Abstract
One of the best-characterized physiological rhythms in plants is the circadian rhythm of CO2 metabolism in Crassulacean acid metabolism (CAM) plants, which is the focus here. The central components of the plant circadian clock have been studied in detail only in Arabidopsis (Arabidopsis thaliana). Full-length cDNAs have been obtained encoding orthologs of CIRCADIAN CLOCK-ASSOCIATED1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1 (TOC1), EARLY FLOWERING4 (ELF4), ZEITLUPE (ZTL), FLAVIN-BINDING KELCH REPEAT F-BOX1 (FKF1), EARLY FLOWERING3 (ELF3), and a partial cDNA encoding GIGANTEA in the model stress-inducible CAM plant, Mesembryanthemum crystallinum (Common Ice Plant). TOC1 and LHY/CCA1 are under reciprocal circadian control in a manner similar to their regulation in Arabidopsis. ELF4, FKF1, ZTL, GIGANTEA, and ELF3 are under circadian control in C3 and CAM leaves. ELF4 transcripts peak in the evening and are unaffected by CAM induction. FKF1 shows an abrupt transcript peak 3 h before subjective dusk. ELF3 transcripts appear in the evening, consistent with their role in gating light input to the circadian clock. Intriguingly, ZTL transcripts do not oscillate in Arabidopsis, but do in M. crystallinum. The transcript abundance of the clock-associated genes in M. crystallinum is largely unaffected by development and salt stress, revealing compensation of the central circadian clock against development and abiotic stress in addition to the well-known temperature compensation. Importantly, the clock in M. crystallinum is very similar to that in Arabidopsis, indicating that such a clock could control CAM without requiring additional components of the central oscillator or a novel CAM oscillator.
In higher plants, a circadian clock controls hypocotyl elongation, daily leaf movements, flowering time, and the rhythm of CO2 fixation in Crassulacean acid metabolism (CAM; McClung, 2001). Some of the molecular components of the central plant clock have been identified. The closely related single Myb-repeat transcription factors, termed CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and the pseudoresponse regulator/CONSTANS-motif protein, TIMING OF CAB EXPRESSION1 (TOC1), form at least part of the central clock (Eriksson and Millar, 2003). CCA1 and LHY are expressed around dawn when they bind to the promoter of TOC1 and repress its expression (Alabadi et al., 2001). As dusk approaches, the levels of CCA1 and LHY decrease, TOC1 expression is derepressed, and TOC1 is expressed during the evening. TOC1 is thought to act as a positive element in the regulation of CCA1 and LHY, but it is clear that this occurs in concert with the action of other genes. Candidates for genes that act in concert with TOC1 to induce CCA1 and LHY include EARLY FLOWERING3 (ELF3), GIGANTEA (GI), and EARLY FLOWERING4 (ELF4; Doyle et al., 2002; Eriksson and Millar, 2003).
A number of other genes have been implicated in the Arabidopsis (Arabidopsis thaliana) clock. ELF3 is a novel protein that has been shown to gate light input to the clock (McWatters et al., 2000). When ELF3 is expressed in the evening, light input to reset the clock is blocked. The ZEITLUPE (ZTL) family of proteins, ZTL, FLAVIN-BINDING KELCH REPEAT F-BOX1 (FKF1), and LIGHT, OXYGEN OR VOLTAGE (LOV), KELCH PROTEIN2 (LKP2), have all been shown to play a role in circadian clock function. An Arabidopsis 35S::LKP2 overexpressing line has arrhythmic clock output in several circadian rhythms (Schultz et al., 2001). ztl is a long-period mutant, whereas fkf1 is a late-flowering mutant in long days and has subtle defects in circadian control of gene expression (Nelson et al., 2000; Somers et al., 2000). Unlike ZTL, FKF1 does not affect circadian period and has been demonstrated to act downstream of the clock to control the photoperiodic switch to flowering (Imaizumi et al., 2003). The FKF1 LOV domain can bind to a flavin mononucleotide chromophore and undergo light-induced photochemistry, suggesting that FKF1 could directly sense blue light (Imaizumi et al., 2003). ZTL has recently been shown to interact directly with TOC1 and target it for degradation via a proteasome-dependent pathway (Mas et al., 2003). Furthermore, the abundance of the ZTL protein itself is under circadian control with the turnover involving the proteasome, and ZTL forms part of an SCF complex in vivo (Kim et al., 2003; Han et al., 2004).
The physiology and biochemistry of the circadian rhythm of CAM CO2 fixation is relatively well characterized (Wilkins, 1992; Lüttge, 2000, 2003). Phosphoenolpyruvate carboxylase (PEPc) catalyzes primary CO2 fixation in CAM plants. PEPc is activated at night by a phosphorylation event that is driven by a circadian clock. Circadian oscillations in the phosphorylation state of PEPc are due to circadian control of the expression and activity of PEPc kinase (PPCK; Carter et al., 1991; Hartwell et al., 1996, 1999, 2002). Phosphorylation reduces the sensitivity of PEPc to feedback inhibition by malate, and this makes phospho-PEPc more active in vivo at night, allowing malate to accumulate in the vacuole. Interestingly, the circadian control of the CAM pathway and PPCK expression and activity can be overridden by metabolic perturbations that influence the subcellular localization of malate (Borland et al., 1999).
We have a far more limited knowledge of the molecular machinery that underlies the CAM CO2 rhythms. One proposal considers that the movement of malate into and out of the vacuole, with the tonoplast functioning as a discrete hysteresis switch, could itself form the basis of the CAM circadian oscillator (Wilkins, 1992; Lüttge, 2000, 2003). Computer modeling of this biophysical oscillator has demonstrated that it could sustain robust rhythmicity of CAM over a range of temperatures (Lüttge, 2000). However, Wyka et al. (2004) found that preventing nocturnal malate accumulation in Kalanchoë daigremontiana did not cause the predicted phase delays generated by the aforementioned computer model. The authors conclude that their results rule out vacuolar malic acid accumulation as the central pacemaking process in Kalanchoë (Wyka et al., 2004). The current paradigm is that the central circadian oscillator responsible for all rhythmic outputs consists of a discrete suite of genes, which form an autoregulatory negative feedback loop. A testable hypothesis assumes that a similar clock could provide the circadian control of the CAM pathway and that the movement of malate in and out of the vacuole responds to a molecular clock. Conversely, the metabolic signals provided by the operation of CAM (e.g. high cytosolic malate during the day or high starch at dusk) could be involved in entraining the central clock genes or in modulating the output pathways from the clock. To test these hypotheses, we have identified orthologs of the molecular components of the plant circadian oscillator from the model plant, Mesembryanthemum crystallinum (Common Ice Plant), in which CAM is inducible. These genes allow us to address fundamental questions about the molecular basis of the circadian control of the CAM pathway.
We report the cloning and characterization of orthologs of seven Arabidopsis circadian clock-associated genes from M. crystallinum, hereafter referred to as McCCA1/McLHY, McTOC1, McELF4, McZTL, McFKF1, McGI, and McELF3. Here, we characterize all of these clock-associated genes in a plant species other than Arabidopsis and, importantly, we demonstrate central clock operation in a CAM species. We identify McZTL as a clock-associated gene whose regulation has changed during the divergence of M. crystallinum and Arabidopsis from a common ancestor. Also, it is established that the central clock operates robustly throughout development, regardless of stress conditions.
RESULTS
Identification of Central Circadian Clock Genes in M. crystallinum
Database searches using the TBLASTN search algorithm against the M. crystallinum gene index of expressed sequence tags (ESTs; Kore-eda et al., 2004) revealed EST sequences with similarity to AtCCA1/AtLHY, AtELF4, AtZTL, AtGI, and AtELF3. We used these EST sequences to design the reverse transcription (RT)-PCR primers in Table I that specifically target the M. crystallinum orthologs of these genes. The RT-PCR products were cloned and sequenced to confirm that the amplified product was gene specific. We also cloned a fragment of McZTL and McFKF1 via degenerate PCR, using primers to the conserved domains of plant ZTL family proteins (see “Materials and Methods”). A fragment of the McTOC1 gene was isolated using primers specific to sugar beet (Beta vulgaris) EST sequences (GenBank accession nos. BI543444 and BI543434), with high similarity to AtTOC1. As a member of the order Caryophyllales, sugar beet is in the same taxonomic order as M. crystallinum. We reasoned, therefore, that there was a strong likelihood that the McTOC1 gene would be closely related to the sugar beet TOC1 gene. The identity of the resulting 300-bp McTOC1 PCR fragment was confirmed by cloning and sequencing.
Table I.
Primers used for RT-PCR analyses and cloning
Primers with “F” at end of name are forward; primers with “R” at end of name are reverse.
| Primer Name | Sequence | Product Size | Optimal Cycle No. |
|---|---|---|---|
| bp | |||
| McCCA1/LHYF | 5′-GCAAAATGCAACAGAAACCA-3′ | 350 | 22 |
| McCCA1/LHYR | 5′-ATACTTGCTGTGGCCAAGGT-3′ | ||
| McTOC1F | 5′-TTCATTGATCGAAGTAAAGTCAG-3′ | 300 | 28 |
| McTOC1R | 5′-CCAGCCTCAAGCACTTTACA-3′ | ||
| McELF4F | 5′-ATGTGGCGATCATTCAGGA-3′ | 199 | 25 |
| McELF4R | 5′-CCCTTCATATTCAATCCACCA-3′ | ||
| McZTLF | 5′-GTGTGGCGAGAAATTCCAGT-3′ | 350 | 25 |
| McZTLR | 5′-TCCTCAGTTGGGTCAAGAAGA-3′ | ||
| McFKF1F | 5′-GCAAACTTAGGTGGCTGACC-3′ | 400 | 25 |
| McFKF1R | 5′-CGGCTTGGAGAGAGATTCCT-3′ | ||
| McGIF | 5′-ATTCCCCCACTCACCATACA-3′ | 302 | 29 |
| McGIR | 5′-GCAAGAGGGCAAACTATCCA-3′ | ||
| McELF3F | 5′-CTCGTAGAGAAGGCGAGCTG-3′ | 426 | 25 |
| McELF3R | 5′-ACACAAGGCTCCAGCAAGAT-3′ | ||
| McCAB2F | 5′-TCGCCATCCTACCTTACCG-3′ | 849 | 18 |
| McCAB2R | 5′-TCATTCATTTGGCAACAACG-3′ | ||
| McCCR1/2F | 5′-ATTCGGCACGAGTTTCAAT-3′ | 315 | 26 |
| McCCR1/2R | 5′-CTCGTTGACGGTGATTTGAC-3′ | ||
| McPPCKF | 5′-TCTACTCAGGCAAGGGATTTG-3′ | 699 | 19 |
| McPPCKR | 5′-GAACAAGAACTGGCCCAGAA-3′ | ||
| McUBQF | 5′-TGTTGCATGGTCTGGTTGTT-3′ | 211 | 18 |
| McUBQR | 5′-CGGAAAGAAAAACTTTGATTCAC-3′ |
Based on the EST and PCR fragment sequences corresponding to each gene, sequence-specific primers were designed for 5′ and 3′ RACE-PCR of the corresponding full-length cDNAs. The 3′ sequences of McCCA1/LHY, McZTL, and McELF3 were obtained by completing the sequence of existing partial cDNAs. The 5′ end of the clock genes was amplified from cDNA synthesized using mRNA known to possess each transcript at high abundance (Figs. 2–4). Similarly, the 3′ ends of McTOC1, McELF4, and McFKF1 were amplified. The RACE-PCR products were either TOPO cloned and sequenced to confirm the identity of the corresponding full-length cDNA sequences for each gene or sequenced directly by primer walking (McCCA1/LHY).
Figure 2.
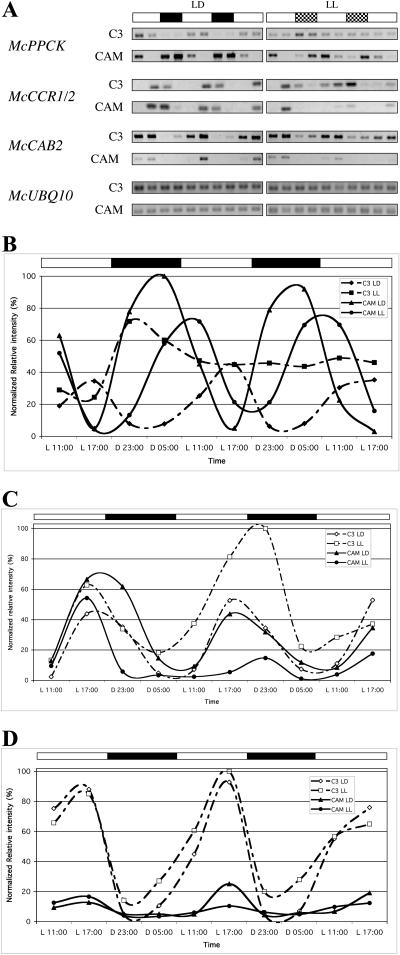
CAM induction mediates alterations in the diurnal and circadian control of McPPCK and McCAB2. C3- and CAM-induced M. crystallinum were entrained in LD (12/12) for 29 d (C3) or 65 d (CAM). CAM-induced plants had been salt stressed for 19 d to ensure complete induction of CAM. At the start of the experiment, one-half of the plants were maintained in LD while one-half were transferred to LL. Leaf samples were collected in duplicate at the indicated time points and RNA was isolated. Semiquantitative RT-PCR was performed on the RNA and the resulting band intensities were normalized to the McUBQ10 loading control. Diamonds, C3 in LD; squares, C3 in LL; triangles, CAM in LD; and circles, CAM in LL. A, Gel images for McPPCK, McCCR1/2, McCAB2, and McUBQ10. B, Normalized data for McPPCK transcript abundance. C, Normalized data for McCCR1/2 transcript abundance. D, Normalized data for McCAB2 transcript abundance.
Figure 3.
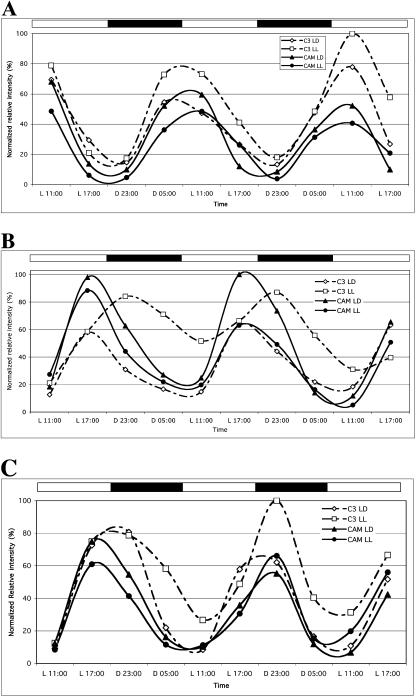
McCCA1/LHY, McTOC1, and McELF4 are clock controlled in both C3 and CAM leaves. M. crystallinum plants were entrained in LD (12/12) for 29 d (C3) or 65 d (CAM). CAM-induced plants had been salt stressed for 19 d to ensure complete induction of CAM. At the start of the experiment, one-half of the plants were maintained in LD while one-half were transferred to LL. Leaf samples were collected at the indicated time points and RNA was isolated. Semiquantitative RT-PCR was performed on the RNA and the resulting band intensities were normalized to the McUBQ10 loading control. Diamonds, C3 in LD; squares, C3 in LL; triangles, CAM in LD; and circles, CAM in LL. A, McCCA1/LHY transcript abundance. B, McTOC1 transcript abundance. C, McELF4 transcript abundance.
Figure 4.
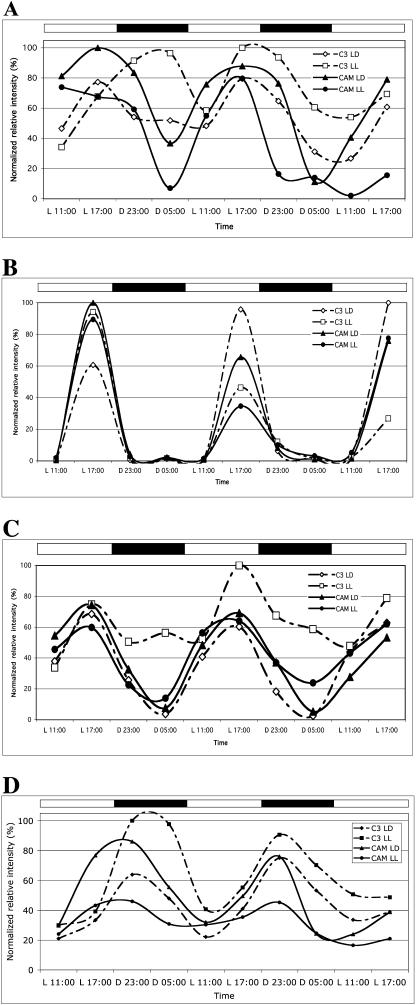
McZTL is a clock-controlled gene in C3 and CAM leaves of M. crystallinum. C3- and CAM-induced M. crystallinum plants were entrained in LD (12/12) for 29 d (C3) or 65 d (CAM). CAM-induced plants had been salt stressed for 19 d to ensure complete induction of CAM. At the start of the experiment, one-half of the plants were maintained in LD while one-half were transferred to LL. Leaf samples were collected at the indicated time points and RNA was isolated. Semiquantitative RT-PCR was performed on the RNA and the resulting band intensities were normalized to the McUBQ10 loading control. Diamonds, C3 in LD; squares, C3 in LL; triangles, CAM in LD; and circles, CAM in LL. A, McZTL transcript abundance. B, McFKF1 transcript abundance. C, McGI transcript abundance. D, McELF3 transcript abundance.
Confirmation of Clock Gene Identity
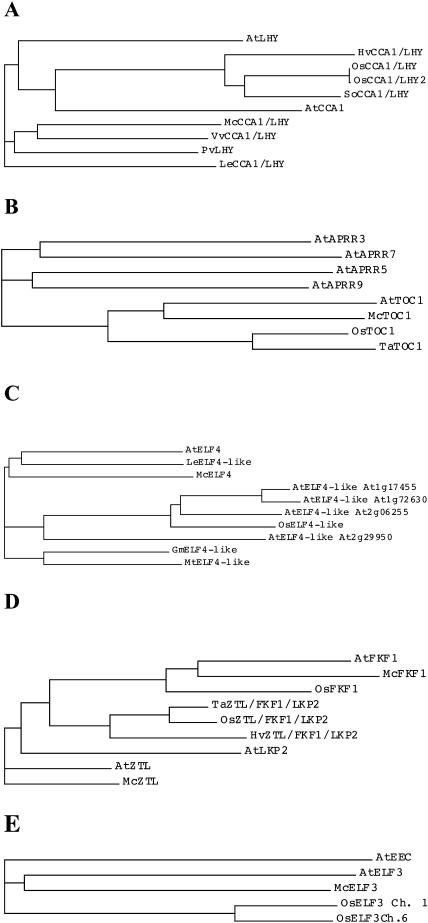
To confirm that we had cloned bona fide M. crystallinum orthologs of the Arabidopsis clock genes, we performed multiple sequence alignments between the M. crystallinum sequences and the confirmed sequences for the corresponding Arabidopsis genes. We also included the available annotated full-length rice (Oryza sativa) orthologs of each clock gene (deduced from the rice genome sequence and The Institute for Genomic Research [TIGR] rice gene index) in the multiple alignments, plus a number of other full-length orthologs identified as tentative consensus (TC) sequences in the TIGR plant gene indexes. Phylogenetic trees generated using the Vector NTI suite AlignX program confirmed the identity of all M. crystallinum clock genes (Fig. 1). McCCA1/McLHY (AY371287) shows a high degree of similarity with AtCCA1 (37.5% identity) and AtLHY (42.7% identity), particularly in the single Myb repeat at the N terminus. Specifically, the Myb repeat of McCCA1/LHY shares 82.2% identity with AtLHY and 83.2% identity with AtCCA1. The phylogenetic tree demonstrates that McCCA1/LHY is most closely related to the VvCCA1/LHY gene from Vitis vinifera and that both of these sequences are embedded on a branch of the tree that includes the PvCCA1/LHY gene from Phaseolus vulgaris (AJ420902; Kaldis et al., 2003; Fig. 1A). The tree does not resolve whether McCCA1/LHY is most closely related to AtCCA1 or AtLHY; hence, we maintained its dual name. However, the tree does reveal that the available full-length monocot CCA1/LHY sequences from rice (OsCCA1/LHY and OsCCA1/LHY2), sugarcane (Saccharum officinarum [SoCCA1/LHY]), and barley (Hordeum vulgare [HvCCA1/LHY]) are most closely related to AtCCA1, suggesting that the rice gene, which has been named OsLHY elsewhere (Izawa et al., 2003), might be termed OsCCA1. At present, we have not detected a rice LHY ortholog in the publicly available rice genome and EST databases.
Figure 1.
Phylogenetic tree analysis reveals that the M. crystallinum clock genes are the orthologs of the Arabidopsis clock genes. A, McCCA1/LHY tree. McCCA1/LHY (GenBank accession no. AY371287), AtCCA1 (U28422), AtLHY (AJ006404), PvLHY (common bean, AJ420902), LeCCA1/LHY (tomato, BT012912), VvCCA1/LHY (grape, TIGR TC32028), HvCCA1/LHY (barley, TC121173), SoCCA1/LHY (sugarcane, TC13152), OsCCA1/LHY (rice, TC231874) and OsCCA1/LHY2 (TC231873). B, McTOC1 tree. McTOC1 (GenBank accession no. AY371288), AtTOC1 (AF272039), AtAPRR3 (AB046956), AtAPRR5 (AB046955), AtAPRR7 (AB046954), AtAPRR9 (AB046953), OsTOC1 (rice, TIGR rice pseudomolecules ch. 2 9630.t03778, TC235603) and TaTOC1 (wheat, TC150832). C, McELF4 tree. McELF4 (AY371289), AtELF4 (At2g40080), AtELF4-like (At1g17455), AtELF4-like (At1g72630), AtELF4-like (At2g29950), AtELF4-like (At2g06255), OsELF4-like (rice, AF119222, AF161269 ch. 11), GmELF4-like (soybean, TIGR TC166607), LeELF4-like (tomato, TC118553), and MtELF4-like (barrel medic; TC80100). D, McZTL and McFKF1 tree. McZTL (AY371290), McFKF1 (AY371291), AtZTL (AF254413), AtFKF1 (AF216523), AtLKP2 (AB038797), OsFKF1 (rice, TIGR rice pseudomolecules ch. 11 9639.t03046), OsZTL/FKF1/LKP2 (rice; TIGR pseudomolecules ch. 2 9630.t00468, TC217395), TaZTL/FKF1/LKP2 (wheat, TC174398), and HvZTL/FKF1/LKP2 (barley, TC111655). E, McELF3 tree. McELF3 (AY371291), AtELF3 (At2g25930), AtEEC (At3g21320), OsELF3 Ch.1 (rice, AP000399), and OsELF3 Ch. 6 (rice, AP003296). ch, Chromosome.
Casein kinase II can phosphorylate AtCCA1 and AtLHY in vitro and is necessary for its circadian oscillator function (Daniel et al., 2004). Using mass spectrometry, these authors have determined that S-5 and S-6 plus 1 or more of S-431, S-432, S-433, and S-484 are phosphorylated by CK2. These sites are conserved in McCCA1/LHY.
The phylogenetic tree based on the alignment of the McTOC1 gene (AY371288) with the family of 5 Arabidopsis pseudoresponse regulator genes confirms that we have identified a M. crystallinum gene that is most closely related to AtTOC1/AtAPRR1 (44.5% identity; Fig. 1B). The wheat (Triticum aestivum) and rice TOC1 orthologs (TaTOC1 and OsTOC1) form a separate branch of the tree but are clearly more closely related to AtTOC1 than AtAPRR3, AtAPRR5, AtAPRR7, and AtAPRR9. McELF4 (AY371289) and AtELF4 show 41% identity overall and the phylogenetic tree indicates that McELF4 is most closely related to AtELF4 and LeELF4-like when compared to the 4 Arabidopsis ELF4-like genes (Fig. 1C). The rice ELF4-like gene falls among AtELF4-like genes on a separate branch of the tree to AtELF4. We also identified 2 ELF4-like sequences from the model legumes barrel medic (Medicago truncatula) and soybean (Glycine max; MtELF4-like and GmELF4-like), and these form a third branch on the ELF4-tree.
The phylogenetic tree in Figure 1D provides good support for McZTL (AY371290; 72% identity with AtZTL) and McFKF1 (AY371291; 80.4% identity with AtFKF1) being the closest orthologs of the respective Arabidopsis genes. The rice, wheat, and barley sequences all have highest similarity to AtFKF1. A rice gene was identified on chromosome 11 and termed OsFKF1 because it clusters with AtFKF1 and McFKF1 (Fig. 1D). The analysis precluded a precise placement of the other monocot sequences in relation to ZTL, FKF1, or LKP2. The alignment of McELF3 (AY371292) with AtELF3 (34.3% identity) was restricted to conserved blocks identified previously in alignments of AtELF3 with a number of homologous ESTs (Hicks et al., 2001; Liu et al., 2001). In other regions of the protein, McELF3 shows limited similarity to AtELF3 or to the OsELF3 orthologs. McELF3 is most similar to AtELF3 rather than to AtEEC, which lies on a separate branch of the tree (Fig. 1E). The two rice OsELF3-like sequences are highly similar to one another and are orthologs of AtELF3.
Temporal Transcript Abundance Profiles of Central Clock Genes in C3 and CAM M. crystallinum Leaves in Light/Dark Cycles and Free-Running Conditions
Transcript abundance profiles for the M. crystallinum clock gene orthologs were established in the leaves of both young C3 and older CAM-induced plants using semiquantitative RT-PCR. We examined the transcript abundance of each gene in both 12-h-light/12-h-dark (LD) cycles and continuous light (LL) to determine whether each gene oscillates in response to either light/dark cycles or a circadian clock. Abundance of McUBQ10 (TIGR TC4886) was used as a loading control and all data were normalized to the McUBQ10 signal (Fig. 2A). As controls for CAM induction and evening- and day-expressed clock-controlled genes (CCG), we monitored the transcript abundance of McPPCK (AF158091) as a known CAM-induced CCG, a McCCR1/2 ortholog (TC6352) as a known evening-expressed CCG, and McCAB2 (AF003128) as a known day-expressed CCG (Fig. 2). While McPPCK has previously been reported to be a CCG in CAM-induced leaves, with transcript abundance peaking in the night under the control of the circadian clock, our data show that McPPCK transcripts are not under the control of the clock in C3 M. crystallinum (Taybi et al., 2000; Dodd et al., 2003; Fig. 2B). In fact, in C3 leaves illuminated at a light intensity of 500 to 550 μE m−2 s−1, McPPCK transcript levels are light-induced and in C3, LL McPPCK transcripts plateau at a high, light-induced signal with no clear evidence of clock control. These data demonstrate a clear phase shift in the transcript peak of McPPCK with the transition of CAM leaves from LD to LL. In LD cycles, McPPCK transcript levels peak predawn, while in LL conditions, the transcript levels peak 6 h later in the midmorning. This demonstration of circadian-controlled McCCR1/2 and McCAB2 expression in M. crystallinum is noteworthy. Furthermore, the peak expression of McCCR1/2 in the evening (Fig. 2C) and McCAB2 in the day (Fig. 2D) matches the expression pattern for the corresponding genes in Arabidopsis. In LD cycles, the peak level of McCCR1/2 transcripts in the evening is largely unaffected by CAM induction (Fig. 2C), while the peak level of McCAB2 transcripts is strongly repressed (approximately 5-fold) by CAM induction (Fig. 2D).
The transcript abundance of McCCA1/LHY displays robust oscillations in C3 and CAM leaves (Fig. 3A). There was little difference between the maximum level of transcript detected in the C3 and CAM leaf samples. This indicates that the clock control of McCCA1/LHY is not affected by CAM induction (Fig. 3A). The peak of McCCA1/LHY transcripts occurs after dawn, but expression is also high predawn, indicating a broad dawn-phased peak of transcript (Fig. 3A). This phased expression of McCCA1/LHY is in keeping with the circadian control of CCA1/LHY in Arabidopsis (Wang and Tobin, 1998). In contrast, McTOC1 transcript levels peak at 5 pm when McCCA1/LHY levels decline rapidly (Fig. 3B). One notable feature of C3 McTOC1 transcript abundance is that in LL, the peak of McTOC1 transcripts is phase delayed 6 h to a peak at 11 pm. Furthermore, there is evidence that the steady-state transcript abundance of McTOC1 increased in the CAM leaves (compare C3 LD with CAM LD in Fig. 3B). The transcript abundance profiles for McCCA1/LHY and McTOC1 are consistent with reciprocal regulation, as has also been reported for Arabidopsis (Fig. 3, A and B; Alabadi et al., 2001). When McCCA1/LHY transcripts are high in the morning, McTOC1 transcripts reach their trough, and when McTOC1 transcripts peak in the late afternoon, McCCA1/LHY declines rapidly toward an evening trough (Fig. 3, A and B).
The transcript abundance of McELF4 displays robust oscillations in both C3 and CAM leaves of M. crystallinum under LD and LL (Fig. 3C). McELF4 transcript levels reach their peak around subjective dusk and their trough around subjective dawn. This abundance profile is compatible with the proposed role of AtELF4 as part of the evening-expressed mechanism of the central clock (Doyle et al., 2002).
The transcript abundance profiles of the 2 ZEITLUPE family genes McZTL and McFKF1 were found to be under circadian control in C3 and CAM M. crystallinum (Fig. 4, A and B). In C3 leaves, McZTL transcript levels peak in the afternoon and reach their trough in the morning. In CAM-induced leaves, McZTL transcripts also peak in the afternoon, but McZTL transcripts are high between the morning and the evening in CAM leaves, reaching a sharp trough before subjective dawn (Fig. 4A). It is clear that the circadian transcript abundance profile of McZTL changes with CAM induction with a more prolonged period of high transcript abundance in the CAM leaves. McFKF1 transcripts peak sharply in the afternoon and are low to undetectable throughout the remainder of the 24-h cycle (Fig. 4B). In Arabidopsis, FKF1 transcript levels peak 7 to 10 h after subjective dawn, while McFKF1 transcripts peak 9 h after subjective dawn (Nelson et al., 2000; Fig. 4B).
McGI transcript levels oscillate in C3 and CAM leaves with peak transcript levels occurring in the afternoon (Fig. 4C). McGI transcripts reach their trough before subjective dawn. Again, this is consistent with the circadian transcript profile of GI in Arabidopsis. In Arabidopsis, AtGI transcript levels peak in the evening, between 8 and 12 h after dawn, while in M. crystallinum, McGI transcript levels peak 9 h after dawn (Fowler et al., 1999; Park et al., 1999; Fig. 4C).
The transcript level of McELF3 is under circadian control in C3 and CAM leaves (Fig. 4D). Transcripts of this gene increase in the latter part of the day and remain on throughout the night. This is consistent with the McELF3 gene playing a role in gating the input of light signals to reset the clock in M. crystallinum, as has been demonstrated for the Arabidopsis ELF3 gene (McWatters et al., 2000). In both C3 and CAM M. crystallinum leaves, McELF3 transcripts reach their trough in the morning after subjective dawn.
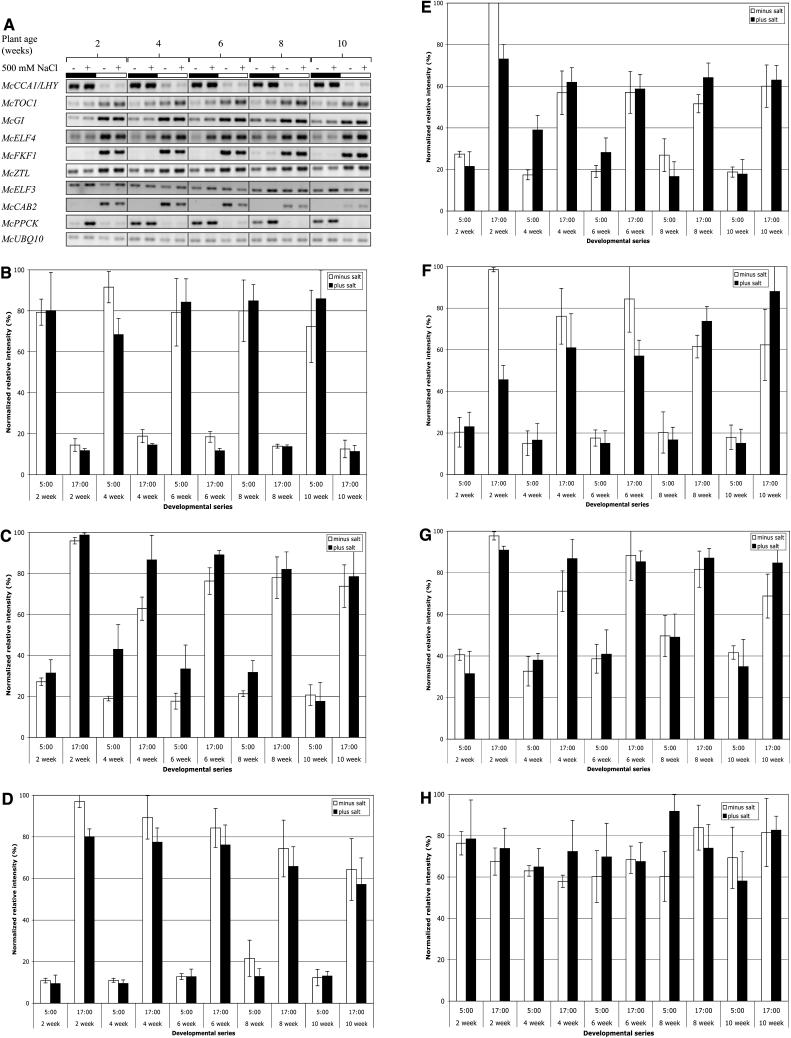
Clock Operation during Development and in Response to Salt Stress
For the transcript abundance analysis shown in Figures 2 to 4, we used 29- to 31-d-old plants (measured from the day of seed sowing) as the C3 plants, and 65- to 67-d-old mature plants that had been salt stressed with 500 mm NaCl for almost 3 weeks as the CAM plants. These plants were chosen to directly compare fully C3 leaves with fully CAM-induced leaves. This raises the possibility that the changes in the expression of McTOC1 and McZTL that we identified in the experiment in Figures 2 to 4 may be due to plant development or salt stress or the interaction of both factors. To investigate whether the regulation of the components of the central circadian clock is influenced by development or salt stress, we sampled M. crystallinum plants every 2 weeks between 2 and 10 weeks of age. For each age group of plants, one-half were salt stressed with 500 mm NaCl for 1 week prior to sampling, while one-half of the plants were maintained well watered as a control. It should be noted that we grew the plants in 1-L pots and thus we cannot rule out the possibility that the well-watered plants were subjected to mild water stress as their root volume increased over time. We collected leaf samples at 5 am (3 h prior to dawn) and 5 pm (3 h prior to dusk) and isolated RNA. We then performed semiquantitative RT-PCR to determine the steady-state transcript abundance of each of the clock-associated genes throughout development with and without salt stress. This analysis revealed that the transcript abundance of the majority of the M. crystallinum clock-associated genes is remarkably constant throughout development and in response to salt stress (Fig. 5, A–H). Unlike the other clock genes, which all show a good differential in transcript abundance between 5 am and 5 pm, there is very little difference in the transcript abundance of McELF3 between 5 am and 5 pm in Figure 5H, despite the fact that McELF3 is clearly a CCG (Fig. 4D). Comparison of the 5 am and 5 pm time points in Figure 4D reveals that McELF3 transcript levels differ less at these 2 time points in LD cycles. At 5 am, McELF3 levels are decreasing to their trough at 11am, while at 5 pm, McELF3 levels are increasing to their peak at 11 pm.
Figure 5.
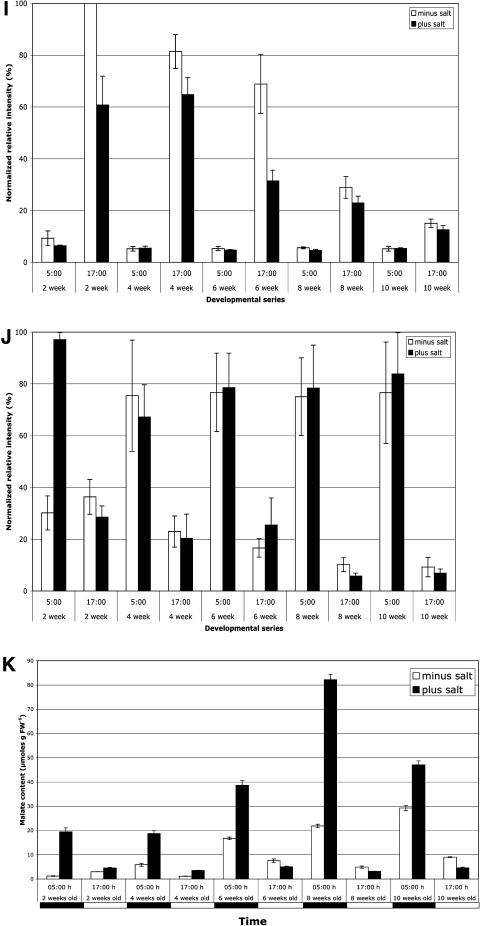
The transcript abundance of clock-associated genes shows compensation against development and salt stress. M. crystallinum plants were grown in 12/12 LD cycles. Salt-stressed plants were irrigated with 500 mm NaCl for 1 week prior to sampling. Leaf samples were collected at 5 am, 3 h prior to dawn, and at 5 pm, 3 h prior to dusk, from plants that were 2, 4, 6, 8, and 10 weeks old, and RNA was isolated. Semiquantitative RT-PCR was performed on the RNA. McUBQ10 was amplified from the same RNA samples to check the RNA loading. A, Typical gel image for the clock-associated genes plus McPPCK and McCAB2. B to J, Average normalized relative intensity of triplicate biological replica experiments. The error bars represent the sd. B, CCA1. C, TOC1. D, GI. E, ELF4. F, FKF1. G, ZTL. H, ELF3. I, CAB2. J, PPCK. K, Leaf malate content was determined for the same leaf samples as an indicator of nocturnal malate accumulation due to the operation of CAM.
CAB genes have previously been reported to be repressed by salt stress in M. crystallinum (Kore-eda et al., 2004). The 5 pm peak in the transcript abundance of McCAB2 is salt repressed in 2-, 4-, and 6-week-old plants (Fig. 5I). There is also a significant developmental decrease in McCAB2 transcript levels. Even though salt stress causes a repression of McCAB2 in 2-, 4-, and 6-week-old plants, there is still an additional developmental repression of McCAB2 as the plants age (Fig. 5I). In 10-week-old plants, peak levels of McCAB2 are low with and without salt stress (Fig. 5I).
The regulation of the transcript level of McPPCK throughout development, with and without salt stress, is very interesting (Fig. 5J). This gene is generally regarded as a salt-induced CAM-specific gene (Taybi et al., 2000; Dodd et al., 2003). However, only in the 2-week-old unstressed plants are McPPCK transcript levels low both predawn and predusk. A week of salt stress induces McPPCK levels very strongly in the dark at 5 am even in these 2-week-old plants. We did not see light induction of McPPCK in the 2-week-old unstressed plants used in this experiment, unlike the light induction observed in the C3 plants used in the experiment in Figure 2. This is most likely due to the fact that the plants for the experiment shown in Figures 2 to 4 were grown at 500 to 550 μmol photons m−2 s−1, while the plants used in the experiment in Figure 5 were grown at 300 to 330 μmol photons m−2 s−1. It is well known that the magnitude of the light induction of PPCK activity and PPCK transcripts in C3 and C4 plants is dependent on the light intensity (Nimmo et al., 1987; Bakrim et al., 1992; Fontaine et al., 2002; Marsh et al., 2003). Our data suggest this is also true for McPPCK in unstressed leaves of M. crystallinum that are performing C3 photosynthesis. In all the other plants, from 4 to 10 weeks old, McPPCK transcripts are present in the dark even in the well-watered control plants. Salt stress has little effect on the McPPCK level in these older plants. It is clear that the unstressed nocturnal level of McPPCK in the 4- to 10-week-old plants is substantial. This suggests that a key molecular component of the circadian control of CAM is nocturnally expressed in unstressed plants from 4 weeks old onward. Examination of the malate content of the same leaf samples used for RNA isolation revealed that malate levels were higher at 5 am than at 5 pm, indicative of nocturnal CO2 fixation via PEPc associated with CAM, in all but the 2-week-old unstressed plants in which the malate level is higher at 5 pm and McPPCK levels are low at both 5 am and 5 pm (Fig. 5K). Although McPPCK transcripts are high at 5 am in the 4-week-old unstressed plants, the amount of malate accumulated is low (6 μmol g−1 fresh weight) compared to the level in the 4-week-old salt-stressed leaves (19 μmol g−1 fresh weight) in which the level of McPPCK transcripts at 5 am is very similar to that in the unstressed leaves. Clearly, the 4-week-old unstressed leaves are performing only a little nocturnal malate accumulation. Examination of the transcript levels for several other CAM-associated genes (e.g. PEPc, malic enzyme, and enolase), which are known to be induced by salt stress during the induction of CAM, revealed that these genes were not induced in the 4-week-old unstressed leaves, suggesting that even if the plants had experienced mild water limitation due to their limited 1-L pot size, this was not sufficient to induce CAM (data not shown). Thus, the nocturnal induction of McPPCK in 4-week-old unstressed plants is not indicative of the leaves performing significant CAM as judged by malate accumulation. Examination of the regulation of McPPCK transcript levels throughout the dark period in 4-week-old unstressed leaves and 8-week-old stressed leaves revealed that, while McPPCK transcripts are high in the 4-week-old unstressed leaves at the 5 am time point reported here and at 8 am, they are low earlier in the night at 8 pm, 11 pm, and 2 am (data not shown). However, the 8-week-old salt-stressed leaves have high McPPCK transcript levels at 11 pm, 2 am, 5 am, and 8 am, revealing a significantly prolonged peak of McPPCK transcript abundance in these older stressed leaves (data not shown). Thus, the substantially greater malate accumulation at 5 am in the 8-week-old stressed leaves correlates with a much broader or more prolonged peak of McPPCK levels. This important result indicates that it is not the quantity of McPPCK at a particular time point during the night that is important for determining the capacity for nocturnal malate accumulation, but the duration of the McPPCK transcript peak throughout the night.
DISCUSSION
Circadian Clock Genes in M. crystallinum
We have cloned the M. crystallinum orthologs corresponding to seven plant circadian clock-associated genes. All seven of the M. crystallinum genes reported here were cloned and characterized from the model C3 plant Arabidopsis. This report of the cloning and characterization of all clock-associated genes in a second plant species not only documents evolutionary conservation and adaptation of the clock function, but also provides information about clock functioning in a CAM plant and compensation of the clock against development and abiotic stress.
Our phylogenetic analysis of the available full-length CCA1/LHY sequences reveals some very interesting points. First, Arabidopsis is the only species that possesses 2 genes belonging to the CCA1/LHY family. The two rice sequences in our phylogenetic tree represent transcripts from a single gene. A BLAST search against the TIGR rice genome database (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1&database=Genes_in_TIGR_Rice_Pseudomolecules) revealed only 1 rice OsCCA1/LHY-related sequence on chromosome 8, and this is the gene that encodes OsCCA1/LHY and OsCCA1/LHY2. Furthermore, the TIGR TC sequences from which we derived the 2 rice genes in Figure 1A align perfectly, except for a small number of base changes (23 out of 3,499 bases for OsCCA1/LHY compared to OsCCA1/LHY2) and 5 gaps (30–160 bp in length), suggesting that OsCCA1/LHY and OsCCA1/LHY2 are splice variants from the same gene. OsCCA1/LHY contains 4 unspliced introns that are not present in OsCCA1/LHY2. However, it is noteworthy that the unspliced introns do not create premature stop codons and thus the unspliced version of the OsCCA1/LHY transcript could encode a functional protein. The rice OsCCA1/LHY gene clearly merits further investigation in relation to alternative splicing. Second, all of the available monocot CCA1/LHY sequences are most closely related to AtCCA1, revealing that AtCCA1 itself has evolved relatively slowly since the monocot-dicot split. To improve our understanding of the evolution of the plant circadian clock, it will be important to determine whether other dicot species possess 2 genes in the CCA1/LHY family as the Arabidopsis paradigm might suggest.
The TOC1 phylogenetic tree shows that McTOC1 is the nearest neighbor to AtTOC1 (Fig. 1B). There are numerous plant ESTs that encode fragments of TOC1 genes from other species, but only ESTs from rice and wheat assemble into full-length TOC1 sequences (Fig. 1B). As in AtTOC1, in McTOC1 the 2 Asp residues that are essential for phosphorelay in response regulator proteins are changed to Glu (Asp-1) and Asn (Asp-2; Strayer et al., 2000). This implies that these residues may themselves play an important role in the function of the pseudoresponse regulator domain of TOC1 because they are conserved between the Aizoaceae and the Brassicaceae. The C-terminal CONSTANS domain of AtTOC1 is also well conserved in McTOC1, and C terminal to this motif is a region rich in acidic residues. Overall, the C termini of the 2 proteins (from residue 512 of AtTOC1 and 444 of McTOC1 to the C terminus) are 61% identical. The C-terminal acidic region is less highly conserved in the other Arabidopsis pseudoresponse regulator proteins APRR3, APRR5, APRR7, and APRR9. For example, the C terminus of McTOC1 (from residue 444 to the C terminus) shares between 33% and 42% identity with the C terminus of AtAPRR3, AtAPRR5, AtAPRR7, and AtAPRR9. Such an acidic region is a common feature of many transcriptional activators, and the conservation of this region among the TOC1 genes implies they may act as transcription factors.
McELF4 is clearly a good ortholog of AtELF4 (Fig. 1C). The ELF4 genes encode very small, nuclear localized proteins of around 110 amino acid residues with no known functional domains (Doyle et al., 2002; Khanna et al., 2003). Only 22 residues are identical between all of the available ELF4 and ELF4-like sequences.
McZTL and McFKF1 were named based on their phylogenetic relationships revealed in the tree in Figure 1D. The gene duplication event that generated these two genes clearly predates the separation of the Aizoaceae and the Brassicaceae from their common ancestor. AtLKP2 sits alone on a separate branch of the tree, suggesting that there could be a McLKP2 gene in the M. crystallinum genome. The ZEITLUPE family of proteins in Arabidopsis contains 3 conserved protein motifs: a LOV-type PER ARNT SIM domain near the N terminus, a 40-amino acid F-box in the center of the protein, and 6 C-terminal kelch repeats. All 3 of these motifs are conserved in McZTL and McFKF1, indicating that the proteins are likely to function in a similar manner in M. crystallinum.
McELF3 is most closely related to AtELF3. ELF3 is a novel protein that lacks identifiable protein motifs. However, ELF3 does align with short ESTs from a range of plant species, and this has allowed a number of ELF3 conserved domains to be identified (Hicks et al., 2001; Liu et al., 2001). All of these domains are conserved in McELF3. AtELF3 is nuclear localized, but the putative nuclear localization signal is poorly conserved in McELF3 (2 out of 7 residues identical; Hicks et al., 2001). Furthermore, only a single residue of the putative AtELF3 nuclear localization signal is conserved in the rice ELF3 orthologs. However, McELF3 is predicted to be nuclear localized based on its generally basic sequence (PSORTII; Reinhardt's method gives a 94.1% reliability that McELF3 is nuclear localized; k-NN prediction gives a 65.2% probability that McELF3 localizes to the nucleus).
McPPCK and McCCR1/2 Regulation in M. crystallinum
McPPCK transcript levels show a 6-h phase delay in their peak in LL conditions in CAM leaves (Fig. 2B). This phase delay in LL has not been reported previously and is important in terms of understanding CAM CO2 fixation rhythms in LL. None of the central clock genes that we have studied display a similar phase delay in CAM leaves in LL. This suggests that if the CCA1/LHY and TOC1 oscillator is providing the temporal information that coordinates McPPCK during CAM, then the output pathway that links the central clock to McPPCK must include an element that undergoes a phase delay relative to the underlying oscillator when CAM leaves are transferred to LL. Alternatively, McPPCK control may occur via a different oscillator that is phase delayed in LL.
Our analysis of the regulation of McCCR1/2 cycling in LL (Fig. 2, A and C) reveals that, while this gene oscillates robustly in C3 leaves, the oscillations damp out very rapidly in CAM-induced leaves. However, McCCR1/2 transcript levels do cycle in CAM leaves under driven LD conditions, so it is clear that McCCR1/2 may be specifically uncoupled from its clock in CAM-induced leaves. In Arabidopsis, AtCCR2 expression is controlled in part by its own protein in a suboscillator feedback loop (Heintzen et al., 1997). This makes it possible that the autoregulation of McCCR1/2 is lost in salt-stressed, CAM-induced leaves even though the central clock itself is largely compensated against stress and continues to operate robustly in CAM-induced leaves.
The M. crystallinum Clock Genes Are Themselves Clock Controlled
Our analysis of the regulation of the clock-associated genes in C3 and CAM leaves allowed us to discern two key points about the control of the transcript level of these genes in M. crystallinum. First, the transcript abundance of all seven clock-associated genes not only oscillated in LD, but also continued to oscillate in LL. Second, we identified McZTL as the one gene whose transcript abundance profile differs from its Arabidopsis ortholog. AtZTL transcripts do not oscillate in LD or LL (Somers et al., 2000), but we have demonstrated that McZTL transcripts oscillate in LD and LL in both C3 and CAM leaves.
The amplitude of the oscillations in the relative transcript abundance of McCCA1/LHY changes little with CAM induction. This supports the hypothesis that these single Myb-repeat transcription factors form part of the central oscillator in M. crystallinum. If the transcript abundance of these genes changed markedly with the C3-to-CAM switch, it would suggest that the clock includes other components that are more important for maintaining robust rhythmicity throughout the life of the plant. It is of particular interest in this context to note that the relative transcript abundance for three isoforms of the light-harvesting chlorophyll a/b-binding protein (McCAB) oscillated in both C3 and CAM M. crystallinum (Fig. 2D; data not shown). Although rhythmicity of the McCAB transcript oscillations is maintained between C3 and CAM, the amplitude of McCAB rhythms decreases significantly between C3 leaves and CAM leaves because McCAB genes are repressed by CAM induction (Fig. 2D). The AtCCA1/LHY transcription factors bind to the promoter of AtCAB genes and mediate both the acute response of these genes to light and their circadian regulation (Wang et al., 1997; Wang and Tobin, 1998). It is therefore significant that, although McCAB gene transcript levels are repressed after CAM induction, McCCA1/LHY levels are relatively unchanged between C3 and CAM. This suggests the existence of a repressor protein binding to the McCAB promoters to down-regulate their expression in CAM-induced leaves. Alternatively, these transcripts are globally repressed as a result of stress treatment and/or are selectively targeted for turnover following stress.
Our data on the transcript profile of McZTL in C3 and CAM M. crystallinum demonstrate that, during plant evolution, the control of the ZTL gene has altered between a member of the Aizoaceae and a member of the Brassicaceae. Conservative estimates, based on the fossil record, of the dates of divergence of the major eudicot clades put the date of divergence of the Brassicales (Arabidopsis) at 89.5 million years ago and the Caryophyllales (Mesembryanthemum) at 83 million years ago (http://www.flmnh.ufl.edu/deeptime). Thus, a change has occurred in the regulation of the transcript abundance of ZTL between Arabidopsis and Mesembryanthemum sometime in the last 80 to 90 million years. Recently, AtZTL has been reported to be regulated posttranscriptionally via different circadian phase-specific degradation rates (Kim et al., 2003). It will be interesting to see whether McZTL is also regulated posttranscriptionally via proteasome-dependent proteolytic degradation in M. crystallinum, in addition to the circadian control of its relative transcript abundance reported here. Our findings suggest that there will be many subtle differences in the machinery of the plant circadian clock that become apparent as more plant species are studied with respect to their clock genes and the regulatory interactions that constitute the central plant clock. Studying the evolution of the plant clock will undoubtedly provide substantial insight into which genes are the most fundamental components of the central plant clock and which genes are more peripheral and therefore have been subjected to greater or lesser selection pressure during evolution.
The Plant Circadian Clock Is Largely Compensated against Development and Abiotic Stress
Our data examining the influence of development and salt stress on the operation of the central clock (Fig. 5) provide insight into environmental compensation within the plant circadian oscillator. While it is well established that the clock is temperature compensated to permit robust rhythmicity over a broad temperature range (Somers et al., 1998), compensation of the plant clock against other environmental signals has not been examined previously. Our data reveal that the plant clock is largely compensated against severe abiotic stress in the form of 500 mm NaCl. Furthermore, the circadian control of the plant clock genes also changes little during the life of the plant in the absence of salt stress. The fact that the central clock genes operate robustly, showing only relatively small changes throughout the life cycle of M. crystallinum even in the face of severe salt stress, is further evidence of just how important the clock is to the competitive success of a plant. By maintaining consistent clock function in a diverse range of conditions, the plant can ensure that it still optimizes the temporal aspects of its metabolism that are vital to reproductive success. This latter point is particularly relevant in a stress-inducible CAM plant, where it is clear that coordination of the CAM pathway by the circadian clock avoids potentially catastrophic futile cycles between malate synthesis and decarboxylation.
The identification of the clock genes reported here in a model inducible CAM plant that can switch rapidly from C3 to CAM sets the stage for future work aimed at a detailed understanding of the molecular basis of the circadian control of the CAM pathway. We now have the necessary molecular tools to manipulate the expression of these genes in transgenic CAM plants and examine the effect these perturbations have on the circadian control of CO2 fixation via the CAM pathway. These experiments will allow us to finally resolve the nature of the circadian oscillator that controls CAM.
MATERIALS AND METHODS
Plant Material
Mesembryanthemum crystallinum plants were grown from seed in vermiculite irrigated with half-strength Hoagland solution in a growth chamber on a 12-h-light (23°C)/12-h-dark (18°C) cycle. The photoperiod was for 12 h from 8 am until 8 pm. Fluorescent lighting provided a photon flux density of 500 to 550 μmol photons m−2 s−1. Young plants aged 29 d were used for the collection of C3 leaves and plants that were 65 d old and had been stressed with 500 mm NaCl for 19 d were used for the CAM leaves. One-half of the plants were in LD cycles and one-half were transferred to LL and constant temperature (23°C) at the beginning of the experiment (8 am, lights on). Duplicate leaf samples were collected every 6 h for a total of 60 h at the following times: 11 am, 5 pm, 11 pm, and 5 am, giving a total of 10 time points for each treatment (C3 LD, C3 LL, CAM LD, and CAM LL). For the experiment to examine the role of development and salt stress in clock gene control (Fig. 5), the plants were grown in 1-L pots at 300 to 330 μmol photons m−2 s−1, and the salt-stressed plants were irrigated with 500 mm NaCl for 1 week prior to sampling. All samples were frozen in liquid nitrogen and stored at −80°C until use.
RNA Isolation
Frozen leaf samples were ground in liquid nitrogen using a mortar and pestle, and total RNA was isolated from the frozen powder using a cetyl-trimethyl-ammonium bromide-based RNA extraction procedure (Hartwell et al., 1996). Total RNA samples were treated with DNase (DNA-free kit; Ambion., Austin, TX) to eliminate contaminating genomic DNA. Many of the primers used for the amplification of specific genes flanked introns, and contamination by genomic DNA was never detected in any of the RT-PCR reactions. Following DNase treatment, the quantity and purity of the total RNA were determined spectrophotometrically as described (Sambrook et al., 1989). All RNA samples were diluted to a concentration of 0.2 μg/μL prior to use in RT reactions.
Semiquantitative RT-PCR
The total RNA samples (2 μg) were mixed with 1 μg of anchored oligo(dT) (5′-AAGCTTTTTTTTTTTTTTTV-3′) and incubated at 95°C for 2 min and immediately cooled on ice. RT was carried out in a reaction mixture (40 μL) containing the denatured RNA plus oligo(dT), 1× Stratascript RT buffer (Stratagene, La Jolla, CA), 1 mm dNTPs (Invitrogen, Carlsbad, CA), and 40 units of Stratascript RT (Stratagene). The reaction was incubated at 37°C for 90 min, followed by 95°C for 5 min.
PCR reactions were performed using 1 μL of each reverse transcribed cDNA sample in a reaction mixture (10 μL) containing 1× Sigma ReadyMix REDTaq PCR reaction mix with MgCl2 (Sigma, St. Louis) and 1 μm of each forward and reverse primer. The primer sequences and the predicted product sizes are indicated in Table I. PCR reactions were conducted in a programmable thermocycler (PTC200 DNA engine; MJ Research, Watertown, MA), and the optimal number of PCR cycles for each gene is indicated in Table I. Standard PCR cycles were 95°C for 2 min, a gene-specific number of cycles of 55°C/30 s, 72°C/1 min, 95°C/30 s, and a final extension step of 55°C for 30 s followed by 72°C for 7 min. All products were separated on a 1% agarose gel in 1× Tris-acetate EDTA and stained with ethidium bromide. Gels were visualized using a Bio-Rad gel documentation system and band intensities were quantified using Quantity One software (Bio-Rad Laboratories, Hercules, CA). An M. crystallinum polyubiquitin gene (McUBQ10; TIGR TC4886) with high homology to Arabidopsis (Arabidopsis thaliana) UBQ10 was used as the loading control for the RT-PCR. The McUBQ10 primer sequences used are given in Table I. The quantified RT-PCR signals for all the clock genes examined here were normalized to the McUBQ10 signal to correct for minor variations in the loading of RNA into the RT reactions and/or the efficiency of the RT reactions. All experiments were carried out in duplicate; in each case, similar trends were observed. The data presented in Figures 2 to 4 are from individual experiments that are representative of the results obtained. The data in Figure 5, B–J, show the mean normalized relative intensity from three biological replicates and the error bars represent the sd. The gel in Figure 5A shows the raw data for a single replicate.
The gene-specific primers for McCCA1/LHY, McELF4, McGI, and McELF3 were designed to target partial 3′ EST sequences corresponding to fragments of the M. crystallinum orthologs. The ESTs were identified from the M. crystallinum gene index (http://www.tigr.org/tdb/tgi/mcgi; Kore-eda et al., 2004) using the TBLASTN search algorithm and the Arabidopsis CCA1/LHY, ELF4, GI, and ELF3 protein sequences as input sequences. The FKF1 ESTs have GenBank accession numbers AI026317, AA791399, AA791403, and AI822178. The CCA1/LHY ESTs have accession numbers BF479609, BM300086, and BM034918. The ELF3 EST has accession number BF479720, the ELF4 EST has accession number CA838873, and the GI EST has accession number CA838115.
Degenerate PCR to Clone the M. crystallinum ZTL and FKF1 Orthologs
To isolate the M. crystallinum cDNA sequences corresponding to the Arabidopsis clock-associated genes ZTL and FKF1, a degenerate PCR strategy was employed. The degenerate PCR primers ZTLF 5′ CAWGGNGADYTDYTNAAYTTY 3′ (corresponding to the conserved amino acid motif QGELLNF), and ZTLR1 5′ RCTHGCYTRDGADARYTCATG 3′ and ZTLR2 5′ RCTYGCYAARCAHARYTCATG 3′ (corresponding to the conserved amino acid motifs HELSLAS found in AtZTL and HELCLAS found in AtFKF1, respectively) were used. The conserved regions of the ZEITLUPE family of proteins were identified by aligning EST sequences from Arabidopsis, tomato (Lycopersicum esculentum), Sorghum bicolor, soybean (Glycine max), wheat (Triticum aestivum), barley (Hordeum vulgare), rice (Oryza sativa), Lotus japonicus, Zea mays, M. crystallinum, and barrel medic (Medicago truncatula).
Cloning a 5′ Fragment of the M. crystallinum TOC1 Gene
We identified two ESTs from sugar beet (Beta vulgaris; accession nos. BI543444 and BI543434) in the GenBank EST database with homology to the 5′ end of the Arabidopsis TOC1 gene. Sugar beet, like M. crystallinum, is a member of the taxonomic order Caryophyllales, which justified primer design to conserved regions between the sugar beet and Arabidopsis TOC1 orthologs. The primers BvTOC1F 5′-TTCATTGATCGAAGTAAAGTCAG-3′ and BvTOC1R 5′-CCAGCCTCAAGCACTTTACA-3′ were used to amplify a 311-bp fragment of the M. crystallinum TOC1 cDNA via RT-PCR.
Cloning and Sequencing of RT-PCR and RACE-PCR Products
All RT-PCR and RACE-PCR products were gel purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA) and cloned into the TA vector (pCR4-TOPO) using the TOPO TA cloning kit for sequencing (Invitrogen) according to the manufacturer's protocols. The CCA1/LHY 5′ RACE-PCR product was sequenced directly by primer walking. Cloned inserts in pCR4-TOPO were sequenced fully on both strands by the in-house DNA-sequencing units at the University of Arizona, Tucson, and the University of Nevada, Reno, to confirm the gene specificity of the RT-PCR reactions. All PCR reactions were found to have targeted the gene of interest.
Cloning the Full-Length cDNAs for CCA1/LHY, TOC1, ELF4, ZTL, FKF1, and ELF3 Orthologs from M. crystallinum Using RACE-PCR
The 5′ and 3′ ends of the M. crystallinum CCA1/LHY, TOC1, ZTL, FKF1, ELF4, and ELF3 cDNAs were amplified using the SMART RACE-PCR kit (CLONTECH, Palo Alto, CA) according to the manufacturer's protocols. SMART cDNA was synthesized from total RNA corresponding to the time of the transcript peak for each clock gene as determined with semiquantitative RT-PCR analysis. The gene-specific primer for cloning the 5′ end of CCA1/LHY was 5′-CTTGCTGTGGCCAAGGTTTCCCTAGC-3′; for the 5′ end of ZTL, 5′-AGGAGCAACACCTCCAGGGTTTCCAG-3′; for the 3′ end of ZTL, 5′-CAAGCACAGTTCATGCACGCTCGATG-3′; for the 5′ end of FKF1, 5′-GCAAACTTAGGTGGCTGACCGGGAAC-3′; for the 5′ end of ELF3, 5′-CTGCCGTCTTTCTGCTTGTATTGACTGG-3′; for the 5′ end of TOC1, 5′-TCCCTCGGCATTAAGTGCATCAATAACCTG-3′; for the 3′ end of TOC1, 5′-TGTGATAACGATTCCAAGAGCTGCGAGGAG-3′; for the 5′ end of ELF4, 5′-CCCTTCATATTCAATCCACCATTCTCCC-3′; for the 3′ end of ELF4, 5′-TGAAGAATGTGGCGATCATTCAGGAATTG-3′. The 3′ end of CCA1/LHY, FKF1, and ELF3 was obtained by sequencing the 3′ end of ESTs in the database.
Malate Determination
Samples (500 mg) of the same leaves that were used for RNA isolation (including the duplicate samples) were ground in liquid nitrogen and extracted in 7 mL of 80% methanol at 70°C. The methanol extracts were dried down and resuspended in 0.5 mL 200 mm Bicine, pH 7.8. The concentration of malate was determined using the enzymatic method described by Möllering (1974). Each extract was assayed in triplicate.
DNA and Protein Sequence Analysis
DNA sequence data were analyzed using Vector NTI Suite for MacOSX (Informax, Frederick, MD). Database searches were conducted using the National Center for Biotechnology Information (NCBI) network version of BLAST 2.0 (Altschul et al., 1997). Multiple sequence alignments and phylogenetic trees were generated using the AlignX program within the Vector NTI Suite for MacOSX. AlignX uses a modified ClustalW algorithm to align the amino acid sequences and builds phylogenetic trees using the neighbor-joining method (Saitou and Nei, 1987).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to requisite permission from any third-party owners of all parts of the material. Obtaining any permissions will be the responsibility of the requester.
Sequence data from this article have been deposited in the GenBank/EMBL data libraries under the following accession numbers: McCCA1/LHY, AY371287; McTOC1, AY371288; McELF4, AY371289; McZTL, AY371290; McFKF1, AY371291; and McELF3, AY371292.
Acknowledgments
J.H. thanks Susie Boxall for working on this project voluntarily. We thank Christine B. Michalowski for growing some of the plants used in this study.
This work was supported by a Biotechnology and Biological Sciences Research Council, UK, David Phillips Fellowship (grant no. JF14818 to J.H.), and in part by the National Science Foundation (grant no. DBI–9813360 to H.J.B.). J.C.C. acknowledges support from the National Science Foundation (grant nos. IBN–0196070 and DBI–9813360) and the Nevada Agricultural Experiment Station (publication no. 03043029).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.054577.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakrim N, Echevarria C, Cretin C, Cretin C, Arriodupont M, Pierre JN, Vidal J, Chollet R, Gadal P (1992) Regulatory phosphorylation of Sorghum leaf phosphoenolpyruvate carboxylase. Identification of the protein-serine kinase and some elements of the signal-transduction cascade. Eur J Biochem 204: 821–830 [DOI] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999) Metabolite control overrides circadian regulation of phosphoenolpyruvate carboxylase kinase and CO2 fixation in Crassulacean acid metabolism. Plant Physiol 121: 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PJ, Nimmo HG, Fewson CA, Wilkins MB (1991) Circadian rhythms in the activity of a plant protein kinase. EMBO J 10: 2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Griffiths H, Taybi T, Cushman JC, Borland AM (2003) Integrating diel starch metabolism with the circadian and environmental regulation of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Planta 216: 789–797 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Millar AJ (2003) The circadian clock. A plant's best friend in a spinning world. Plant Physiol 132: 732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25: 115–122 [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE (2004) Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 333–342 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (2002) Probing the circadian control of phosphoenolpyruvate carboxylase kinase expression in Kalanchoë fedtschenkoi. Funct Plant Biol 29: 663–668 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071–1078 [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6: 113–120 [DOI] [PubMed] [Google Scholar]
- Kaldis A-D, Kousidis P, Kesanopoulus K, Prombona A (2003) Light and circadian regulation in the expression of LHY and Lhcb genes in Phaseolus vulgaris. Plant Mol Biol 52: 981–997 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kikis EA, Quail PH (2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133: 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Geng R, Somers DE (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kore-eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman JC (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene 341: 83–92 [DOI] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U (2000) Tonoplast functioning as the master switch for circadian regulation of Crassulacean acid metabolism. Planta 211: 761–769 [DOI] [PubMed] [Google Scholar]
- Lüttge U (2003) Circadian rhythmicity: Is the “biological clock” hardware or software? Prog Bot 64: 277–319 [Google Scholar]
- Marsh JT, Sullivan S, Hartwell J, Nimmo HG (2003) Structure and expression of phosphoenolpyruvate carboxylase kinase genes in Solanaceae. A novel gene exhibits alternative splicing. Plant Physiol 133: 2021–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim W-Y, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McClung CR (2001) Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol 52: 139–162 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Möllering H (1974) L-malate: determination with malate dehydrogenase and glutamate-oxaloacetate transaminase. In HU Bergmeyer, ed, Methods of Enzymatic Analysis, Vol 3. Verlag Chemie, Weinheim, Germany, pp 1589–1593
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Nimmo GA, McNaughton GAL, Fewson CA, Wilkins MB, Nimmo HG (1987) Changes in the kinetic properties and phosphorylation state of phosphoenolpyruvate carboxylase in Zea mays leaves in response to light and dark. FEBS Lett 213: 18–22 [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in Crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wilkins MB (1992) Circadian rhythms: their origin and control. New Phytol 121: 347–375 [DOI] [PubMed] [Google Scholar]
- Wyka TP, Bohn A, Duarte HM, Kaiser F, Lüttge UE (2004) Perturbations of malate accumulation and the endogenous rhythms of gas exchange in the Crassulacean acid metabolism plant Kalanchoë daigremontiana: testing the tonoplast-as-oscillator model. Planta 219: 705–713 [DOI] [PubMed] [Google Scholar]