ABSTRACT
Invasions by eukaryotes dependent on environmentally acquired bacterial mutualists are often limited by the ability of bacterial partners to survive and establish free-living populations. Focusing on the model legume-rhizobium mutualism, we apply invasion biology hypotheses to explain how bacteriophages can impact the competitiveness of introduced bacterial mutualists. Predicting how phage-bacteria interactions affect invading eukaryotic hosts requires knowing the eco-evolutionary constraints of introduced and native microbial communities, as well as their differences in abundance and diversity. By synthesizing research from invasion biology, as well as bacterial, viral, and community ecology, we create a conceptual framework for understanding and predicting how phages can affect biological invasions through their effects on bacterial mutualists.
KEYWORDS: bacteriophages, microbial communities, rhizosphere-inhabiting microbes, invasion ecology, multi-trophic interactions, plant-microbe interactions
MUTUALIST AVAILABILITY DRIVES HOST INVASIONS
Biological invasion is a fascinating and troublesome phenomenon: it causes major ecological and economic costs but also provides important ecological insights (1 – 3). An invasion occurs when a species introduced to a new range proliferates there and becomes pestiferous (4 – 6). The ability to invade is strongly influenced by the biota, either native or introduced, with which an introduced species interacts in the new range (7, 8).
Many eukaryotes depend on bacterial mutualists that are horizontally transmitted [Table 1 (9 – 13)]. These bacteria do not disperse with host propagules and instead infect the host from a free-living stage. Thus, for mutualist-dependent eukaryotes to invade, these horizontally transmitted symbionts must arrive independently and survive as free-living bacteria (4, 14 – 19). Bacteriophages, viruses that infect bacteria, strongly shape bacterial community composition (20 – 29). Thus, as a eukaryotic mutualist moves into a new range, its fate could hinge on the ecological and evolutionary outcomes of bacterium-bacteriophage interactions encountered there by the free-living bacterial symbionts on which it depends. Here, we explore how rhizobiophages, bacteriophages that specialize on rhizobia, could affect range expansions by legumes and rhizobia (Fig. 1), which is a well-studied model of mutualist-dependent invasion. To do so, we introduce invasion biology and then use it to predict how phages could influence an invasion by a mutualist-dependent eukaryote.
TABLE 1.
Examples of environmentally acquired mutualistic bacteria that associate with invasive eukaryotic hosts
| Invasive eukaryote | Mutualistic bacterium | References |
|---|---|---|
| Alnus glutinosa (European alder) | Frankia sp. | Schwob et al. (30) |
| Casuarina cunninghamiana (River oak) | Frankia sp. | Zimpfer et al. (31) |
| Gunnera tinctoria (Chilean rhubarb) | Nostoc sp. | Gioria and Osborne (32) |
| Hedera helix (European ivy) | Bacillus amyloliquefaciens | Soares et al. (33) |
| Myrica faya (firetree) | Frankia sp. | Burleigh and Dawson; Vitousek et al.; Walker and Vitousek (34 – 36) |
| Various legumes | Rhizobia (e.g., Bradyrhizobium sp.) | La Pierre et al.; Rodríguez-Echeverría et al.; Stepkowski et al. (37 – 39) |
| Agrilus mali (apple buprestid) | Pantoea sp. and Pseudomonas orientalis | Bozorov et al. (40) |
| Agrilus planipennis (emerald ash borer) | Streptomyces sp., Erwinia sp., and Burkholderia cepacia | Vasanthakumar et al. (41) |
| Riptortus pedestris (a species of broad-headed bug) | Burkholderia sp. | Kikuchi et al.; Kikuchi et al. (42, 43) |
| Sirex noctilio (sirex woodwasp) | Streptomyces sp. and γ-proteobacteria | Adams et al. (44) |
| Various coreoid and lygaeoid stinkbugs | Burkholderia sp. | Kaltenpoth and Flórez; Kikuchi et al. (45, 46) |
| Various insects | e.g., gut bacteria | Lu et al. (15) |
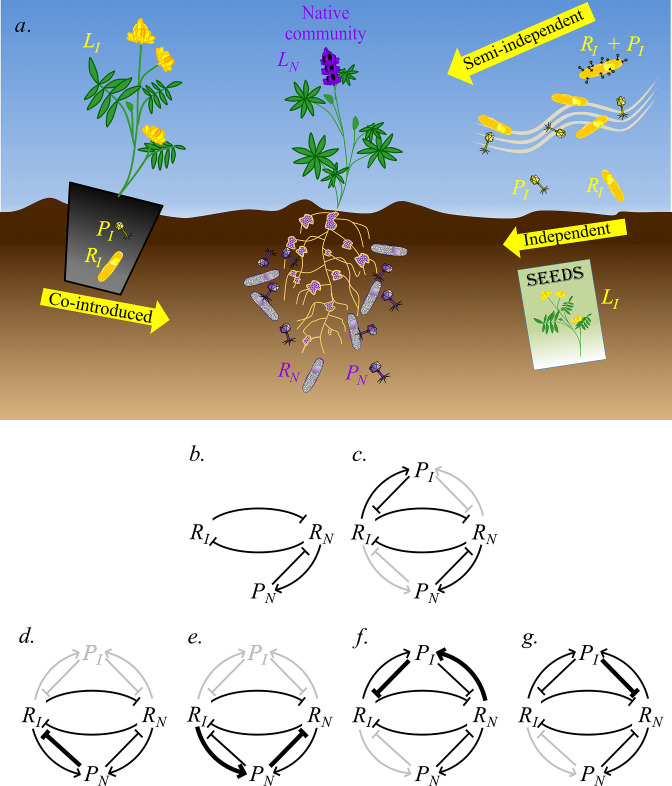
FIG 1.
(a) This cartoon depicts different paths along which an invaded community might assemble. In the center, the native community, depicted in cool colors, includes the native legume (LN ), nodulated by its native rhizobium symbiont (RN ), which also lives free in the soil and hosts native phage (PN ). Biological entities from a hypothetical distant home range community, depicted in warm colors, can arrive via several pathways. On the left, a potted legume (LI ) is co-introduced with rhizobium (RI ) and phage (PI ) from its home range. On the lower right, the introduced legume (LI ) arrives as seed, with neither symbiont nor phage. On the top right, rhizobium from the distant home range is co-introduced with phage (RI + PI ). Alternatively, in the center right, phage (PI ) and/or rhizobium (RI ) arrive independently. (b–g) When introduced rhizobia (RI ) and native rhizobia (RN ) compete, the various ways they might interact with phages lead to different invasion biology hypotheses, as described below. (b) Introduced rhizobia (RI ) arrive without their familiar home range phages (PI ), are not infected by native phages (PN ), and compete with native rhizobia (RN ) (“enemy escape”). (c) Both rhizobia and phages are introduced from the same distant population. If each rhizobium is preyed upon only by its familiar phage (gray lines absent), then each rhizobium might be regulated by its familiar phage; however, the relative magnitudes of the dark lines determine if invasion can occur. If either rhizobium is attacked by an unfamiliar phage, the gray lines between that pair are present (dark). Again, several outcomes are possible, as described below. (d) Native phages affect introduced rhizobia more negatively than native rhizobia (“biotic resistance” via “reverse spillover”). (e) Native phages are strongly amplified by introduced rhizobia but more negatively affect native rhizobia (“enemy spillback”). (f) Introduced phages are strongly amplified by native rhizobia but more negatively affect introduced rhizobia (biotic resistance via “subsidized pathogen”). (g) Introduced phages more negatively affect native rhizobia than introduced rhizobia (“enemy spillover”). Note that panels d and e can occur, regardless of the presence of introduced phages, and that panels f and g can occur, regardless of whether native phages can attack introduced rhizobia. Lines indicate the direction of the interaction effect. Arrowheads indicate interactions that increase fitness of the biological entity at which the arrowheads point, while a flathead indicates interactions that decrease fitness of the receiving partner. Interactions and partners depicted in gray might be either present or absent, as described above. Arrow thickness indicates the magnitude of the interaction effect.
A MODEL SYSTEM: LEGUMES, RHIZOBIA, AND RHIZOBIOPHAGES
Legumes and rhizobia
Soil-dwelling rhizobia infect legume roots, populate intercellular spaces, and stimulate production of specialized organs called nodules (47). In each nodule, a subset of the intercellular rhizobium population is engulfed by host cells and encapsulated within an intracellular symbiosome. There, some rhizobia differentiate into specialized endosymbiotic cells called bacteroids, which reduce atmospheric di-nitrogen (N2) in mutualistic exchange for photosynthates (48, 49). Within the nodule, symbiotic rhizobia can reach high cell densities (50 – 52), but once released from senescing nodules, rhizobia must survive the abiotic and biotic stresses of soil (53 – 62).
Free-living rhizobia survive as saprotrophs or persister cells in bulk soil [53, 54, 63, but see reference (64)], which is not penetrated by roots and holds few resources and inhabitants (65, 66). Fortunate cells eventually encounter a rhizosphere (67, 68), the ecologically complex habitat surrounding roots (69 – 72). Rhizobia can proliferate in rhizospheres of both legumes and non-legumes (73, 74), sometimes surviving for years without a legume host (75). The rhizosphere community is rich in both competitors (57, 76 – 82) and natural enemies, such as rhizobiophages (83, 84).
Rhizobia can facilitate legume invasions
Legumes (Fabaceae) are potent invaders (85). They endanger native plants directly by competition and indirectly by increasing soil nitrogen (N) concentration, which hinders habitat restoration (86, 87) and facilitates non-leguminous invasive plants (88 – 91). A legume species usually partners with particular groups of rhizobia (69, 92 – 96) and obtains greater benefit from familiar, co-evolved rhizobia (37, 97 – 102). However, rhizobia and plants disperse independently (Fig. 1a), with senescing nodules releasing reproductive rhizobia into adjacent soil (103). Rhizobia passively disperse long distances by dust storms (104 – 107) and can also be moved with soil or co-transported with the roots of adult legume hosts (37 – 39, 108 – 116). Nevertheless, rhizobium genotypes are not cosmopolitan and often exhibit a significant biogeographic structure at various spatial scales (37, 117 – 121). Since novel habitats lack familiar rhizobia, establishing legume crops onto new continents requires inoculation with compatible rhizobia (122 – 125). Lacking such deliberate inoculation, range expansion by rhizobium-dependent wild legumes requires that familiar rhizobia either co-disperse or arrive independently (75, 76, 93, 99, 126). How most symbiotic bacteria disperse remains poorly understood (127, 128).
For some legumes, greater soil mineral N can reduce the need for rhizobia (129), but for many legumes, successful invasion depends on the presence or introduction of compatible rhizobia (16, 76, 130 – 132). Despite this dependence, there are multiple cases in which rhizobial symbionts have apparently co-invaded with rhizobium-dependent legumes (37 – 39, 108, 112, 115), with legumes representing almost 10% of the invasive plants recorded for North America (85).
Rhizobiophages
Surprisingly little is known about rhizobiophage diversity. Early studies classified rhizobiophages by morphology into at least three families (Siphoviridae, Myoviridae, and Podoviridae), all within the order Caudovirales (83, 133). Next-generation sequencing has suddenly increased information about rhizobiophage genomes, revealing a broader taxonomic diversity (84, 134 – 143). However, owing to limited research, the number and diversity of described rhizobiophage genomes available on GenBank comprise only a fraction of the recorded genomes of their rhizobium hosts [e.g., see references (144 – 147)].
The spatial structure of bacteriophage diversity is poorly described in general and known primarily from aquatic ecosystems (148 – 152). It is typically thought that the distribution of a bacteriophage is limited only by the presence of its host, though evidence to support this claim is still missing (153). Bacteriophages can passively move short distances in soil (23) [reviewed in reference (154)], and some phages, either as virions or as prophages, might disperse long distances with wind-borne dust [Fig. 1a (155)] or as stowaways in transported soil. Accordingly, some phages are widely distributed (156 – 158). However, many phage communities are spatially structured (29, 134, 150, 151, 159 – 162), and phage community composition in soil can vary immensely even across small spatial scales [>10 m (163 – 165)]. Communities of rhizobiophages differ strongly among nearby (<10 km distant) legume populations: phages from different agricultural fields of the same host legume rarely showed more than 88% average nucleotide identity (134), and an unpublished analysis of 141 genome sequences of Bradyrhizobium spp. from different continents found that all of the 31 detected prophages were unique (J. Van Cauwenberghe, unpublished data). These observations suggest that rhizobiophages disperse poorly over longer distances, but they might nonetheless accompany deliberately applied rhizobium inoculum. Sharma and colleagues (83) detected compatible rhizobiophages in locations where rhizobia were intentionally inoculated onto legumes introduced for afforestation and soil rehabilitation. Often, however, rhizobia being developed for agricultural inoculum are screened for lysogeny (166, 167), and such efforts have been further facilitated by genomic methods (168).
Phage predation may affect rhizobium success
Mutualism theory predicts that when individual hosts interact with many symbionts, selection favors hosts that can choose the most cooperative symbionts (169 – 171). For example, legumes can constrain infection by compatible but less beneficial rhizobia (70, 92, 172 – 178). However, legumes seldom control which genotypes nodulate them (118, 131, 179 – 182), and legume choice cannot overcome rhizosphere effects (53, 172, 180, 183, 184). For example, crop nodules are rarely occupied by the most effective nitrogen fixers (125, 185, 186) because those genotypes fail to compete in the rhizosphere (54, 186, 187). Instead, the nodulation chances of a rhizobium genotype increases with its cell density in the rhizosphere (184, 188, 189), which means it must compete effectively (125, 190) and survive natural enemies in the rhizosphere (191, 192).
Rhizobiophages are abundant in soils (193 – 195), especially in legume rhizospheres (133, 196, 197), where they can reduce rhizobium nodulation rates and plant growth (198, 199). Phage density is correlated with the decline of free-living (saprophytic) rhizobia in soil (200), and rhizobiophage infection can strongly regulate rhizobium populations (201 – 209). Applying particular rhizobiophages can improve legume crop production by controlling highly competitive rhizobium genotypes that are inefficient N2 fixers (202, 203). Rhizobiophages might similarly influence the relative competitive success of native versus introduced rhizobia.
APPLYING INVASION BIOLOGY THEORIES TO LEGUME-RHIZOBIUM-RHIZOBIOPHAGE SYSTEMS
As with infectious disease epidemics, complex ecological interactions drive the fates of biotic invasions. After an infectious agent is introduced to a host population, the agent can either disappear, lodge as a commensal, or spread. Similarly, depending on the ecological interactions it encounters, an introduced species could immediately disappear, quietly persist with no apparent effect on the native community, or become pestiferous, disrupting the native community, either ecologically or economically or both.
Lytic bacteriophages influence bacterial community composition by causing heavy mortality on specific bacteria (20 – 27, 29, 210, 211). Temperate phages following the lysogenic pathway produce more complex effects. They can confer benefits to their hosts, such as superinfection exclusion (212 – 214) and auxiliary metabolic genes (215, 216), but still turn lethal when they activate their lytic pathway. Thus, bacteriophages might alter the success or failure of bacterial symbionts that can drive the population expansion of introduced eukaryotic hosts. Invasion biology theory helps analyze the many paths along which rhizobiophages could indirectly influence legume invasions (Fig. 1b through g).
Invasion theory (217) proposes mechanisms by which biotic interactions might either facilitate invasion (Fig. 1b, e, and g) or produce “biotic resistance,” i.e., the ability of a native community to resist exotic invasion (Fig. 1c, d, and f). For example, suppressive soils rich in phages that infect Ralstonia solanacearum can resist establishment by that plant pathogen (218). Similar mechanisms might be responsible for the aforementioned failure of inoculated rhizobia to competitively occupy either soil (54, 186, 187) or nodule communities (125, 185, 186). We hope this paper stimulates testing of the hypotheses described below.
The earliest invasion biology hypothesis derives from an assumption underlying classical biological control of crop pests (219); i.e., organisms proliferate when introduced in a new range because they arrived without the natural enemies that controlled them in the home range [“enemy escape” or the “enemy release hypothesis” (4, 7, 220 – 222); Fig. 1b]. Eukaryotic hosts commonly proliferate after dispersing over long distances without viral enemies (223). For example, plant species introduced to the U.S. are infected with 24% fewer virus species (224) than in their European home ranges. If eukaryotic hosts arrive and associate with mutualistic bacteria that have dispersed without bacteriophage enemies, the host and bacteria might similarly co-proliferate in the new range. Thus, introduced rhizobia that have escaped compatible rhizobiophages from their home range might outcompete native rhizobia, which remain regulated by their own rhizobiophage enemies, thereby facilitating a legume invasion.
Regardless of whether they escape home range phages, introduced bacteria also encounter “unfamiliar phages”. If evolutionary pressures (e.g., ongoing local adaptation and negative frequency-dependent selection) overcome constraints (e.g., genetic distance and fitness trade-offs), phage host ranges might evolve to encompass previously unfamiliar bacteria (e.g., Fig. 1d through g). Thus, enemy release could be fleeting [e.g., see references (4, 225 – 228)], with the fate of introduced rhizobia depending on how they and native rhizobia interact with phages.
Native phages that can infect introduced rhizobia might hamper co-proliferation of introduced legumes and rhizobia (biotic resistance via “reverse spillover”; Fig. 1d). Alternatively, native phages could facilitate invasion by spilling back onto native bacteria from introduced bacteria [(229, 230) Fig. 1e]. Such “enemy spillback” (231, 232), also called “local pathogen accumulation” (233), could occur if native phages only rarely infect introduced rhizobia (thereby producing little change in the density of introduced rhizobia) but persistently achieve unusually large burst sizes when they do. Because spillback from introduced rhizobia might increase phage density, the phenomenon might be detected by comparing phage abundance in the presence versus absence of introduced rhizobia. Both reverse spillover and enemy spillback can occur in either the presence or the absence of introduced phages (hence the gray lines in Fig. 1d and e)
Non-native phages co-introduced with rhizobia could also either deter or promote co-proliferation and invasion of introduced rhizobia and legumes. Introduced phages might deter invasion simply by continuing to specialize on and regulate co-introduced rhizobia (Fig. 1c). Introduced phages could also deter rhizobium invasion by proliferating more luxuriantly on occasionally infected native rhizobia but most negatively affecting the density of introduced rhizobia (biotic resistance via “subsidized pathogen” [(14) Fig. 1f]. This subsidy of the introduced pathogen could arise if native rhizobia are either more abundant or because they produce comparatively larger burst sizes than introduced rhizobia. We know of no examples of this phenomenon.
Alternatively, introduced phages that infect native bacteria (either rhizobia or other competitors) could facilitate legume-rhizobium invasion by spilling over onto and decimating competing native bacterial communities [“enemy spillover,” a form of “apparent competition” (234 – 236); Fig. 1g]. Although not yet documented for rhizobia, this phenomenon has been observed in other microbial introductions (212, 230, 237, 238). Enemy spillover can occur, e.g., when introduced bacteria carry a prophage, which allows them to outcompete native bacteria that lack resistance to this phage [i.e., phage-mediated allelopathy (239)]. A prophage that is induced in only a few of its lysogenic hosts might continue to replicate lytically on competing susceptible hosts, which could then be eliminated, while protecting its lysogenic hosts via superinfection exclusion (212 – 214). In a recent study simulating bacterial invasions in vitro, bacteria dispersing to nearby patches could outcompete native bacteria only when carrying phages to which the latter were susceptible (240).
Thus, regardless of the path along which an invaded community assembles (Fig. 1a), phages can influence invasion by mutualist bacteria. In some scenarios, phages facilitate legume invasion (Fig. 1b, e, and g), whereas in others, they hamper invasion (Fig. 1c, d, and f). Key questions, then, are (i) how often do rhizobia disperse to a new range without their co-evolved rhizobiophages? (ii) how likely is it that rhizobiophage host ranges include or acquire novel rhizobia? (iii) which rhizobia (native or introduced) will be most negatively affected by phages? and (iv) how will rhizobiophage effects on a rhizobium community cascade onto host legume populations?
Whether a phage will affect a novel host bacterium (e.g., spillover) more negatively than its original host (e.g., spillback) depends largely on the relative effectiveness of mechanisms involved in the various stages of infection. These mechanisms include the ability of the phage to attach to each host [e.g., as quantified by adsorption rates (241)], the effectiveness of rhizobium intracellular defense mechanisms, such as restriction-modification systems, CRISPR-Cas systems, abortive infection, and assembly interference [reviewed in references (242 – 244)], or resistance conferred by prophages [i.e. superinfection exclusion (242, 245)], and the phage’s ability to overcome these defenses [reviewed in reference (246)]. How phages will influence outcomes of bacterial competition also depends on population and community-level processes, as discussed below.
ECO-EVOLUTIONARY FACTORS INFLUENCE HOW RHIZOBIOPHAGES AFFECT LEGUME-RHIZOBIUM INVASION
Rhizobiophages might influence legume invasions by lowering rhizobium density. Indeed, early experiments using single-strain inoculation found that adding rhizobiophages sometimes reduced rhizobium density (205) but not always (206). However, we think rhizobiophages are more likely to influence legume invasions by altering the composition of rhizobium communities (201 – 203, 205 – 207, 209). Accordingly, in experiments creating tripartite microbial communities containing rhizobiophages that infect one of two competing rhizobia, phage-resistant strains often occupy a higher percentage of nodules than phage-sensitive strains (202, 203, 206, 207). For example, a phage specialized on Bradyrhizobium japonicum USDA 117 altered the competitive outcome between USDA 117 and B. japonicum USDA 110 by reducing in-soil population size and nodule occupancy of its USDA 117 host (202, 203). Such results suggest that rhizobiophages alter apparent competition among rhizobium taxa. Indeed, simple mathematical models of bacterial interactions with lytic phages produce numerical dynamics akin to other predator-prey models (247). However, overlapping temporal scales of evolutionary and numerical dynamics can drive continuing fluctuations following initial community assembly, necessitating a new conceptual framework (24, 28, 248).
Below, we outline some of the decisive factors known to determine how phage communities affect the structure of bacterial communities and suggest, given these principles, which rhizobium community, introduced or native, will be most negatively affected by novel phages. We also consider how these effects might cascade up to affect the invasion potential of a host legume.
Coevolution
The interdependence of bacterial and phage fitness often produces a co-evolutionary arms race (24, 249, 250): bacteria experience selection for various defensive traits [e.g., alterations to receptors by which bacteriophages attach or mechanisms that recognize and degrade phage DNA or block bacteriophage replication (244, 251)], but bacteriophage populations evolve the ability to use different attachment sites or evade recognition by bacterial hosts (246, 252). A co-evolving partner that fails to counter-adapt quickly faces extinction [i.e., the red queen hypothesis (253)]. Phages typically evolve faster and become locally adapted: more infective on sympatric than allopatric bacteria [e.g., see references (254, 255)]. Thus, naturally occurring phage-bacteria interaction networks usually consist of modules involving local phages adapted to related bacteria (136, 256 – 258) from nearby locations (24, 134, 254, 255, 259). Since the genetic distance between familiar versus recently encountered hosts influences whether a bacteriophage can infect unfamiliar hosts (258, 260) and introduced rhizobia often occupy genetic clusters distinct from native rhizobia (37, 116), phages might not infect unfamiliar rhizobia.
Specialization
Both phage host range (i.e., the number of types of bacteria a phage can infect and lyse) and the breadth of bacterial resistance (i.e., the number of types of phages a bacterium can resist) are measures of specialization which strongly influence microbial community composition. Generalists, i.e., phages that can infect and lyse more types of bacteria or bacteria that can resist infection by more types of phages, should be more successful than specialists, unless generalization involves trade-offs (261, 262). For example, most ways by which bacteria prevent phage infection are costly to bacterial growth (263 – 266), which limits how many types of phage a bacterium can resist and could also cause bacteria to lose resistance to other phages (267). Similarly, specialist phages might infect and lyse few types of bacteria but obtain larger burst sizes or higher adsorption rates than do generalist phages attacking those same bacteria (268, 269). Thus, the fitness benefits a phage obtains from each bacterium type trade off with the number of bacterial types it can infect and the phylogenetic distances among them [e.g., see references (269 – 274)].
Accordingly, phages are usually specialized within a locality. For example, some rhizobiophages associated with common bean rhizobia were extreme specialists, infecting less than 1% of tested rhizobia (134). However, phages within a community can vary in host range: some rhizobiophages are generalists, infecting more than 90% of local, closely related hosts (134). Phages infecting via more conserved surface receptors may infect a more phylogenetically diverse range of hosts (275, 276). However, even generalist phages are rarely able to infect more than a few taxa and, if so, would only infect certain strains within each taxon (261, 277, 278).
The genetic distance between familiar versus recently encountered bacteria reduces the likelihood a phage can infect such unfamiliar hosts (258, 260). Adapting to new bacteria is most difficult for phages highly specialized on distantly related hosts (258, 260, 271). However, even minor mutations (271, 279, 280) can add new host species or genera (225, 270, 281). Thus, phages might be maladapted only during the initial encounter with novel bacteria (14, 282), e.g., in plant pathogens (232, 283). Native rhizobiophages might adapt to a rapidly expanding population of introduced rhizobia, or introduced rhizobiophages might adapt to the numerically dominant native rhizobia.
Relative abundance
Frequency-dependent selection favors phages that adopt abundant hosts (284), which causes those hosts to decline. Multiple studies have documented this “kill-the-winner” process (26, 285 – 287). Thus, the relative effect of phages on introduced or native rhizobium communities may depend largely on the initial relative abundances of both communities. Newly introduced rhizobia are likely to be rare (288), which selects for phages that can infect and drive down the abundance of native rhizobia relative to introduced rhizobia. These phages could be either introduced phages, with shifted host ranges, or native phages. Indeed, if introduced phages evolve to infect the more abundant members of the native rhizobium community, they might disrupt the community enough to benefit introduced rhizobia (spillover). As introduced rhizobia proliferate and become invasive, however, selection on phages would reverse. Thus, delayed eco-evolutionary feedback could yield fluctuating-selection dynamics (289).
Diversity
Rhizobium communities are highly diverse (145, 290, 291), comprising strains with various phage resistance profiles (134), but the diversity of rhizobiophages is still poorly known (136). It is unclear whether and how bacterial diversity predicts how novel phages might structure a host community comprising both familiar and novel bacteria. It is also poorly known how bacterial diversity affects phage evolution [but see reference (274)] and how such evolution could feed back to affect the host community. In kill-the-winner dynamics, “winning” phages can increase bacterial diversity by functioning as keystone predators (292), but a more diverse host community may be more likely to contain bacteria that can survive a greater variety of phages [sampling effect (293)]. Nonetheless, a less diverse bacterial community comprising generalists with relatively broad phage resistance [e.g., with few phage receptors or with effective broad spectrum phage-defense systems (294 – 299)] might still be more resilient to more different phages than is a more diverse community of specialists, each resistant to a different phage (e.g., due to more specialized phage-defense systems).
In general, richer phage communities can better control microbial communities (300, 301), either by including phages with larger or more rapidly expanding host ranges (300 – 303) or by including a greater diversity of specialized phages, each of which attacks different bacterial hosts [sampling effect (293)]. Experiments using phages, either to modify bacterial communities in marine and freshwater systems or to combat pathogenic bacteria in medicine and agriculture (27, 154, 304 – 307), generally find that cocktails of multiple phages provide broader and more durable (i.e., reduced rate at which phage resistance evolves in bacterial hosts) bacterial control than obtained by deploying phages individually (27, 305, 308). However, bacteria are more likely to evolve generalized resistance to a more diverse community of phages (309, 310). For example, Betts et al. (310) found that more diverse phage communities caused selective sweeps of lipopolysaccharide (LPS) synthesis gene mutations, which conferred broad resistance. Nevertheless, introducing even a low-diversity phage community might sufficiently disturb the competitive balance within a bacterial community to compromise its resistance to invasion. Bacterial communities are generally composed of a few dominant genotypes and many rarer genotypes (311, 312), so the decline or eradication of a single dominant genotype via a kill-the-winner process could create a dynamic cascade in which previously rarer genotypes become dominant (26, 287).
Dispersal affects diversity
The diversity of introduced communities of phages and bacteria depends on their respective large-scale population structure and their introduction histories. Communities and populations usually become genetically depauperate as they disperse over long distances (313, 314), suggesting that native communities of phage and bacteria are likely to be more diverse than those established by a single, small introduction. However, if either phages and/or bacteria have been introduced multiple times from multiple locations, then the introduced communities might be very diverse (313, 315 – 318). Rhizobia co-invading with legume hosts often exhibit evidence of multiple introductions (108, 116), possibly by accompanying more than one species of congeneric legume hosts (39, 116, 319, 320). It would be interesting to compare the diversity of rhizobiophages in such communities with those found in rhizobium communities formed by single introductions.
Pleiotropic effects of phage resistance in rhizobia
Rhizobiophages can potentially influence legume fate when they select rhizobia with phage resistance traits that share pleiotropic effects with symbiosis or mutualism traits. Phage resistance traits in rhizobia may trade off with their abilities to engage with legumes such as rhizosphere colonization (321), nodulation (322, 323), or nitrogen fixing efficiency (205, 322). Some bacteriophage-resistant mutants of Bradyrhizobium japonicum (324) have alterations in cell surface LPSs, which are also common sites of phage attachment (323). Defective LPS prevents nodulation by disabling communication between legumes and rhizobia (190, 325, 326). Alternatively, phage resistance can be associated with more and larger nodules, higher nitrogenase activity (327, 328), and enhanced host nitrogen content (329). Finally, such pleiotropic effects are not always observable (330). Thus, if rhizobia evolve resistance to novel phages, any pleiotropic effects of these traits could either disrupt or improve their cooperation with familiar legumes, depending on the magnitude and direction of pleiotropy.
CONCLUSIONS AND FUTURE DIRECTIONS
Dependency in rhizobial mutualists appears to be an Achilles’ heel for many invading legumes (16, 37, 93, 112). Thus, legume invasions might be either foiled or promoted by the evolutionary and ecological effects of native and/or co-introduced bacteriophage enemies. The probability of enemy escape is initially determined by the likelihood that the enemy arrives in the new range and subsequently by the adaptive potential of the phages and bacteria, which determines whether introduced rhizobia might be hampered by new enemies or can enjoy the benefits provided by enemy spillover and enemy spillback (Fig. 1).
Our ability to predict the relative probabilities of these various scenarios, both in this system and among other eukaryotes dependent on mutualists infectiously acquired from the environment, is hampered by how little is known about these processes (enemy escape, spillover, biotic resistance via reverse spillover, biotic resistance via subsidized pathogen, and spillback) in this and other bacterium-bacteriophage systems. Progress in this area depends upon identifying bacteriophage communities associated with rhizobia and legumes, and characterizing the ecological and evolutionary interactions among these populations in both native and non-native habitats. Invasions of other eukaryotes dependent on infectiously acquired bacterial mutualists should receive similar attention.
ACKNOWLEDGMENTS
NSF grants DEB-1457508 and IOS-1759048 to E.L.S. funded the work and supported J.V.C. J.V.C. was partially supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany ´s Excellence Strategy – EXC 2051 – Project-ID 390713860, the Alexander von Humboldt Foundation in the context of an Alexander von Humboldt-Professorship founded by German Federal Ministry of Education and Research, and the European Union’s Horizon 2020 research and innovation program, under the Marie Skłodowska-Curie Actions Innovative Training Networks grant agreement no. 955974 (VIROINF).
E.L.S. and J.V.C. developed the ideas, prepared the figures, reviewed the literature, and drafted and organized the manuscript. Both authors provided critical review before giving approval for submission of the final version.
Contributor Information
Jannick Van Cauwenberghe, Email: jannick.van.cauwenberghe@uni-jena.de.
Jacob Yount, The Ohio State University, Columbus, Ohio, USA .
Katarzyna Danis-Wlodarczyk, The Ohio State University, Columbus, Ohio, USA .
REFERENCES
- 1. Diagne C, Leroy B, Vaissière A-C, Gozlan RE, Roiz D, Jarić I, Salles J-M, Bradshaw CJA, Courchamp F. 2021. High and rising economic costs of biological invasions worldwide. Nature 592:571–576. doi: 10.1038/s41586-021-03405-6 [DOI] [PubMed] [Google Scholar]
- 2. Vaissière AC, Courtois P, Courchamp F, Kourantidou M, Diagne C, Essl F, Kirichenko N, Welsh M, Salles JM. 2022. The nature of economic costs of biological invasions. Biol Invasions 24:2081–2101. doi: 10.1007/s10530-022-02837-z [DOI] [Google Scholar]
- 3. Vitousek PM, D’Antonio CM, Loope LL, Westbrooks R. 1996. Biological invasions as global environmental change. Am Sci 84:468–478. doi:https://www.jstor.org/stable/29775751 [Google Scholar]
- 4. Mitchell CE, Agrawal AA, Bever JD, Gilbert GS, Hufbauer RA, Klironomos JN, Maron JL, Morris WF, Parker IM, Power AG, Seabloom EW, Torchin ME, Vázquez DP. 2006. Biotic interactions and plant invasions. Ecol Lett 9:726–740. doi: 10.1111/j.1461-0248.2006.00908.x [DOI] [PubMed] [Google Scholar]
- 5. Mallon CA, Elsas J van, Salles JF. 2015. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol 23:719–729. doi: 10.1016/j.tim.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 6. Litchman E. 2010. Invisible invaders: non-pathogenic invasive microbes in aquatic and terrestrial ecosystems. Ecol Lett 13:1560–1572. doi: 10.1111/j.1461-0248.2010.01544.x [DOI] [PubMed] [Google Scholar]
- 7. Catford JA, Jansson R, Nilsson C. 2009. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers Distrib 15:22–40. doi: 10.1111/j.1472-4642.2008.00521.x [DOI] [Google Scholar]
- 8. Cavieres LA, Callaway R. 2021. Facilitation and the invasibility of plant communities. J Ecol 109:2019–2028. doi: 10.1111/1365-2745.13627 [DOI] [Google Scholar]
- 9. Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of symbiodinium. Annu Rev Ecol Evol Syst 34:661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417 [DOI] [Google Scholar]
- 10. Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992 [DOI] [PubMed] [Google Scholar]
- 11. Gomes SIF, Kielak AM, Hannula SE, Heinen R, Jongen R, Keesmaat I, De Long JR, Bezemer TM. 2020. Microbiomes of a specialist caterpillar are consistent across different habitats but also resemble the local soil microbial communities. Anim Microbiome 2:37. doi: 10.1186/s42523-020-00055-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hannula SE, Zhu F, Heinen R, Bezemer TM. 2019. Foliar-feeding insects acquire microbiomes from the soil rather than the host plant. Nat Commun 10:1254. doi: 10.1038/s41467-019-09284-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957 [DOI] [PubMed] [Google Scholar]
- 14. Dickie IA, Bufford JL, Cobb RC, Desprez-Loustau ML, Grelet G, Hulme PE, Klironomos J, Makiola A, Nuñez MA, Pringle A, Thrall PH, Tourtellot SG, Waller L, Williams NM. 2017. The emerging science of linked plant–fungal invasions. New Phytol 215:1314–1332. doi: 10.1111/nph.14657 [DOI] [PubMed] [Google Scholar]
- 15. Lu M, Hulcr J, Sun J. 2016. The role of symbiotic microbes in insect invasions. Annu Rev Ecol Evol Syst 47:487–505. doi: 10.1146/annurev-ecolsys-121415-032050 [DOI] [Google Scholar]
- 16. Simonsen AK, Dinnage R, Barrett LG, Prober SM, Thrall PH. 2017. Symbiosis limits establishment of legumes outside their native range at a global scale. Nat Commun 8:14790. doi: 10.1038/ncomms14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfe BE, Klironomos JN. 2005. Breaking new ground: soil communities and exotic plant invasion. BioScience 55:477–487. doi: 10.1641/0006-3568(2005)055[0477:BNGSCA]2.0.CO;2 [DOI] [Google Scholar]
- 18. Traveset A, Richardson DM. 2014. Mutualistic interactions and biological invasions. Annu Rev Ecol Evol Syst 45:89–113. doi: 10.1146/annurev-ecolsys-120213-091857 [DOI] [Google Scholar]
- 19. Richardson DM, Allsopp N, D’Antonio CM, Milton SJ, Rejmánek M. 2000. Plant invasions — the role of mutualisms. Biol Rev Camb Philos Soc 75:65–93. doi: 10.1017/s0006323199005435 [DOI] [PubMed] [Google Scholar]
- 20. Bohannan BJM, Lenski RE. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Letters 3:362–377. doi: 10.1046/j.1461-0248.2000.00161.x [DOI] [Google Scholar]
- 21. Fuhrman JA, Noble RT. 1995. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr 40:1236–1242. doi: 10.4319/lo.1995.40.7.1236 [DOI] [Google Scholar]
- 22. Hennes KP, Simon M. 1995. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl Environ Microbiol 61:333–340. doi: 10.1128/aem.61.1.333-340.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimura M, Jia ZJ, Nakayama N, Asakawa S. 2008. Ecology of viruses in soils: past, present and future perspectives. Soil Sci Plant Nutr 54:1–32. doi: 10.1111/j.1747-0765.2007.00197.x [DOI] [Google Scholar]
- 24. Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38:916–931. doi: 10.1111/1574-6976.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathias CB, Kirschner AKT, Velimirov B. 1995. Seasonal variations of virus abundance and viral control of the bacterial production in a backwater system of the danube river. Appl Environ Microbiol 61:3734–3740. doi: 10.1128/aem.61.10.3734-3740.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morella NM, Gomez AL, Wang G, Leung MS, Koskella B. 2018. The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol Ecol 27:2025–2038. doi: 10.1111/mec.14542 [DOI] [PubMed] [Google Scholar]
- 27. Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, Shen Q, Friman VP. 2019. Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol 37:1513–1520. doi: 10.1038/s41587-019-0328-3 [DOI] [PubMed] [Google Scholar]
- 28. Chevallereau A, Pons BJ, van Houte S, Westra ER. 2022. Interactions between bacterial and phage communities in natural environments. Nat Rev Microbiol 20:49–62. doi: 10.1038/s41579-021-00602-y [DOI] [PubMed] [Google Scholar]
- 29. Braga LPP, Spor A, Kot W, Breuil MC, Hansen LH, Setubal JC, Philippot L. 2020. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome 8:52. doi: 10.1186/s40168-020-00822-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwob G, Roy M, Pozzi AC, Herrera-Belaroussi A, Fernandez MP. 2018. In planta sporulation of frankia spp. as a determinant of alder-symbiont interactions. Appl Environ Microbiol 84:e01737-18. doi: 10.1128/AEM.01737-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zimpfer JF, Kennedy GJ, Smyth CA, Hamelin J, Navarro E, Dawson JO. 1999. Localization of Casuarina-infective Frankia near Casuarina Cunninghamiana trees in Jamaica. Can J Bot 77:1248–1256. doi: 10.1139/b99-063 [DOI] [Google Scholar]
- 32. Gioria M, Osborne BA. 2013. Biological flora of the British isles: Gunnera tinctoria. J Ecol 101:243–264. doi: 10.1111/1365-2745.12022 [DOI] [Google Scholar]
- 33. Soares MA, Li H-Y, Bergen M, da Silva JM, Kowalski KP, White JF. 2016. Functional role of an endophytic Bacillus amyloliquefaciens in enhancing growth and disease protection of invasive English ivy (Hedera helix L.). Plant Soil 405:107–123. doi: 10.1007/s11104-015-2638-7 [DOI] [Google Scholar]
- 34. Burleigh SH, Dawson JO. 1994. Occurrence of Myrica-nodulating Frankia in Hawaiian volcanic soils. Plant Soil 164:283–289. doi: 10.1007/BF00010080 [DOI] [Google Scholar]
- 35. Vitousek PM, Walker LR, Whiteaker LD, Mueller-Dombois D, Matson PA. 1987. Biological invasion by myrica faya alters ecosystem development in Hawaii. Science 238:802–804. doi: 10.1126/science.238.4828.802 [DOI] [PubMed] [Google Scholar]
- 36. Walker LR, Vitousek PM. 1991. An invader alters germination and growth of native dominant tree in Hawai’i. Ecology 72:1449–1455. doi: 10.2307/1941117 [DOI] [Google Scholar]
- 37. La Pierre KJ, Simms EL, Tariq M, Zafar M, Porter SS. 2017. Invasive legumes can associate with many mutualists of native legumes, but usually do not. Ecol Evol 7:8599–8611. doi: 10.1002/ece3.3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodríguez-Echeverría S, Crisóstomo JA, Nabais C, Freitas H. 2009. Belowground mutualists and the invasive ability of acacia longifolia in coastal dunes of Portugal. Biol Invasions 11:651–661. doi: 10.1007/s10530-008-9280-8 [DOI] [Google Scholar]
- 39. Stepkowski T, Moulin L, Krzyzanska A, Mcinnes A, Law IJ, Howieson J. 2005. European origin of bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl Environ Microbiol 71:7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bozorov TA, Rasulov BA, Zhang D. 2019. Characterization of the gut microbiota of invasive agrilus mali matsumara (coleoptera: buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci Rep 9:4923. doi: 10.1038/s41598-019-41368-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vasanthakumar A, Handelsman JO, Schloss PD, Bauer LS, Raffa KF. 2008. Gut microbiota of an invasive subcortical beetle, agrilus planipennis fairmaire, across various life stages. Environ Entomol 37:1344–1353. doi: 10.1603/0046-225x(2008)37[1344:gmoais]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 42. Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U S A 109:8618–8622. doi: 10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl Environ Microbiol 77:4075–4081. doi: 10.1128/AEM.00358-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, Currie CR, Raffa KF. 2011. Cellulose-degrading bacteria associated with the invasive woodwasp sirex noctilio. ISME J 5:1323–1331. doi: 10.1038/ismej.2011.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaltenpoth M, Flórez LV. 2020. Versatile and dynamic symbioses between insects and burkholderia bacteria. Annu Rev Entomol 65:145–170. doi: 10.1146/annurev-ento-011019-025025 [DOI] [PubMed] [Google Scholar]
- 46. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between burkholderia gut symbionts and their heteropteran hosts. ISME J 5:446–460. doi: 10.1038/ismej.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dilworth MJ, James EK, Sprent JI, Newton WE. 2008. Nitrogen-fixing leguminous symbioses. In Nitrogen fixation: origins, applications, and research progress. Vol. 7. Springer, Dordrecht. doi: 10.1007/978-1-4020-3548-7 [DOI] [Google Scholar]
- 48. Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805. doi: 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- 49. Poole P, Ramachandran V, Terpolilli J. 2018. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16:291–303. doi: 10.1038/nrmicro.2017.171 [DOI] [PubMed] [Google Scholar]
- 50. West SA, Kiers ET, Pen I, Denison RF. 2002. Sanctions and mutualism stability: when should less beneficial mutualists be tolerated?. J Evol Biol 15:830–837. doi: 10.1046/j.1420-9101.2002.00441.x [DOI] [Google Scholar]
- 51. Simms EL, Taylor DL. 2002. Partner choice in nitrogen-fixation mutualisms of legumes and rhizobia. Integr Comp Biol 42:369–380. doi: 10.1093/icb/42.2.369 [DOI] [PubMed] [Google Scholar]
- 52. Bergersen FJ. 1982. Root nodules of legumes: structure and functions. Research Studies Press, Chichester. [Google Scholar]
- 53. Brockwell J, Bottomley PJ, Thies JE. 1995. Manipulation of rhizobia microflora for improving legume productivity and soil fertility: a critical assessment. Plant Soil 174:143–180. doi: 10.1007/BF00032245 [DOI] [Google Scholar]
- 54. Denton MD, Reeve WG, Howieson JG, Coventry DR. 2003. Competitive abilities of common field isolates and a commercial strain of Rhizobium leguminosarum bv. trifolii for clover nodule occupancy. Soil Biol Biochem 35:1039–1048. doi: 10.1016/S0038-0717(03)00146-9 [DOI] [Google Scholar]
- 55. Brockwell J, Gault RR, Zorin M, Roberts MJ. 1982. Effects of environmental variables on the competition between inoculum strains and naturalized populations of Rhizobium trifolii for nodulation of Trifolium subterraneum L. and on rhizobia persistence in the soil. Aust J Agric Res 33:803–815. doi: 10.1071/AR9820803 [DOI] [Google Scholar]
- 56. Chatel DL, Parker CA. 1973. Survival of field-grown rhizobia over the dry summer period in Western Australia. Soil Biol Biochem 5:415–423. doi: 10.1016/0038-0717(73)90068-0 [DOI] [Google Scholar]
- 57. Corich V, Giacomini A, Vendramin E, Vian P, Carlot M, Concheri G, Polone E, Casella S, Nuti MP, Squartini A. 2007. Long term evaluation of field-released genetically modified rhizobia. Environ Biosafety Res 6:167–181. doi: 10.1051/ebr:2007006 [DOI] [PubMed] [Google Scholar]
- 58. Giongo A, Ambrosini A, Vargas LK, Freire JRJ, Bodanese-Zanettini MH, Passaglia LMP. 2008. Evaluation of genetic diversity of bradyrhizobia strains nodulating soybean [Glycine max (L.) Merrill] isolated from South Brazilian fields. Appl Soil Ecol 38:261–269. doi: 10.1016/j.apsoil.2007.10.016 [DOI] [Google Scholar]
- 59. Richardson AE, Simpson RJ. 1989. Acid-tolerance of rhizobium growing in an acid soil. Soil Biol Biochem 21:87–95. doi: 10.1016/0038-0717(89)90016-3 [DOI] [Google Scholar]
- 60. Slattery JF, Pearce DJ, Slattery WJ. 2004. Effects of resident rhizobial communities and soil type on the effective nodulation of pulse legumes. Soil Biol Biochem 36:1339–1346. doi: 10.1016/j.soilbio.2004.04.015 [DOI] [Google Scholar]
- 61. Vlassak KM, Vanderleyden J. 1997. Factors influencing nodule occupancy by inoculant rhizobia. CRC Crit Rev Plant Sci 16:163–229. doi: 10.1080/07352689709701948 [DOI] [Google Scholar]
- 62. Keeler AM, Rafferty NE. 2022. Legume germination is delayed in dry soils and in sterile soils devoid of microbial Mutualists: species-specific implications for upward range expansions. Ecol Evol 12:e9186. doi: 10.1002/ece3.9186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ratcliff WC, Denison RF. 2011. Bacterial persistence and bet hedging in. PLoS Biol 4:98–100. doi: 10.4161/cib.14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hartel PG, Alexander M. 1983. Growth and survival of cowpea rhizobia in acid, aluminum-rich soils. Soil Sci Soc Am 47:502–506. doi: 10.2136/sssaj1983.03615995004700030021x [DOI] [Google Scholar]
- 65. Starkey RL. 1931. Some influences of the development of higher plants upon the microorganisms in the soil: IV. Influence of proximity to roots on abundance and activity of microorganisms. Soil Sci 32:367–393. doi: 10.1097/00010694-193111000-00003 [DOI] [Google Scholar]
- 66. Rengel Z, Marschner P. 2005. Nutrient availability and management in the rhizosphere: exploiting genotypic differences. New Phytol 168:305–312. doi: 10.1111/j.1469-8137.2005.01558.x [DOI] [PubMed] [Google Scholar]
- 67. Denison RF, Kiers ET. 2011. Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr Biol 21:R775–R785. doi: 10.1016/j.cub.2011.06.018 [DOI] [PubMed] [Google Scholar]
- 68. Masson-Boivin C, Sachs JL. 2018. Symbiotic nitrogen fixation by rhizobia — the roots of a success story. Curr Opin Plant Biol 44:7–15. doi: 10.1016/j.pbi.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 69. Graham PH. 2008. Ecology of the root-Nodule bacteria of legumes, p 23–58. In Dilworth MJ, James EK, Sprent JI, Newton WE (ed), Nitrogen-fixing leguminous symbioses. Springer, The Netherlands, Dordrecht. doi: 10.1007/978-1-4020-3548-7 [DOI] [Google Scholar]
- 70. Rangin C, Brunel B, Cleyet-Marel J-C, Perrineau M-M, Béna G. 2008. Effects of medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural sinorhizobium species community. Appl Environ Microbiol 74:5653–5661. doi: 10.1128/AEM.01107-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T. 2015. Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53:403–424. doi: 10.1146/annurev-phyto-082712-102342 [DOI] [PubMed] [Google Scholar]
- 72. Ndungu SM, Messmer MM, Ziegler D, Gamper HA, Mészáros É, Thuita M, Vanlauwe B, Frossard E, Thonar C. 2018. Cowpea (Vigna unguiculata L. Walp) hosts several widespread bradyrhizobial root nodule symbionts across contrasting agro-ecological production areas in Kenya. Agric Ecosyst Environ 261:161–171. doi: 10.1016/j.agee.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nautiyal CS. 1997. Rhizosphere competence of Pseudomonas sp. NBRI9926 and rhizobium sp. NBRI9513 involved in the suppression of chickpea (Cicer arietinum L.) pathogenic fungi. FEMS Microbiol Ecol 23:145–158. doi: 10.1111/j.1574-6941.1997.tb00398.x [DOI] [Google Scholar]
- 74. Velázquez E, Carro L, Flores-Félix JD, Martínez-Hidalgo P, Menéndez E, Ramírez-Bahena M-H, Mulas R, González-Andrés F, Martínez-Molina E, Peix A. 2017. The legume nodule microbiome: a source of plant growth-promoting bacteria BT, p 41–70. In Kumar V, Kumar M, Sharma S, Prasad R (ed), Probiotics and plant health. Springer, Singapore. doi: 10.1007/978-981-10-3473-2 [DOI] [Google Scholar]
- 75. Narożna D, Pudełko K, Króliczak J, Golińska B, Sugawara M, Mądrzak CJ, Sadowsky MJ. 2015. Survival and competitiveness of bradyrhizobium japonicum strains 20 years after introduction into field locations in Poland. Appl Environ Microbiol 81:5552–5559. doi: 10.1128/AEM.01399-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lopez ZC, Friesen ML, Von Wettberg E, New L, Porter S. 2021. Microbial mutualist distribution limits spread of the invasive legume medicago polymorpha. Biol Invasions 23:843–856. doi: 10.1007/s10530-020-02404-4 [DOI] [Google Scholar]
- 77. Brockwell J, Roughley RJ, Herridge DF. 1987. Population dynamics of Rhizobium japonicum strains used to inoculate three successive crops of soybean. Aust J Agric Res 38:61–74. doi: 10.1071/AR9870061 [DOI] [Google Scholar]
- 78. Bushby HVA. 1993. Colonization of rhizospheres by bradyrhizobium sp. in relation to strain persistence and nodulation of some pasture legumes. Soil Biol Biochem 25:597–605. doi: 10.1016/0038-0717(93)90199-L [DOI] [Google Scholar]
- 79. Ferreira MC, Hungria M. 2002. Recovery of soybean inoculant strains from uncropped soils in Brazil. Field Crops Research 79:139–152. doi: 10.1016/S0378-4290(02)00119-3 [DOI] [Google Scholar]
- 80. Kuykendall LD. 1989. Influence of glycine max nodulation on the persistence in soil of a genetically marked bradyrhizobium japonicum strain. Plant Soil 116:275–277. doi: 10.1007/BF02214558 [DOI] [Google Scholar]
- 81. Murray BBR, Thrall PH, Woods MJ. 2001. Acacia species and rhizobial interactions: implications for restoration of native vegetation. Ecol Manage Restor 2:213–219. doi: 10.1046/j.1442-8903.2001.00086.x [DOI] [Google Scholar]
- 82. Reyes VG, Schmidt EL. 1979. Population densities of rhizobium japonicum strain 123 estimated directly in soil and rhizospheres. Appl Environ Microbiol 37:854–858. doi: 10.1128/aem.37.5.854-858.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sharma RS, Mohmmed A, Babu CR. 2002. Diversity among rhizobiophages from rhizospheres of legumes inhabiting three ecogeographical regions of India. Soil Biol Biochem 34:965–973. doi: 10.1016/S0038-0717(02)00030-5 [DOI] [Google Scholar]
- 84. Cubo MT, Alías-Villegas C, Balsanelli E, Mesa D, de Souza E, Espuny MR. 2020. Diversity of sinorhizobium (Ensifer) meliloti bacteriophages in the rhizosphere of medicago marina: myoviruses, filamentous and N4-like podovirus. Front Microbiol 11:22. doi: 10.3389/fmicb.2020.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ehrenfeld JG. 2003. Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 6:503–523. doi: 10.1007/s10021-002-0151-3 [DOI] [Google Scholar]
- 86. Corbin JD, D’Antonio CM. 2004. Can carbon addition increase competitiveness of native grasses? A case study from California. Restor Ecol 12:36–43. doi: 10.1111/j.1061-2971.2004.00299.x [DOI] [Google Scholar]
- 87. WallisDeVries MF, Bobbink R. 2017. Nitrogen deposition impacts on biodiversity in terrestrial ecosystems: mechanisms and perspectives for restoration. Biol Conserv 212:387–389. doi: 10.1016/j.biocon.2017.01.017 [DOI] [Google Scholar]
- 88. Liu G, Yang YB, Zhu ZH. 2018. Elevated nitrogen allows the weak invasive plant Galinsoga quadriradiata to become more vigorous with respect to inter-specific competition. Sci Rep 8:1–8. doi: 10.1038/s41598-018-21546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maron JL, Connors PG. 1996. A native nitrogen-fixing shrub facilitates weed invasion. Oecologia 105:302–312. doi: 10.1007/BF00328732 [DOI] [PubMed] [Google Scholar]
- 90. Van Riper LC, Larson DL, Larson JL. 2010. Nitrogen-limitation and invasive sweetclover impacts vary between two great plains plant communities. Biol Invasions 12:2735–2749. doi: 10.1007/s10530-009-9678-y [DOI] [Google Scholar]
- 91. Maron JL, Jefferies RL. 1999. Bush lupine mortality, altered resource availability, and alternative vegetaion states. Ecology 80:443–454. doi: 10.1890/0012-9658(1999)080[0443:BLMARA]2.0.CO;2 [DOI] [Google Scholar]
- 92. Ehinger M, Mohr TJ, Starcevich JB, Sachs JL, Porter SS, Simms EL. 2014. Specialization-generalization trade-off in a bradyrhizobium symbiosis with wild legume hosts. BMC Ecol 14:8. doi: 10.1186/1472-6785-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Harrison TL, Simonsen AK, Stinchcombe JR, Frederickson ME. 2018. More partners, more ranges: generalist legumes spread more easily around the globe. Biol Lett 14:20180616. doi: 10.1098/rsbl.2018.0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang D, Yang S, Tang F, Zhu H. 2012. Symbiosis specificity in the legume - rhizobial mutualism. Cell Microbiol 14:334–342. doi: 10.1111/j.1462-5822.2011.01736.x [DOI] [PubMed] [Google Scholar]
- 95. Liu WYY, Ridgway HJ, James TK, Premaratne M, Andrews M. 2012. Characterisation of rhizobia nodulating Galega officinalis (goat’s rue) and Hedysarum coronarium (sulla). New Zeal Plant Prot 65:192–196. doi: 10.30843/nzpp.2012.65.5365 [DOI] [Google Scholar]
- 96. Young NR, Mytton LR. 1983. The response of white clover to different strains of Rhizobium trifolii in hill land reseeding. Grass and Forage Sci 38:13–19. doi: 10.1111/j.1365-2494.1983.tb01615.x [DOI] [Google Scholar]
- 97. Komatsu KJ, Simms EL. 2020. Invasive legume management strategies differentially impact mutualist abundance and benefit to native and invasive hosts. Restor Ecol 28:378–386. doi: 10.1111/rec.13081 [DOI] [Google Scholar]
- 98. Parker MP. 1995. Plant fitness variation caused by different mutualist genotypes. Ecology 76:1525–1535. doi: 10.2307/1938154 [DOI] [Google Scholar]
- 99. Rodríguez-Echeverría S, Fajardo S, Ruiz-Díez B, Fernández-Pascual M. 2012. Differential effectiveness of novel and old legume-rhizobia mutualisms: implications for invasion by exotic legumes. Oecologia 170:253–261. doi: 10.1007/s00442-012-2299-7 [DOI] [PubMed] [Google Scholar]
- 100. Shelby N, Duncan RP, Putten WH, McGinn KJ, Weser C, Hulme PE, Austin A. 2016. Plant mutualisms with rhizosphere microbiota in introduced versus native ranges. J Ecol 104:1259–1270. doi: 10.1111/1365-2745.12609 [DOI] [Google Scholar]
- 101. Cauwenberghe JV, Visch W, Michiels J, Honnay O. 2016. Selection mosaics differentiate rhizobium–host plant interactions across different nitrogen environments. Oikos 125:1755–1761. doi: 10.1111/oik.02952 [DOI] [Google Scholar]
- 102. Wandrag EM, Birnbaum C, Klock MM, Barrett LG, Thrall PH, Cheng L. 2020. Availability of soil mutualists may not limit non-native Acacia invasion but could increase their impact on native soil communities. J Appl Ecol 57:786–793. doi: 10.1111/1365-2664.13577 [DOI] [Google Scholar]
- 103. Sprent JI, Sutherland JM, Faria SM. 1987. Some aspects of the biology of nitrogen-fixing organisms. Phil Trans R Soc Lond B 317:111–129. doi: 10.1098/rstb.1987.0051 [DOI] [Google Scholar]
- 104. Vinuesa P, Silva C, Werner D, Martínez-Romero E, Martı E. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54. doi: 10.1016/j.ympev.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 105. Reche I, D’Orta G, Mladenov N, Winget DM, Suttle CA. 2018. Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J 12:1154–1162. doi: 10.1038/s41396-017-0042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weil T, De Filippo C, Albanese D, Donati C, Pindo M, Pavarini L, Carotenuto F, Pasqui M, Poto L, Gabrieli J, Barbante C, Sattler B, Cavalieri D, Miglietta F. 2017. Legal immigrants: invasion of alien microbial communities during winter occurring desert dust storms. Microbiome 5:32. doi: 10.1186/s40168-017-0249-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Šantl-Temkiv T, Amato P, Casamayor EO, Lee PKH, Pointing SB. 2022. Microbial ecology of the atmosphere. FEMS Microbiol Rev 46:fuac009. doi: 10.1093/femsre/fuac009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Andrus AD, Andam C, Parker MA. 2012. American origin of cupriavidus bacteria associated with invasive mimosa legumes in the Philippines. FEMS Microbiol Ecol 80:747–750. doi: 10.1111/j.1574-6941.2012.01342.x [DOI] [PubMed] [Google Scholar]
- 109. Warrington S, Ellis A, Novoa A, Wandrag EM, Hulme PE, Duncan RP, Valentine A, Le Roux JJ. 2019. Cointroductions of Australian acacias and their rhizobial mutualists in the Southern hemisphere. J Biogeogr 46:1519–1531. doi: 10.1111/jbi.13602 [DOI] [Google Scholar]
- 110. Weir BS, Turner SJ, Silvester WB, Park D-C, Young JM. 2004. Unexpectedly diverse mesorhizobium strains and rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by bradyrhizobium species unexpectedly diverse mesorhizobium strains and rhizobium leguminosa. Appl Environ Microbiol 70:5980–5987. doi: 10.1128/AEM.70.10.5980-5987.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Crisóstomo JA, Rodríguez-Echeverría S, Freitas H. 2013. Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume acacia saligna, an intercontinental study. Applied Soil Ecology 64:118–126. doi: 10.1016/j.apsoil.2012.10.005 [DOI] [Google Scholar]
- 112. Horn K, Parker IM, Malek W, Rodríguez-Echeverría S, Parker MA. 2014. Disparate origins of Bradyrhizobium symbionts for invasive populations of Cytisus scoparius (Leguminosae) in North America. FEMS Microbiol Ecol 89:89–98. doi: 10.1111/1574-6941.12335 [DOI] [PubMed] [Google Scholar]
- 113. Lemaire B, Van Cauwenberghe J, Chimphango S, Stirton C, Honnay O, Smets E, Muasya AM. 2015. Recombination and horizontal transfer of nodulation and ACC deaminase (acdS) genes within alpha- and betaproteobacteria nodulating legumes of the cape fynbos biome. FEMS Microbiol Ecol 91:fiv118. doi: 10.1093/femsec/fiv118 [DOI] [PubMed] [Google Scholar]
- 114. Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, Boatwright JS, Honnay O, Smets E, Sprent J, James EK, Muasya AM. 2015. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the core cape subregion (South Africa). FEMS Microbiol Ecol 91:1–17. doi: 10.1093/femsec/fiu024 [DOI] [PubMed] [Google Scholar]
- 115. Ndlovu J, Richardson DM, Wilson JRU, Le Roux JJ, Ladiges P. 2013. Co-invasion of South African ecosystems by an Australian legume and its rhizobial symbionts. J Biogeogr 40:1240–1251. doi: 10.1111/jbi.12091 [DOI] [Google Scholar]
- 116. Porter SS, Faber-Hammond JJ, Friesen ML. 2018. Co-invading symbiotic mutualists of medicago polymorpha retain high ancestral diversity and contain diverse accessory genomes. FEMS Microbiol Ecol 94:1–12. doi: 10.1093/femsec/fix168 [DOI] [PubMed] [Google Scholar]
- 117. Bay SK, McGeoch MA, Gillor O, Wieler N, Palmer DJ, Baker DJ, Chown SL, Greening C. 2020. Soil bacterial communities exhibit strong biogeographic patterns at fine taxonomic resolution. mSystems 5:e00540-20. doi: 10.1128/mSystems.00540-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dove R, Wolfe ER, Stewart NU, Ballhorn DJ. 2020. Ecoregion - rather than sympatric legumes - influences symbiotic bradyrhizobium associations in invasive scotch broom (Cytisus scoparius) in the Pacific Northwest. Northwest Sci 94:142–159. doi: 10.3955/046.094.0205 [DOI] [Google Scholar]
- 119. Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. 2012. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. doi: 10.1038/nrmicro2795 [DOI] [PubMed] [Google Scholar]
- 120. Van Cauwenberghe J, Michiels J, Honnay O. 2015. Effects of local environmental variables and geographical location on the genetic diversity and composition of rhizobium leguminosarum nodulating vicia cracca populations. Soil Biol Biochem 90:71–79. doi: 10.1016/j.soilbio.2015.08.001 [DOI] [Google Scholar]
- 121. van der Gast CJ. 2015. Microbial biogeography: the end of the ubiquitous dispersal hypothesis Environ Microbiol 17:544–546. doi: 10.1111/1462-2920.12635 [DOI] [PubMed] [Google Scholar]
- 122. Kirchner O. 1895. Die wurzelknollchen der sojabohne. Cohn, Beiträge zur Biol der Pflanz VII:213–223. [Google Scholar]
- 123. Gangulee N. 1926. The organism forming nodules on crotalaria juncea. Ann Appl Bio XIII:256–259. doi: 10.1111/j.1744-7348.1926.tb04270.x [DOI] [Google Scholar]
- 124. Brockwell J, Fettell NA, Bowman AM, Smith W, Sweeney G, Charman N, Ballard RA. 2008. Symbiotic competence of rose clover (Trifolium hirtum All). Aust J Agric Res 59:802–813. doi: 10.1071/AR07469 [DOI] [Google Scholar]
- 125. Mendoza-Suárez M, Andersen SU, Poole PS, Sánchez-Cañizares C. 2021. Competition, nodule occupancy, and persistence of inoculant strains: key factors in the rhizobium-legume symbioses. Front Plant Sci 12:690567. doi: 10.3389/fpls.2021.690567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bamba M, Nakata S, Aoki S, Takayama K, Núñez-Farfán J, Ito M, Miya M, Kajita T. 2016. Wide distribution range of rhizobial symbionts associated with pantropical sea-dispersed legumes. Antonie van leeuwenhoek. Int J Gen Mol Microbiol 109:1605–1614. doi: 10.1007/s10482-016-0761-y [DOI] [PubMed] [Google Scholar]
- 127. Choudoir MJ, Barberán A, Menninger HL, Dunn RR, Fierer N. 2018. Variation in range size and dispersal capabilities of microbial taxa. Ecology 99:322–334. doi: 10.1002/ecy.2094 [DOI] [PubMed] [Google Scholar]
- 128. Walters KE, Capocchi JK, Albright MBN, Hao Z, Brodie EL, Martiny JBH. 2022. Routes and rates of bacterial dispersal impact surface soil microbiome composition and functioning. ISME J 16:2295–2304. doi: 10.1038/s41396-022-01269-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Menge DNL, Wolf AA, Funk JL. 2015. Diversity of nitrogen fixation strategies in mediterranean legumes. Nature Plants 1:15064. doi: 10.1038/nplants.2015.64 [DOI] [PubMed] [Google Scholar]
- 130. Parker MA. 2001. Mutualism as a constraint on invasion success for legumes and rhizobia. Divers Distrib 7:125–136. doi: 10.1046/j.1472-4642.2001.00103.x [DOI] [Google Scholar]
- 131. Parker MA, Malek W, Parker IM. 2006. Growth of an invasive legume is symbiont limited in newly occupied habitats. Divers Distrib 12:563–571. doi: 10.1111/j.1366-9516.2006.00255.x [DOI] [Google Scholar]
- 132. Delavaux CS, Weigelt P, Magnoli SM, Kreft H, Crowther TW, Bever JD. 2022. Nitrogen-fixing symbiotic bacteria act as a global filter for plant establishment on Islands. Commun Biol 5:1–6. doi: 10.1038/s42003-022-04133-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Werquin M, Ackermann H-W, Levesque RC. 1988. A study of 33 bacteriophages of Rhizobium meliloti. Appl Environ Microbiol 54:188–196. doi: 10.1128/aem.54.1.188-196.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Van Cauwenberghe J, Santamaría RI, Bustos P, Juárez S, Ducci MA, Fleming TF, Etcheverry AV, González V. 2021. Spatial patterns in phage-rhizobium coevolutionary interactions across regions of common bean domestication. ISME J 15:2092–2106. doi: 10.1038/s41396-021-00963-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Santamaría RI, Bustos P, Van Cauwenberghe J, González V. 2022. Hidden diversity of double-stranded DNA phages in symbiotic rhizobium species. Philos Trans R Soc Lond B Biol Sci 377:20200468. doi: 10.1098/rstb.2020.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Van Cauwenberghe J, Santamaría RI, Bustos P, González V. 2022. Novel lineages of single-stranded DNA phages that coevolved with the symbiotic bacteria Rhizobium. Front Microbiol 13:1–14. doi: 10.3389/fmicb.2022.990394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Brewer TE, Stroupe ME, Jones KM. 2014. The genome, proteome and phylogenetic analysis of Sinorhizobium meliloti phage Φm12, the founder of a new group of T4-superfamily phages. Virology 450–451:84–97. doi: 10.1016/j.virol.2013.11.027 [DOI] [PubMed] [Google Scholar]
- 138. Dziewit L, Oscik K, Bartosik D, Radlinska M. 2014. Molecular characterization of a novel temperate Sinorhizobium bacteriophage, ФLM21, encoding DNA methyltransferase with CcrM-like specificity. J Virol 88:13111–13124. doi: 10.1128/JVI.01875-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ford S, Moeskjær S, Young P, Santamaría RI, Harrison E. 2021. Introducing a novel, broad host range temperate phage family infecting Rhizobium leguminosarum and beyond. Front Microbiol 12:765271. doi: 10.3389/fmicb.2021.765271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gunathilake KMD, Halmillawewa AP, MacKenzie KD, Perry BJ, Yost CK, Hynes MF. 2021. A bacteriophage infecting mesorhizobium species has a prolate capsid and shows similarities to a family of caulobacter crescentus phages. Can J Microbiol 67:147–160. doi: 10.1139/cjm-2020-0281 [DOI] [PubMed] [Google Scholar]
- 141. Halmillawewa AP, Restrepo-Córdoba M, Yost CK, Hynes MF. 2015. Genomic and phenotypic characterization of Rhizobium gallicum phage vB_RglS_P106B. Microbiology (Reading) 161:611–620. doi: 10.1099/mic.0.000022 [DOI] [PubMed] [Google Scholar]
- 142. Johnson MC, Tatum KB, Lynn JS, Brewer TE, Lu S, Washburn BK, Stroupe ME, Jones KM. 2015. Sinorhizobium meliloti phage ΦM9 defines a new group of T4-superfamily phages with unusual genomic features, but a common T =16 capsid. J Virol 89:10945–10958. doi: 10.1128/JVI.01353-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Santamaría RI, Bustos P, Sepúlveda-Robles O, Lozano L, Rodríguez C, Fernández JL, Juárez S, Kameyama L, Guarneros G, Dávila G, González V. 2014. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl Environ Microbiol 80:446–454. doi: 10.1128/AEM.02256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kumar N, Lad G, Giuntini E, Kaye ME, Udomwong P, Shamsani NJ, Young JPW, Bailly X. 2015. Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol 5:140133. doi: 10.1098/rsob.140133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Pérez Carrascal OM, VanInsberghe D, Juárez S, Polz MF, Vinuesa P, González V. 2016. Population genomics of the symbiotic plasmids of sympatric nitrogen-fixing rhizobium species associated with phaseolus vulgaris. Environ Microbiol 18:2660–2676. doi: 10.1111/1462-2920.13415 [DOI] [PubMed] [Google Scholar]
- 146. Porter SS, Chang PL, Conow CA, Dunham JP, Friesen ML. 2017. Association mapping reveals novel serpentine adaptation gene clusters in a population of symbiotic mesorhizobium. ISME J 11:248–262. doi: 10.1038/ismej.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Fields B, Moffat EK, Harrison E, Andersen SU, Young JPW, Friman V-P. 2022. Genetic variation is associated with differences in facilitative and competitive interactions in the Rhizobium leguminosarum species complex. Environ Microbiol 24:3463–3485. doi: 10.1111/1462-2920.15720 [DOI] [PubMed] [Google Scholar]
- 148. Hurwitz BL, Sullivan MB. 2013. The Pacific ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One 8:e57355. doi: 10.1371/journal.pone.0057355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wietz M, Millán-Aguiñaga N, Jensen PR. 2014. CRISPR-Cas systems in the marine actinomycete Salinispora: linkages with phage defense, microdiversity and biogeography. BMC Genomics 15:936. doi: 10.1186/1471-2164-15-936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Huang S, Zhang S, Jiao N, Chen F, Wommack KE. 2015. Marine cyanophages demonstrate biogeographic patterns throughout the global ocean. Appl Environ Microbiol 81:441–452. doi: 10.1128/AEM.02483-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hanson CA, Marston MF, Martiny JBH. 2016. Biogeographic variation in host range phenotypes and taxonomic composition of marine cyanophage isolates. Front. Microbiol 7:983. doi: 10.3389/fmicb.2016.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Garin-Fernandez A, Pereira-Flores E, Glöckner FO, Wichels A. 2018. Marine genomics the North sea goes viral: occurrence and distribution of North sea bacteriophages. Mar Genomics 41:31–41. doi: 10.1016/j.margen.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 153. Dennehy JJ. 2014. What ecologists can tell virologists. Annu Rev Microbiol 68:117–135. doi: 10.1146/annurev-micro-091313-103436 [DOI] [PubMed] [Google Scholar]
- 154. Svircev AM, Lehman SM, Sholberg P, Roach D, Castle AJ. 2011. Phage biopesticides and soil bacteria: multilayered and complex interactions, p 215–235. In Witzany G (ed), Biocommunication in soil microorganisms. Soil biology. Springer, Berlin, Heidelberg. doi: 10.1007/978-3-642-14512-4 [DOI] [Google Scholar]
- 155. Whon TW, Kim M-S, Roh SW, Shin N-R, Lee H-W, Bae J-W. 2012. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol 86:8221–8231. doi: 10.1128/JVI.00293-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Breitbart M, Miyake JH, Rohwer F. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236:249–256. doi: 10.1016/j.femsle.2004.05.042 [DOI] [PubMed] [Google Scholar]
- 157. Jameson E, Mann NH, Joint I, Sambles C, Mühling M. 2011. The diversity of cyanomyovirus populations along a North-South Atlantic ocean transect. ISME J 5:1713–1721. doi: 10.1038/ismej.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GGZ, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, Felts B, Dinsdale EA, Mokili JL, Edwards RA. 2014. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5:4498. doi: 10.1038/ncomms5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Winter C, Matthews B, Suttle CA. 2013. Effects of environmental variation and spatial distance on bacteria, archaea and viruses in sub-polar and arctic waters. ISME J 7:1507–1518. doi: 10.1038/ismej.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Jung MJ, Kim MS, Yun JH, Lee JY, Kim PS, Lee HW, Ha JH, Roh SW, Bae JW. 2018. Viral community predicts the geographical origin of fermented vegetable foods more precisely than bacterial community. Food Microbiol 76:319–327. doi: 10.1016/j.fm.2018.06.010 [DOI] [PubMed] [Google Scholar]
- 161. Finke JF, Suttle CA. 2019. The environment and cyanophage diversity: insights from environmental sequencing of DNA polymerase. Front Microbiol 10:167. doi: 10.3389/fmicb.2019.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Taboada B, Isa P, Gutiérrez-Escolano AL, Del Ángel RM, Ludert JE, Vázquez N, Tapia-Palacios MA, Chávez P, Garrido E, Espinosa AC, Eguiarte LE, López S, Souza V, Arias CF. 2018. The geographic structure of viruses in the cuatro ciénegas basin, a unique oasis in Northern Mexico, reveals a highly diverse population on a small geographic scale. Appl Environ Microbiol 84:1–25. doi: 10.1128/AEM.00465-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Santos-Medellín C, Estera-Molina K, Yuan M, Pett-Ridge J, Firestone MK, Emerson JB. 2022. Spatial turnover of soil viral populations and genotypes overlain by cohesive responses to moisture in grasslands. Proc Natl Acad Sci U S A 119:1–11. doi: 10.1073/pnas.2209132119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Roux S, Emerson JB. 2022. Diversity in the soil virosphere: to infinity and beyond? Trends Microbiol 30:1025–1035. doi: 10.1016/j.tim.2022.05.003 [DOI] [PubMed] [Google Scholar]
- 165. Durham DM, Sieradzki ET, ter Horst AM, Santos-Medellín C, Bess CWA, Geonczy SE, Emerson JB. 2022. Substantial differences in soil viral community composition within and among four Northern California habitats. ISME Commun 2:1–5. doi: 10.1038/s43705-022-00171-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bergersen FJ, Brockwell J, Gibson AH, Schwinghamer EA. 1971. Studies of natural populations and mutants of rhizobium in the improvement of legume inoculants. Plant Soil 35:3–16. doi: 10.1007/BF02661831 [DOI] [Google Scholar]
- 167. Safronova VI, Novikova NI. 1996. Comparison of two methods for root nodule bacteria preservation: lyophilization and liquid nitrogen freezing. J Microbiol Methods 24:231–237. doi: 10.1016/0167-7012(95)00042-9 [DOI] [Google Scholar]
- 168. Roumiantseva ML, Vladimirova ME, Saksaganskaia AS, Muntyan VS, Kozlova AP, Afonin AM, Baturina OA, Simarov BV. 2022. Ensifer meliloti L6-Ak89, an effective inoculant of medicago lupulina varieties: phenotypic and deep-genome screening. Agronomy 12:766. doi: 10.3390/agronomy12040766 [DOI] [Google Scholar]
- 169. Bull JJ, Rice WR. 1991. Distinguishing mechanisms for the evolution of co-operation. J Theor Biol 149:63–74. doi: 10.1016/s0022-5193(05)80072-4 [DOI] [PubMed] [Google Scholar]
- 170. Sachs JL, Mueller UG, Wilcox TP, Bull JJ. 2004. The evolution of cooperation. Q Rev Biol 79:135–160. doi: 10.1086/383541 [DOI] [PubMed] [Google Scholar]
- 171. Stoy KS, Gibson AK, Gerardo NM, Morran LT. 2020. A need to consider the evolutionary genetics of host–symbiont mutualisms. J Evol Biol 33:1656–1668. doi: 10.1111/jeb.13715 [DOI] [PubMed] [Google Scholar]
- 172. Aguilar OM, Riva O, Peltzer E. 2004. Analysis of rhizobium etli and of its symbiosis with wild phaseolus vulgaris supports coevolution in centers of host diversification. Proc Natl Acad Sci U S A 101:13548–13553. doi: 10.1073/pnas.0405321101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Batstone RT, Dutton EM, Wang D, Yang M, Frederickson ME. 2017. The evolution of symbiont preference traits in the model legume medicago truncatula. New Phytol 213:1850–1861. doi: 10.1111/nph.14308 [DOI] [PubMed] [Google Scholar]
- 174. Gubry-Rangin C, Garcia M, Béna G. 2010. Partner choice in medicago truncatula-sinorhizobium symbiosis. Proc Biol Sci 277:1947–1951. doi: 10.1098/rspb.2009.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Jones DG, Hardarson G. 1979. Variation within and between white clover varieties in their preference for strains of Rhizobium trifolii. Ann Appl Biol 92:221–228. doi: 10.1111/j.1744-7348.1979.tb03867.x [DOI] [Google Scholar]
- 176. Heath KD, Tiffin P. 2007. Context dependence in the coevolution of plant and rhizobial mutualists. Proc Biol Sci 274:1905–1912. doi: 10.1098/rspb.2007.0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. 2010. Host control over infection and proliferation of a cheater symbiont. J Evol Biol 23:1919–1927. doi: 10.1111/j.1420-9101.2010.02056.x [DOI] [PubMed] [Google Scholar]
- 178. Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs JL, Urbina M, Tausczik Y. 2006. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc Biol Sci 273:77–81. doi: 10.1098/rspb.2005.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Crook MB, Lindsay DP, Biggs MB, Bentley JS, Price JC, Clement SC, Clement MJ, Long SR, Griffitts JS. 2012. Rhizobial plasmids that cause impaired symbiotic nitrogen fixation and enhanced host invasion. MPMI 25:1026–1033. doi: 10.1094/MPMI-02-12-0052-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Boyle JA, Simonsen AK, Frederickson ME, Stinchcombe JR. 2021. Priority effects alter interaction outcomes in a legume-rhizobium mutualism. Proc R Soc B 288:20202753. doi: 10.1098/rspb.2020.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Pahua VJ, Stokes PJN, Hollowell AC, Regus JU, Gano-Cohen KA, Wendlandt CE, Quides KW, Lyu JY, Sachs JL. 2018. Fitness variation among host species and the paradox of ineffective rhizobia. J Evol Biol 31:599–610. doi: 10.1111/jeb.13249 [DOI] [PubMed] [Google Scholar]
- 182. Vuong HB, Thrall PH, Barrett LG, Shefferson R. 2017. Host species and environmental variation can influence rhizobial community composition. J Ecol 105:540–548. doi: 10.1111/1365-2745.12687 [DOI] [Google Scholar]
- 183. Van Cauwenberghe J, Lemaire B, Stefan A, Efrose R, Michiels J, Honnay O. 2016. Symbiont abundance is more important than pre-infection partner choice in a rhizobium–legume mutualism. Syst Appl Microbiol 39:345–349. doi: 10.1016/j.syapm.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 184. Westhoek A, Field E, Rehling F, Mulley G, Webb I, Poole PS, Turnbull LA. 2017. Policing the legume-rhizobium symbiosis: a critical test of partner choice. Sci Rep 7:1419. doi: 10.1038/s41598-017-01634-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Triplett EW, Sadowsky MJ. 1992. Genetics of competition for nodulation of legumes. Annu Rev Microbiol 46:399–428. doi: 10.1146/annurev.mi.46.100192.002151 [DOI] [PubMed] [Google Scholar]
- 186. Lardi M, de Campos SB, Purtschert G, Eberl L, Pessi G. 2017. Competition experiments for legume Infection Identify Burkholderia phymatum as a highly competitive β-Rhizobium. Front Microbiol 8:1527. doi: 10.3389/fmicb.2017.01527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Triplett EW, Barta TM. 1987. Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol 85:335–342. doi: 10.1104/pp.85.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Weaver RW, Frederick LR. 1974. Effect of inoculum rate on competitive nodulation of Glycine max L. Merrill. I. greenhouse studies. Agron J 66:229–232. doi: 10.2134/agronj1974.00021962006600020014x [DOI] [Google Scholar]
- 189. Pinochet X, Arnaud F, Cleyet-Marel JC. 1993. Competition for nodule occupancy of introduced bradyrhizobium japonicum strain SMGS1 in French soils already containing bradyrhizobium japonicum strain G49. Can. J. Microbiol 39:1022–1028. doi: 10.1139/m93-155 [DOI] [Google Scholar]
- 190. Wheatley RM, Ford BL, Li L, Aroney STN, Knights HE, Ledermann R, East AK, Ramachandran VK, Poole PS. 2020. Lifestyle adaptations of rhizobium from rhizosphere to symbiosis. Proc Natl Acad Sci U S A 117:23823–23834. doi: 10.1073/pnas.2009094117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Fields B, Friman VP. 2022. Microbial ECO-evolutionary dynamics in the plant rhizosphere. Curr Opin Microbiol 68:102153. doi: 10.1016/j.mib.2022.102153 [DOI] [PubMed] [Google Scholar]
- 192. Burghardt LT, GC . 2023. The evolutionary ecology of rhizobia: multiple facets of competition before, during, and after symbiosis with legumes. Curr Opin Microbiol 72:102281. doi: 10.1016/j.mib.2023.102281 [DOI] [PubMed] [Google Scholar]
- 193. Appunu C, Dhar B. 2008. Isolation and symbiotic characteristics of two Tn5-derived phage-resistant bradyrhizobium japonicum strains that nodulate soybean. Curr Microbiol 57:212–217. doi: 10.1007/s00284-008-9176-y [DOI] [PubMed] [Google Scholar]
- 194. Hatfull GF. 2008. Bacteriophage genomics. Curr Opin Microbiol 11:447–453. doi: 10.1016/j.mib.2008.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. 2008. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol 159:349–357. doi: 10.1016/j.resmic.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 196. Turska-Szewczuk A, Pietras H, Pawelec J, Mazur A, Russa R. 2010. Morphology and general characteristics of bacteriophages infectious to robinia pseudoacacia mesorhizobia. Curr Microbiol 61:315–321. doi: 10.1007/s00284-010-9613-6 [DOI] [PubMed] [Google Scholar]
- 197. Wdowiak S, Małek W, Grzadka M. 2000. Morphology and general characteristics of phages specific for astragalus cicer rhizobia. Curr Microbiol 40:110–113. doi: 10.1007/s002849910021 [DOI] [PubMed] [Google Scholar]
- 198. Allam Y, Amin G, Fattah GA, Hashem A, Egamberdieva D, Abd_Allah EF, El–Didamony G. 2022. The use of rhizobium and mycorrhizae in soil containing rhizobiophage to improve growth and nodulation of cowpea. Sci Agric 79:e20210110. doi: 10.1590/1678-992x-2021-0110 [DOI] [Google Scholar]
- 199. Liu JJ, Liu ZX, Yu H, Yao Q, Yu ZH, Wang GH. 2019. Biological characteristics of Bacteriophages infecting three Typic Rhizobia of legume. Ying yong sheng tai xue bao = J Appl Ecol 30:2775–2782. doi: 10.13287/j.1001-9332.201908.029 [DOI] [PubMed] [Google Scholar]
- 200. Vidor C, Miller RH. 1980. Relative saprophytic competence of rhizobium japonicum strains in soils as determined by the quantitative fluorescent antibody technique (FA). Soil Biol Biochem 12:483–487. doi: 10.1016/0038-0717(80)90084-X [DOI] [Google Scholar]
- 201. Schwinghamer EA, Brockwell J. 1978. Competitive advantage of bacteriocin and phage-producing strains of Rhizobium trifolii in mixed culture. Soil Biol Biochem 10:383–387. doi: 10.1016/0038-0717(78)90062-7 [DOI] [Google Scholar]
- 202. Hashem FM, Angle JS. 1990. Rhizobiophage effects on nodulation, nitrogen fixation, and yield of field-grown soybeans (Glycine max L. Merr.). Biol Fertil Soils 9:330–334. doi: 10.1007/BF00634110 [DOI] [Google Scholar]
- 203. Hashem F. 1988. Rhizobiophage effects on bradyrhizobium japonicum, nodulation and soybean growth. Soil Biol Biochem 20:69–73. doi: 10.1016/0038-0717(88)90128-9 [DOI] [Google Scholar]
- 204. Kankila J, Lindström K. 1994. Host range, morphology and DNA restriction patterns of bacteriophage isolates infecting rhizobium leguminosarum bv. trifolii. Soil Biol Biochem 26:429–437. doi: 10.1016/0038-0717(94)90174-0 [DOI] [Google Scholar]
- 205. Evans J, Barnet YM, Vincent JM. 1979. Effect of a bacteriophage on colonisation and nodulation of clover roots by paired strains of Rhizobium trifolii. Can J Microbiol 25:974–978. doi: 10.1139/m79-149 [DOI] [PubMed] [Google Scholar]
- 206. Barnet YM. 1980. The effect of rhizobiophages on populations of Rhizobium trifolii in the root zone of clover plants. Can J Microbiol 26:572–576. doi: 10.1139/m80-101 [DOI] [PubMed] [Google Scholar]
- 207. Eklund E. 1984. Behaviour of selected Finnish red clover rhizobium inoculants under field conditions, p 336. In Veeger C, Newton WE (ed), In advances in nitrogen fixation research. Nijhoff, The Hague and Pudoc, Wageningen. doi: 10.1007/978-94-009-6923-0 [DOI] [Google Scholar]
- 208. Basit HA, Angle JS, Salem S, Gewaily EM. 1992. Phage coating of soybean seed reduces nodulation by indigenous soil bradyrhizobia. Can J Microbiol 38:1264–1269. doi: 10.1139/m92-208 [DOI] [Google Scholar]
- 209. Ahmad MH, Morgan V. 1994. Characterization of a cowpea (Vigna unguiculata) rhizobiophage and its effect on cowpea nodulation and growth. Biol Fert Soils 18:297–301. doi: 10.1007/BF00570632 [DOI] [Google Scholar]
- 210. Lafferty KD, Dobson AP, Kuris AM. 2006. Parasites dominate food web links. Proc Natl Acad Sci U S A 103:11211–11216. doi: 10.1073/pnas.0604755103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Liu R, Han G, Li Z, Cun S, Hao B, Zhang J, Liu X. 2022. Bacteriophage therapy in aquaculture: current status and future challenges. Folia Microbiol (Praha) 67:573–590. doi: 10.1007/s12223-022-00965-6 [DOI] [PubMed] [Google Scholar]
- 212. Brown SP, Le Chat L, De Paepe M, Taddei F. 2006. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol 16:2048–2052. doi: 10.1016/j.cub.2006.08.089 [DOI] [PubMed] [Google Scholar]
- 213. Burns N, James CE, Harrison E. 2015. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol Appl 8:346–351. doi: 10.1111/eva.12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Li XY, Lachnit T, Fraune S, Bosch TCG, Traulsen A, Sieber M. 2017. Temperate phages as self-replicating weapons in bacterial competition. J R Soc Interface 14:1–7. doi: 10.1098/rsif.2017.0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Mann NH, Cook A, Millard A, Bailey S, Clokie M. 2003. Bacterial Photosynthesis genes in a virus. Nature 424:741. doi: 10.1038/424741a [DOI] [PubMed] [Google Scholar]
- 216. Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe JA, Chisholm SW. 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U S A 108:E757–E764. doi: 10.1073/pnas.1102164108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Elton CS. 1958. The Ecology of invasions by animals and plants. Springer, Boston, MA. doi: 10.1007/978-1-4899-7214-9 [DOI] [Google Scholar]
- 218. Yang K, Wang X, Hou R, Lu C, Fan Z, Li J, Wang S, Xu Y, Shen Q, Friman VP, Wei Z. 2023. Rhizosphere phage communities drive soil suppressiveness to bacterial wilt disease. Microbiome 11:1–18. doi: 10.1186/s40168-023-01463-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Riley CV. 1893. Parasitic and Predaceous insects in applied Entomology. Govt. print. off, Washington. doi: 10.5962/bhl.title.22474 [DOI] [Google Scholar]
- 220. Callaway RM, Thelen GC, Rodriguez A, Holben WE. 2004. Soil biota and exotic plant invasion. Nature 427:731–733. doi: 10.1038/nature02322 [DOI] [PubMed] [Google Scholar]
- 221. Keane RM, Crawley MJ. 2002. Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol 17:164–170. doi: 10.1016/S0169-5347(02)02499-0 [DOI] [Google Scholar]
- 222. Meijer K, Schilthuizen M, Beukeboom L, Smit C. 2016. A review and meta-analysis of the enemy release hypothesis in plant-herbivorous insect systems. PeerJ 4:e2778. doi: 10.7717/peerj.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Blumenthal D, Mitchell CE, Pys P. 2009. Synergy between pathogen release and resource availability. Proc Natl Acad Sci USA 106:7899–7904. doi: 10.1073/pnas.081260710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Mitchell CE, Power AG. 2003. Release of invasive plants from fungal and viral pathogens. Nature 421:625–627. doi: 10.1038/nature01317 [DOI] [PubMed] [Google Scholar]
- 225. Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. 2007. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci 86:2536–2540. doi: 10.3382/ps.2007-00250 [DOI] [PubMed] [Google Scholar]
- 226. Diagne C, Granjon L, Gueye MS, Ndiaye A, Kane M, Niang Y, Tatard C, Brouat C. 2020. Association between temporal patterns in helminth assemblages and successful range expansion of exotic mus musculus domesticus in Senegal. Biol Invasions 22:3003–3016. doi: 10.1007/s10530-020-02304-7 [DOI] [Google Scholar]
- 227. Duffy MA, Hall SR. 2008. Selective Predation and rapid evolution can jointly dampen effects of virulent parasites on daphnia populations. Am Nat 171:499–510. doi: 10.1086/528998 [DOI] [PubMed] [Google Scholar]
- 228. Phillips BL, Kelehear C, Pizzatto L, Brown GP, Barton D, Shine R. 2010. Parasites and pathogens lag behind their host during periods of host range advance. Ecology 91:872–881. doi: 10.1890/09-0530.1 [DOI] [PubMed] [Google Scholar]
- 229. Colautti RI, Ricciardi A, Grigorovich IA, MacIsaac HJ. 2004. Is invasion success explained by the enemy release hypothesis. Ecol Letters 7:721–733. doi: 10.1111/j.1461-0248.2004.00616.x [DOI] [Google Scholar]
- 230. Strauss A, White A, Boots M, Perkins S. 2012. Invading with biological weapons: the importance of disease-mediated invasions. Funct Ecol 26:1249–1261. doi: 10.1111/1365-2435.12011 [DOI] [Google Scholar]
- 231. Kelly DW, Paterson RA, Townsend CR, Poulin R, Tompkins DM. 2009. Parasite spillback: a neglected concept in invasion ecology? Ecology 90:2047–2056. doi: 10.1890/08-1085.1 [DOI] [PubMed] [Google Scholar]
- 232. Flory SL, Clay K, Thrall P. 2013. Pathogen accumulation and long-term dynamics of plant invasions. J Ecol 101:607–613. doi: 10.1111/1365-2745.12078 [DOI] [Google Scholar]
- 233. Eppinga MB, Rietkerk M, Dekker SC, De Ruiter PC, Van der Putten WH, Van der Putten WH. 2006. Accumulation of local pathogens: a new hypothesis to explain exotic plant invasions. Oikos 114:168–176. doi: 10.1111/j.2006.0030-1299.14625.x [DOI] [Google Scholar]
- 234. Borremans B, Faust C, Manlove KR, Sokolow SH, Lloyd-Smith JO. 2019. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Phil Trans R Soc B 374:20180344. doi: 10.1098/rstb.2018.0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Daszak P, Cunningham AA, Hyatt AD. 2000. Emerging infectious diseases of wildlife - threats to biodiversity and human health. Science 287:443–449. doi: 10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- 236. Prenter J, Macneil C, Dick JTA, Dunn AM. 2004. Roles of parasites in animal invasions. Trends Ecol Evol 19:385–390. doi: 10.1016/j.tree.2004.05.002 [DOI] [PubMed] [Google Scholar]
- 237. Joo J, Gunny M, Cases M, Hudson P, Albert R, Harvill E. 2006. Bacteriophage-mediated competition in bordetella bacteria. Proc Biol Sci 273:1843–1848. doi: 10.1098/rspb.2006.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Tompkins DM, White AR, Boots M. 2003. Ecological replacement of native red squirrels by invasive greys driven by disease. Ecol Letters 6:189–196. doi: 10.1046/j.1461-0248.2003.00417.x [DOI] [Google Scholar]
- 239. Davies EV, James CE, Kukavica-Ibrulj I, Levesque RC, Brockhurst MA, Winstanley C. 2016. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J 10:2553–2555. doi: 10.1038/ismej.2016.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. You X, Kallies R, Kühn I, Schmidt M, Harms H, Chatzinotas A, Wick LY. 2022. Phage co-transport with hyphal-riding bacteria fuels bacterial invasion in a water-unsaturated microbial model system. ISME J 16:1275–1283. doi: 10.1038/s41396-021-01155-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Heineman RH, Bull JJ. 2007. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution 61:1695–1709. doi: 10.1111/j.1558-5646.2007.00132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 243. Hampton HG, Watson BNJ, Fineran PC. 2020. The arms race between bacteria and their phage foes. Nature 577:327–336. doi: 10.1038/s41586-019-1894-8 [DOI] [PubMed] [Google Scholar]
- 244. Rostøl JT, Marraffini L. 2019. Ph)Ighting phages: how bacteria resist their parasites. Cell Host Microbe 25:184–194. doi: 10.1016/j.chom.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Susskind MM, Botstein D, Wright A. 1974. Superinfection exclusion by P22 prophage in lysogens of salmonella typhimurium. III. failure of superinfecting phage DNA to enter sieA+ lysogens. Virology 62:350–366. doi: 10.1016/0042-6822(74)90398-5 [DOI] [PubMed] [Google Scholar]
- 246. Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 247. Krysiak-Baltyn K, Martin GJO, Gras SL. 2018. Computational modelling of large scale phage production using a two-stage batch process. Pharmaceuticals 11:1–14. doi: 10.3390/ph11020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Blazanin M, Turner PE. 2021. Community context matters for bacteria-phage ecology and evolution. ISME J 15:3119–3128. doi: 10.1038/s41396-021-01012-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Buckling A, Rainey PB. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci 269:931–936. doi: 10.1098/rspb.2001.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Common J, Morley D, Westra ER, van Houte S. 2019. CRISPR-CAS immunity leads to a coevolutionary arms race between Streptococcus thermophilus and lytic phage. Philos Trans R Soc Lond B Biol Sci 374:20180098. doi: 10.1098/rstb.2018.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Stern A, Sorek R. 2011. The phage-host arms-race: shaping the evolution of microbes. Bioessays 33:43–51. doi: 10.1002/bies.201000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Blower TR, Short FL, Fineran PC, Salmond GPC. 2012. Viral molecular mimicry circumvents abortive infection and suppresses bacterial suicide to make hosts permissive for replication. Bacteriophage 2:234–238. doi: 10.4161/bact.23830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Van Valen L. 1977. The red queen. Am Nat 111:809–810. doi: 10.1086/283213 [DOI] [Google Scholar]
- 254. Koskella B, Thompson JN, Preston GM, Buckling A. 2011. Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria. Am Nat 177:440–451. doi: 10.1086/658991 [DOI] [PubMed] [Google Scholar]
- 255. Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325:833. doi: 10.1126/science.1174173 [DOI] [PubMed] [Google Scholar]
- 256. Rybniker J, Kramme S, Small PL. 2006. Host range of 14 mycobacteriophages in mycobacterium ulcerans and seven other mycobacteria including mycobacterium tuberculosis - application for identification and susceptibility testing. J Med Microbiol 55:37–42. doi: 10.1099/jmm.0.46238-0 [DOI] [PubMed] [Google Scholar]
- 257. Loessner MJ, Calendar R. 2006. The listeria bacteriophages, p 593. In Calendar R (ed), The bacteriophages. Oxford University Press, New York. [Google Scholar]
- 258. Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Modlin RL, Hendrix RW, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics And Evolutionary Science Sea-Phages Program . 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi: 10.1016/j.virol.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Flores CO, Valverde S, Weitz JS. 2013. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. ISME J 7:520–532. doi: 10.1038/ismej.2012.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Wendling CC, Goehlich H, Roth O. 2018. The structure of temperate phage – bacteria infection networks changes with the phylogenetic distance of the host bacteria. Biol Lett 14:20180320. doi: 10.1098/rsbl.2018.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5:806–823. doi: 10.3390/v5030806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE. 2019. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27:51–63. doi: 10.1016/j.tim.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 263. Bohannan BJM, Kerr B, Jessup CM, Hughes JB, Sandvik G. 2002. Trade-offs and coexistence in microbial microcosms. Antonie van leeuwenhoek. Int J Gen Mol Microbiol 81:107–115. doi: 10.1023/A:1020585711378 [DOI] [PubMed] [Google Scholar]
- 264. Brockhurst MA, Buckling A, Rainey PB. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci 272:1385–1391. doi: 10.1098/rspb.2005.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Meaden S, Paszkiewicz K, Koskella B. 2015. The cost of phage resistance in a plant pathogenic bacterium is context-dependent. Evolution 69:1321–1328. doi: 10.1111/evo.12652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Wang J, Gao Y, Zhao F. 2016. Phage–bacteria interaction network in human oral microbiome. Environ Microbiol 18:2143–2158. doi: 10.1111/1462-2920.12923 [DOI] [PubMed] [Google Scholar]
- 267. Avrani S, Schwartz DA, Lindell D. 2012. Virus-host swinging party in the oceans. Mob Genet Elements 2:88–95. doi: 10.4161/mge.20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Keen EC. 2014. Tradeoffs in bacteriophage life histories. Bacteriophage 4:e28365. doi: 10.4161/bact.28365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Ford BE, Sun B, Carpino J, Chapler ES, Ching J, Choi Y, Jhun K, Kim JD, Lallos GG, Morgenstern R, Singh S, Theja S, Dennehy JJ, van Raaij MJ. 2014. Frequency and fitness consequences of bacteriophage 6 host range mutations. PLoS ONE 9:e113078. doi: 10.1371/journal.pone.0113078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Duffy S, Turner PE, Burch CL. 2006. Pleiotropic costs of niche expansion in the RNA bacteriophage Φ6. Genetics 172:751–757. doi: 10.1534/genetics.105.051136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Hall JPJ, Harrison E, Brockhurst MA. 2013. Viral host-adaptation: insights from evolution experiments with phages. Curr Opin Virol 3:572–577. doi: 10.1016/j.coviro.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 272. Molina F, Menor-Flores M, Fernández L, Vega-Rodríguez MA, García P. 2022. Systematic analysis of putative phage-phage interactions on minimum-sized phage cocktails. Sci Rep 12:1–12. doi: 10.1038/s41598-022-06422-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. 2008. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 62:1–11. doi: 10.1111/j.1558-5646.2007.00260.x [DOI] [PubMed] [Google Scholar]
- 274. Sant DG, Woods LC, Barr JJ, McDonald MJ. 2021. Host diversity slows bacteriophage adaptation by selecting generalists over specialists. Nat Ecol Evol 5:350–359. doi: 10.1038/s41559-020-01364-1 [DOI] [PubMed] [Google Scholar]
- 275. Takeuchi I, Osada K, Azam AH, Asakawa H, Miyanaga K, Tanji Y. 2016. The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal twort-like phages. Appl Environ Microbiol 82:5763–5774. doi: 10.1128/AEM.01385-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Vasquez I, Retamales J, Parra B, Machimbirike V, Robeson J, Santander J. 2023. Comparative genomics of a polyvalent escherichia-salmonella phage fp01 and in silico analysis of its receptor binding protein and conserved enterobacteriaceae phage receptor. Viruses 15:379. doi: 10.3390/v15020379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. 2011. Statistical structure of host – phage interactions. Proc Natl Acad Sci U S A 108:E288–E297. doi: 10.1073/pnas.1101595108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Kauffman KM, Chang WK, Brown JM, Hussain FA, Yang J, Polz MF, Kelly L. 2022. Resolving the structure of phage-bacteria interactions in the context of natural diversity. Nat Commun 13:372. doi: 10.1038/s41467-021-27583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. 2011. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog 7:e1002260. doi: 10.1371/journal.ppat.1002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Strobel HM, Horwitz EK, Meyer JR. 2022. Viral protein instability enhances host-range evolvability. PLoS Genet 18:e1010030. doi: 10.1371/journal.pgen.1010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Hall AR, Scanlan PD, Buckling A. 2011. Bacteria-phage coevolution and the emergence of generalist pathogens. Am Nat 177:44–53. doi: 10.1086/657441 [DOI] [PubMed] [Google Scholar]
- 282. Faillace CA, Morin PJ. 2016. Evolution alters the consequences of invasions in experimental communities. Nat Ecol Evol 1:13. doi: 10.1038/s41559-016-0013 [DOI] [PubMed] [Google Scholar]
- 283. Flory SL, Alba C, Clay K, Holt RD, Goss EM. 2018. Emerging pathogens can suppress invaders and promote native species recovery. Biol Invasions 20:5–8. doi: 10.1007/s10530-017-1438-9 [DOI] [Google Scholar]
- 284. Bono LM, Gensel CL, Pfennig DW, Burch CL. 2013. Competition and the origins of novelty: experimental evolution of niche-width expansion in a virus. Biol Lett 9:20120616. doi: 10.1098/rsbl.2012.0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Thingstad TF, Lignell R. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13:19–27. doi: 10.3354/ame013019 [DOI] [Google Scholar]
- 286. Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, Felts B, Haynes M, Liu H, Lipson D, Mahaffy J, Martin-Cuadrado AB, Mira A, Nulton J, Pasić L, Rayhawk S, Rodriguez-Mueller J, Rodriguez-Valera F, Salamon P, Srinagesh S, Thingstad TF, Tran T, Thurber RV, Willner D, Youle M, Rohwer F. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J 4:739–751. doi: 10.1038/ismej.2010.1 [DOI] [PubMed] [Google Scholar]
- 287. Erkus O, de Jager VCL, Spus M, van Alen-Boerrigter IJ, van Rijswijck IMH, Hazelwood L, Janssen PWM, van Hijum SAFT, Kleerebezem M, Smid EJ. 2013. Multifactorial diversity sustains microbial community stability. ISME J 7:2126–2136. doi: 10.1038/ismej.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Erdman LW, Walker RH. 1927. Occurrence of the various groups of legume bacteria in Iowa soils. Proc Iowa Acad Sci 34:53–57. doi:https://scholarworks.uni.edu/pias/vol34/iss1/6/ [Google Scholar]
- 289. Hall AR, Scanlan PD, Morgan AD, Buckling A. 2011. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett 14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x [DOI] [PubMed] [Google Scholar]
- 290. Porter SS, Faber-Hammond J, Montoya AP, Friesen ML, Sackos C. 2019. Dynamic genomic architecture of mutualistic cooperation in a wild population of mesorhizobium. ISME J 13:301–315. doi: 10.1038/s41396-018-0266-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Cavassim MIA, Moeskjær S, Moslemi C, Fields B, Bachmann A, Vilhjálmsson BJ, Schierup MH, W Young JP, Andersen SU. 2020. Symbiosis genes show a unique pattern of introgression and selection within a rhizobium leguminosarum species complex. Microb Genom 6:e000351. doi: 10.1099/mgen.0.000351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Leibold MA. 1996. A graphical model of Keystone predators in food webs: trophic regulation of abundance, incidence, and diversity patterns in communities. The American Naturalist 147:784–812. doi: 10.1086/285879 [DOI] [Google Scholar]
- 293. Huston MA. 1997. Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 110:449–460. doi: 10.1007/s004420050180 [DOI] [PubMed] [Google Scholar]
- 294. Ali Y, Koberg S, Heßner S, Sun X, Rabe B, Back A, Neve H, Heller KJ. 2014. Temperate streptococcus thermophilus phages expressing superinfection exclusion proteins of the Ltp type. Front Microbiol 5:98. doi: 10.3389/fmicb.2014.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Adler BA, Hessler T, Cress BF, Lahiri A, Mutalik VK, Barrangou R, Banfield J, Doudna JA. 2022. Broad-spectrum CRISPR-Cas13a enables efficient Phage genome editing. Nat Microbiol 7:1967–1979. doi: 10.1038/s41564-022-01258-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Jiang S, Chen K, Wang Y, Zhang Y, Tang Y, Huang W, Xiong X, Chen S, Chen C, Wang L. 2023. A DNA phosphorothioation-based Dnd defense system provides resistance against various phages and is compatible with the Ssp defense system. mBio, June:e0093323. doi: 10.1128/mbio.00933-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, Yaakov G, Doron S, Sorek R. 2018. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3:90–98. doi: 10.1038/s41564-017-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial Genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Millman A, Bernheim A, Stokar-Avihail A, Fedorenko T, Voichek M, Leavitt A, Oppenheimer-Shaanan Y, Sorek R. 2020. Bacterial retrons function in anti-phage defense. Cell 183:1551–1561. doi: 10.1016/j.cell.2020.09.065 [DOI] [PubMed] [Google Scholar]
- 300. Pires DP, Melo LDR, Vilas Boas D, Sillankorva S, Azeredo J. 2017. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol 39:48–56. doi: 10.1016/j.mib.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 301. Pires DP, Costa AR, Pinto G, Meneses L, Azeredo J. 2020. Current challenges and future opportunities of phage therapy. FEMS Microbiol Rev 44:684–700. doi: 10.1093/femsre/fuaa017 [DOI] [PubMed] [Google Scholar]
- 302. Sulakvelidze A, Alavidze Z, Morris JGJ. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Wright RCT, Friman VP, Smith MCM, Brockhurst MA. 2021. Functional diversity increases the efficacy of phage combinations. Microbiology 167:001110. doi: 10.1099/mic.0.001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Harada LK, Silva EC, Campos WF, Del Fiol FS, Vila M, Dąbrowska K, Krylov VN, Balcão VM. 2018. Biotechnological applications of bacteriophages: state of the art. Microbiol Res 212–213:38–58. doi: 10.1016/j.micres.2018.04.007 [DOI] [PubMed] [Google Scholar]
- 305. Sieiro C, Areal-Hermida L, Pichardo-Gallardo Á, Almuiña-González R, de Miguel T, Sánchez S, Sánchez-Pérez Á, Villa TG. 2020. A hundred years of bacteriophages: can phages replace antibiotics in agriculture and aquaculture? Antibiotics 9:1–30. doi: 10.3390/antibiotics9080493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Yang Y, Shen W, Zhong Q, Chen Q, He X, Baker JL, Xiong K, Jin X, Wang J, Hu F, Le S. 2020. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front Microbiol 11:1–12. doi: 10.3389/fmicb.2020.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Meaden S, Koskella B. 2013. Exploring the risks of phage application in the environment. Front Microbiol 4:358. doi: 10.3389/fmicb.2013.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Zhao Y, Ye M, Zhang X, Sun M, Zhang Z, Chao H, Huang D, Wan J, Zhang S, Jiang X, Sun D, Yuan Y, Hu F. 2019. Comparing polyvalent bacteriophage and bacteriophage cocktails for controlling antibiotic-resistant bacteria in soil-plant system. Sci Total Environ 657:918–925. doi: 10.1016/j.scitotenv.2018.11.457 [DOI] [PubMed] [Google Scholar]
- 309. Betts Alex, Gifford DR, MacLean RC, King KC. 2016. Parasite diversity drives rapid host dynamics and evolution of resistance in a bacteria-phage system. Evolution 70:969–978. doi: 10.1111/evo.12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Betts A, Gray C, Zelek M, MacLean RC, King KC. 2018. High parasite diversity accelerates host adaptation and diversification. Science 360:907–911. doi: 10.1126/science.aam9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored ‘“rare biosphere’. Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Lynch MDJ, Neufeld JD. 2015. Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13:217–229. doi: 10.1038/nrmicro3400 [DOI] [PubMed] [Google Scholar]
- 313. Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x [DOI] [PubMed] [Google Scholar]
- 314. Nei M, Maruyama T, Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x [DOI] [PubMed] [Google Scholar]
- 315. Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG. 2001. The population biology of invasive species. Annu Rev Ecol Syst 32:305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037 [DOI] [Google Scholar]
- 316. López R, Asensio C, Gilbertson RL. 2006. Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204–1213. doi: 10.1094/PHYTO-96-1204 [DOI] [PubMed] [Google Scholar]
- 317. Ghabooli S, Shiganova TA, Zhan A, Cristescu ME, Eghtesadi-Araghi P, MacIsaac HJ. 2011. Multiple introductions and invasion pathways for the invasive ctenophore mnemiopsis leidyi in Eurasia. Biol Invasions 13:679–690. doi: 10.1007/s10530-010-9859-8 [DOI] [Google Scholar]
- 318. Chown SL, Hodgins KA, Griffin PC, Oakeshott JG, Byrne M, Hoffmann AA. 2015. Biological invasions, climate change and genomics. Evol Appl 8:23–46. doi: 10.1111/eva.12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Klonowska A, Chaintreuil C, Tisseyre P, Miché L, Melkonian R, Ducousso M, Laguerre G, Brunel B, Moulin L. 2012. Biodiversity of mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in new caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol 81:618–635. doi: 10.1111/j.1574-6941.2012.01393.x [DOI] [PubMed] [Google Scholar]
- 320. McGinn KJ, van der Putten WH, Duncan RP, Shelby N, Weser C, Hulme PE. 2016. Trifolium species associate with a similar richness of soil-borne mutualists in their introduced and native ranges. J. Biogeogr 43:944–954. doi: 10.1111/jbi.12690 [DOI] [Google Scholar]
- 321. Evans TJ, Ind A, Komitopoulou E, Salmond GPC. 2010. Phage-selected lipopolysaccharide mutants of pectobacterium atrosepticum exhibit different impacts on virulence. J Appl Microbiol 109:505–514. doi: 10.1111/j.1365-2672.2010.04669.x [DOI] [PubMed] [Google Scholar]
- 322. Kleczkowska J. 1950. A study of phage-resistant mutants of Rhizobium trifolii. J Gen Microbiol 4:298–310. doi: 10.1099/00221287-4-3-298 [DOI] [PubMed] [Google Scholar]
- 323. Stacey G, Pocratsky LA, Puvanesarajah V. 1984. Bacteriophage that can distinguish between wild-type rhizobium japonicum and a non-nodulating mutant. Appl Environ Microbiol 48:68–72. doi: 10.1128/aem.48.1.68-72.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Stacey G, Paau AS, Noel KD, Maier RJ, Silver LE, Brill WJ. 1982. Mutants of rhizobium japonicum defective in nodulation. Arch Microbiol 132:219–224. doi: 10.1007/BF00407954 [DOI] [Google Scholar]
- 325. Bourassa DV, Kannenberg EL, Sherrier DJ, Buhr RJ, Carlson RW. 2017. The lipopolysaccharide lipid a long-chain fatty acid is important for rhizobium leguminosarum growth and stress adaptation in free-living and nodule environments. Mol Plant Microbe Interact 30:161–175. doi: 10.1094/MPMI-11-16-0230-R [DOI] [PubMed] [Google Scholar]
- 326. Thoms D, Liang Y, Haney CH. 2021. Maintaining symbiotic homeostasis: how do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol Plant Microbe Interact 34:462–469. doi: 10.1094/MPMI-11-20-0318-FI [DOI] [PubMed] [Google Scholar]
- 327. Mishra A, Dhar B, Singh RM. 2005. Symbiotic behaviour of phage-resistant mutants of pigeonpea [Cajanus cajan (L.) cv. BaharJ rhizobial strain IHP-195 with the main and alternate hosts. Indian J Genet Plant Breed 65:131–132. [Google Scholar]
- 328. Anand A, Jaiswal SK, Dhar B, Vaishampayan A. 2012. Surviving and thriving in terms of symbiotic performance of antibiotic and phage-resistant mutants of bradyrhizobium of soybean. Curr Microbiol 65:390–397. doi: 10.1007/s00284-012-0166-8 [DOI] [PubMed] [Google Scholar]
- 329. Gupta D, Ramkrishna K. 2001. Symbiotic properties of phage resistant mutants of rhizobium sp. of Vigna aconitifolia. Ann Arid Zo 40:179–184. [Google Scholar]
- 330. Bruch CW, Allen ON. 1955. Description of two bacteriophages active against lotus rhizobia. Soil Sci Soc Am J 19:175–179. doi: 10.2136/sssaj1955.03615995001900020016x [DOI] [Google Scholar]