Abstract
Background & Aims:
Multi-target stool DNA testing (MT-sDNA) is now FDA-approved for average-risk CRC screening. Trials leading to its approval used blinded colonoscopy as the reference standard. In the post-approval screen setting, the clinical performance and impact of MT-sDNA on unblinded colonoscopy has not been described. We measured the impact that knowledge of a positive MT-sDNA test result has on colonoscopy yield and quality.
Methods:
The unblinded group comprised all patients with positive MT-sDNA results on screening from 9/1/14–9/30/15 at a single tertiary center. Off-label test patients were excluded. The blinded group included all MT-sDNA positive participants in a pre-approval screening study from the same center. Detailed colonoscopy findings and withdrawal times were recorded.
Results:
There were 172 MT-sDNA positive patients in the unblinded group and 72 in the blinded group. More total adenomatous/sessile serrated polyps (70% vs 53%, p=0.013) and advanced neoplasms (28% vs 21%, p=0.27) were detected in unblinded than blinded groups. Median numbers of polyps detected were 2 (interquartile range, 1 – 4) and 1 (0 – 2) in unblinded and blinded groups, respectively (p=0.0007). Among polyps detected, flat or slightly raised lesions in the right colon were proportionately more frequent with unblinded (40%) than blinded exams (9%) (p=0.0017). Median withdrawal time was 19 min (13 – 29) in the unblinded group compared to 13 min (10 – 20) in the blinded group (p=0.0001).
Conclusions:
Knowledge of a positive MT-sDNA result appears to have a beneficial impact on the diagnostic yield and quality of subsequent colonoscopy.
Keywords: Colorectal Neoplasms/prevention and control, Feces/analysis, Colonoscopy/standards
Introduction
Colorectal cancer (CRC) ranks second only to lung cancer in mortality with roughly 50,000 deaths annually.1 Both incidence and death rate have fallen over the past three decades,1 with evidence suggesting that this is largely due to increased screening rates and early detection of both precancers and curable-stage CRC.2–6 CRC screening is cost-effective and highly recommended for the general population.7–9
For those participating in programmatic screening, colonoscopy remains the most frequently used modality5, 10 and is supported by observational evidence demonstrating a reduction in CRC incidence and mortality.11, 12 However, there is also evidence that colonoscopy does not yield the same mortality benefit for cancers proximal to the splenic flexure as compared to those in the distal colorectum,4, 12, 13 and missed proximal lesions account for a disproportionate number of interval cancers.14, 15 Furthermore, studies of colonoscopy performance show substantial operator variability in precancer detection rates,16 raising risk of interval cancers.17, 18 More polyps and fewer interval cancers are observed with increasing colonoscope withdrawal time19–21, an important surrogate of examination quality.22–24
Survey data show that one in three individuals in the eligible U. S. population remains unscreened by any modality;6, 10, 25 and the actual participation rate may be just one in two based on record review to mitigate over-reporting bias.26 Randomized controlled observations show that CRC screening participation rates and programmatic adherence is improved when patients are given choices among screening tests.27, 28 Further, survey data suggest that, when given a choice, patients prefer methods that are convenient, non-invasive, and safe.6, 29, 30
Multi-target stool DNA testing (MT-sDNA) was developed to overcome barriers to screening, while maintaining high accuracy, independent of lesion site. Cologuard™ (Exact Sciences, Madison WI) is a clinically-available MT-sDNA test, recently approved by both the United States Food and Drug Administration and Centers for Medicare and Medicaid Services. Cross-sectional screening studies have demonstrated high detection rates of CRC and large precancers, and sensitivity was superior to that of fecal immunohistochemical testing (FIT) for curable-stage CRC and advanced precancers.31, 32 Early observations suggest that the availability of MT-sDNA is attracting new patient participation to CRC screening.33
Previous screen-setting studies of MT-sDNA performance used blinded colonoscopy as the reference standard. We hypothesized that endoscopist knowledge of a positive MT-sDNA result would improve the yield and quality of diagnostic colonoscopy in clinical practice, but this has not been assessed to date. Therefore, our aim was to compare colonoscopy findings and withdrawal times among a post-approval cohort, in which endoscopists were informed of positive MT-sDNA results, to those obtained in an earlier clinical trial at the same institution, in which endoscopists were blinded to MT-sDNA results.
Ethical Considerations
This study was approved by the Mayo Clinic Institutional Review Board.
Materials and Methods
Overall study design
We compared colonoscopic findings and withdrawal times between two groups of patients. The first was a cohort of retrospectively identified patients who underwent colonoscopy at Mayo Clinic (Rochester, MN) for the indication of a positive MT-sDNA test administered in routine clinical practice for average-risk CRC screening; for these patients, the endoscopist was aware of the test result (unblinded group). The second cohort included the subset of patients with a positive MT-sDNA test during participation in a multicenter pre-approval screening study (ClinicalTrials.gov Identifier: NCT01397747). Among all eligible participants of this Multi-Target Colorectal Cancer Screening Test for the Detection of Colorectal Advanced Adenomatous Polyps and Cancer (DeeP-C) study,31 we identified those who completed the protocol at one of three Mayo Clinic sites (Rochester, MN; Jacksonville, FL; Scottsdale, AZ); per protocol, patients underwent a multi-target stool DNA test followed by screening colonoscopy in which endoscopists were blinded to the stool test results (blinded group).
Patients
For the unblinded group, patients with a positive MT-sDNA result from September 1, 2014 through September 30, 2015 were identified from the electronic medical record. Patients at average risk for CRC, those with a history of hyperplastic polyps, adenomas more than 60 months before MT-sDNA, or segmental sigmoid colon resection for benign disease were included. In the event that MT-sDNA was ordered for the evaluation of gastrointestinal symptoms, anemia, or above-average-CRC risk (contrary to FDA-approved labelling) results were excluded from the primary analysis but recorded and described. Above-average-risk for CRC was defined as having inflammatory bowel disease, a prior history of advanced colorectal neoplasia (CRN) (includes adenomatous or sessile serrated adenoma/polyps (SSA/P) ≥10 mm, polyps with any >25% villous component or any high-grade dysplasia, or CRC), any adenomas within 60 months of positive MT-sDNA, colorectal surgery for CRN, or patients with a first degree family member with CRC before age 60 years. Those with prior history of extensive colon resection or known active malignancy of the lungs or gastrointestinal tract were excluded as were all who did not complete diagnostic colonoscopy by December 31, 2015.
Inclusion and exclusion criteria of the blinded cohort comprising DeeP-C clinical trial participants have been previously described.31 Briefly, these were individuals age 50 to 84 years scheduled for average risk screening colonoscopy. Similar to those included in the unblinded cohort (above), patients were excluded from the clinical trial if they had a personal history of CRN, digestive tract cancer, inflammatory bowel disease, previous colon resection other than for sigmoid diverticula, overt rectal bleeding in the previous 30 days, or had a family history of early-onset or syndromic CRC. Patients were also excluded from the trial if they had CRC screening by colonoscopy in the previous 9 years, or barium enema or computed tomographic colonography or flexible sigmoidoscopy in the previous 5 years.
Data Abstraction
Patient records were reviewed by a single examiner (D.H.J.) and entered into a secure centralized database. Colonoscopy procedural details were abstracted from individual endoscopy reports (ProVation, Wolters Kluwer, Alphen aan den Rijn, Netherlands) contained in the electronic record including quality of colon preparation (inadequate, poor, fair, good, excellent), neoplastic and non-neoplastic findings, and withdrawal time. For each colonoscopy, the total number of polypoid lesions, and the size and morphology (by Paris classification34) of the largest or highest grade lesion in each side of the colorectum were recorded. Lesion size was obtained from the pathology report in cases of en bloc resection, and from endoscopic size estimates if treated by piecemeal removal. Histologic diagnoses were abstracted from the clinical pathology report. To record the anatomic locations of CRN, the right hemicolon was defined as the splenic flexure and all proximal segments, and the left colon included all colon segments distal to the splenic flexure. Paris classification was assigned by the reviewer based on endoscopic photographs, and when not available, text description provided in the endoscopy report. For colonoscopies performed outside our institution in the unblinded group, data were abstracted from scanned primary source documents.
Stool DNA testing
Stool assays were performed by technicians who were blinded to clinical findings. Full details of the MT-sDNA including stool collection, sample processing and molecular analysis have been previously described.31 Briefly, patients were given a collection device, containing EDTA based-buffer solution. After receipt of specimen at the clinical laboratory, buffered stool samples were homogenized, separated into aliquots, and frozen at −80°C. For the blinded group, stool aliquots were subsequently sent in batches to one of three laboratories: Exact Sciences (Madison, WI), Mayo Medical Laboratory (Rochester, MN), and Molecular Pathology Laboratory Network (Knoxville, TN). For the unblinded group, all samples were processed and analyzed at Exact Sciences.
The MT-sDNA (now Cologuard) uses quantitative allele-specific realtime target and signal amplification (QuARTS) assays for aberrant methylation of the BMP3 and NDRG4 gene promoter regions, for KRAS gene mutations, and for quantification of β-actin (a reference gene for human DNA quantity); as well as an immunochemical assay for human hemoglobin. Quantitative measurements of each marker were incorporated into a validated logistic-regression algorithm, with a pre-specified cutoff to indicate test positivity. The algorithm and molecular methods have been previously published.31
Statistical Analysis
The primary study endpoint was the rate of detection of CRN. Though the event rate among DeeP-C participants at our institution was unknown prior to this study, CRN was found in 37% of all 9989 total clinical trial participants.31 Assuming equal sizes, approximately 95 patients in each of the blinded and unblinded groups provided 80% power to demonstrate a 20% difference in adenomatous CRN detection in a two-sided test at the 5% significance level. Differences in baseline characteristics were assessed by the Wilcoxon rank-sum test for continuous variables; proportions were assessed by chi-squared or Fisher’s exact test, where appropriate.
Results
From 1908 MT-sDNA tests performed clinically on patients at Mayo Clinic during the post-approval period, 225 (11.8%) were positive. Of the 225 positive tests, 16 were ordered for off label indications (Supplemental Table 1), 8 patients were symptomatic at the time of stool collection, and 29 patients did not undergo colonoscopy by the end of 2015 (Figure 1). From the central clinical trial database of 9989 eligible participants, 356 were identified as having completed the DeeP-C study at Mayo Clinic between June 2011 and November 2012. Of these, 72 (20%) had positive multi-target stool DNA test results. Thus, for the primary analysis, there were 172 patients in the unblinded group and 72 patients in the blinded group.
Figure 1.
Study flow diagram. CRC, colorectal cancer; CRN, colorectal neoplasia
Patient and procedure characteristics
There were no significant differences in age or sex between the unblinded and the blinded groups (Table 1).
Table 1.
Demographics and colonoscopy findings of patients with positive MT-sDNA
| Unblinded | Blinded | p-value | |
|---|---|---|---|
|
Patients N |
172 | 72 | |
| Median age, years (interquartile range) | 69 (61, 75) | 70 (65, 75) | 0.37 |
| Male (%) | 38 | 46 | 0.32 |
| Colonoscopic Findings | |||
| Median number of polyps, n (IQR) | 2 (1, 4) | 1(0, 2) | 0.0007 |
| HP only, n (%) | 14 (8%) | 5 (7%) | 1.0 |
| Any SSA/P or adenomatous polyp, n (%) | 120 (70%) | 38 (53%) | 0.013 |
| Adv CRN*, n (%) | 48 (28%) | 15 (21%) | 0.27 |
| CRC/HGD, n (%) | 4 (2.3%) | 3 (4.2%) | 0.68 |
| Any SSA/P, n (%) | 36 (21%) | 8 (11%) | 0.10 |
| Non-neoplastic hemorrhagic lesionsα, n (%) | 9 (5%) | 6 (8%) | 0.39 |
Includes bleeding hemorrhoids, solitary rectal ulcer, new inflammatory bowel disease, ileitis, and new radiation proctopathy
Includes CRC, any lesion ≥10mm, HGD any size, and TVA any size
CRC, colorectal cancer; CRN, colorectal neoplasia; HGD, high-grade dysplasia; HP, hyperplastic polyp; SSA/P, sessile serrated adenoma/polyp
Over the study period no colonoscopists were lost from the 2012 practice, and fourteen additional colonoscopists were new to the practice in the 2014–15 period. Colon preparation data were available for 170 patients in the unblinded group and all 72 patients in the blinded group. There was no difference in the proportion of patients with inadequate or poor preparations in the unblinded or blinded groups; 4% (6/170) and 6% (4/72) respectively (p = 0.49). In the unblinded group, a total of five patients had diagnostic colonoscopy performed outside our institution. Of these, three were negative for neoplasia. For both patients with colonoscopies positive for neoplasia, source documents were scanned in our electronic medical record.
Colorectal neoplasia
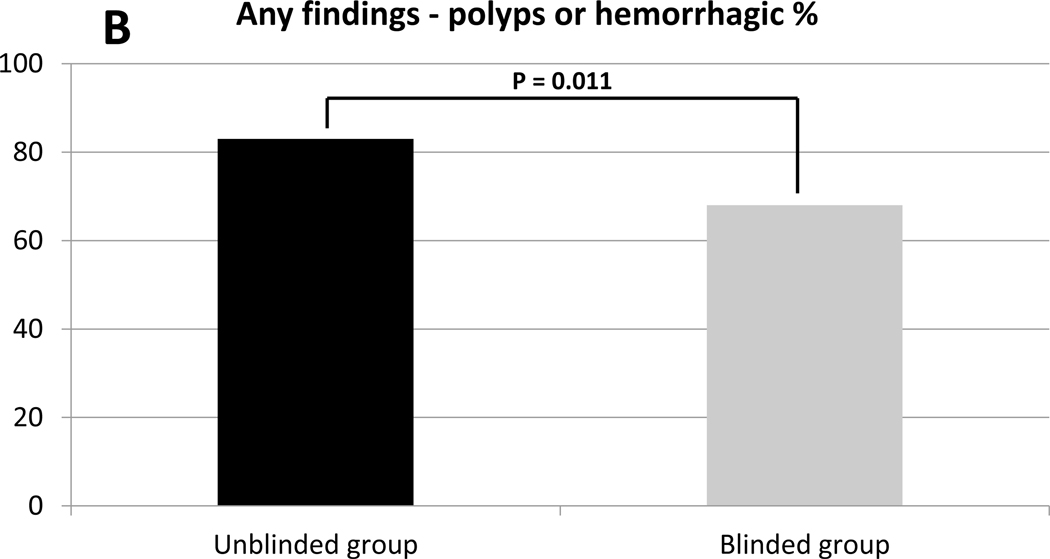
Any polypoid lesions including hyperplastic polyps (HP), were more common in the unblinded group 78% (134/172) than the blinded group 60% (43/72) (p = 0.0047) (Table 1, Figure 2A). In the unblinded group, there were a median of 2 polyps (IQR 1 – 4) detected per colonoscopy compared to a median of 1 polyp (IQR 0–2) (p = 0.0007) in the blinded group (Table 1, Figure 2B). This difference was not attributable to HP lesions as the proportion of patients in whom HPs were the only neoplastic finding were not different between the unblinded and blinded groups at 8% (14/172) and 7% (5/72) respectively (p = 1.0) (Table 1).
Figure 2.
Colonoscopy yield in MT-sDNA positive patients: findings in unblinded and blinded groups. (A) Proportions of patients with any polypoid lesion, including hyperplastic polyps. (B) Number of polypoid lesions per patient, including hyperplastic polyps. (C) Any sessile serrated adenoma/polyp (SSA/P) or adenoma.
SSA/Ps and Adenomas:
The finding of at least one SSA/P or adenomatous lesion of any size was significantly more common in the unblinded group than the blinded group at 70% (120/172) and 53% (38/72) respectively (p = 0.013) (Table 1, Figure 2C). Sessile serrated polyps of any size were found in 21% (36/172) of unblinded examinations and of 11% (8/72) of blinded ones (p=0.10). Advanced CRN was found in 28% (48/172) and 21% (15/72) of the respective groups (p=0.27). Numbers of CRC or HGD were very few in both groups (Table 1). The majority of patients with adenomatous CRN in each group had at least one lesion in the right colon: 78% (93/120) in the unblinded group and 84% (32/38) in the blinded group (p=0.49). Among patients with advanced CRN, the majority in each group had a right-sided lesion; this included 65% (31/48) in the unblinded and 66% (10/15) in the blinded group.
Polyp morphology:
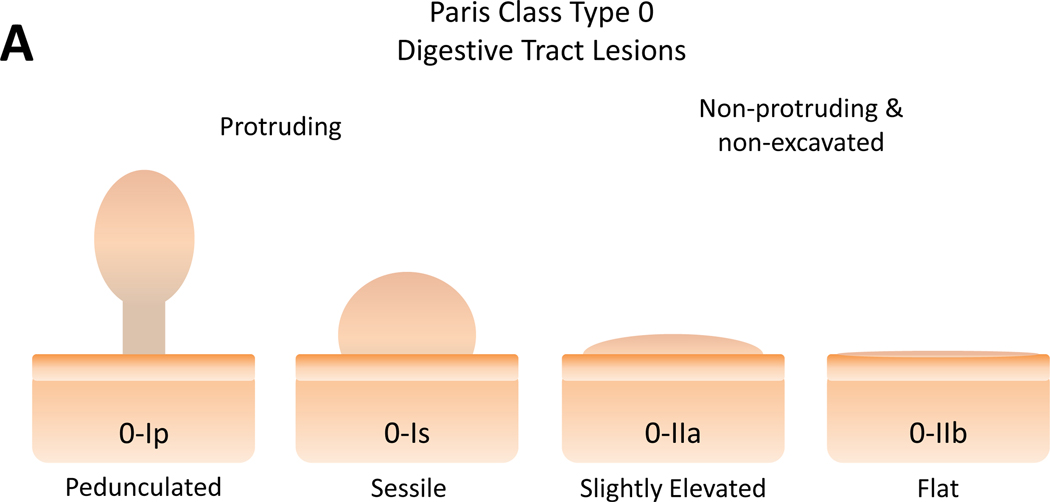
Polyp morphology according to the Paris classification is illustrated in Figure 3A. Among patients with SSA/P or adenomatous polyps in the right colon, 40% (39/93) in the unblinded group had flat or slightly raised (Paris class 0-IIa or 0-IIb) lesions versus 9% (3/32) in the blinded group (p=0.0017)(Figure 3B). Of patients that had a left-sided adenoma, there was no difference between groups with respect to with type 0-IIa morphology lesions (Figure 3C); there were no class 0-IIb lesions in the left colon. While more polyps were photographed in the unblinded group than the blinded group, 88% (161/183) and 66% (40/61) respectively (p=0.002); there was no significant difference in the proportion of photographed polyps classified as Paris class 0-IIa or 0-IIb 24% (38/161) and 10% (4/40) in the unblinded and blinded groups respectively (p=0.08).
Figure 3.
Polyp morphology and distribution of flat polyps among MT-sDNA positive patients. (A) Paris classification of superficial colon polyps. Modified and used with permission from the Endoscopic Classification Review Group: Update on the Paris classification of superficial neoplastic lesions in the digestive tract Endoscopy. June 2005;37(6):570–578. Percent of detected adenomas or SSA/P with Paris Class 0-IIa and 0-IIb morphology in the right (B) and left (C) hemicolon.
Incidental findings
When patients met inclusion criteria but the colonoscopy was negative for a polypoid lesion, 24% (9/38) of unblinded group patients had non-neoplastic hemorrhagic findings that may have explained test positivity; this rate was similar in the blinded group 21% (6/29) (Table 1). The details of these findings are outlined in Supplemental Table 2.
Colonoscope Withdrawal Time
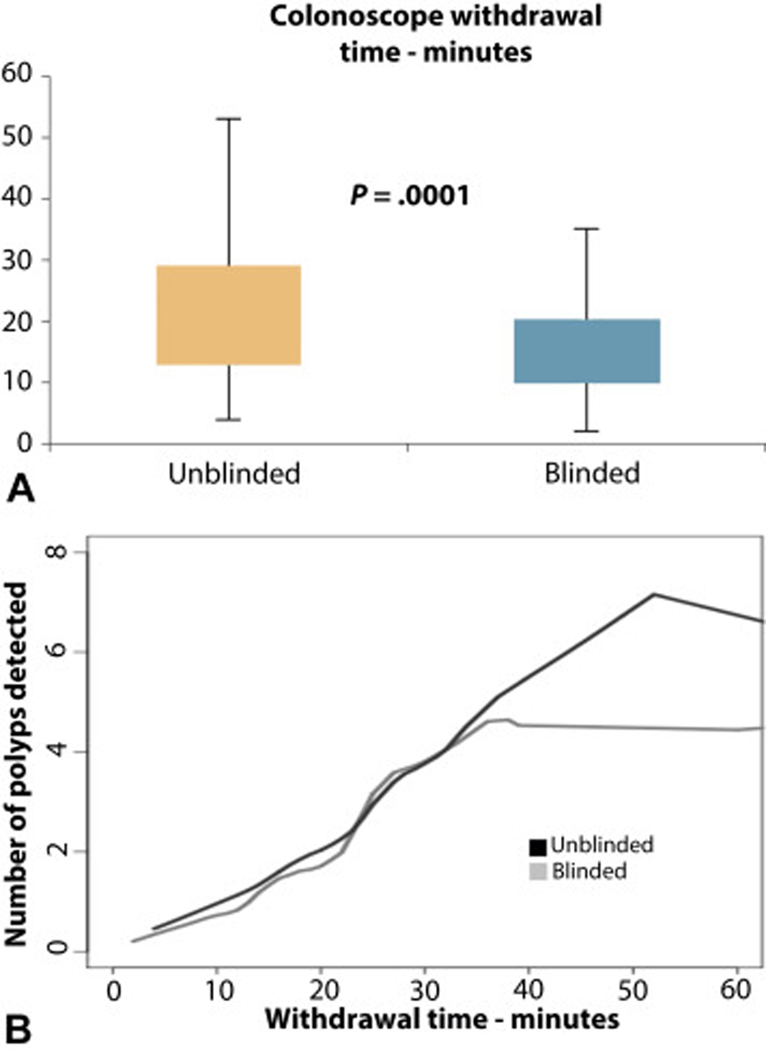
Colonoscopy withdrawal times were available for 129 patients in the unblinded group and 71 in the blinded group. These were significantly longer in the unblinded group at 19 minutes (IQR 13 – 29) compared to the blinded group median time of 13 minutes (IQR 10 – 20) (p=0.0001)(Figure 4A). Overall, number of polyps detected correlated with withdrawal time (Figure 4B).
Figure 4.
Colonoscopy withdrawal times. (A) Comparison of withdrawal times between unblinded and blinded groups. (B) Correlation between withdrawal times and number of polyps detected.
Discussion
Based on our findings, knowledge of a positive screening MT-sDNA test result appears to improve both the diagnostic yield and quality of the subsequent colonoscopic examination. Significantly more patients with pre-cancers and more polyps per patient were found when endoscopists were informed of MT-sDNA results compared to findings in the blinded group. Further, median endoscope withdrawal time, was significantly longer in the unblinded group.
The proportion of patients with any adenomatous CRN, including SSA/P, was significantly higher in the unblinded group and in excess of double the rate of adenoma detection set by major societies as a measure of colonoscopy quality.24 Proximal serrated polyps were detected in 21% of our unblinded MT-sDNA positive patients, which is greater than the 13% prevalence of proximal serrated polyps reported in a screening population undergoing colonoscopy.16 The marked increase in detection rates of often inconspicuous right sided flat lesions (Paris class 0-IIa and 0-IIb) by unblinded endoscopists further supports improved quality resulting from knowledge of a positive MT-sDNA test result.
Advanced CRN was found in 28% of patients in the unblinded group. For context, the historical prevalence of advanced CRN reported in the general population is roughly three to four times less.36–43 This indirect comparison underscores the potential for enrichment of disease at diagnostic colonoscopy following positive MT-sDNA screening. We also observed a higher than expected proportion of advanced CRN in the right hemicolon as well; over half of advanced CRN in both groups was found in the right colon. The present study was not powered to measure a difference in the rate of advanced CRN between the blinded and unblinded groups.
When endoscopists are informed of a positive MT-sDNA result, withdrawal times are significantly longer than in those blinded to the test result suggesting that such knowledge enhances quality of inspection. Increased quality is also supported by the increased number of polyps per patient found in the unblinded group. It cannot be determined from study observations whether the increased number of polyps detected in the unblinded group was the cause or the consequence of the longer withdrawal times in that group. Longer withdrawal times in the unblinded group may reflect a slower and more fastidious inspection resulting in more polyps found, or a more mindful inspection may have simply led to detection of more polyps which then required more time for subsequent removal. In either case, what is most relevant is the increased number of polyps detected and removed by unblinded endoscopists. As advocated by some, measuring the number of adenomas per colonoscopy may be a more accurate measure of inspection quality than the conventional approach measuring the rate of patients with an adenoma.44 As endoscopists’ knowledge of MT-sDNA positivity was shown to increase yield of neoplastic findings over those from blinded examinations, this functionally increases the true positive rate, and as a consequence, MT-sDNA performance estimates of both sensitivity and specificity would be higher than those derived from studies based on blinded endoscopies.
We acknowledge several potential limitations to this study. Patients undergoing screening in routine clinical practice may differ in unmeasured ways from those who participated in the DeeP-C trial; however, inclusion and exclusion criteria for both cohorts were similar by design, and there were not significant differences in their age or sex distributions. MT-sDNA positivity rates appear lower in the post-approval cohort, compared to the blinded pre-approval group. While there were no changes to the test assays or analytic algorithm between the two time periods, we hypothesize there may be potential differences in exposure to prior colorectal cancer screening between the two groups. Examination of this phenomenon will require further study. Colonoscopy withdrawal time is a measure of endoscopy quality due to positive correlation with adenoma detection rate and decreased interval CRC.18, 45 While this is an important clinical practice metric,24, 46 it is a surrogate marker with possible effect saturation after 9 minutes.19, 45 Therefore, we did not use this as the primary outcome measure. In the present study, withdrawal time measurements could not be sub-divided into elements of inspection time and therapy time. We did demonstrate that the number of polyps detected per unit of withdrawal time was similar between groups. Given this shared relationship, it follows that the longer withdrawal times in the unblinded group would be associated with more polyps found.
We took great care to estimate baseline endoscopist performance in our practice over the study period using measurements from procedures performed with a diagnosis of average-risk CRC screening. There are likely unmeasured differences in colonoscopy practice between 2012 and 2014–15. We internally examined our institutional polyp detection rate (PDR) and withdrawal time (WT) over the study period and found no significant change (data not shown). Thus, any change in practice at our institution between years does not explain the increase in yield in the unblinded group. Also, the practice of photodocumentation during colonoscopy in our practice has changed during the study period as demonstrated by the lower number of polyps photographed in the blinded group. This is likely related to a transition in documentation software, as ProVation was instituted in our Rochester practice in late 2012. The observed differences in photographed subtle lesions is likely due lower photography rate, but also that these lesions may have been missed endoscopically.
Not all patients with a positive MT-sDNA completed diagnostic colonoscopy by the end of the study period. While missing data from these patients could potentially have impacted the outcome measures, the overall follow-up rate in our practice was excellent; 87% (196/225) of patients complied with recommended colonoscopy. This is consistent with a previous report observing greater adherence to surveillance colonoscopy over screening colonoscopy, suggesting the patient’s knowledge of increased risk may improve adherence.47
Finally, we were surprised that a study similar to ours has not been reported examining what effect that knowledge of a fecal occult blood test result has on colonoscopy quality or yield. For the present study, we had the unique opportunity to access a large number of clinical outcomes on patients with positive MT-sDNA results following colonoscopies by both blinded and unblinded endoscopists. Furthermore, procedures in this single center study allowed similar patient groups examined by the same endoscopy group and within a similar time frame.
In conclusion, endoscopist knowledge of MT-sDNA positivity in practice may improve both the yield and quality of the colonoscopic examination. These data suggest a potential additive benefit of MT-sDNA to current screening colonoscopy practices, which may extend to individual endoscopist performance and to overall CRC screening adherence, areas which warrant further study.
Supplementary Material
Grant support:
This research was supported by a grant from the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma and the Paul Calabresi Program in Clinical-Translational Research (NCI CA90628), the Carol M. Gatton endowment for Digestive Diseases Research, and Mayo Clinic.
Presented in part at the 116th Annual Meeting of the American Gastroenterological Association, May 22, 2016, San Diego, CA.
Abbreviations:
- CRC
colorectal cancer
- CRN
colorectal neoplasia
- FIT
fecal immunohistochemical test
- HP
hyperplastic polyp
- MT-sDNA
multi-target stool DNA
- QuARTS
quantitative allele-specific realtime target and signal amplification
- SSA/P
sessile serrated adenoma/polyp
Footnotes
Disclosures:
Mayo Clinic is a minor equity holder in Exact Sciences (Madison WI) and has an intellectual property agreement with Exact Sciences under which inventors, such as Drs. Kisiel and Ahlquist and Mr. Mahoney could receive royalties in accordance with Mayo Clinic policy. Dr. Johnson, Dr. Sweetser, Mrs. Burger and Ms. Devens have no relevant disclosures.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Ward EM, Jemal A. Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992–2008. Cancer Epidemiol Biomarkers Prev 2012;21:411–6. [DOI] [PubMed] [Google Scholar]
- 3.Hewett DG, Kahi CJ, Rex DK. Does colonoscopy work? J Natl Compr Canc Netw 2010;8:67–76; quiz 77. [DOI] [PubMed] [Google Scholar]
- 4.Baxter NN, Warren JL, Barrett MJ, et al. Association between colonoscopy and colorectal cancer mortality in a US cohort according to site of cancer and colonoscopist specialty. J Clin Oncol 2012;30:2664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klabunde CN, Cronin KA, Breen N, et al. Trends in colorectal cancer test use among vulnerable populations in the United States. Cancer Epidemiol Biomarkers Prev 2011;20:1611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinwachs D, Allen JD, Barlow WE, et al. National Institutes of Health state-of-the-science conference statement: Enhancing use and quality of colorectal cancer screening. Ann Intern Med 2010;152:663–7. [DOI] [PubMed] [Google Scholar]
- 7.Rex DK, Johnson DA, Anderson JC, et al. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. Am J Gastroenterol 2009;104:739–50. [DOI] [PubMed] [Google Scholar]
- 8.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008;134:1570–95. [DOI] [PubMed] [Google Scholar]
- 9.Screening for Colorectal Cancer: U.S. Preventive Services Task Force Recommendation StatementScreening for Colorectal Cancer. Annals of Internal Medicine 2008;149:627–637. [DOI] [PubMed] [Google Scholar]
- 10.Steele CB, Rim SH, Joseph DA, et al. Colorectal cancer incidence and screening - United States, 2008 and 2010. MMWR Surveill Summ 2013;62 Suppl 3:53–60. [PubMed] [Google Scholar]
- 11.Kahi CJ, Imperiale TF, Juliar BE, et al. Effect of screening colonoscopy on colorectal cancer incidence and mortality. Clin Gastroenterol Hepatol 2009;7:770–5; quiz 711. [DOI] [PubMed] [Google Scholar]
- 12.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013;369:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxter NN, Goldwasser MA, Paszat LF, et al. Association of colonoscopy and death from colorectal cancer. Ann Intern Med 2009;150:1–8. [DOI] [PubMed] [Google Scholar]
- 14.Robertson DJ, Lieberman DA, Winawer SJ, et al. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut 2014;63:949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010;138:2088–100. [DOI] [PubMed] [Google Scholar]
- 16.Kahi CJ, Hewett DG, Norton DL, et al. Prevalence and variable detection of proximal colon serrated polyps during screening colonoscopy. Clin Gastroenterol Hepatol 2011;9:42–6. [DOI] [PubMed] [Google Scholar]
- 17.Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski MF, Regula J, Kraszewska E, et al. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med 2010;362:1795–803. [DOI] [PubMed] [Google Scholar]
- 19.Butterly L, Robinson CM, Anderson JC, et al. Serrated and adenomatous polyp detection increases with longer withdrawal time: results from the New Hampshire Colonoscopy Registry. Am J Gastroenterol 2014;109:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barclay RL, Vicari JJ, Doughty AS, et al. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. N Engl J Med 2006;355:2533–41. [DOI] [PubMed] [Google Scholar]
- 21.Benson ME, Reichelderfer M, Said A, et al. Variation in colonoscopic technique and adenoma detection rates at an academic gastroenterology unit. Dig Dis Sci 2010;55:166–71. [DOI] [PubMed] [Google Scholar]
- 22.Rex DK. Colonoscopic withdrawal technique is associated with adenoma miss rates. Gastrointest Endosc 2000;51:33–6. [DOI] [PubMed] [Google Scholar]
- 23.Rex DK, Petrini JL, Baron TH, et al. Quality indicators for colonoscopy. Gastrointest Endosc 2006;63:S16–28. [DOI] [PubMed] [Google Scholar]
- 24.Rex DK, Schoenfeld PS, Cohen J, et al. Quality indicators for colonoscopy. Am J Gastroenterol 2015;110:72–90. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian S, Klosterman M, Amonkar MM, et al. Adherence with colorectal cancer screening guidelines: a review. Prev Med 2004;38:536–50. [DOI] [PubMed] [Google Scholar]
- 26.Ferrante JM, Ohman-Strickland P, Hahn KA, et al. Self-report versus medical records for assessing cancer-preventive services delivery. Cancer Epidemiol Biomarkers Prev 2008;17:2987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inadomi JM, Vijan S, Janz NK, et al. Adherence to colorectal cancer screening: a randomized clinical trial of competing strategies. Arch Intern Med 2012;172:575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang PS, Wheat CL, Abhat A, et al. Adherence to Competing Strategies for Colorectal Cancer Screening Over 3 Years. Am J Gastroenterol 2016;111:105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moawad FJ, Maydonovitch CL, Cullen PA, et al. CT colonography may improve colorectal cancer screening compliance. AJR Am J Roentgenol 2010;195:1118–23. [DOI] [PubMed] [Google Scholar]
- 30.Gluecker TM, Johnson CD, Harmsen WS, et al. Colorectal cancer screening with CT colonography, colonoscopy, and double-contrast barium enema examination: prospective assessment of patient perceptions and preferences. Radiology 2003;227:378–84. [DOI] [PubMed] [Google Scholar]
- 31.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 32.Redwood DG, Asay ED, Blake ID, et al. Stool DNA Testing for Screening Detection of Colorectal Neoplasia in Alaska Native People. Mayo Clin Proc 2016;91:61–70. [DOI] [PubMed] [Google Scholar]
- 33.Berger BM, Hooker A, Bethke L, et al. Colorectal Cancer Screening With Multi-target Stool DNA-based Testing: Previous Screening History of the Initial Patient Cohort. Am J Gastroenterol 2015;110:S607. [Google Scholar]
- 34.Update on the paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy 2005;37:570–8. [DOI] [PubMed] [Google Scholar]
- 35.Francis DL, Rodriguez-Correa DT, Buchner A, et al. Application of a conversion factor to estimate the adenoma detection rate from the polyp detection rate. Gastrointest Endosc 2011;73:493–7. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman DA, Williams JL, Holub JL, et al. Race, ethnicity, and sex affect risk for polyps >9 mm in average-risk individuals. Gastroenterology 2014;147:351–8; quiz e14–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman DA, Holub J, Eisen G, et al. Prevalence of polyps greater than 9 mm in a consortium of diverse clinical practice settings in the United States. Clin Gastroenterol Hepatol 2005;3:798–805. [DOI] [PubMed] [Google Scholar]
- 38.Lieberman DA, Holub JL, Moravec MD, et al. Prevalence of colon polyps detected by colonoscopy screening in asymptomatic black and white patients. JAMA 2008;300:1417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B, Holub J, Peters D, et al. Prevalence of colon polyps detected by colonoscopy screening of asymptomatic Hispanic patients. Dig Dis Sci 2012;57:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regula J, Rupinski M, Kraszewska E, et al. Colonoscopy in colorectal-cancer screening for detection of advanced neoplasia. N Engl J Med 2006;355:1863–72. [DOI] [PubMed] [Google Scholar]
- 41.Schroy PC 3rd, Coe A, Chen CA, et al. Prevalence of advanced colorectal neoplasia in white and black patients undergoing screening colonoscopy in a safety-net hospital. Ann Intern Med 2013;159:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner H, Hoffmeister M, Arndt V, et al. Protection from right- and left-sided colorectal neoplasms after colonoscopy: population-based study. J Natl Cancer Inst 2010;102:89–95. [DOI] [PubMed] [Google Scholar]
- 43.Saini SD, Kim HM, Schoenfeld P. Incidence of advanced adenomas at surveillance colonoscopy in patients with a personal history of colon adenomas: a meta-analysis and systematic review. Gastrointest Endosc 2006;64:614–26. [DOI] [PubMed] [Google Scholar]
- 44.Kahi CJ, Vemulapalli KC, Johnson CS, et al. Improving measurement of the adenoma detection rate and adenoma per colonoscopy quality metric: the Indiana University experience. Gastrointest Endosc 2014;79:448–54. [DOI] [PubMed] [Google Scholar]
- 45.Shaukat A, Rector TS, Church TR, et al. Longer Withdrawal Time Is Associated With a Reduced Incidence of Interval Cancer After Screening Colonoscopy. Gastroenterology 2015;149:952–7. [DOI] [PubMed] [Google Scholar]
- 46.Vavricka SR, Sulz MC, Degen L, et al. Monitoring colonoscopy withdrawal time significantly improves the adenoma detection rate and the performance of endoscopists. Endoscopy 2016. [DOI] [PubMed] [Google Scholar]
- 47.Greenspan M, Chehl N, Shawron K, et al. Patient Non-adherence and Cancellations Are Higher for Screening Colonoscopy Compared with Surveillance Colonoscopy. Dig Dis Sci 2015;60:2930–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.