosothiol content was quantified according to a standard curve created with GSNO.
Gel Electrophoresis and Western-Blot Analysis
Proteins were separated by SDS-PAGE on 12% polyacrylamide gels (Laemmli, 1970), were transferred onto polyvinylidene difluoride membranes and blocked with 1% nonfat milk powder and 1% bovine serum albumin. The blots were incubated with anti-biotin mouse monoclonal antibody conjugated with alkaline phosphatase at a dilution of 1:10,000 for 1 h. Antibodies raised against PSII oxygen-evolving complex 33, α-ATPase, β-ATPase, and α-tubulin were incubated for 1 h, followed by incubation with goat anti-rabbit IgG conjugated to alkaline phosphatase. Cross-reacting protein bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium as substrates.
Biotin Labeling of S-Nitrosylated Proteins
Frozen Arabidopsis cells were ground under liquid nitrogen to a fine powder. Proteins were extracted by adding 12 mL of HEN buffer (25 mm HEPES, pH 7.7, 1 mm EDTA, and 0.1 mm neocuproine) puffer to 10 g of cell powder. Cell debris was removed by centrifugation (20,000g, 10 min, 4°C) and protein concentration was determined according to Bradford (1976) with bovine serum albumin as standard.
The in vitro S-nitrosylation and subsequent biotinylation of S-nitrosylated proteins were done as described by Jaffrey (2001) with minor modifications. After treating the supernatant with GSNO for 20 min at room temperature (RT), the proteins were incubated with 20 mm MMTS and 2.5% SDS at 50°C for 20 min with frequent vortexing for blocking nonnitrosylated free Cys residues. Residual MMTS was removed by precipitation with 2 volumes of −20°C acetone and the proteins were resuspended in 0.1 mL of HENS buffer (HEN buffer containing 1% SDS)/mg protein. Biotinylation was achieved by adding 2 mm biotin-HPDP and 1 mm ascorbate and incubation at RT for 1 h.
Purification of Biotinylated Proteins
After removing biotin-HPDP, the precipitated proteins were resuspended in 0.1 mL of HENS buffer/mg of protein and 2 volumes of neutralization buffer (20 mm HEPES, pH 7.7, 100 mm NaCl, 1 mm EDTA, and 0.5% Triton X-100). A total of 15 μL of neutravidin-agarose/mg of protein were added and incubated for 1 h at RT. The matrix was washed extensively with 20 volumes of washing buffer (600 mm NaCl in neutralization buffer) and bound proteins were eluated with 100 mm β-mercaptoethanol in neutralization buffer.
nanoLC/MS/MS Analyses
Proteins were dissolved in 50 μL of 0.1 m NH4HCO3/10% acetonitrile and digested with 3 μg of trypsin at 37°C overnight. Bands from stained SDS gels were cut out, washed, and treated with trypsin according to Shevchenko et al. (1996). Digested peptides were extracted by vortexing for 3 h with 100 μL of 5% formic acid. All tryptic peptide samples were dried and redissolved in 50 μL 0.1% trifluoroacetic acid and 5% acetonitrile. Peptides were separated by reversed-phase chromatography using an UltiMate Capillary/Nano liquid chromatography system (LC Packings, Amsterdam). Portions of 20-μL sample were loaded to a precolumn (300 μm × 5 mm, 5 μm C18, 100 Å, PepMap, LC Packings) and eluted and fractionated on a self-packed analytical column (75 μm × 120 mm packed with YMC-Gel ODS-A (3 μm C18; YMC, Kyoto) with a gradient of 5% to 55% acetonitrile at a flow rate of 150 nL/min in 40 min. Eluted peptides were continuously delivered to a Q-Tof Ultima mass spectrometer (Waters/Micromass, Manchester, UK) by electrospray and analyzed by MS/MS employing data-dependent analysis (3 most abundant ions in each cycle; 0.3 s MS m/z 400–2,000 and maximum 4.8 s MS/MS m/z 50–3,000, continuum mode, 60 s dynamic exclusion). The MS/MS raw data were processed and converted into Micromass pkl-format using MassLynx 4.0 ProteinLynx. The resulting pkl-files were used for searching the NCBInr (Viridiplantae) protein database by Mascot search program (Matrixscience, London) with 0.2 D mass tolerance. Only matches calculated as significant by the Mascot search algorithm were considered in the protein identification.
GAPDH Activity Assay
The activity of GAPDH was determined according to Mohr et al. (1996) with some modifications. Crude extracts of Arabidopsis cell suspension cultures (in 50 mm Tris-Cl, pH 7.5) were reduced with 10 mm DTT for 20 min at room temperature. Residual DTT was removed with Sephadex G-25M columns. A total of 150 to 300 μg protein were incubated with 50 μm arsenate and 100 μg/mL 3-phosphogylcerinaldehyde and were adjusted to 950 μL with 50 mm Tris-Cl, pH 7.5. The reaction was initiated by adding 50 μL of 10 mm NAD+ and the reduction of NAD+ to NADH was monitored at 340 nm.
Inhibition assays were done as described in (Mohr et al., 1999) with minor modifications. In brief, after DTT treatment and desalting, reduced cell culture extracts were incubated with different concentrations of GSNO, SNP, or GSH at room temperature. After 2 min, the treated cell culture extracts were added to the assay mixture and GAPDH activity was determined subsequently as described above.
Supplementary Material
Acknowledgments
We thank Dr. Anna Sokolenko (LMU, Munich) for supplying antibodies against PSII oxygen-evolving complex 33, α-ATPase, and β-ATPase.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1110 Innate Immunity) and by Bayerisches Staatsministerium für Umwelt, Gesundheit, und Verbraucherschutz.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058719.
References
- Aniya Y, Anders MW (1989) Activation of rat liver microsomal glutathione S-transferase by reduced oxygen species. J Biol Chem 264: 1998–2002 [PubMed] [Google Scholar]
- Bethke PC, Badger MR, Jones RL (2004) Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16: 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C (2001) Nitric oxide and the regulation of gene expression. Trends Cell Biol 11: 66–75 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 77: 248–254 [DOI] [PubMed] [Google Scholar]
- Carlberg I, Rintamaki E, Aro EM, Andersson B (1999) Thylakoid protein phosphorylation and the thiol redox state. Biochemistry 38: 3197–3204 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Huckelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Giustarini D, Rossi R, Colombo R, Milzani A (2003) Reversible S-glutathionylation of Cys 374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic Biol Med 34: 23–32 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Milzani A, Giustarini D, Di Simplicio P, Colombo R, Rossi R (2000) S-NO-actin: S-nitrosylation kinetics and the effect on isolated vascular smooth muscle. J Muscle Res Cell Motil 21: 171–181 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Desimone M, Henke A, Wagner E (1996) Oxidative stress induces partial degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase in isolated chloroplasts of barley. Plant Physiol 111: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Achkor H, Titarenko E, Martinez MC (2003) The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett 543: 136–139 [DOI] [PubMed] [Google Scholar]
- Durner J, Gow AJ, Stamler JS, Glazebrook J (1999) Ancient origins of nitric oxide signaling in biological systems. Proc Natl Acad Sci USA 96: 14206–14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton P, Jones ME, McGregor E, Dunn MJ, Leeds N, Byers HL, Leung KY, Ward MA, Pratt JR, Shattock MJ (2003) Reversible cysteine-targeted oxidation of proteins during renal oxidative stress. J Am Soc Nephrol 14: S290–S296 [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Sandermann H Jr (1997) Maize glutathione-dependent formaldehyde dehydrogenase cDNA: a novel plant gene of detoxification. Plant Mol Biol 34: 843–854 [DOI] [PubMed] [Google Scholar]
- Foster MW, Stamler JS (2004) New insights into protein S-nitrosylation: mitochondria as a model system. J Biol Chem 279: 25891–25897 [DOI] [PubMed] [Google Scholar]
- Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, Bonetto V, Mengozzi M, Duffieux F, Miclet E, et al (2002) Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci USA 99: 3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratelli M, Demol H, Puype M, Casagrande S, Villa P, Eberini I, Vandekerckhove J, Gianazza E, Ghezzi P (2003) Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics 3: 1154–1161 [DOI] [PubMed] [Google Scholar]
- Garcia-Mata C, Lamattina L (2003) Abscisic acid, nitric oxide and stomatal closure: Is nitrate reductase one of the missing links? Trends Plant Sci 8: 20–26 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J (2004) Nitric oxide is induced by wounding and influences jasmonic acid signalling in Arabidopsis thaliana. Planta 218: 938–946 [DOI] [PubMed] [Google Scholar]
- Huang X, von Rad U, Durner J (2002) Nitric oxide induces the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Huber SC, Hardin SC (2004) Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol 7: 1–5 [DOI] [PubMed] [Google Scholar]
- Ito H, Iwabuchi M, Ogawa K (2003) The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol 44: 655–660 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3: 193–197 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: PL1. [DOI] [PubMed] [Google Scholar]
- Ji Y, Toader V, Bennett BM (2002) Regulation of microsomal and cytosolic glutathione S-transferase activities by S-nitrosylation. Biochem Pharmacol 63: 1397–1404 [DOI] [PubMed] [Google Scholar]
- Jia L, Bonaventura C, Bonaventura J, Stamler JS (1996) S-Nitrosohaemoglobin: a dynamic activity of blood involved in vascular control. Nature 380: 221–226 [DOI] [PubMed] [Google Scholar]
- Klatt P, Pineda Molina E, Perez-Sala D, Lamas S (2000) Novel application of S-nitrosoglutathione-Sepharose to identify proteins that are potential targets for S-nitrosoglutathione-induced mixed-disulphide formation. Biochem J 349: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge I, Gutteck-Amsler U, Zollinger M, Do KQ (1997) S-Nitrosoglutathione in rat cerebellum: identification and quantification by liquid chromatography-mass spectrometry. J Neurochem 69: 2599–2607 [DOI] [PubMed] [Google Scholar]
- Koh YH, Suzuki K, Che W, Park YS, Miyamoto Y, Higashiyama S, Taniguchi N (2001) Inactivation of glutathione peroxidase by NO leads to the accumulation of H2O2 and the induction of HB-EGF via c-Jun NH2-terminal kinase in rat aortic smooth muscle cells. FASEB J 15: 1472–1474 [DOI] [PubMed] [Google Scholar]
- Kuncewicz T, Sheta EA, Goldknopf IL, Kone BC (2003) Proteomic analysis of s-nitrosylated proteins in mesangial cells. Mol Cell Proteomics 2: 156–163 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lawler JM, Song W (2002) Specificity of antioxidant enzyme inhibition in skeletal muscle to reactive nitrogen species donors. Biochem Biophys Res Commun 294: 1093–1100 [DOI] [PubMed] [Google Scholar]
- Leshem Y, Haramaty E (1996) The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. foliage. J Plant Physiol 148: 258–263 [Google Scholar]
- Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, Cotgreave IA (2002) Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys 406: 229–240 [DOI] [PubMed] [Google Scholar]
- Liu L, Enright E, Sun P, Tsai SY, Mehta P, Beckman DL, Terrian DM (2002) Inactivation of annexin II tetramer by S-nitrosoglutathione. Eur J Biochem 269: 4277–4286 [DOI] [PubMed] [Google Scholar]
- Marcus Y, Altman-Gueta H, Finkler A, Gurevitz M (2003) Dual role of cysteine 172 in redox regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase activity and degradation. J Bacteriol 185: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Lamas S (2004) Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch Biochem Biophys 423: 192–199 [DOI] [PubMed] [Google Scholar]
- Mehta RA, Fawcett TW, Porath D, Mattoo AK (1992) Oxidative stress causes rapid membrane translocation and in vivo degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase. J Biol Chem 267: 2810–2816 [PubMed] [Google Scholar]
- Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B (1999) Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem 274: 9427–9430 [DOI] [PubMed] [Google Scholar]
- Mohr S, Stamler JS, Brune B (1996) Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem 271: 4209–4214 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C (1995) Natural resistance and nitric oxide. Cell 82: 873–876 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wendehenne D, Durner J, Noad R, Klessig DF (2000) Nitric oxide modulates the activity of two tobacco enzymes aconitase. Plant Physiol 122: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot 53: 1237–1247 [PubMed] [Google Scholar]
- Padgett CM, Whorton AR (1995) S-Nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol 269: C739–C749 [DOI] [PubMed] [Google Scholar]
- Perez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM (1999) Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J Biol Chem 274: 17075–17079 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127: 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M (1999) Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci 4: 136–141 [DOI] [PubMed] [Google Scholar]
- Ruiz F, Corrales FJ, Miqueo C, Mato JM (1998) Nitric oxide inactivates rat hepatic methionine adenosyltransferase In vivo by S-nitrosylation. Hepatology 28: 1051–1057 [DOI] [PubMed] [Google Scholar]
- Saville B (1958) A scheme for the colorimetric determination of microgram amounts of thiols. Analyst 83: 670–672 [Google Scholar]
- Sha S, Minakuchi K, Higaki N, Sato K, Ohtsuki K, Kurata A, Yoshikawa H, Kotaru M, Masumura T, Ichihara K, et al (1997) Purification and characterization of glutaredoxin (thioltransferase) from rice (Oryza sativa L.). J Biochem (Tokyo) 121: 842–848 [DOI] [PubMed] [Google Scholar]
- Shenton D, Grant CM (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Sies H, Dafre AL, Ji Y, Akerboom TP (1998) Protein S-thiolation and redox regulation of membrane-bound glutathione transferase. Chem Biol Interact 111–112: 177–185 [DOI] [PubMed] [Google Scholar]
- Song JJ, Lee YJ (2003) Differential role of glutaredoxin and thioredoxin in metabolic oxidative stress-induced activation of apoptosis signal-regulating kinase 1. Biochem J 373: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Pupillo P, Trost P (2002) The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J Biol Chem 277: 44946–44952 [DOI] [PubMed] [Google Scholar]
- Stamler JS (1994) Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78: 931–936 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J (1992. a) Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci USA 89: 7674–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC (2001) Nitrosylation. The prototypic redox-based signaling mechanism. Cell 106: 675–683 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Singel DL, Loscalzo J (1992. b) Biochemistry of nitric oxide and its redox-activated forms. Science 258: 1898–1902 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ (1997) (S)NO signals: translocation, regulation, and a consensus motif. Neuron 18: 691–696 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yamasaki H (2002) Reversible inhibition of photophosphorylation in chloroplasts by nitric oxide. FEBS Lett 512: 145–148 [DOI] [PubMed] [Google Scholar]
- Tsikas D, Sandmann J, Holzberg D, Pantazis P, Raida M, Frolich JC (1999) Determination of S-nitrosoglutathione in human and rat plasma by high-performance liquid chromatography with fluorescence and ultraviolet absorbance detection after precolumn derivatization with o-phthalaldehyde. Anal Biochem 273: 32–40 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Durner J, Klessig DF (2004) Nitric oxide signalling and plant defense. Curr Opin Plant Biol 7: 449–455 [DOI] [PubMed] [Google Scholar]
- Wippermann U, Fliegmann J, Bauw G, Langebartels C, Maier K, Sandermann H Jr (1999) Maize glutathione-dependent formaldehyde dehydrogenase: protein sequence and catalytic properties. Planta 208: 12–18 [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol 45: 18–27 [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J (2004) Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Kallis RP, Ewy RG, Portis AR Jr (2002) Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform. Proc Natl Acad Sci USA 99: 3330–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Portis AR Jr (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA 96: 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.