Abstract
Caffeoyl coenzyme A 3-O-methyltransferases (CCoAOMTs) are S-adenosyl-l-methionine-dependent O-methyltransferases (OMTs) involved in lignin biosynthesis. Plant CCoAOMTs belong to a distinct family of OMTs, more closely related to the mammalian catechol OMTs than to other plant OMTs. The crystal structure of alfalfa (Medicago sativa) CCoAOMT in complex with the reaction products S-adenosine-l-homocysteine and feruloyl/sinapoyl CoAs presented here belong to a structurally and mechanistically distinct family of plant small molecule OMTs. These structures provide a new understanding of the substrate preferences and the catalytic mechanism accompanying CCoAOMT-mediated O-methylation of CoA-linked phenylpropanoid substrates.
In plant cells, methylated hydroxycinnamic acid derivatives are important precursors in the biosynthesis of cell wall esterified phenolic compounds, soluble sinapate esters, dimeric lignans, and the extensively cross-linked polymer lignin. Lignin and esterified phenolic compounds provide structural support for the cell walls of conducting tissues and tracheary elements and were critical in the evolution of higher plants from their aquatic progenitors. Additionally, lignin serves as an inducible physical barrier against pathogen infections (Vance et al., 1980), and after cellulose is the second most abundant natural substance found on earth.
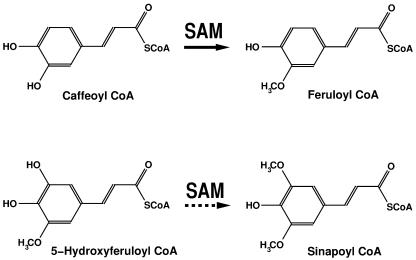
Two S-adenosyl-l-Met (SAM)-dependent O-methyltransferases (OMTs) are involved in the methylation of monolignol precursors: caffeic acid 3-OMTs (COMTs; Zubieta et al., 2002) and caffeoyl CoA OMTs (CCoAOMTs). CCoAOMT has been isolated and characterized from a number of different plants, including alfalfa (Medicago sativa; Inoue et al., 1998), zinnia (Zinnia elegans; Ye et al., 1994), carrot (Daucus carota; Kühnl et al., 1989), parsley (Petroselinum crispum; Pakusch et al., 1989), poplar (Populus spp.; Meyermans et al., 2000), pine (Pinus taeda; Li et al., 1999), and tobacco (Nicotiana tabacum; Martz et al., 1998). Currently, the methylation steps in the lignin biosynthetic pathway are believed to traverse two main branches dependent on COMT and CCoAOMT activities, respectively. In vitro, COMT methylates caffeoyl and 5-hydroxyferuloyl free acids, aldehydes, and alcohols, with a preference for the 5-hydroxylated compounds, whereas CCoAOMT methylates caffeoyl and 5-hydroxyferuloyl CoA thioesters (Fig. 1) with an in vitro kinetic preference for caffeoyl CoA (Parvathi et al., 2001).
Figure 1.
CCoAOMT-catalyzed methylation reaction. CCoAOMT methylates caffeoyl and 5-hydroxyferuloyl CoA thioesters with an in vitro kinetic preference for caffeoyl CoA. The dotted arrow emphasizes this possible substrate selectivity. SAM is the methyl donor in both cases.
Transgenic experiments that down-regulated COMT activity severely reduced the syringyl monomer content of the resultant lignin but did not decrease the overall lignin content of the transgenic plants (Atanassova et al., 1995; Doorsselaere et al., 1995; Hopkins et al., 2001). These results strongly implicate COMT as the primary 5-hydroxyl methyltransferase present in plants. In contrast, anti-sense down-regulation of tobacco CCoAOMT yielded plants with a reduction in both guaiacyl and syringyl monolignol content and a strong alteration of their development fate (Zhong et al., 1998; Pincon et al., 2001). Curiously, unlike experiments conducted in tobacco, down-regulation of alfalfa CCoAOMT primarily affected guaiacyl content (Guo et al., 2000). These experiments suggest that, at least in some plants, CCoAOMT is the primary 3-hydroxyl methyltransferase. Moreover, these results also imply that there may be little functional redundancy in vivo between CCoAOMT and COMT (Humphreys and Chapple, 2002). Thus, rather than operating through a metabolic grid, the biosynthesis of lignin may operate through a series of sequential steps where CCoAOMT and COMT play equally important yet distinct roles.
Primary sequence analysis and structural comparison with other plant OMTs demonstrate that COMT and CCoAOMT belong to two distinct families of OMTs. The family to which COMT (Zubieta et al., 2002) belongs includes two other structurally characterized plant OMTs: alfalfa isoflavone OMT (IOMT; 13% sequence identity with alfalfa CCoAOMT, but 42% sequence identity with alfalfa COMT) and alfalfa chalcone OMT (ChOMT; 11% sequence identity with alfalfa CCoAOMT, but 56% identity with alfalfa COMT; Zubieta et al., 2001). A second family of plant OMTs only includes enzymes that exhibit a high degree of specificity for CoA thioesters, with sequence identity ranging from 50% to 90% with CCoAOMT. It should be noted though that the complete functional characterization of the CCoAOMT family of related plant enzymes has not been carried out, suggesting that the CCoAOMT family may include members displaying specificities for substrates other than CoA-linked molecules. In fact, an enzyme related to CCoAOMT from ice plant (Mesembryanthemum crystallinum) was recently characterized and shown to methylate various flavonoid and phenylpropanoid conjugates (Vogt, 2004).
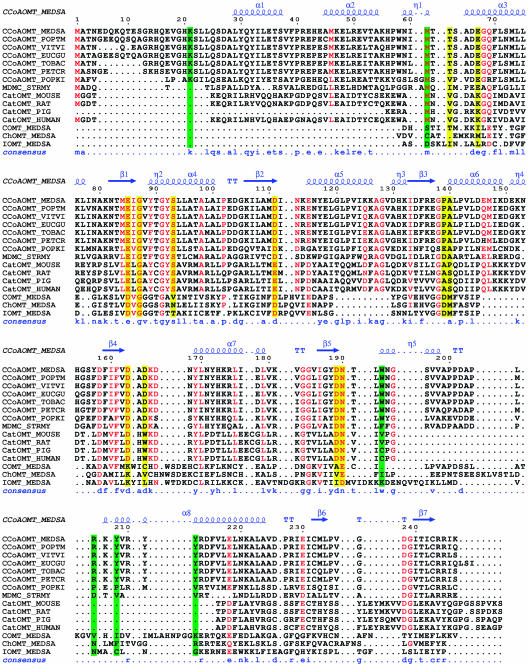
Based on sequence identity, the closest non-CoA OMT to this CCoAOMT family is the group of animal catechol OMTs (Fig. 2). The soluble portion of catechol OMT from rat (19% sequence identity with CCoAOMT) has been structurally characterized (Vidgren et al., 1994) and bears a high degree of structural similarity to CCoAOMT. Another group of methyltransferases closely related to plant CCoAOMTs includes enzymes responsible for the methylation of antibiotics synthesized by bacteria (Pospiech et al., 1996).
Figure 2.
Sequence alignment of OMTs. Sequence alignment of alfalfa CCoAOMT with related OMTs, including 6 plant OMTs that exhibit a preference for CoA-linked thioesters, one macrolide 4-OMT (MDMC_STRMY or Ma40MT) from Streptomyces mycarofaciens, the soluble part of 4 animal catechol OMTs, including the structurally characterized rat catechol OMT (Protein Data Bank accession no. 1VID), and 3 structurally characterized plant OMTs (alfalfa COMT, ChOMT, and IOMT; Protein Data Bank accession nos. 1KYZ, 1FPQ, and 1FPX, respectively) truncated to the region that aligns with CCoAOMT. The secondary structure of alfalfa CCoAOMT is displayed above and the consensus sequence for the CCoAOMT family below the alignment. Highly conserved residues between CCoAOMT and catechol OMT families are red. Residues involved in divalent metal ion and cofactor binding are depicted on a yellow background. Residues involved in substrate recognition are depicted on a green background. This figure was prepared with ESPript (Gouet et al., 1999) and corrected by hand to closely match structural alignments when such structures are available.
The crystal structures of alfalfa CCoAOMT in complex with the reaction products S-adenosyl-l-homocysteine (SAH) and feruloyl/sinapoyl CoAs presented here belong to a structurally and mechanistically distinct family of plant small molecule OMTs. These structures provide a new understanding of the substrate preferences and the catalytic mechanism accompanying CCoAOMT mediated O-methylation of CoA-linked substrates.
RESULTS
Structural Elucidation and Description
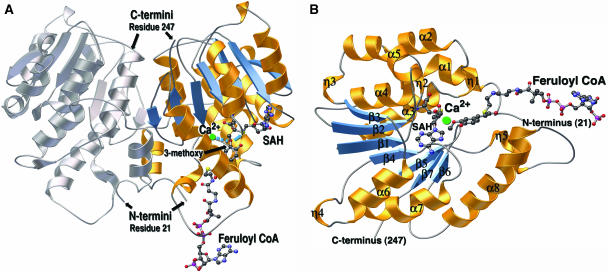
Alfalfa CCoAOMT is a 28-kD protein consisting of 247 amino acids (EC 2.1.1.104; Inoue et al., 1998) that forms a homodimer in solution. The crystallographic structures obtained at 2.45 Å and 2.65 Å resolution with CoA-linked substrates bound reveal the architecture of the dimeric form of CCoAOMT (Fig. 3A). Due to the lack of the N-terminal dimerization domain present in previously characterized plant OMTs (Zubieta et al., 2001, 2002), the interaction between monomers in the CCoAOMT dimer is novel. The dimerization interface includes residues 26 to 40, 66 to 78, and 212 to 226 and involves mostly hydrophobic interactions. The first 20 amino acid residues are not observed in the electronic density.
Figure 3.
Ribbon diagrams of the CCoAOMT three-dimensional structure. A, Ribbon diagram of the alfalfa CCoAOMT homodimer, with one monomer colored (helices are gold and β-strands are blue) and the other monomer in gray. The dyad axis is oriented vertically in the plane of the page. The SAH and feruloyl CoA molecules are depicted as colored ball and sticks (carbon is black; nitrogen is blue; oxygen is red; and sulfur is yellow). The Ca2+ ion is in green. A coordination bond from the Ca2+ ion to the 3-methoxy oxygen of feruloyl CoA is shown as a dashed blue line. A blue arrow points from the same oxygen to the sulfur position on SAH that is attached to the reactive methyl group in SAM. The N-terminal and C-terminal amino acid residues visible in the final electron density map are numbered. B, Ribbon diagram of the CCoAOMT monomer. The SAH and feruloyl CoA molecules are depicted as ball and sticks and color-coded as in A. Secondary structures are numbered according to Figure 2. α is α-helix, β is β-strand, and η is 3/10 helix. Ribbon diagrams were produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org).
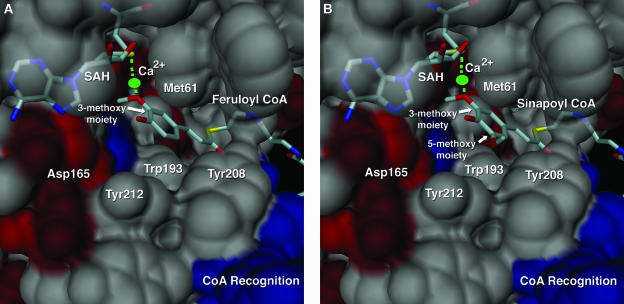
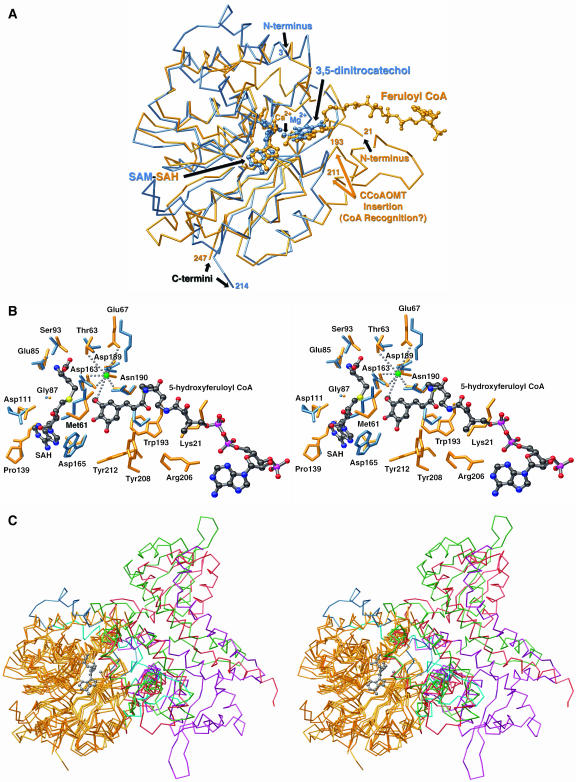
Each monomer (Fig. 3B) consists of a single catalytic domain. This domain exhibits a core α/β Rossmann fold that provides the binding site for SAM/SAH (Rossmann et al., 1974). All structurally characterized SAM-dependent OMTs share this architectural motif. The adenine portion of SAM/SAH forms hydrogen bonds with Asp-165 and the backbone nitrogen of Ala-140. Hydrogen bonds also form between the Rib hydroxyls and the carboxyl group of Asp-111. For SAH, the terminal-amino group hydrogen bonds with Gly-87 and Ser-93 and forms a salt bridge with Glu-85. The terminal carboxylate of SAH participates in a hydrogen-bonding network with Ser-93 and a well-ordered water molecule. Weaker electron density was observed in the active site for crystals soaked in the presence of caffeoyl CoA and 5-hydroxyferuloyl CoA. In every crystal examined, electron density was observed only in one protein molecule out of the four protein molecules present in the asymmetric unit. This observation could be due to limited access in the crystalline state to the active site of some protein chains or may be due to the higher intrinsic flexibility or disorder of some protein chains in the asymmetric unit. Nevertheless, the electron density associated with one active site was sufficient to model the CoA-linked phenolic substrates (Fig. 4, A and B). A single orientation for the CoA tail was not discernable, and instead the CoA conjugate appears somewhat disordered, extending out of the active site and residing in multiple conformations. This lack of defined density for the CoA tail of substrates is partly due to the intrinsic instability of the CoA-linked thioester bond upon prolonged incubations with crystals resulting in a lower effective concentration of substrate in solution. Nevertheless, the electron density is of sufficient quality to allow the CoA tail to be modeled into the enzyme-active site. The orientation used in the model shown suggests that the surface residues Lys-21, near the N terminus, and Arg-206, on the CCoAOMT insertion element (Fig. 5, A and B), interact with the negatively charged 3′-phosphate group of the adenosine 3′-5′-diphosphate moiety of CoA. The phenolic moiety of the molecule is more ordered in the active site and is sequestered by a number of aromatic residues including Tyr-208, Tyr-212, and Trp-193. Met-61, Asp-163, and Asn-190 complete the rest of the substrate-binding pocket.
Figure 4.
Close-up molecular surface view of the CCoAOMT active site. A, The active site view includes a Ca2+ ion (green), with SAH and feruloyl CoA shown as color-coded bonds. Residues surrounding the phenylpropanoid portion of feruloyl CoA are illustrated as molecular surfaces and labeled accordingly. Surfaces are color coded according to their properties (acidic residues in red, basic residues in blue, neutral or hydrophobic residues in gray). B, The same view as in A for the sinapoyl CoA complex. Both the 3-methoxy and 5-methoxy portions of the sinapoyl product are labeled. This figure was produced with DINO (Visualizing Structural Biology, 2002, http://www.dino3d.org).
Figure 5.
Structural superposition of CCoAOMT with representative methyltransferases. A, Three-dimensional superposition of alfalfa CCoAOMT (blue) and rat catechol OMT (gold) structures. The nature of the bound cofactor (SAM in catechol OMT and SAH in CCoAOMT) is color coded as for the backbone depiction. Residues 193 to 211 in alfalfa CCoAOMT appear as an insertion in the core fold as defined by catechol OMT and appear to form a structural extension involved in weakly tethering the CoA-linked tail of CCoAOMT substrates. This figure was produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org). B, Close-up view of the alfalfa CCoAOMT active site (residues in gold) compared to the catechol OMT active site (residues in blue) shown in stereo. Residue numbers correspond to the numbers for alfalfa CCoAOMT. The figure was produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org). C, Superposition of a subset of structurally characterized and architecturally distinct small molecule OMTs. Three-dimensional backbone superposition shown in stereo of some structurally characterized SAM-dependent OMTs known to methylate small molecules. The Rossmann fold, common to all of these OMTs, is colored gold. The variable parts are color coded as follows: alfalfa CCoAOMT (magenta), rat catechol OMT (blue), alfalfa COMT (green), alfalfa ChOMT (violet), and alfalfa IOMT (red). This figure was produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org).
Unlike previously characterized plant OMTs, which most likely utilize general base catalysis for activation of the methyl accepting hydroxyl group (Zubieta et al., 2001, 2002), CCoAOMT contains a well-ordered active-site divalent metal-binding site suggestive of a metal-dependent catalytic mechanism (Vidgren et al., 1994; Ibrahim et al., 1998). Specifically, a calcium ion is observed in the active site (derived from the crystallization solution) surrounded by an octahedral arrangement of hydroxyl and carboxyl ligands. The side chain oxygens of Thr-63, Glu-67, Asp-163, Asp-189, and Asn-190 are involved in the chelation of the calcium ion, with the coordination complex completed by the 3-methoxy oxygen atom of feruloyl CoA (or sinapoyl CoA) and a water molecule. This calcium ion, present at high concentration in the crystallization solution, substitutes for the expected physiological ligand, Mg2+, the latter of which is present in the catechol OMT structure previously reported (Vidgren et al., 1994). In CCoAOMT, the metal ion mediates the deprotonation of the caffeoyl 3-hydroxyl group and maintains the resultant oxoanion in close proximity to the reactive methyl group of SAM (approximately 3Å) allowing for facile transmethylation to occur.
Metal-Dependent Transmethylation
By stripping the metal ion from CCoAOMT with EDTA and then replacing the putative Ca2+ ion with Mg2+, Mn2+, or Zn2+, the role of the divalent metal ion was examined. The EDTA-treated enzyme displayed a specific activity only 15% the level of untreated enzyme. Addition of Mg2+, Ca2+, and Zn2+ restored activity to 100% the level of untreated CCoAOMT. Curiously, the divalent cation Mn2+ only restored activity to 35% of untreated CCoAOMT, most likely due to the higher ligand-binding energy of Mn2+ versus the other divalent cations tested (Table I).
Table I.
Activity of CCoAOMT in the presence of different metal cations
Row 1 refers to CCoAOMT untreated by EDTA, row 2 CCoAOMT dialyzed overnight with 5 mm EDTA, and rows 3 to 6 after EDTA treatment (EDTA was depleted and 2 mm of the appropriate metal cation was added). All assays used 0.1 mm caffeoyl CoA and 0.1 mm 14C-labeled SAM. Activity expressed as counts per microgram of CCoAOMT for untreated CCoAOMT assayed in the presence of 2 mm Mg2+ was set to 100% and all other reactions were then referenced to this value. Specific activities for untreated CCoAOMT were as reported previously (Inoue et al., 1998).
| Activity (% of Wild Type) | |
|---|---|
| CCoAOMT | 100 |
| CCoAOMT + 5 mm EDTA | 15 |
| CCoAOMT + 1 mm Ca2+ | 100 |
| CCoAOMT + 1 mm Mg2+ | 100 |
| CCoAOMT + 1 mm Zn2+ | 100 |
| CCoAOMT + 1 mm Mn2+ | 35 |
DISCUSSION
Three-dimensional superposition of the CCoAOMT and catechol OMT structures (Fig. 5, A and B) shows a high level of similarity in the core of the enzyme, despite poor sequence similarity. Based on a structure alignment performed with the Dali server, the root mean square deviation between the 183 aligned alpha carbons is 2.4 Å. The Rossmann fold, encompassing the catalytic divalent cation and the SAM/SAH-binding sites, is highly conserved. The overall architectural similarity, the substrate-binding site, and the role of a divalent cation in the catalytic mechanism, structurally, functionally, and evolutionarily relate plant CCoAOMTs and mammalian catechol OMTs. However, the two taxonomically distinct enzyme families adopt different oligomerization states. While CCoAOMT is dimeric, the soluble portion of mammalian catechol OMT is monomeric, and no evidence of dimerization exists for the full-length rat catechol OMT. Unlike previously characterized plant OMTs (Zubieta et al., 2001, 2002) in which the dimeric state is critical to phenolic substrate recognition, the CCoAOMT dimer does not appear to be necessary for substrate recognition and transmethylation. Indeed, each substrate and cofactor interacts with only one monomer of the CCoAOMT dimer, unlike other plant OMTs whose N-terminal dimerization domains contribute to the active site of the dyad-related monomer (Zubieta et al., 2001, 2002).
With regard to the mechanistically and structurally related catechol OMTs of mammalian origin, other differences between CCoAOMT and catechol OMT occur in the region lying near the CoA-binding site of CCoAOMT, including the N-terminal residues, a Pro-rich loop spanning residues 193 to 211 of CCoAOMT, as well as the loop between positions 237 and 239. This latter loop is shortened with respect to the structurally equivalent loop in catechol OMT that includes Glu-199 involved in substrate recognition in catechol OMT. Remarkably, when superimposing the structures, Glu-199 of catechol OMT resides at an equivalent spatial position to Lys-21 of CCoAOMT. In CCoAOMT, the N-terminal extension and the 193 to 211 and 237 to 239 loops form part of the substrate-binding pocket and likely determine the specificity of CCoAOMT for the CoA-linked substrates. Notably, the regiospecificity and kinetic activity of the recently described CCoAOMT family member characterized from ice plant appears to be influenced by the presence or absence of the amino terminal region, which clearly in CCoAOMT plays a role in recognition of the phenylpropanoid conjugate (Vogt, 2004).
Considering the structure and its three-dimensional comparison with catechol OMT, we can suggest a catalytic mechanism based on the metal-mediated deprotonation of the caffeoyl 3-hydroxyl group followed by transmethylation due to the juxtapositioning of a reactive phenolic oxyanion near the reactive methyl group of SAM. The environment around the caffeoyl 3-hydroxyl moiety of the substrate is electropositive due to the positively charged sulfur of SAM and the Ca2+ ion. Most likely, the caffeoyl 3-hydroxyl exists as an oxyanion in order to balance charge in the active site region. Chelation to the metal ion further positions the 3-oxyanion moiety in close proximity to the reactive methyl group of SAM (approximately 3Å). This metal-dependent catalytic mechanism is also postulated for mammalian catechol OMT and related methyltransferases (Vidgren et al., 1994; Ibrahim et al., 1998).
The role of the residues in CCoAOMT, previously suggested to be important for enzyme activity (Hoffmann et al., 2001), can now be reanalyzed based on the experimentally determined structure of CCoAOMT. The conclusions drawn from this structure stand in stark contrast to those obtained based on a sequence alignment with rat catechol OMT and subsequent homology modeling (Hoffmann et al., 2001), illustrating the utility of experimentally determined structures even when homology exists to related and structurally characterized protein family members. In this previously published structure-function study of CCoAOMT, the observed reduction of activity upon mutation of Asp-66 and Gln-69 (numbering according to the alfalfa CCoAOMT sequence) was attributed to the possible role of these residues in substrate binding (Hoffmann et al., 2001).
More specifically, based upon a homology model computed based upon low sequence identity with mammalian catechol OMT, Asp-66 was hypothesized to make contact with the adenine moiety of CoA, whereas Gln-69 was hypothesized to bind either the adenine or the phosphate group. The experimental structure of CCoAOMT actually shows no evidence of such an interaction for Asp-66 and reveals that Gln-69 points in the opposite direction to the adenine and CoA phosphate, making any role of Gln-69 in CoA binding unlikely. The importance of these residues in the enzyme activity is most likely due to their proximity to Thr-63, which is involved in the divalent metal ion coordination, and to Met-61, which is involved in substrate binding. The loss of all activity previously observed upon mutating Arg-228 to Thr can be attributed only indirectly to catalysis through structural stabilization and not to a direct role in substrate recognition as previously suggested (Hoffmann et al., 2001).
The comparison of the CCoAOMT structure with other known OMT structures (rat catechol OMT, alfalfa IOMT, alfalfa ChOMT, and alfalfa COMT; Fig. 5C) demonstrates, despite the differences related to substrate specificity, a high degree of structural similarity in the SAM-binding region of methyltransferases exists within and between kingdoms. This implies the fundamental conservation of the SAM-binding protein fold throughout evolution. The high degree of sequential divergence between plant OMTs, animal catechol OMTs, and bacterial OMTs does not impact the overall fold but does result in altered oligomerization states, altered modes of recognizing hydroxyl bearing substrates, and most notably, altered catalytic mechanisms accompanying catalytic transfer of a methyl moiety from SAM to a suitably placed methyl acceptor.
MATERIALS AND METHODS
Materials
Ni2+-nitrilotriacetic acid agarose (NTA) resin was purchased from Qiagen (Valencia, CA). Benzamidine-Sepharose and Superdex-200 FPLC columns were obtained from Amersham Biosciences (Piscataway, NJ). Cryoloops were purchased from Hampton Research (Laguna Niguel, CA).
Protein Expression and Purification
Alfalfa (Medicago sativa) CCoAOMT was cloned into the Escherichia coli expression vector pET-15b (Novagen, Madison, WI). The CCoAOMT construct was transformed into E. coli BL21(DE3). Transformed E. coli were grown at 37°C in terrific broth containing 100 μg/mL ampicillin until A600 = 1.0. After induction with 0.5 mm isopropyl-β-1-thio-galactopyranoside, the cultures were grown for 6 h at 25°C. Cells were pelleted, harvested, and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0; 500 mm NaCl; 20 mm imidazole, pH 8.0; 20 mm β-mercaptoethanol; 10% [v/v] glycerol; and 1% [v/v] Tween 20). After sonication and centrifugation, the supernatant was passed over a Ni2+-NTA column (Qiagen), washed with 10 bed volumes of lysis buffer, 10 bed volumes of wash buffer (50 mm Tris-HCl, pH 8.0; 500 mm NaCl; 20 mm imidazole, pH 8.0; 20 mm β-mercaptoethanol; and 10% [v/v] glycerol), and the His-tagged protein was eluted with elution buffer (50 mm Tris-HCl, pH 8.0; 500 mm NaCl; 250 mm imidazole, pH 8.0; 20 mm β-mercaptoethanol; and 10% [v/v] glycerol). Incubation with thrombin during dialysis for 24 h at 4°C against 25 mm HEPES, pH 7.5; 100 mm NaCl; and 1 mm dithiothreitol (DTT) removed the N-terminal His-tag. Dialyzed protein was reloaded onto a Ni2+-NTA column to remove cleaved His-tag followed by thrombin depletion using a benzamidine-Sepharose column (Pharmacia Biotech, Piscataway, NJ). Gel filtration of a Superdex-200 column (Amersham Biosciences) equilibrated with 25 mm HEPES, pH 7.5; 100 mm NaCl; and 1 mm DTT provide >98% pure protein. Fractions containing the protein of interest were pooled and concentrated to approximately 10 mg/mL in the presence of 1 mm SAM and stored at −80°C. Selenomethionine-derivatized protein was obtained from E. coli grown in minimal media with appropriate amino acids and selenoMet (Sigma-Aldrich, St. Louis) added (Doublie, 1997). Expression and purification steps were carried out as described for unsubstituted wild-type enzyme.
Metal Dependence
Purified CCoAOMT was dialyzed against 25 mm HEPES, 100 mm NaCl, and 5 mm EDTA for 12 h. The protein was then concentrated to approximately 2 mg/mL. Typical reactions were run in 50-μL volumes containing 250 mm HEPES, pH 7.5; 50 mm NaCl; 0.1 mm adenosyl-l-Met-14C-methyl; 0.1 mm caffeoyl CoA; approximately 50 μg of CCoAOMT; and no added metal or 2 mm Ca2+, Mg2+, Zn2+, or Mn2+. CCoAOMT protein untreated with EDTA was assayed using 50 mm NaCl, 0.1 mm adenosyl-l-Met-14C-methyl, 0.1 mm caffeoyl CoA, approximately 50 μg of CCoAOMT, and 2 mm Mg2+. Reactions were allowed to proceed for 1 h at room temperature after which time 10 μL of 3 m NaOH was added to hydrolyze the thioester bond. After vortexing the solution, 40 μL of 1 m HCl was added to neutralize the NaOH and to protonate the liberated caffeic and ferulic acids. Finally, the 14C-labeled products were extracted with 200 μL of ethyl acetate and subjected to scintillation counting. Specific activity expressed as counts per microgram of CCoAOMT for untreated CCoAOMT assayed in the presence of 2 mm Mg2+ was set to 100% and all other reactions were then referenced to this value.
Protein X-Ray Crystallography
Crystals of CCoAOMT were grown by vapor diffusion in hanging drops consisting of a 1:1 mixture of protein and crystallization buffer (CCoAOMT: 12.5% [w/v] polyethylene glycol 8000; 0.05 m 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid, pH 8.5; 0.20 m calcium acetate; and 2 mm DTT; at 15°C). Cryo protectants consisted of 20% (v/v) glycerol, 20% (w/v) polyethylene glycol 8000, 0.2 m calcium acetate, and 0.05 m 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid, pH 8.5. Crystals were soaked in 1 mm of SAM prior to freezing.
The CCoAOMT structure was determined using x-ray diffraction data collected at 105 K on beamline FIP-BM30A (Roth et al., 2002) at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Data reduction was accomplished using the HKL suite (Otwinowski and Minor, 1997) in combination with PROW (Bourgeois, 1999) for the two-dimensional deconvolution of overlapping reflections due to the unusually large unit cell. For crystals with a significant mosaicity (caffeoyl CoA), the deconvolution procedure was less successful, and therefore data completeness was reduced. This occurred mostly along the third axis of the reciprocal lattice. Finally, due to the structural differences with other known OMTs, including mammalian catechol OMT, structure determination by the molecular replacement method was not possible. Therefore, a dataset at 3.1 Å resolution was first collected with a selenomethionine derivative crystal near the Se absorption edge to maximize the anomalous signal. The space group was C2221 (a = 60.84 Å, b = 136.69 Å, c = 335.13 Å) with potentially 4 molecules/asymmetric unit. Twenty-seven of the 32 Se atom sites were found by direct methods with the Shake-and-Bake program (Weeks and Miller, 1999). These sites were refined with MLPHARE (Computational Collaborative Project Number 4, 1979). After solvent flattening with the DM program (Cowtan, 1994), the resultant electron density map was sufficient for the unambiguous modeling of residues 21 to 247 of CCoAOMT and a SAH molecule of protein chain A with the O molecular modeling package (Jones et al., 1993). Two other protein chains (B and C) were then located with the AMoRe program (Navaza, 1994) and the last protein chain (polypeptide chain D), which is highly disordered, by manual matching of the remaining sites, using this first model as a template. Molecule A forms a dimer with molecule C, related by a noncrystallographic 2-fold axis, and molecules B and D form physiological dimers with crystallographic 2-fold axes. A 2.45-Å native dataset was collected at the Stanford Synchrotron Radiation Laboratory (SSRL, Menlo Park, CA) and reduced with the HKL suite. This dataset was used for refinement of the initial model by simulated annealing with the CNS program (Brunger et al., 1998). Two datasets at 2.65- to 2.7-Å resolution were also collected at ESRF on crystals soaked in 1 mm caffeoyl CoA and 1 mm 5-hydroxyferuloyl CoA for 2 and 24 h, respectively, in addition to SAM. Data reduction was accomplished with the HKL suite and refinement with CNS. In all refinements, the four molecules in the asymmetric unit were treated independently. Density was observed in the Fo-Fc map in which the reaction product (feruloyl CoA and sinapoyl CoA, respectively) were modeled (pdb id: 1sui and 1sus, respectively). Due to the activity of CCoAOMT in solution and in crystals, SAM has undergone methyl transfer, resulting in the appearance of SAH in all of the structures examined thus far. Whereas comparative refinement of SAM and SAH in the active site did not help to discriminate at this resolution, weak positive density observed in the Fo-Fc map close to the caffeoyl moiety indicated that the methyl group is indeed transferred from SAM to the substrate. No significant structural differences were observed between native and feruloyl/sinapoyl CoA complexes (0.25–0.29 Å root mean square displacement) with the most significant structural differences located in the N-terminal portion of CCoAOMT and the long loop between Val-197 and Lys-207, demonstrating unperturbed secondary and tertiary structures upon binding of the hydroxyl-bearing substrate (Table II).
Table II.
Crystallographic data, phasing, and refinement statistics for alfalfa CCoAOMT
The native dataset was collected at SSRL on beamline 9.2. The Se anomalous and caffeoyl (product feruoyl) CoA datasets were collected at ESRF on beamline FIP-BM30A. Data in parentheses are for the last resolution shell.
| Dataset | ||||
|---|---|---|---|---|
| Data Collection | ||||
| Sample | Native | Se Anomalous Dataset | Caffeoyl CoA | 5-Hydroxyferuloyl CoA |
| Wavelength (≈) | 1.08 | 0.979 | 0.979 | 0.979 |
| Resolution (≈) | 2.45 | 3.04 | 2.65 | 2.70 |
| Total reflections | 156,080 | 68,152 | 81,794 | 75,450 |
| Unique reflections | 48,750 | 49,777 | 34,905 | 35,644 |
| Completeness (%) | 94.6 (97.7) | 92.7 (79.0) | 85.7 (43.0) | 94.2 (77.0) |
| I/σ | 20.1 (3.0) | 10.0 (1.36) | 22.3 (1.03) | 19.6 (1.36) |
| Rsym (%)a | 6.0 (37.0) | 7.0 (14.8) | 6.3 (24.9) | 7.6 (21.8) |
| Redundancy | 3.2 (1.83) | 1.4 (1.05) | 2.34 (0.68) | 2.11 (1.27) |
| Refinement | ||||
| Rcryst (%)b | 26.0 | – | 23.6 | 24.3 |
| Rfree (%)c | 30.1 | – | 28.8 | 29.1 |
| Protein atoms | 7,210 | – | 7,210 | 7,210 |
| Ligand atoms | 112 | – | 170 | 171 |
| Water molecules | 66 | – | 66 | 66 |
| Rmsd bond lengths (≈) | 0.008 | – | 0.009 | 0.008 |
| Rmsd bond angles (o) | 1.4 | – | 1.5 | 1.4 |
| <B-factor> protein (≈2) | 52.8 | – | 63.1 | 55.5 |
Rsym = Σ|Ih − <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalent reflections.
Rcryst = Σ|Fobs − Fcalc|/ΣFobs, where summation is over the data used for refinement.
Rfree was calculated using 5% of data excluded from refinement.
Accession Numbers
The accession numbers for the sequences shown in Figure 2 are as follows: CCoAOMTs from alfalfa (AAC28973.1), Populus tremuloides (AAA80651.1), Vitis vinifera (CAA90969.1), Eucalyptus gunnii (CAA72911.1), tobacco (Nicotiana tabacum; AAC49913.1), parsley (Petroselinum crispum; CAA90894.1), and Populus kitakamiensis (BAA19102.1); Ma4OMT from Streptomyces mycarofaciens (Q00719); the soluble part of four animal catechol OMTs from Mus musculus (AAC33334.1), Rattus norvegicus (CAA78276.1), Sus scrofa (Q99028), Homo sapiens (AAH11935.1); and COMT (AAB46623.1), ChOMT (AAB48059.1), and IOMT (AAC49928.1) from alfalfa. The other genes referred to in the text are QOMT from Myxococcus xanthus (AAC44130.1) and Ma8OMT from Streptomyces glaucescens (P39896).
Acknowledgments
We thank the staff of the SSRL macromolecular crystallography team, members of the Noel laboratory for technical assistance, and Lynn Gregory at the Institut de Biologie Structurale for her careful proofreading. We also acknowledge the ESRF for provision of synchrotron radiation facilities. Finally, we thank Thomas Vogt of Leibniz-Institute of Plant Biochemistry (Halle) and Milton Stubbs of Martin-Luther-Universität Halle-Wittenberg for communicating their results prior to publication.
This work was supported by the National Science Foundation (grant no. 0236027 to J.P.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.048751.
References
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier M-T, Monties B, Fritig B, Legrand M (1995) Altered lignin composition in transgenic tobacco expressing O-methlytransferase sequences in sense and antisense orientation. Plant J 8: 465–477 [Google Scholar]
- Bourgeois D (1999) New processing tools for weak and/or spatially overlapped macromolecular diffraction patterns. Acta Crystallogr D Biol Crystallogr 55: 1733–1741 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, et al (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Computational Collaborative Project Number 4 (1979) A suite of programs for protein crystallography. SERC, Daresbury Laboratory, Warrington, UK
- Cowtan K (1994) dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31: 34–38 [Google Scholar]
- Doorsselaere JV, Baucher M, Chognot E, Chabbert B, Tollier MT, Petit-Conil M, Leplé JC, Pilate G, Cornu D, Monties B, et al (1995) A novel lignin in poplar trees with a reduced caffeic acid/ferulic acid O-methyltransferase activity. Plant J 8: 855–864 [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2000) Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa (Medicago sativa L.): impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Bergdoll M, Thion L, Erard M, Legrand M (2001) Identification of the enzymatic active site of tobacco caffeoyl-coenzyme A O-methyltransferase by site-directed mutagenesis. J Biol Chem 276: 36831–36838 [DOI] [PubMed] [Google Scholar]
- Hopkins DW, Webster EA, Chudek JA, Halpin C (2001) Decomposition in soil of tobacco plants with genetic modifications to lignin biosynthesis. Soil Biol Chem 33: 1455–1462 [Google Scholar]
- Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5: 224–229 [DOI] [PubMed] [Google Scholar]
- Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36: 1–10 [DOI] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Balance GM, Ni W, Stürzer C, Dixon RA (1998) Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1993) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 49: 148–157 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E (1989) Elicitor induced S-adenosyl-1-methionine: caffeoyl-CoA 3-O-methyltransferase from carrot cell suspension cultures. Plant Sci 60: 21–25 [Google Scholar]
- Li L, Osakabe Y, Chandrashekhar PJ, Chiang VL (1999) Secondary xylem-specific expression of caffeoyl-coenzyme A 3-O-methyltransferase plays an important role in the methylation pathway associated with lignin biosynthesis in loblolly pine. Plant Mol Biol 40: 555–565 [DOI] [PubMed] [Google Scholar]
- Martz F, Maury S, Pincon G, Legrand M (1998) cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyltransferase, a lignin biosynthetic enzyme. Plant Mol Biol 36: 427–437 [DOI] [PubMed] [Google Scholar]
- Meyermans H, Morrell K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devresse B, Van Beeumen J, Marita JM, et al (2000) Modifications in lignin and accumulation of phenolic glycosides in poplar xylem upon down-regulation of caffeoyl coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Otwinowski Z, Minor W (1997) Methods in enzymology 276. In CW Carter Jr, RM Sweet, eds, Macromolecular Crystallography, Part A. Academic Press, London, pp 307–326
- Pakusch AE, Kneusel RE, Matern U (1989) S-Adenosyl-1-methionine: trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys 271: 488–494 [DOI] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Pincon G, Maury S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2001) Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Pospiech A, Bietenhader J, Schupp T (1996) Two multifunctional peptide synthetases and an O-methyltransferase are involved in the biosynthesis of the DNA-binding antibiotic and antitumour agent saframycin Mx1 from Myxococcus xanthus. Microbiol 142: 741–746 [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW (1974) Chemical and biological evolution of nucleotide-binding protein. Nature 250: 194–199 [DOI] [PubMed] [Google Scholar]
- Roth M, Carpentier P, Kaïkati O, Joly J, Charrault P, Pirocchi M, Kahn R, Fanchon E, Jacquamet L, Borel F, et al (2002) FIP: a highly automated beamline for multiwavelength anomalous diffraction experiments. Acta Crystallogr D Biol Crystallogr 58: 805–814 [DOI] [PubMed] [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT (1980) Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18: 259–288 [Google Scholar]
- Vidgren J, Svensson LA, Liljas A (1994) Crystal structure of catechol O-methyltransferase. Nature 368: 354–357 [DOI] [PubMed] [Google Scholar]
- Vogt T (2004) Regiospecificity and kinetic properties of a plant natural product O-methyltransferase are determined by its N-terminal domain. FEBS Lett 561: 159–162 [DOI] [PubMed] [Google Scholar]
- Weeks CM, Miller R (1999) The design and implementation of SnB v2.0. Appl Cryst D 32: 120–124 [Google Scholar]
- Ye ZH, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Iii WH, Negrel J, Ye ZH (1998) Dual methylation pathways in lignin biosynthesis. Plant Cell 10: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, He X-Z, Dixon RA, Noel JP (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer J-L, Dixon RA, Noel JP (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]