Abstract
Drought is a major abiotic stress affecting all levels of plant organization and, in particular, leaf elongation. Several experiments were designed to study the effect of water deficits on maize (Zea mays) leaves at the protein level by taking into account the reduction of leaf elongation. Proteomic analyses of growing maize leaves allowed us to show that two isoforms of caffeic acid/5-hydroxyferulic 3-O-methyltransferase (COMT) accumulated mostly at 10 to 20 cm from the leaf point of insertion and that drought resulted in a shift of this region of maximal accumulation toward basal regions. We showed that this shift was due to the combined effect of reductions in growth and in total amounts of COMT. Several other enzymes involved in lignin and/or flavonoid synthesis (caffeoyl-CoA 3-O-methyltransferase, phenylalanine ammonia lyase, methylenetetrahydrofolate reductase, and several isoforms of S-adenosyl-l-methionine synthase and methionine synthase) were highly correlated with COMT, reinforcing the hypothesis that the zone of maximal accumulation corresponds to a zone of lignification. According to the accumulation profiles of the enzymes, lignification increases in leaves of control plants when their growth decreases before reaching their final size. Lignin levels analyzed by thioacidolysis confirmed that lignin is synthesized in the region where we observed the maximal accumulation of these enzymes. Consistent with the levels of these enzymes, we found that the lignin level was lower in leaves of plants subjected to water deficit than in those of well-watered plants.
Drought is one of the major environmental factors decreasing plant productivity. It affects various metabolic processes but has scarcely been related to lignin biosynthesis. Lignin is a major polymer of plant cell walls that confers hydrophobicity and mechanical strength to the walls of conducting vessels to withstand the negative pressure generated by transpiration. Its variability has been comprehensively reviewed (Boudet et al., 1995; Campbell and Sederoff, 1996; Baucher et al., 1998, 2003; Whetten et al., 1998; Anterola and Lewis, 2002; Boerjan et al., 2003). Lignin is composed of p-hydroxyphenyl (H), guaiacyl (G), and/or syringyl (S) units, the proportions of which vary according to botanical and cytological criteria (Lewis and Yamamoto, 1990). In addition, grass lignin contains p-coumaric acid (pCA) and ferulic acid (FA; Higuchi, 1997). The complex pathway of lignin biosynthesis was recently rewritten by Humphreys and Chapple (2002), and the in vivo enzymatic reactions were highlighted. Caffeic acid/5-hydroxyferulic 3-O-methyltransferase (COMT) was initially thought to be a bifunctional enzyme that used both caffeic and 5-hydroxyferulic acids as substrates (Davin and Lewis, 1992). However, its activity toward 5-hydroxyconiferyl aldehyde and alcohol was demonstrated by recent transgenic studies on Arabidopsis (Arabidopsis thaliana; Humphreys and Chapple, 2002), sweetgum (Liquidambar styraciflua; Osakabe et al., 1999), alfalfa (Medicago sativa; Parvathi et al., 2001), and poplar (Populus spp.; Li et al., 2000). As 5-hydroxyconiferyl aldehyde was found to be both the preferred substrate and the inhibitor of caffeate and 5-hydroxyferulate methylation (Zubieta et al., 2001), Li et al. (2000) suggested that COMT be renamed 5-hydroxyconiferyl aldehyde O-methyltransferase.
In maize (Zea mays), a single gene codes for COMT protein (Collazo et al., 1992). Another maize gene, ZRP4, showed sequence similarities with various plant and animal O-methyltransferases (OMTs) but shared only 44% identity with maize COMT. It was proposed to be involved in suberin biosynthesis (Held et al., 1993). The brown midrib 3 (bm3) natural mutation affects the structure and expression of the COMT gene in maize. This mutation induces a reddish-brown pigmentation in the leaf midrib (Neuffer et al., 1968) and affects lignin content and structure (Vignols et al., 1995). Recently, an antisense strategy was applied to down-regulate COMT gene expression in maize. Similar to the bm3 mutants, but in an attenuated manner, COMT-deficient transgenic lines displayed altered lignin content and composition (Piquemal et al., 2002).
The formation of stress lignin has originally been reported under pathogen attack (Vance et al., 1980). Since then, the activation of the phenylpropanoid metabolism has been observed under various stress conditions (Dixon and Paiva, 1995). The expression of COMT and caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) genes was induced in response to infection by tobacco mosaic virus in tobacco (Nicotiana tabacum; Pellegrini et al., 1993; Maury et al., 1999). The induction of CCoAOMT also was observed during the disease resistance response of parsley (Petroselinum crispum; Pakusch et al., 1989; Schmitt et al., 1991). Furthermore, a putative OMT gene from barley was induced by fungal pathogens and UV light (Gregersen et al., 1994). Finally, different sequences of the COMT II promoter were responsible for gene induction by methyl jasmonic acid, UV light, tobacco mosaic virus, or wounding (Toquin et al., 2003).
In a previous proteomic study on maize leaves, we found that COMT accumulated at the basal regions of leaves subjected to drought during their growth (Riccardi et al., 1998). The leaf meristem is located at the base, and there is a gradient of tissue age along the maize leaf (Ben-Haj-Salah and Tardieu, 1995). Drought causes a reduction of growth due to a reduction of meristematic activity (Ben-Haj-Salah and Tardieu, 1997). As a consequence, tissues of the same age are closer to the leaf point of insertion in water-stressed leaves compared to control leaves. It is thus difficult to decipher the respective effects of age and of cellular response to drought. To our knowledge, no study has described the position or age at which maize leaf cells undertake lignification under normal development. In this study, we analyzed segments evenly sampled along the leaf at different stages of development to separate direct effects of water deficit from side effects related to the decrease of growth. Based on the profiles of COMT and of other enzymes involved in lignin synthesis, we drew a hypothesis on the localization of lignin synthesis in leaves and on the effect of drought on lignification. This hypothesis was then further confirmed by analyzing profiles of lignin along the leaf.
RESULTS
Identification of Two Isoforms of COMT Encoded by the Same COMT Gene
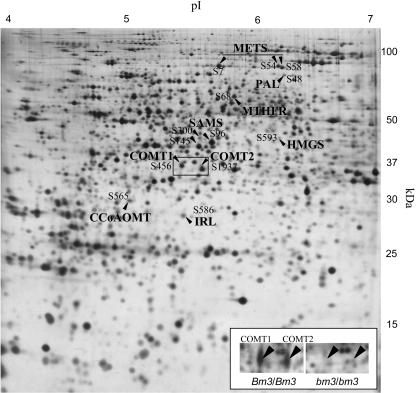
We previously identified a COMT protein in the base of maize leaves (spot 383 in Riccardi et al., 1998; renamed S456 in this study). The relative amount of this protein increased when plants were subjected to water deficit. To check the COMT identity, we compared line Io to a near-isogenic line bearing the bm3 mutation (bm3/bm3; Vignols et al., 1995). Spot S456 was absent in the isogenic lines bearing the bm3 mutation. Additionally, we also observed the absence of another spot with an apparent molecular mass of 37 kD and a pI slightly more acidic (Fig. 1, inset). The relative amount of this protein also increased significantly in leaves subjected to water deficit in both F2 and Io inbred lines (spot 91 in Riccardi et al., 1998; renamed S1937 in this study). An analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) provided evidence that it was also a COMT. We named the more acidic protein COMT1 (S456, pI ≈ 5.4) and the less acidic one COMT2 (S1937, pI ≈ 5.6). Because the two isoforms disappeared in a line known to bear a deficient COMT gene (Vignols et al., 1995) and were identified as COMT according to LC-MS/MS analysis, they most likely correspond to two different products of the same gene, differing by a posttranslational modification, the nature of which remains to be elucidated.
Figure 1.
Identification of two isoforms of COMT in maize leaves. Arrows indicate the proteins identified by LC-MS/MS and correlated with COMT1, CCoAOMT, and PAL on a silver-stained 2D gel of leaf proteins from the F2 inbred line of maize. The inset shows an enlargement of the framed region in the Io wild-type line (Bm3/Bm3) and in the near-isogenic line bearing the bm3 mutation (bm3/bm3). COMT1 and COMT2 are present in Bm3/Bm3 and absent in bm3/bm3.
Shift of COMT Peaks toward the Base of Water-Stressed Maize Leaves and Relation with Tissue Age
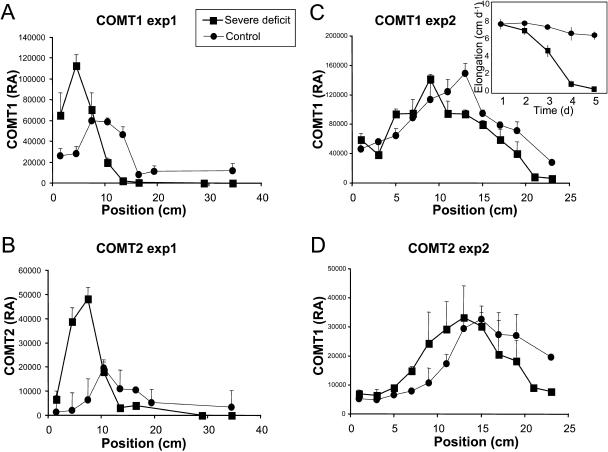
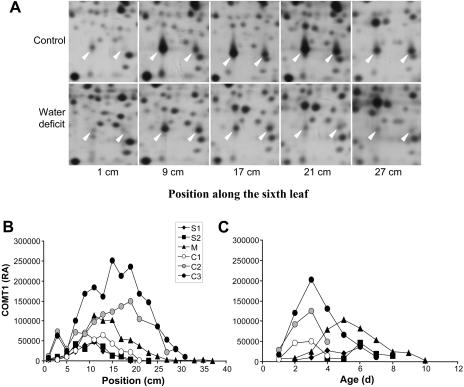
We next examined expression profiles at the protein level of both COMTs in leaves of water-stressed plants. We first grew Io plants in perlite (experiment 1) as was done previously by Riccardi et al. (1998). We found that COMT1 accumulated maximally at an 8- to 14-cm distance from the point of insertion of control leaves (Fig. 2A). When plants were not watered for 10 d, the maximal amount of COMT1 was located at 2 to 8 cm, indicating a shift toward the point of insertion. COMT2 accumulated at a slightly higher position than COMT1 in both conditions, the maximal accumulation being at 10 to 16 and 4 to 10 cm in control and water-stressed leaves, respectively (Fig. 2B).
Figure 2.
COMT1 and COMT2 profiles in response to water deficit. A and B, Relative amounts of COMT1 and COMT2 along leaf 6 of Io plants in experiment 1. C and D, Relative amounts of COMT1 and COMT2 along leaf 6 of Io plants in experiment 2 at day 3. The inset in C shows the elongation rate of leaf 6 in experiment 2. Vertical bars show se (n = 3 in experiment 1; n = 2 in experiment 2). RA, Relative amount.
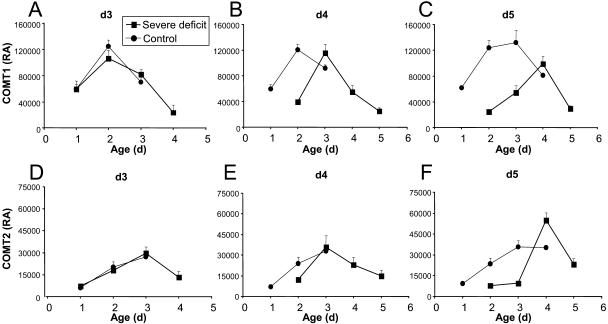
To look at the effect of water deficit on the profiles of COMT accumulation as a function of time and leaf growth, a second experiment was then performed in which leaf growth was measured daily and samples were harvested after 3 to 5 d of water deprivation. Io plants were cultivated in a soil-sand mixture to investigate the effect of delayed water deficit. The growth of leaf 6 stopped after 5 d without watering, whereas it stayed at a constant rate in control plants (Fig. 2C, inset). After 3 d of no watering (day 3), the region of maximal accumulation of COMT1 and COMT2 shifted toward the base of the leaf in response to drought (Fig. 2, C and D). In the following days (days 4 and 5), no notable quantitative or position change was observed in control leaves for COMT1, whereas the position of maximal accumulation of COMT2 moved 2 cm down in drought-stressed leaves and its quantity increased. Because of the reduction of cell division and elongation in the elongation zone at the base of the leaf under drought conditions, tissues at the same position have different ages in control and water-stressed leaves. The shift of maximal COMT accumulation could therefore be due to a drought-induced growth reduction. We thus expressed the relative amount of COMTs as a function of computed tissue ages (Fig. 3). In control leaves harvested on days 3 and 4, COMT1 mostly accumulated in 2-d-old tissues. On day 5, maximal expression of COMT1 was mostly distributed between 2- and 3-d-old tissues. In water-stressed leaves, maximal accumulation of COMT1 was found in 2-, 3-, and 4-d-old tissues at days 3, 4, and 5, respectively (Fig. 3, A–C). COMT2 profiles showed no maximum in well-watered leaves, probably because the analyzed segments were not old enough. In water-stressed leaves, the maximal accumulation of COMT2 was observed in 3-d-old tissues at both days 3 and 4 (Fig. 3, D and E) and in 4-d-old tissues at day 5 (Fig. 3F). Taken together, these results showed that the age of maximal accumulation was delayed at least for COMT1 in drought-stressed leaves. It was also observed that the maximal accumulation of COMT2 occurred at least 1 d after the maximal accumulation of COMT1 in control leaves.
Figure 3.
Relative amounts of COMT isoforms in function of tissue age in Io leaves of experiment 2. Relative amounts of COMT1 in leaves harvested at day 3 (A), day 4 (B), and day 5 (C) are shown. Relative amounts of COMT2 in leaves harvested at day 3 (D), day 4 (E), and day 5 (F) also are shown. Vertical bars show se (n = 4 to 11). RA, Relative amount.
Variation of Amount of COMT with Leaf Development
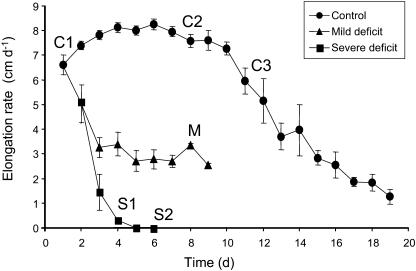
A third experiment was performed to compare the effect of water deficit with the effect of leaf growth decrease in the absence of stress and to study two levels of water deficit. F2 plants were cultivated in a soil:compost (1:1) mixture. Harvesting dates did not correspond to treatment duration as in experiment 2 (number of days) but to particular stages of leaf growth. Control leaves were collected at three different stages: C1 and C2 at the beginning and the end of the period of maximal growth (8 cm d−1), respectively, and C3 during growth decrease. Mildly drought-stressed leaves (M) were harvested when growth reduction had stabilized (approximately 4 cm d−1). Severely water-stressed leaves were collected at two stages: S1 just before growth stopped (<1 cm d−1) and S2 2 d later. Plants reached C1, C2, C3, M, S1, and S2 stages at 1, 8, 11, 8, 4, and 6 d, respectively (Fig. 4). Under control conditions (C1 to C3), COMT1 profiles showed a steady increase during leaf development as well as a shift of maximal expression levels toward the tip (Fig. 5, A and B). In C1 leaves, maximal accumulation occurred from 9 to 15 cm from the point of insertion. In C3 leaves, the area of maximal expression was located farther from the base (15–19 cm from the point of insertion), and the relative amount reached its highest level. Under mild water-deficit conditions (M), the strongest accumulation of COMT1 was observed at 11 to 15 cm from the point of insertion, and its relative amount was lower than that observed for C2 or C3. Under severe water deficit (S1 and S2), the relative COMT1 expression level was considerably reduced, as compared with controls, and its profile was very similar in S1 and S2 leaves (Fig. 5B). It showed a maximal accumulation at 11 cm.
Figure 4.
Elongation rate of leaf 6 of F2 plants in experiment 3. Average days of harvesting are indicated for controls (C1 to C3), and plants subjected to mild (M) and severe (S1 and S2) water deficit. Vertical bars show se (n = 3).
Figure 5.
Variation of relative amounts of COMT with leaf development. A, Silver-stained 2D gel of leaf proteins from the F2 inbred line in experiment 3. Arrowheads indicate COMT1 (left) and COMT2 (right). B, Relative amount of COMT1 along the leaves of F2 plants in experiment 3. C, Relative amount of COMT1 in function of tissue age of F2 leaves in experiment 3.
Quantification was less accurate for COMT2 than for COMT1 because of the presence of a close spot (identified as a malate dehydrogenase) that occasionally merged with it (Fig. 5A). Nevertheless, several observations could be made. Similar to COMT1, the level of accumulation and the position of maximal accumulation of COMT2 were lower in C1 than in C2 and C3, but there was no clear decrease of the amount of COMT2 in response to water deficit. As observed in experiments 1 and 2, the region of maximal accumulation of COMT2 was slightly higher in the leaf than that of COMT1, suggesting a timing delay between COMT1 and COMT2 accumulation.
For both COMTs, the position of maximal accumulation under mild water deficit was intermediate between control and severe water-deficit treatments. COMT1 was mostly expressed in 3-d-old tissues of control leaves, whatever the stage, in 5-d-old tissues of mildly stressed leaves (M), and in 6-d-old tissues of severely water-stressed leaves (S1 and S2; Fig. 5C). In agreement with experiment 2, tissues showing maximal accumulation were older in water-stressed leaves, although they were at a lower position than in control leaves.
Phenylpropanoid Metabolism
More than 300 of the proteins detected on two-dimensional (2D) gels were identified by LC-MS/MS (D. Vincent, H. Corti, M. Davanture, L. Negroni, and M. Zivy, unpublished data). Two of them were directly related to lignin biosynthesis: a CCoAOMT and a Phe ammonia lyase (PAL; EC 4.3.1.5), which controls the first step of the phenylpropanoid pathway (Fig. 1A). COMT1, CCoAOMT, and PAL showed similar profiles (data not shown), the correlation between their relative amounts in the different samples varying between 0.59 and 0.77.
In order to find other proteins behaving similarly, we looked for proteins highly correlated with these three proteins (Fig. 1; Table I; Supplemental Fig. 1). Among the highest correlated proteins (r > 0.60), we found three S-adenosyl-l-Met (SAM) synthases (SAMS; EC 2.5.1.6). SAMS catalyzes the conversion of ATP and l-Met into SAM, which is a donor of the methyl group known to be involved in methylation occurring in monolignol and/or flavonoid biosynthesis, as those catalyzed by COMT. Among the other highly correlated proteins were all the identified Met synthases (METS; EC 2.1.1.14), which produce Met by transferring a methyl group from 5-methyltetrahydrafolate (5-CH3-THF) to homo-Cys (hCys), and a methylenetetrahydrofolate reductase (MTHFR; EC 1.5.1.20 and 1.7.99.5), which generates 5-CH3-THF (Roje et al., 1999). Thus, MTHFR and METS contribute to the production of Met, from which SAM is generated. The other identified proteins found to be highly correlated with COMT, CCoAOMT, and PAL were an isoflavone reductase-like protein (IRL; EC 1.3.1.-) and a hydroxymethylglutaryl-CoA synthase (HMGS; EC 4.1.3.5) involved in the synthesis of mevalonate (Alex et al., 2000).
Table I.
Identified proteins highly correlated with COMT1, CCoAOMT, and PAL
Only proteins showing a correlation coefficient greater than 0.60 with at least one of these three enzymes are shown.
| Spot | Correlation to COMT1 | Correlation to CCoAOMT | Correlation to PAL | |
|---|---|---|---|---|
| COMT1 | S456 | 1.00 | 0.60 | 0.77 |
| CCoAOMT | S565 | 0.60 | 1.00 | 0.59 |
| PAL | S48 | 0.77 | 0.59 | 1.00 |
| METS | S7 | 0.56 | 0.45 | 0.60 |
| METS | S54 | 0.71 | 0.56 | 0.91 |
| METS | S58 | 0.72 | 0.52 | 0.82 |
| MTHR | S68 | 0.60 | 0.55 | 0.57 |
| SAMS | S96 | 0.78 | 0.60 | 0.82 |
| SAMS | S145 | 0.77 | 0.42 | 0.71 |
| SAMS | S300 | 0.64 | 0.55 | 0.61 |
| IRL | S586 | 0.83 | 0.63 | 0.70 |
| HMGS | S593 | 0.61 | 0.35 | 0.64 |
Relation between COMT and Lignification Profiles
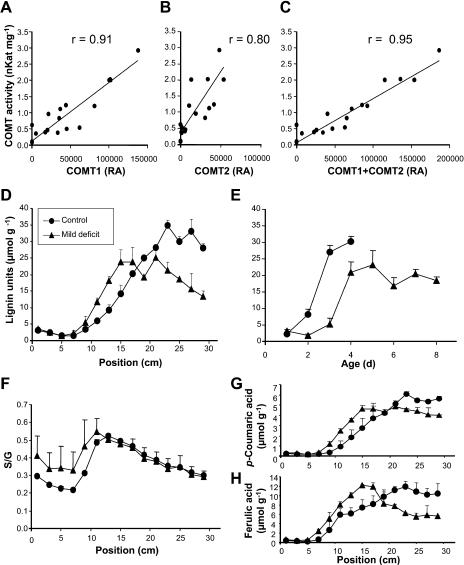
To check whether the variations in relative amounts of COMT determined by proteomic analysis could be correlated with changes in its activity, in vitro measurements of COMT activity were carried out on samples of leaves used for 2D analysis in experiment 1. We found that the activity was significantly correlated with protein amounts of COMT1 and COMT2 (Fig. 6). The best correlation (r = 0.95) was observed when COMT1 and COMT2 protein amounts were summed (Fig. 6C). This result shows that both enzymes could be active.
Figure 6.
Relation between COMT and lignification profiles. A to C, In vitro activity of COMT in function of relative amounts of COMT1 and COMT2 measured on 2D gels. r, Correlation coefficient. D, Measurement of uncondensed lignin units (H + G + 5OHG + S) along leaf 6 in experiment 3. E, Measurement of uncondensed lignin units in function of tissue age. F, S:G ratio along leaf 6. G, Measurement of FA along leaf 6 in experiment 3. Vertical bars show se (n = 2 in D, F, and G; n = 3–8 in E). RA, Relative amount.
We then performed lignin analyses to determine whether the relative amounts of COMT, CCoAOMT, and PAL correlate with lignification along the leaves. Lignin determination by the standard Klason gravimetric method could not be applied to 2-cm-long leaf segments (about 20 mg of dry matter). Indeed, this method not only requires a preliminary solvent extraction step to eliminate proteins and other interfering compounds, but also is sample demanding (100–300 mg of an extract-free sample is necessary to recover a final lignin amount that can be weighted with some accuracy, this sample demand increasing when the lignin amount is low). As the alternative spectrometric methods of lignin determination would suffer interference drawbacks from nonlignin phenolics, the only way to unequivocally characterize native lignin in such samples remains by the analysis of specific lignin-derived compounds, namely, thioacidolysis monomers. We used thioacidolysis for the determination of uncondensed lignin H, G, and S units (see “Materials and Methods”) in the 2-cm-long dried leaf fragments collected from the sixth leaf of the control (C2) and moderately drought-stressed (M) plants (experiment 3). The total yield in (H + G + S) lignin-derived monomers ranged between 2 and 35 μmol g−1 of dried leaf samples (Fig. 6D). G and S monomers were the main lignin-derived compounds, whereas H monomers were recovered in low or trace amounts (relative amount ranging between 1% and 6% of the [H + G + S] total). If we compare these yields to those obtained for maize stem lignins (Méchin et al., 2000), we approximate that the total lignin amount of the dried leaves ranges between 0.2% and 3.5% by weight. This is an approximation, as thioacidolysis yields from leaf and stem lignins probably differ.
Duplicate experiments from two different analogous leaf samples gave fairly close thioacidolysis yields, with ses ranging between 5% and 20% for most samples. As shown in Figure 6D, thioacidolysis yield, which herein is the only tool that can approximate lignin amount, was very low in the first 7 cm of the control leaf, then a steady increase occurred before stabilization at 22 cm from the point of insertion. In the mildly drought-stressed leaves, thioacidolysis yield stabilized nearer the base (15 cm from the point of insertion) at a lower level (20 μmol g−1), which suggests a lower lignin amount and/or a lower proportion of uncondensed lignin units. Similar to the COMT profile, the lignification profile, as reflected by thioacidolysis yield, shifted toward the base of the leaf. When expressed as a function of computed tissue age, lignification was found to be both delayed and down-regulated by water deficit (Fig. 6E). Thus, drought stress altered COMT1 levels and lignification in a consistent manner. In addition, we found that thioacidolysis yield decreased in the upper parts of the drought-stressed leaf (from 21–30 cm from the point of insertion; Fig. 6D). Because a decrease in lignin content as the tissue ages is unlikely, this trend may be due to the fact that cell wall lignin is more cross-linked in this area by thioacidolysis-resistant bonds. Thioacidolysis yield can also increase with the frequency of the S lignin units, as these S units are more involved in uncondensed bonds. Thus, a variation of the S:G thioacidolysis ratio could bias thioacidolysis-based comparisons of lignin levels between controls and plants subjected to water deficit. However, the S:G ratio stayed at similarly low values (0.2–0.5) whatever the plant (control or water stressed; Fig. 6F).
pCA and FA were recovered from the thioacidolysis of dried leaf samples together with the lignin-derived G and S monomers. As thioacidolysis is not aimed at the quantitative cleavage of ester bonds, the amounts of pCA and FA were only considered comparatively. In grass cell walls, most p-coumarate accretion occurs with lignification (Musel et al., 1997; MacAdam and Grabber, 2002). In our analyses, the thioacidolysis-released pCA ranged between 0.3 and 6 μmol g−1 of dried leaf samples (Fig. 6G). This amount displayed a very strong relationship to the amount of lignin-derived monomers (r2 = 0.99 and 0.94 for the control leaf series and the drought-stressed leaf series, respectively), and the water stress both reduced the yield of thioacidolysis-released pCA and of lignin-derived monomers. This result confirms that thioacidolysis-released pCA may be a convenient, albeit indirect, indicator of lignification in grass samples. The efficiency of thioacidolysis in FA extraction is negatively related to the level of tissue lignification; thus, profiles of thioacidolysis-released FA do not reflect profiles of total FA, and concentrations cannot be compared between controls and stressed plants. Nevertheless, the shapes of thioacidolysis-released FA profiles (Fig. 6H) show similarities with those of uncondensed lignin units.
The reduced amounts of lignin units under mild water deficit further confirm the overall down-regulation of lignification in maize leaves subjected to drought stress, as has been observed at the enzymatic level.
DISCUSSION
This study was aimed at deciphering COMT variations along maize leaves through development and water deficits and relating them to the lignification process. Two COMT isoforms (COMT1 and COMT2) were identified, and both disappeared in a bm3/bm3 genotype, i.e. in the presence of a deficient COMT gene (Vignols et al., 1995). This strongly suggests that both isoforms are the products of the same COMT gene, which is present as a single copy in the maize genome. The predicted protein sequence, composed of 364 amino acids, has an estimated pI of 6.03 and an estimated molecular mass of 39.572 kD (Collazo et al., 1992). Both COMTs have similar molecular mass (37 kD) but different pIs (5.4 and 5.6 for COMT1 and COMT2, respectively). The putative posttranslational modification causing the pI difference was not identified. Because both proteins have similar molecular mass, a proteolysis process is unlikely. In addition, it is unlikely that it occurred during protein extraction because the proportions of both proteins changed along the leaf. Because COMT2 accumulated and decreased after COMT1, we could hypothesize that COMT2 is an intermediate form of COMT before degradation. However, the relatively large proportion of COMT2 and the better correlation of in vitro enzymatic activity with the sum of both isoforms than with each of them are not consistent with this hypothesis (Fig. 6). The putative posttranslational modification may reflect distinct substrate specificities because COMT can methylate various substrates (Davin and Lewis, 1992; Maury et al., 1999; Osakabe et al., 1999; Li et al., 2000; Parvathi et al., 2001; Zubieta et al., 2002).
In the three experiments we performed, the profiles of COMT repeatedly showed a maximal accumulation in a region of the leaf located above the zone of elongation, which is 6 to 8 cm long (Ben-Haj-Salah and Tardieu, 1995). This feature is consistent with the hypothesis that cells enter in a zone of lignification after the completion of divisions and elongation. The area where the COMT concentration increases could correspond to the beginning of this zone. COMT could then be actively degraded when the cells have reached a sufficient level of lignification. Thus, the peak of COMT accumulation could occur in the region where lignification takes place in leaves. Collazo et al. (1992) showed that the levels of COMT transcripts were undetectable in maize root tips, high in the elongation zone, and low in the differentiating zone. Transcript expression is thus maximal in the elongation zone of maize roots, whereas, in our study, protein maximal expression occurred beyond the elongation zone of maize leaves. This is consistent with a lag between transcript and protein synthesis.
By computing an estimation of tissue age, we showed that the highest level of COMT accumulation was reached in tissues that were older in water-stressed leaves than in well-watered leaves. If COMT accumulation was not repressed during drought and was synthesized mostly between days 2 and 3, like in controls, the position of maximal accumulation would go down progressively toward the base of the leaf just by the effect of growth decrease on the gradient of tissue age. On the contrary, if COMT accumulation abruptly ceased, maximal accumulation should stay in tissues that were 2 to 3 d old at the beginning of water deficit. These tissues stop their progression toward leaf end when growth is arrested. Our results on plants subjected to severe water deficit are consistent with the last hypothesis. The position of maximal accumulation did not continuously progress toward the leaf base but was stabilized after a few centimeters. The estimated age of maximal accumulation in S1 was 6 d (Fig. 5), whereas S1 samples were collected on average at day 4 (Fig. 4). Accordingly, tissues showing maximal accumulation of COMT were formed approximately 2 d before day 0. It is thus likely that most of the amount of COMT observed in samples subjected to severe drought actually corresponded to the rest of COMT that was synthesized in control conditions. The observed shift of the maximal accumulation toward older tissues would thus result from the total (S1 and S2 leaves) or partial (M leaves) repression of COMT synthesis. The reduction of COMT amount in M, S1, and S2 leaves could also be explained by the absence (or reduction) of COMT synthesis, which could no longer compensate for the protein turnover.
We also showed that COMT levels increased during maize leaf development up to a stage where growth decreases (stage C3; Fig. 5, B and C). This increase is temporary because both COMT isoforms disappeared from control leaves after the end of growth (data not shown). Several studies showed that COMT activity increases in the successive internodes of alfalfa (Inoue et al., 1998, 2000) and tobacco (Maury et al., 1999). In various growing organs, COMT transcript levels gradually increased during alfalfa development (Gowri et al., 1991). A perfect correlation was obtained between transcript levels and activity of COMT during the growing season of aspen (Meng and Campbell, 1998). Therefore, development over time modifies COMT expression in leaves as well as in other organs.
Quantitative variations were not identical, although the shape of COMT profiles and the effect of drought on the position of maximal expression were conserved among the three experiments in different environmental conditions and with different genotypes. Indeed, the relative amount of COMT1 decreased in drought-stressed leaves in experiments 2 and 3, while an increase was noted by Riccardi et al. (1998) and in experiment 1. The increase observed by Riccardi et al. (1998) can be explained by the shift of maximal accumulation toward the leaf base since samples were only taken in this part of the leaf. As the level of COMT accumulation changes widely according to the stage of leaf development, the increase observed in water-stressed leaves of experiment 1 might be due to differences in the stage of development that were not precisely controlled in this experiment, where leaf growth was not measured. However, it cannot be excluded that the amount of COMT is at least temporarily increased by drought when plants are cultivated in perlite. Several studies showed the activation of COMT expression in response to various stresses but not to drought.
COMT1 was highly correlated with CCoAOMT, PAL, SAMS, METS, MTHFR, IRL, and HMGS. CCoAOMT and PAL directly contribute to the synthesis of monolignols, whereas SAMS, METS, and MTHFR contribute to the synthesis of SAM. SAM is synthesized by SAMS from Met, which is synthesized by METS from hCys and 5-CH3-THF, the latter being provided by MTHFR. The coordinated expression of several of these genes has already been reported. Transcription of PAL, SAMS, and COMT genes was induced in elicited cells of alfalfa (Gowri et al., 1991; Ni et al., 1996). Increased accumulation of SAMS transcripts or protein has been repeatedly observed in tissues where lignification occurs, i.e. vascular tissues (Peleman et al., 1989) and wood-forming tissues (Van der Mijnsbrugge et al., 2000; Whetten et al., 2001). Coordinated accumulation of SAMS and METS was observed during seed germination in Arabidopsis (Gallardo et al., 2002). SAM is the primary methyl group donor, and it is used in the methylation reactions catalyzed by COMT and CCoAOMT in monolignol biosynthesis. It is also involved in the methylation of flavonoids, DNA, proteins, quaternary ammonium compounds, and many other metabolites. In addition, SAM is the precursor of ethylene, polyamines, and nicotianamine (Moffatt and Weretilnyk, 2001). Thus, SAM plays a major role in different metabolisms. Nevertheless, since many enzymes involved in its synthesis correlate with COMT, CCoAOMT, and PAL, our data suggest that, in this case, their variation is linked to the synthesis of monolignols. No other protein of the lignin biosynthesis pathway was identified in the 2D gels. Some of them might be present but not identified so far. Others could have their molecular mass or pI outside the ranges used for these 2D gels, or their quantity could be lower than the detection threshold.
IRL acts in isoflavonoid metabolism, which suggests that this secondary metabolism is also down-regulated by drought. A correlation between isoflavone reductase and PAL transcript accumulation also was observed in elicited cell suspension cultures of alfalfa (Ni et al., 1996). No direct relation was found between cell wall formation and the last protein showing high correlation with COMT1, CCoAOMT, and PAL, namely, HMGS. HMGS is involved in the synthesis of mevalonate, which is a precursor for various isoprenoid compounds, including sterols. Interestingly, it has been reported that HMGS is down-regulated by osmotic stress, whereas it is induced by wounding in Brassica juncea (Alex et al., 2000).
The effect of drought on the phenylpropanoid pathway and other proteins involved in cell wall formation is not well documented. CCoAOMTs, but not COMT, were found to be induced by drought in needles of maritime pine (Pinus pinaster; Costa et al., 1998) and in roots of Arabidopsis (Bianchi et al., 2002). SAMS genes were up-regulated under salt stress conditions in tomato seedlings (Espartero et al., 1994) and in loblolly pine (Pinus taeda) roots (Chang et al., 1995), but osmotic stress substantially decreased PAL activity in wheat (Triticum aestivum) coleoptiles (Wakabayashi et al., 1997).
To confirm that a lignification zone occurs in the area where COMT and other enzymes possibly involved in lignification accumulate, we performed lignin analyses along the leaf. To unequivocally monitor lignification in small and poorly lignified samples, we determined the lignin-derived thioacidolysis monomers as the specific and sensitive signature of lignin content and structure. Under control conditions, the level of uncondensed lignin units, i.e. the parent structures of thioacidolysis phenolic monomers, remained very low in the elongation zone; it increased from approximately 9 to 23 cm and stabilized above. Thus, the lignification zone is located just beyond the elongation zone. The increase of uncondensed lignin unit amount along the leaf is consistent with the progression of lignin deposition during cell maturation in maize stem internodes (Joseleau and Ruel, 1997) and the deposition of secondary cell wall from 1.5 to 7.5 cm distal to the ligule, i.e. in the maturation zone of elongating tall fescue leaf blades (MacAdam and Nelson, 2002). The relative amount of COMT diminished above 24 cm, while the lignin level remained constant (Fig. 6D). It can be assumed that once the cells are lignified, they do not need COMT activity anymore, provided that lignin is not naturally degraded. When mild water deficit was applied, lignification reached its maximum at a lower position and at a lower level than in control leaves. Profiles in function of tissue age were also consistent with COMT regulation. In water-stressed leaves, tissues in which lignification increased were older than in controls, although their position was lower. Thus, the position of the lignification zone as well as the effect of drought on both lignin content and the position of this zone are consistent with the hypothesis that COMT and other correlated enzymes are involved in lignification and that their profiles of expression in stressed leaves are related to a reduction of lignin accumulation.
No change in lignin composition was observed in leaves subjected to water deficit. Thus, the observed decrease of enzymes involved in methylation reactions should not be interpreted as a specific reduction of these particular reactions but as a marker of a general reduction of monolignol synthesis. The levels of lignin-derived monomers, as well as those of FA and pCA, concomitantly decreased in the upper part of drought-stressed leaves. This could be explained by increased cell wall cross-linking in this area. It could come from cell wall phenolics that might participate in oxidatively mediated cross-links with the formation of condensed bonds in lignins and of diferulic units (Grabber et al., 2000). In agreement with this hypothesis, all peroxidase activities reported so far increased in response to salt stress in wheat root cells (Jbir et al., 2001).
In conclusion, these data provide additional insight into the mechanism of lignification in plants. They allowed the localization of a lignification zone above the elongation zone in which the expression of proteins involved in the metabolism of lignin is maximal. The good correlation between proteomic and lignin measurements lends further support to the notion that lignin accumulation increases when leaf growth decreases in well-watered plants, i.e. when the leaf approaches its final size. In response to water deficit, this lignification zone is shifted toward the leaf base and the level of lignin decreases. The analysis of expression profiles of involved proteins showed that their repression was consistent with both the new position of the lignification zone and the reduction of lignin content. It can be suggested that the decrease of lignification is an adaptative response to drought since the continuous accumulation of lignin in the absence of growth would lead to the lignification of the zone of elongation, which would compromise growth recovery upon rehydration.
MATERIALS AND METHODS
Culture Conditions
Maize (Zea mays) plants were grown in a growth cabinet at a light intensity of 450 μmol m−2 s−1 during the 16-h photoperiod, 25°C day and 18°C night temperatures, and 50% relative humidity. Two lines of maize were used: F2, a flint line from the Institut National de la Recherche Agronomique (France), and Io, an American dent line from the Iodent group.
Three experiments were conducted. In experiment 1, Io inbred-line plants (three replicates) were grown in perlite (one plant per 1-L-capacity plastic pot) and watered with nutrient solution (Hydrocani C2 + Hydroplus Fer H23; HURELARC, Baillet en France, France). A subset of plants was stressed by withholding watering at the fifth-leaf stage (approximately 3-week-old plants). After 10 d of stress, 3-cm-long fragments of the sixth leaf were harvested, immediately frozen in liquid nitrogen, and stored at −196°C.
In experiment 2, Io inbred-line seeds were placed in 1-L-capacity plastic pots and filled with a soil:sand (1:1) mixture. The pots were weighed and watered daily to maintain a substrate humidity of 50% (full irrigation) until the treatment started (day 0). The length of leaves 5 and 6, from pot edge to tip, was measured daily. The control plants were maintained at full irrigation until they were sampled. Drought treatment was initiated by stopping watering when the fifth leaf reached a 30-cm length (day 0), corresponding roughly to 3-week-old plants. Two-centimeter-long fragments of the sixth leaf were harvested from control and water-stressed plants (two replicates), 3, 4, and 5 d after day 0 (days 3, 4, and 5, respectively). Once collected, the leaf samples were immediately frozen in liquid nitrogen and stored at −196°C.
Experiment 3 was conducted the same way as experiment 2 up to day 0, except that the F2 inbred line was used instead of the Io line and the substrate was a soil:compost (1:1) mixture. The control plants were fully irrigated every day until they were sampled. Two regimes of water deficit were tested: watering to stabilize substrate humidity at 30% (mild water deficit) and no watering at all (severe water deficit). Two-centimeter-long fragments of the sixth leaf were sampled at various developmental stages based on leaf elongation rates (see Fig. 4). Three different plants were used for each stage/treatment. Once collected, the leaf samples were immediately frozen in liquid nitrogen and stored at −196°C.
The near-isogenic lines for the bm3 mutation in the genetic background of Io were kindly provided by Dr. Y. Barrière, Institut National de la Recherche Agrnomique (Lusignan, France).
Protein Extraction and 2D-PAGE
A denaturing protein extraction was achieved essentially as described previously (Damerval et al., 1986), except that 60 μL of resolubilization solution was used to resuspend 1 mg of pellet. Each leaf fragment was individually extracted. 2D-PAGE (O'Farrell, 1975) was performed as described previously (Damerval et al., 1986), with a protein load of 30 μL. The 2D gels were silver stained using the method of Damerval et al. (1987) modified by Burstin et al. (1993).
Quantitative Proteomic Analyses
Silver-stained 2D gels were scanned (model 7899; Eikonix, Bedford, MA) with a spatial resolution of 1 pixel/100 μm and an optical density range from 0 to 1.2. Image processing, spot detection, and quantification (integration of pixel value within spot contours) were carried out with the Kepler package (LSB, Rockville, MD). To compensate for staining variation between gels, relative intensities were computed by scaling spot intensities of each gel relative to the sum of spot intensities within the same rectangle.
Spot Identification
COMT2 was previously identified by Edman microsequencing (Riccardi et al., 1998). The other proteins were identified by LC-MS/MS after in-gel digestion by trypsin. HPLC was performed with the Ultimate LC system combined with the Famos autosampler and Switchos II microcolumn switching for preconcentration (LC Packing, Amsterdam). The sample was loaded on the column (PepMAP C18, 5 μm, 75 μm i.d., 15 cm; LC Packing) using a preconcentration step on a micro-precolumn cartridge (300 μm i.d., 5 mm).
Four microliters of the sample were loaded on the precolumn at 5 μL min−1. After 3 min, the precolumn was connected with the separating column and the gradient was started at 200 nL min−1. Buffers were 0.1% HCOOH in water (A) and 0.1% HCOOH in acetonitrile (B). A linear gradient from 5% to 45% B for 25 min was applied. The LCQ Deca XP+ (Thermofinnigan, Les Ulis, France) was used with a nano-electrospray interface. Ionization (1.2–1.4 kV ionization potential) was performed with liquid junction and noncoated capillary probe (New Objective, Cambridge, MA). Proteins were identified by using the SEQUEST package, which compares experimental spectra to the theoretical spectra computed according to database sequences. The criterion used for positive identification of proteins was as follows: At least two peptides of the same protein must be identified with an X-correlation score greater than 1.5, 2.2, and 3.3 for single-, double-, and triple-charged peptide ions, respectively.
Enzyme Activity
Crude protein was extracted from leaf samples as described previously (Van Doorsselaere et al., 1995). The protein content was determined by the Bradford (1976) method, using the Bio-Rad reagent (Bio-Rad Laboratories, Hercules, CA). COMT enzyme assays were conducted with 10 μg of crude protein using 50 mm tritiated S-adenosyl-l-[methyl-3H]Met (NEN, Boston) and 3 mm caffeic acid (Van Doorsselaere et al., 1995). The assays were conducted under steady-state conditions after 10 min of incubation at 30°C.
Lignin Analysis
Lignification of each leaf segment was analyzed by thioacidolysis as described previously (Lapierre, 1993, 1995) using 0.0215 mg mL−1 docosane solution (vehicle, CH2Cl2) as a gas chromatography internal standard. Thioacidolysis of plant tissues coupled to gas chromatography-mass spectrometry of the lignin-derived products makes it possible to study very small amounts of lignin without interference by pCA and FA (Lapierre et al., 1995). The thioethylated H, G, and S monomers originate from the H, G, and S lignin units only involved in labile β–O–4 ether bonds, the most frequent interunit linkages of native lignin. Therefore, the total yield in thioacidolysis monomers reflects the amount of lignin units only involved in β–O–4 ether bonds, referred to as uncondensed lignin units.
Lignin was studied in the sixth leaves from F2 control (at maximal elongation rate C2, experiment 3) and mildly water-stressed plants (stage M, experiment 3). Fifteen consecutive 2-cm-long segments covering the thirty-first centimeter of the leaf were collected. The experiment was repeated on two different leaves from two different control or water-stressed plants.
Estimation of Tissue Age in Leaf Segments
Because of the position of the meristem at the base of the leaf, there is a gradient of age from the base to the top of the leaf. A simple estimation of tissue age was computed by using the daily measurements of leaf growth. Every centimeter gained by the leaf each day was assumed to be caused by the development of the same length of new tissue at the leaf base. This estimation does not take into account the fact that tissue elements actually migrate more slowly while they are in the elongation zone. Thus, an estimation of segmental elongation rate (SER) is necessary. Approximations were attempted by using SER profiles computed by Ben-Haj-Salah and Tardieu (1995). They systematically increased the estimated tissue ages but did not change the differences between treatments (data not shown). This method, although theoretically more exact, relies on parameters difficult to estimate (SER), especially when growth is not constant. In addition, it does not take into account the fact that cell division occurs all along the elongation zone (Tardieu et al., 2000); that is, most cells are younger than the tissue element to which they belong. Thus, the tissue age was roughly estimated by the leaf elongation rates.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requester.
Supplementary Material
Acknowledgments
We thank Hélène Corti and Marlène Davanture for technical assistance, and Sylvie Coursol, Dominique de Vienne, and Grant R. Cramer for careful reading of this paper.
This work was supported by Génoplante.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.050815.
References
- Alex D, Bach TJ, Chye ML (2000) Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J 22: 415–426 [DOI] [PubMed] [Google Scholar]
- Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Baucher M, Halpin C, Petit-Conil M, Boerjan W (2003) Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W (1998) Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci 17: 125–197 [Google Scholar]
- Ben-Haj-Salah H, Tardieu F (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (analysis of the coordination between cell division and cell expansion). Plant Physiol 109: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haj-Salah H, Tardieu F (1997) Control of leaf expansion rate of droughted maize plants under fluctuating evaporative demand (a superposition of hydraulic and chemical messages?). Plant Physiol 114: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M, Damerval C, Vartanian N (2002) Identification of proteins regulated by cross-talk between drought and hormone pathways in Arabidopsis wild-type and auxin-insensitive mutants axr1 and axr2. Funct Plant Biol 29: 55–61 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Boudet AM, Lapierre C, Grima-Pettenati J (1995) Tansley review No. 80. Biochemistry and molecular biology of lignification. New Phytol 129: 203–236 [DOI] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burstin J, Zivy M, de Vienne D, Damerval C (1993) Analysis of scaling methods to minimize experimental variations in two-dimensional electrophoresis quantitative data. Applications to the comparison of maize inbred lines. Electrophoresis 14: 1067–1073 [DOI] [PubMed] [Google Scholar]
- Campbell MM, Sederoff R (1996) Variation in lignin content and composition. Plant Physiol 110: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear JD, Dias MADL, Funkhouser EA, Newton RJ, Cairney J (1995) Gene expression under water deficit in loblolly pine (Pinus taeda L.): isolation and characterization of cDNA clones. Physiol Plant 95: 1–10 [Google Scholar]
- Collazo P, Montoliu L, Puigdomenech P, Rigau J (1992) Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol 20: 857–867 [DOI] [PubMed] [Google Scholar]
- Costa P, Bahrman N, Frigerio JM, Kremer A, Plomion C (1998) Water-deficit-responsive proteins in maritime pine. Plant Mol Biol 38: 587–596 [DOI] [PubMed] [Google Scholar]
- Damerval C, de Vienne D, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7: 52–54 [Google Scholar]
- Damerval C, le Guilloux M, Blaisonneau J, de Vienne D (1987) A simplification of Heukeshoven and Dernick's silver staining of proteins. Electrophoresis 8: 158–159 [Google Scholar]
- Davin LB, Lewis NG (1992) Phenylpropanoid metabolism: biosynthesis of monolignols, lignans and neolignans, lignins and suberins. In HA Stafford, RK Ibrahim, eds, Phenolic Metabolism in Plants. Plenum Press, New York, pp 325–375
- Dixon R, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espartero J, Pintor-Toro JA, Pardo JM (1994) Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol Biol 25: 217–227 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L). X. Molecular cloning and expression of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol 97: 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem 48: 6106–6113 [DOI] [PubMed] [Google Scholar]
- Gregersen L, Christensen AB, Sommer-Knudsen J, Collinge DB (1994) A putative O-methyltransferase from barley is induced by fungal pathogens and UV light. Plant Mol Biol 26: 1797–1806 [DOI] [PubMed] [Google Scholar]
- Held BM, Wang H, John I, Wurtele ES, Colbert JT (1993) An mRNA putatively coding for an O-methyltransferase accumulates preferentially in maize roots and is located predominantly in the region of the endodermis. Plant Physiol 102: 1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T (1997) Biochemistry and Molecular Biology of Wood. Springer-Verlag, Berlin
- Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5: 224–229 [DOI] [PubMed] [Google Scholar]
- Inoue K, Parvathi K, Dixon RA (2000) Substrate preferences of caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferases in developing stems of alfalfa (Medicago sativa L.). Arch Biochem Biophys 375: 175–182 [DOI] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Ballance GM, Ni W, Sturzer C, Dixon RA (1998) Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbir N, Chaibi W, Ammar S, Jemmali A, Ayadi A (2001) Root growth and lignification of two wheat species differing in their sensitivity to NaCl, in response to salt stress. C R Acad Sci III 324: 863–868 [DOI] [PubMed] [Google Scholar]
- Joseleau J-P, Ruel K (1997) Study of lignification by noninvasive techniques in growing maize internodes. An investigation by Fourier transform infrared cross-polarization-magic angle spinning 13C-nuclear magnetic resonance spectroscopy and immunocytochemical transmission electron microscopy. Plant Physiol 114: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C (1993) Application of new methods for the investigation of lignin structure. In HG Jung, DR Buxton, RD Hatfield, J Ralph, eds, Forage Cell Wall Structure and Digestibility. ASA-CSSA-SSSA, Madison, WI, pp 133–166
- Lapierre C, Pollet B, Rolando R (1995) New insights into the molecular architecture of hardwood lignins by chemical degradation methods. Res Chem Intermediat 21: 397–412 [Google Scholar]
- Lewis NG, Yamamoto E (1990) Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41: 455–496 [DOI] [PubMed] [Google Scholar]
- Li L, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- MacAdam JW, Grabber JH (2002) Relationship of growth cessation with the formation of diferulate cross-links and p-coumaroylated lignins in tall fescue leaf blades. Planta 215: 785–793 [DOI] [PubMed] [Google Scholar]
- MacAdam JW, Nelson CJ (2002) Secondary cell wall deposition causes radial growth of fibre cells in the maturation zone of elongating tall fescue leaf blades. Ann Bot (Lond) 89: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury S, Geoffroy P, Legrand M (1999) Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol 121: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin V, Argillier O, Menanteau V, Barrière C, Mila I, Pollet B, Lapierre C (2000) Relationship of wall composition to in vitro digestibility of maize inbred lines. J Sci Food Agric 80: 574–580 [Google Scholar]
- Meng H, Campbell WH (1998) Substrate profiles and expression of caffeoyl coenzyme A and caffeic acid O-methyltransferases in secondary xylem of aspen during seasonal development. Plant Mol Biol 38: 513–520 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Weretilnyk EA (2001) Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells. Physiol Plant 113: 435–442 [Google Scholar]
- Musel G, Schindler T, Bergfeld R, Ruel K, Jacquet G, Lapierre C, Speth V, Schopfer P (1997) Structure and distribution of lignin in primary and secondary cell walls of maize coleoptiles analyzed by chemical and immunological probes. Planta 201: 146–159 [Google Scholar]
- Neuffer MG, Jones L, Zuber MS, editors (1968) The Mutants of Maize. Crop Science Society of America, Madison, WI
- Ni W, Sewalt VJH, Korth KL, Blount JW, Ballance GM, Dixon RA (1996) Stress responses in alfalfa. XXI. Activation of caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase genes does not contribute to changes in metabolite accumulation in elicitor-treated cell-suspension cultures. Plant Physiol 112: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250: 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Tsao C, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA 96: 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakusch A-E, Kneusel RE, Matern U (1989) S-adenosyl-l-methionine:trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys 271: 488–494 [DOI] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler GJ, Seurinck J, Botterman J, Alliote T, Van Montagu M, Inzé D (1989) Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M (1993) Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol 103: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J, Chamayou S, Nadaud I, Beckert M, Barriere Y, Mila I, Lapierre C, Rigau J, Puigdomenech P, Jauneau A, et al (2002) Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi F, Gazeau P, de Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize. Quantitative variation and polypeptide identification. Plant Physiol 117: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S, Wang H, McNeil SD, Raymond RK, Appling DR, Shachar-Hill Y, Bohnert HJ, Hanson AD (1999) Isolation, characterization, and functional expression of cDNAs encoding NADH-dependent methylenetetrahydrofolate reductase from higher plants. J Biol Chem 274: 36089–36096 [DOI] [PubMed] [Google Scholar]
- Schmitt D, Pakusch AE, Matern U (1991) Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem 266: 17416–17423 [PubMed] [Google Scholar]
- Tardieu F, Reymond M, Hamard P, Granier C, Muller B (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J Exp Bot 51: 1505–1514 [DOI] [PubMed] [Google Scholar]
- Toquin V, Grausem B, Geoffroy P, Legrand M (2003) Structure of the tobacco caffeic acid O-methyltransferase (COMT) II gene: identification of promoter sequences involved in gene inducibility by various stimuli. Plant Mol Biol 52: 495–509 [DOI] [PubMed] [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT (1980) Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18: 259–288 [Google Scholar]
- Van der Mijnsbrugge K, Meyermans H, Van Montagu M, Bauw G, Boerjan W (2000) Wood formation in poplar: identification, characterization, and seasonal variation of xylem proteins. Planta 210: 589–598 [DOI] [PubMed] [Google Scholar]
- Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier MT, Petit-Conil M, Leple JC, Pilate G, Cornu D, Monties B, et al (1995) A novel lignin in poplar trees with a reduced caffeic/5-hydroxyferulic acid O-methyltransferase activity. Plant J 8: 855–864 [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Hoson T, Kamisaka S (1997) Osmotic stress suppresses cell wall stiffening and the increase in cell wall-bound ferulic and diferulic acids in wheat coleoptiles. Plant Physiol 113: 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff R (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49: 585–609 [DOI] [PubMed] [Google Scholar]
- Whetten RW, Sun Y-H, Zhang YZ, Sederoff R (2001) Functional genomic and cell wall biosynthesis in loblolly pine. Plant Mol Biol 47: 275–291 [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.