Abstract
Root iron mobilization genes are induced by iron deficiency downstream of an unknown signaling mechanism. The FER gene, encoding a basic helix-loop-helix domain protein and putative transcription factor, is required for induction of iron mobilization genes in roots of tomato (Lycopersicon esculentum). To study upstream regulatory events of FER action, we examined the control of FER gene and FER protein expression in response to iron nutritional status. We analyzed expression of the FER gene and FER protein in wild-type plants, in mutant plants with defects in iron uptake regulation, and in 35S transgenic plants that overexpressed the FER gene. An affinity-purified antiserum directed against FER epitopes was produced that recognized FER protein in plant protein extracts. We found that the FER gene and FER protein were consistently down-regulated in roots after generous (100 μm, physiologically optimal) iron supply compared to low (0.1 μm) and sufficient (10 μm) iron supply. FER gene and FER protein expression were also occasionally down-regulated at sufficient compared to low iron supply. Analysis of FER protein expression in FER overexpression plants, as well as cellular protein localization studies, indicated that FER was down-regulated by high iron at the posttranscriptional level. The FER protein was targeted to plant nuclei and showed transcriptional activation in yeast (Saccharomyces cerevisiae). FER protein regulation in the iron accumulation mutant chloronerva indicated that FER protein expression was not directly controlled by signals derived from iron transport. We conclude that FER is able to affect transcription in the nucleus and its action is controlled by iron supply at multiple regulatory levels.
Iron is an essential component for multiple proteins and enzymes. Limiting iron nutrition may result in severe growth retardation and diverse defects in all organisms. Due to the low solubility of iron in aerobic or alkaline conditions, organisms have developed strategies based on iron reduction or iron chelation to mobilize iron for increased uptake across cellular membranes. Since iron can also have toxic effects, the uptake of iron is tightly regulated in response to iron availability and requirement. Structural genes for iron mobilization have been well characterized in lower and higher organisms and are generally induced upon iron deficiency and down-regulated upon high iron (Crosa, 1997; Andrews et al., 1999; Kaplan, 2002; Van Ho et al., 2002; Curie and Briat, 2003). These findings indicate iron-responsive regulatory mechanisms acting at the transcriptional level.
To date, transcription factors that control iron mobilization genes are mainly characterized in bacteria and lower eukaryotes. In bacteria, the protein FUR is a universal transcription factor that represses the genes for iron transport and iron metabolism, acting both as an intracellular iron sensor and as a regulator (Bagg and Neilands, 1987; Escolar et al., 1999). In yeast (Saccharomyces cerevisiae), the transcription factor Aft1 up-regulates the high-affinity iron transport system upon iron deficiency and is differentially localized in the cell in response to iron (Yamaguchi-Iwai et al., 1995, 1996, 2002). The majority of vertebrate iron regulation research has centered on the posttranscriptional control mediated by the iron regulatory proteins IRP1 and IRP2 (for review, see Pantopoulos, 2004). Transcriptional regulators for important iron deficiency-induced iron transport proteins, such as NRAMP2 in the mammalian duodenum, however, are not described (Kaplan, 2002).
In tomato (Lycopersicon esculentum), the FER gene encoding a basic helix-loop-helix (bHLH) protein and putative transcription factor was found essential for up-regulation of iron mobilization responses in the root (Ling et al., 2002). fer mutant plants are iron deficient and show severe leaf chlorosis (Brown et al., 1971; Brown and Ambler, 1974). Tomato plants mobilize iron through reduction and uptake of Fe2+. FRO2, a membrane-bound iron reductase of the FRO family, and the divalent metal transporter IRT1 are essential components for iron mobilization in dicot plants, as shown for Arabidopsis (Arabidopsis thaliana; Eide et al., 1996; Robinson et al., 1999; Vert et al., 2002). fer mutant plants do not reduce iron and are not able to induce expression of LeFRO1, LeFRO-TC124302, and LeIRT1 in the root, the homologs of AtFRO2 and AtIRT1 (Bereczky et al., 2003; Li et al., 2004; Bauer et al., 2004). fer mutant plants are incapable of mobilizing sufficient iron from the soil into the root. FER gene action was restricted to the root, where it may control mobilization of iron from the soil into the root epidermis and in the vascular cylinder to xylem or phloem (Ling et al., 2002). The mechanisms by which the FER protein regulates iron uptake in response to iron have not been investigated. FER gene expression levels were previously found to be similar in roots grown under deficient (0.1 μm) and sufficient (10 μm) iron supply (Ling et al., 2002). Generous (100 μm) iron supply conditions have not been tested. Induced expression of downstream iron mobilization genes was only found in iron-starved roots, but not generally in roots or leaves of transgenic plants that constitutively expressed FER (Bereczky et al., 2003). It was concluded that the iron regulator FER must be more active for the induction of downstream genes at low iron supply than at high iron supply, and that FER mRNA or FER protein alone was either not stable in leaves, not stable at high iron supply, or not sufficient when alone. From the regulation of FER, it might be possible to deduce the signals mediating iron regulation. To gain further insight into the mechanisms that control the FER gene and FER protein, we investigated their expression at the cellular and whole-organ level. Our results suggest that FER action was controlled at the transcriptional and posttranslational level by iron.
RESULTS
FER Gene Expression in Response to Iron Availability
The FER gene mediates iron mobilization when plants are exposed to iron deficiency but not at high iron supply (Ling et al., 2002). Differential FER gene expression in response to iron availability might be one possibility to control FER action. The FER gene was previously shown to be expressed at a similar level upon low (0.1 μm FeNaEDTA in the medium) and sufficient (10 μm FeNaEDTA in the medium) iron supply, suggesting that FER mRNA might not be regulated at the transcriptional level (Ling et al., 2002). However, the effect of a generous iron supply, such as of 100 μm, on FER gene expression was previously not tested.
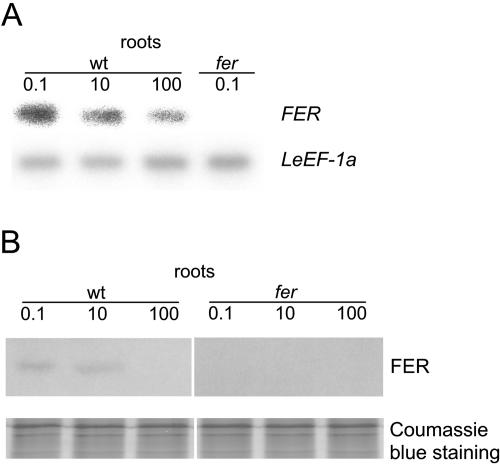
Here, we compared the expression levels of the FER gene in plants grown at low (0.1 μm FeNaEDTA), sufficient (10 μm FeNaEDTA), and generous (100 μm FeNaEDTA) iron supply. We observed that plants grew well in the hydroponic condition when supplied with 10 or 100 μm FeNaEDTA in Hoagland medium, whereas at 0.1 μm FeNaEDTA, plants developed leaf chlorosis (data not shown). Since 100 μm is the regular concentration of iron in multiple plant growth media (Duchefa Biochemie, Haarlem, The Netherlands), this concentration can be regarded as physiologically optimal. We found highest FER expression in response to iron deficiency. At sufficient iron supply, FER gene expression was either decreased compared to low iron supply, in two out of four experiments (Fig. 1A; see also Fig. 6A, wild-type lanes), or at a similar level, in two out of four experiments, as found previously by Ling et al. (2002). At generous iron supply, the FER transcript level was consistently down-regulated (Fig. 1A; see also Fig. 6A). No signal was obtained in the fer mutant plants, regardless of iron supply, due to the presence of a large insertion of approximately 4 kb in between the binding sites for oligonucleotides used in the reverse transcription (RT)-PCR experiments (Fig. 1A; Ling et al., 2002). Taken together, FER mRNA levels responded to different iron availability conditions with a marked down-regulation at generous iron supply.
Figure 1.
Regulation of FER gene and FER protein expression by iron availability in roots. Wild-type and fer mutant plants were grown in the presence of 0.1, 10, or 100 μm FeNaEDTA. A, Semiquantitative RT-PCR analysis of FER mRNA levels in tomato roots. FER expression levels were normalized according to the constitutively expressed LeEF-1a gene. FER signals were absent in the fer plants due to the presence of an insertion within the region to be amplified. B, Western-blot analysis on total protein extracts; 9 μg protein were loaded in each lane. Coomassie Blue staining was used to demonstrate equal loading with proteins.
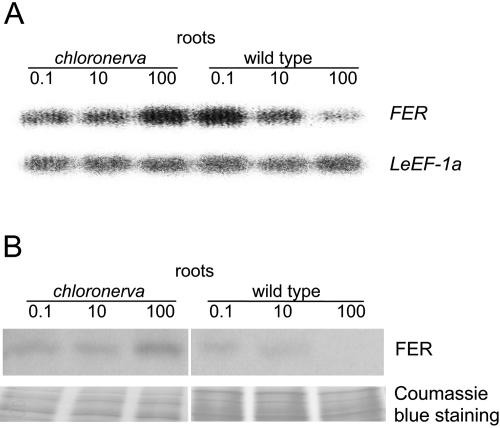
Figure 6.
LeFER expression in chloronerva plants. A, Semiquantitative RT-PCR analysis of FER mRNA levels in roots from chloronerva and wild-type plants grown under deficient (0.1 μm), sufficient (10 μm), and generous (100 μm) iron supply. FER transcript abundance is normalized according to the constitutively expressed LeEF-1a gene. B, Western-blot analysis using anti-N-FER antiserum on total root extracts from chloronerva and wild-type plants grown under 0.1, 10, or 100 μm FeNaEDTA; 9 μg protein were loaded in each lane. Coomassie Blue staining was used to demonstrate equal loading with proteins.
FER Protein Expression in Response to Iron Supply
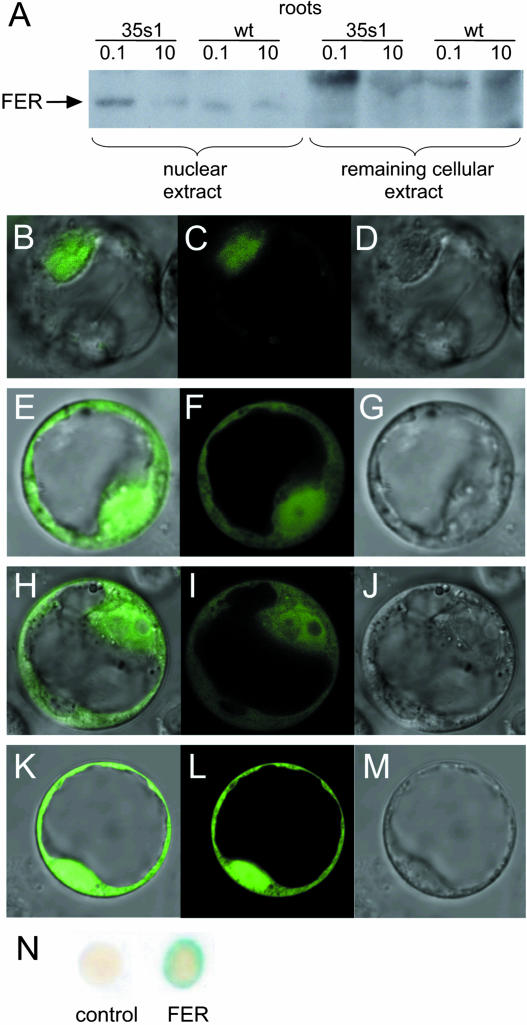
To check whether FER protein levels parallel FER mRNA expression, we developed an affinity-purified polyclonal anti-FER antiserum from rabbit directed against the N-terminal FER peptide, excluding the helix-loop-helix domain (N-FER). This serum was hereafter termed anti-N-FER antiserum. Using western-blot analysis, anti-N-FER antiserum recognized N-FER and the whole intact FER protein when expressed in Escherichia coli (data not shown). Western-blot analysis was subsequently conducted on total root and leaf plant protein extracts. In wild-type root protein extracts, a band of 37 kD was immunologically detectable (Fig. 1B). This band was absent in fer mutant root extracts regardless of iron supply (Fig. 1B). This band was also absent in wild-type leaf protein extracts, but detectable in leaf protein extracts of transgenic plants that ectopically expressed the FER gene in leaves (see Fig. 2C). Since 37 kD was the predicted size of the tomato FER protein, these results indicate that the anti-N-FER antiserum detected FER protein. Tomato plants were grown at low (0.1 μm), sufficient (10 μm), and generous (100 μm) iron supply. In wild-type plants, FER protein levels were either similar (in two experiments out of three) or slightly lower (in one experiment out of three) when plants were grown at sufficient compared to deficient iron supply (Fig. 1B; see also Figs. 2A and 6B). At 100 μm FeNaEDTA supply, the amount of FER protein was consistently undetectable in wild type (Fig. 1B; see also Figs. 2A and 6B). Thus, FER protein expression followed a marked down-regulation at generous iron supply and was induced when iron supply was limiting.
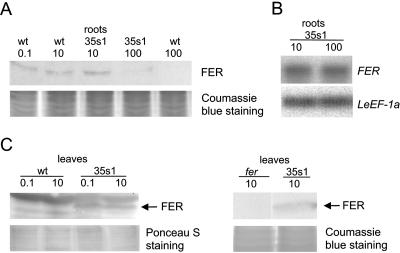
Figure 2.
A, Western-blot analysis using anti-N-FER antiserum on total protein extracts from roots of wild-type and 35s1 transgenic plants that overexpressed the FER gene, grown at 0.1, 10, or 100 μm FeNaEDTA. B, Semiquantitative RT-PCR analysis of FER mRNA levels in tomato roots of 35s1 plants grown at sufficient and high iron supply. FER expression levels were normalized according to the constitutively expressed LeEF-1a gene. C, Western-blot analysis using anti-N-FER antiserum on total protein extracts from leaves of wild-type and 35s1 transgenic plants grown at 0.1 or 10 μm FeNaEDTA (left), and western-blot analysis using anti-FER antiserum (right). FER protein is indicated by an arrow; 9 μg protein were loaded in each lane in A and C. Coomassie Blue or Ponceau S staining was used to demonstrate equal loading with proteins.
FER Gene and FER Protein Expression in Transgenic Plants Constitutively Expressing FER
Previously, we showed functional complementation of transgenic fer mutant plants by overexpression of an intact FER cDNA behind the cauliflower mosaic virus 35S promoter (lines C1-2 = 35s1 and C2-8 = 35s2; Ling et al., 2002; Bereczky et al., 2003). 35s1 plants contained a full-length FER cDNA, whereas that of the 35s2 plants was 21 bp shorter and started with the second ATG start codon, both in the fer mutant background (“Materials and Methods”). 35s1 plants were slightly better complemented than 35s2 plants (Ling et al., 2002). Although the FER gene was expressed constitutively at low and sufficient iron supply in these transgenic plants, molecular iron mobilization responses were stronger at low iron supply than at sufficient iron supply and detectable in roots but not in leaves (Bereczky et al., 2003). These previous results suggested that FER gene action was regulated at the posttranscriptional level, such as via protein stability or protein activation. Here, we investigated this possibility in more detail.
First, we analyzed whether FER mRNA and FER protein were expressed in transgenic 35s1 plants grown upon sufficient and generous iron supply. We found that FER mRNA was produced in 35s1 plant roots regardless of iron supply, as expected from constitutive FER gene expression using the 35S promoter (Fig. 2B). However, the FER protein level was clearly down-regulated at generous versus sufficient iron supply in roots (Fig. 2A).
In leaves of transgenic FER overexpression plants, FER protein was stably expressed (Fig. 2C). Since the anti-N-FER antiserum recognized multiple protein bands in leaf protein extracts, we generated as a control an anti-FER antiserum that was directed against full-length FER protein and affinity purified against C-terminal FER peptides (C-FER). The anti-FER antiserum recognized a single protein band in leaf extracts of 35s1 plants but not of fer mutant plants that corresponded to a 37-kD FER protein (Fig. 2C, right).
Taken together, FER mRNA and FER protein levels were separately regulated in the transgenic 35s FER overexpression plants, indicating control of FER protein at the posttranscriptional level. Moreover, the presence of FER protein was not sufficient for FER action, suggesting additional control at the protein level.
FER Protein Expression in Root Single Nuclei
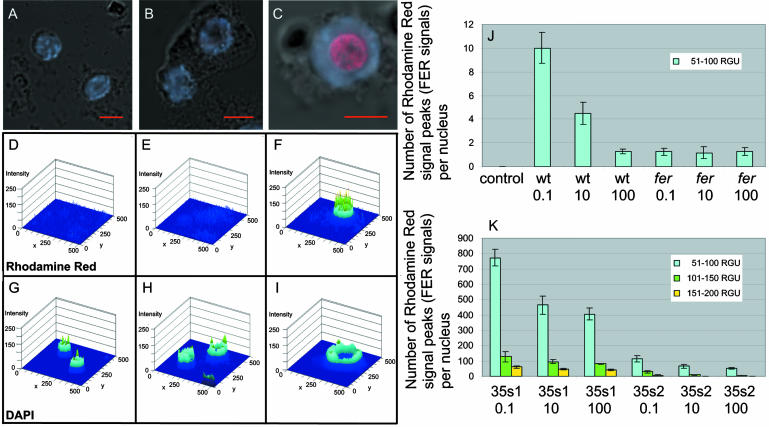
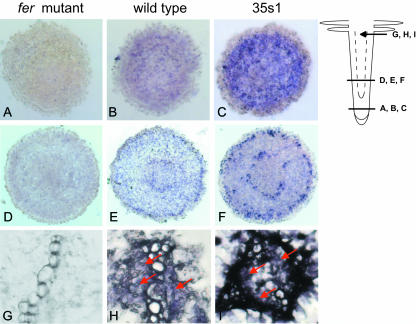
To further analyze FER protein expression, we employed immunolocalization of FER in single root tip nuclei (Houben et al., 1999). Briefly, isolated root tip nuclei were immunolabeled with anti-N-FER antiserum and rhodamine red-labeled secondary antibody. Nuclear genomic DNA was counterstained with 4′,6-diamino-phenylindole (DAPI). The advantage of this method was that immunolocalization signals could be quantified. The intensities of fluorescent signals were examined by laser scanning microscope image software so that fluorescent signal peaks could be counted per nucleus and statistically analyzed (“Materials and Methods”). In fer mutant plants grown at 0.1, 10, and 100 μm FeNaEDTA, fluorescence levels of 1.27, 1.15, and 1.3 signal peaks/nucleus were observed, respectively (Fig. 3, A, D, G, and J). In wild-type plants, low-intensity signal peaks (between 51 and 100 relative gray-scale units; RGU) were observed (Fig. 3, B, E, H, and J). The signals were spread throughout the nucleus without an obvious pattern (Fig. 3B). On average, in nuclei of iron-starved wild-type cells, 10.0 signal peaks were found, in nuclei of iron-sufficient cells, 4.5 signal peaks were found, and in nuclei of generous iron-treated plants, 1.3 signal peaks were found (Fig. 3J). In the 35s1 and 35s2 plants, the numbers of fluorescent signal peaks were about 100 to 400 times higher compared to wild type and of higher intensity (Fig. 3, C, F, I, and K). Signal intensity and number of signals decreased in the nuclei of the transgenic plants when they were exposed to sufficient and generous iron supply (Fig. 3K). For the 35s1 line, signal intensity and number were higher than in the 35s2 line (Fig. 3K). Additionally, the higher abundance of FER in the nuclei of 35s1 and 35s2 roots led to a rearrangement in the subnuclear signal localization. Compared to the diffused signals observed for wild-type nuclei, the transgenic lines exhibited a stronger concentration of the fluorescence signal in the nucleolus (Fig. 3C). In summary, the intensity of FER protein staining in single root tip nuclei suggested an iron-dependent FER expression in the root tips.
Figure 3.
Immunolocalization of FER on single root tip nuclei detected by anti-N-FER antiserum, followed by rhodamine red-coupled anti-rabbit IgG and counterstained with DAPI. A to C, Superimposed confocal images of rhodamine red, DAPI, and differential interference contrast (DIC). D to I, Diagrams, created by the laser scanning microscope 5 image software, presenting the respective rhodamine red (D–F) and DAPI (G–I) fluorescent signal peaks. The images represent the intensities of the fluorescent signals plotted on the same surface as the respective superimposed confocal image in A to C. Different levels of fluorescent signal intensities are represented by different colors: blue, 1 to 50 RGU; blue-green, 51 to 100 RGU; green, 101 to 150 RGU; and yellow, 151 to 200 RGU. The images represent examples for the fer mutant (A, D, and G), wild-type grown at 0.1 FeNaEDTA (B, E, and H), and 35s1 plants grown at 0.1 μm FeNaEDTA (C, F, and I). J, Mean number of fluorescent rhodamine red signal peaks per nucleus for the negative control (secondary antibody omitted), fer mutant, and wild-type plants under all iron supply conditions tested (μm FeNaEDTA). Only signal peaks with intensities between 51 to 100 RGU were counted. Higher signal intensities were not detected for these samples. sd are indicated; n = 10 nuclei. K, Mean number of fluorescent rhodamine red signal peaks per nucleus for 35s1 and 35s2 plants grown under different iron supply conditions (μm FeNaEDTA). Three levels of fluorescent signal intensities were counted, between 51 and 200 RGU. sd are indicated; n = 10 nuclei.
FER Protein Localization in Root Transverse Sections
To check whether FER protein might show differential cellular localization in response to iron supply, we performed immunolocalization of FER in transverse root tip sections (Fig. 4). Wild-type, fer mutant, and transgenic 35s1 plants were grown at deficient, sufficient, and generous iron supply conditions. fer mutant plants displayed no specific FER signals, showing again the specificity of the anti-N-FER antiserum (Fig. 4, A, D, and G). In additional negative controls for secondary antibody specificity, no signals were detected throughout the root sections (data not shown). At generous iron, no signals were detected both in wild-type and in 35s1 plants (data not shown). FER protein expression signals were only detected in wild-type and 35s1 plants grown at sufficient and low iron supply. In these cases, the expression patterns were similar (Fig. 4 shows data for sufficient iron supply). In wild-type plants, FER protein was localized in cells of the root tip except those of the root cap (Fig. 4B). In the root elongation zone, a specific pattern of FER expression was observed, represented by two rings with higher signal concentration (Fig. 4E). The two rings of FER expression signals comprised the cell layer of the epidermis and a cell layer surrounding the vascular cylinder, perhaps the differentiating endodermis. Diffused signals could also be seen in the cortex cells. In the mature root hair zone, the signals were concentrated in the parenchyma cells inside the vascular cylinder (Fig. 4H). The 35s1 roots showed the same pattern of FER staining with more intense signals than the wild-type roots. Despite constitutive expression of FER mRNA in the 35s1 plants (Fig. 2B; for constitutive expression of the 35S promoter in transgenic tomato roots, see also Moghaieb et al., 2004), the FER protein pattern was the same in the 35s1 plants as in wild type (compare Fig. 4, C, F, and I with Fig. 4, B, E, and H). These results suggest that FER protein was expressed in distinct cell types at low and sufficient iron supply, independent of FER mRNA expression. The cellular FER protein expression might be regulated by posttranscriptional in addition to transcriptional control.
Figure 4.
FER immunolocalization using anti-N-FER antiserum on 10-μm paraffin-embedded tomato root cross-sections of fer mutant (A, D, and G), wild-type (B, E, and H), and 35s1 (C, F, and I) plants. A to C, Cross-sections from the meristematic root zone. D to F, Cross-sections from the elongation root zone as indicated on the root scheme. G to I, Magnified views of the central cylinder from cross-sections in the root hair zone. The presence of FER protein was revealed by violet staining from indirect immunolabeling with a secondary antibody coupled to alkaline phosphatase.
Subcellular Localization of FER Protein and Transcriptional Activation
The single-nuclei immunoassays indicated localization of the bHLH domain protein FER in nuclei. We analyzed whether FER protein might show differential localization within the cell in response to iron supply. Therefore, we investigated subcellular localization of FER. Crude nuclear and remaining cellular protein fractions were prepared from root protein extracts of wild-type and 35s1 plants. In western-blot analysis, FER protein was mainly detected in the nuclear, but not in the remaining, cellular fractions of the analyzed lines grown at deficient and sufficient iron supply (Fig. 5A). Therefore, intracellular localization of FER was presumably not dependent on iron concentrations.
Figure 5.
FER protein subcellular localization and transcriptional activation. A, Western-blot analysis using anti-N-FER antiserum on cytosolic and remaining cellular protein fractions from roots of wild-type and 35s1 plants grown at 0.1 or 10 μm FeNaEDTA. The presence of FER protein is indicated by an arrow. B to M, Confocal images of Arabidopsis protoplasts transiently transformed with C-terminal GFP fusion constructs showing GFP fusion protein localization. B to D, Full-length FER∷GFP. E to G, N-FER∷GFP. H to J,C-FER∷GFP. K to M, Free GFP. B, E, H, and K, Superimposed GFP and DIC images. C, F, I, and L, GFP fluorescence. D, G, J, and M, DIC images. N, Yeast one-hybrid assay showing transcription activation capacity of FER. Transcription activation is visualized by a positive LacZ assay (dark color of the colonies). Empty vector was used as a negative control.
To further confirm nuclear localization of FER, we employed a green fluorescent protein (GFP) tagging technique. Arabidopsis protoplasts were transiently transformed with a construct containing 35S∷FER-GFP. The FER∷GFP fusion protein was localized in the nucleus (Fig. 5, B–D). Only very few and light signals were located outside the nucleus. In contrast, free GFP was located in the cytoplasm and the nucleus (Fig. 5, K–M). For the purpose of determining the location of the putative nuclear localization signal in the FER protein, two truncated N- and C-terminal FER∷GFP fusion constructs were tested. Neither of the two protein parts contained the helix-loop-helix domain (N- and C-terminal parts) and was able to trigger GFP localization strictly to the nucleus, as was the case for full-length FER∷GFP (Fig. 5, E–J). Presumably, the presence of a sequence contained in the helix-loop-helix domain was necessary for the proper nuclear localization of the FER protein.
bHLH domain proteins are usually nuclear transcription factors. Since FER was localized to the nucleus, it might act there as a transcription factor. To investigate the potential of FER to activate transcription, we performed a yeast one-hybrid assay. Full-length FER was fused to the GAL4 DNA-binding domain and transferred into yeast cells containing the GAL4-responsive upstream activating sequence fused to a minimal promoter and the lacZ reporter gene. Full-length FER was able to promote reporter gene activity, indicating that FER alone was able to activate transcription in this assay (Fig. 5N). Therefore, FER is presumably able to affect nuclear transcription in plants.
Regulation of FER Protein Expression in chloronerva Mutants
To gain further insight into iron-mediated down-regulation of FER, we examined whether FER mRNA and FER protein expression were influenced by internal or external iron availability. For these experiments, we utilized the chloronerva mutant as a tool. chloronerva plants lack the metal and iron chelator nicotianamine, produced normally by an intact CHLORONERVA gene product (= nicotianamine synthase; Ling et al., 1999). Nicotianamine is required for intracellular and intercellular transport of iron to target components or compartments. Lack of nicotianamine causes local iron deficiencies. Despite sufficient iron supply, chloronerva plants mobilize and take up more iron into the root than wild type (for review, see Scholz et al., 1992). Although extra iron is transported to the shoots, it cannot be delivered to targets in all leaf cells, resulting in intercostal leaf chlorosis. It was previously found that the FER gene was expressed in chloronerva mutant roots (Bereczky et al., 2003). Here, we analyzed iron dependence of FER gene and FER protein expression in chloronerva plants. We observed that at 100 μm iron supply, chloronerva mutant leaves turned green and short-root phenotypes were rescued compared to low iron supply (data not shown). These findings indicate that chloronerva mutants were capable of responding to iron. The wild-type cultivar Bonner Beste (the background of the chloronerva mutant) showed decreased FER mRNA and FER protein expression at generous iron supply (100 μm), similar to the wild-type cultivar Moneymaker (Fig. 6, compare with Fig. 1). In chloronerva mutant plants, however, FER mRNA and FER protein expression levels were both enhanced compared to wild type, which was particularly evident at generous iron supply (Fig. 6). Therefore, external iron supply was not sufficient to down-regulate FER.
DISCUSSION
Here, we analyzed the upstream regulation of the FER gene and FER protein essential for onset of iron mobilization responses at low iron supply. FER protein action is controlled through transcriptional regulation at the mRNA level and posttranscriptional regulation at the protein level, depending on the iron nutritional status. The action of FER is suppressed by high iron, whereas at low iron FER exerts positive control over iron mobilization responses. These findings are in agreement with the evolutionary tendency for negative control of key regulators in cellular processes.
Transcriptional and Posttranscriptional Control of FER
The bHLH domain protein FER is a nuclear protein in plant cells that has transcription factor activity in yeast cells and, presumably, also in plants. As a regulator for iron uptake, FER is supposed to sense the iron nutritional status upstream of its action. We found regulation of FER at different levels. First, the FER gene was regulated at the transcriptional level by iron, whereby gene expression decreased with iron supply. This effect was very consistent when comparing generous iron supply (a physiologically optimal condition) with low or sufficient iron supply conditions. However, iron regulation was not consistent when comparing low and sufficient iron supply. Occasionally, FER mRNA levels were higher at low iron supply versus sufficient iron supply, and, at other times, the levels were similar as was previously described by Ling et al. (2002). Up-regulation of FER mRNA at low and sufficient iron supply compared to generous iron supply suggests that additional upstream iron-regulated transcription factors may control FER gene expression. Second, FER protein was controlled at the posttranscriptional or protein stability level. In wild-type, chloronerva, and fer mutant plants, the amount of FER transcripts correlated well with the amount of FER protein. An exception to this was observed in transgenic lines expressing a functional FER protein using the constitutive 35S promoter in the fer mutant background instead of the natural FER promoter. The 35S promoter was not regulated by iron and resulted in constitutive FER mRNA expression levels. Despite this, FER protein was down-regulated at generous iron in these transgenic lines. Since the transgenic FER cDNA constructs were devoid of the natural 5′ and 3′ untranslated region of the FER gene, down-regulation of FER protein was presumably not the effect of low mRNA stability due to the untranslated regions. Most likely, FER protein was not stable at generous iron supply. Down-regulation of FER protein was not evident only in western-blot experiments. We also observed a discrepancy between previously studied mRNA in situ expression (Ling et al., 2002) and protein in situ expression investigated here. In the undifferentiated root cells of the root tip, FER mRNA in situ signals were detected in all cells in transverse sections, while protein signals were present in all cells except those of the root cap. In the elongating root zone, mRNA signals were mainly present in the epidermis and, to a lesser degree, in cortical cells. However, protein signals were mainly present in the epidermis and in an inner ring of cells, perhaps the differentiating endodermis, surrounding the vascular cylinder, as well as to a lower extent in cortical cells. In the root hair zone, expression of mRNA and protein signals were both confined to parenchymatic cells in the vascular cylinder. Since the same protein expression pattern was observed between plants expressing FER behind its natural promoter and behind the constitutive 35S promoter, we suggest that FER protein was differentially stable in different root tissues. The root cells that express FER protein seem relevant for regulation of iron uptake at the root tip, such as the epidermis, the developing endodermis, and the vascular parenchyma. Third, FER protein was controlled at the level of protein action. Ectopic expression of the FER gene in roots grown upon sufficient iron supply or in leaves did not result in elevated expression of FER gene-dependent LeIRT1 or LeNRAMP1 genes as shown by Bereczky et al. (2003). Here, we showed that, in these cases, FER protein was produced. Despite that, FER was not sufficient for inducing the downstream responses. For its action as a transcription factor, FER might require an additional protein-binding partner. bHLH domain proteins bind DNA as hetero- or homodimers. Alternatively, FER might be activated or inactivated by posttranslational modification.
Control of FER at different levels may allow a rapid and fine-tuned adaptation to changing iron requirements. Levels of active FER protein appear to be controlled more tightly than the levels of FER mRNA. Available FER mRNA may represent a reserve for new protein production even under conditions of generous iron supply, where FER protein is rapidly degraded or not produced. At sufficient iron supply, control of FER protein action seemed more important than control through protein production or stability. Interestingly, protein stability control was also discussed for AtIRT1 and AtFRO2, 2 essential components for iron mobilization in Arabidopsis (Connolly et al., 2002, 2003), and might be a general feature involved in plant iron regulation.
Iron Availability Signals Regulating FER
It was previously hypothesized that nicotianamine may act as a sensor for iron availability in the network of events controlled by FER (Bereczky et al., 2003). Increased FER mRNA and FER protein expression were detected at generous iron supply in the chloronerva mutant. Despite high iron concentrations in the environment, FER protein was stable in the chloronerva mutant. Phenotypic analysis showed that chloronerva mutants responded to iron and were able to overaccumulate iron and metals (for review, see Scholz et al., 1992). Generous iron supply partially rescued the chloronerva plants. Despite iron uptake into chloronerva roots, FER mRNA and FER protein levels were elevated, and FER protein was active in inducing iron mobilization genes (see also Bereczky et al., 2003). FER is therefore not likely controlled by a signaling cascade directly emitted from successful iron transport signals.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum) seedlings were grown in a hydroponic system in Hoagland solution according to Stephan and Prochazka (1989). Twelve days after germination, the plants were transferred into Hoagland solution with different iron concentrations: 0.1 μm FeNaEDTA for limiting iron supply conditions, 10 μm FeNaEDTA for sufficient iron supply, and 100 μm FeNaEDTA for generous iron supply, and grown for an additional 8 d before harvesting for further analyses (100 μm FeNaEDTA is physiologically optimal and recommended for multiple plant growth media (see catalog from Duchefa Biochemie, Haarlem, The Netherlands). Plant lines used were fer mutant (T3238fer), chloronerva mutant, and wild-type cultivars Moneymaker and Bonner Beste. Transgenic lines 35s1 and 35s2 contained an intact FER cDNA starting at the first ATG (position 20; AF437878) and second ATG (position 41; AF437878) start codons, respectively, driven by the constitutive cauliflower mosaic virus 35S promoter in the fer mutant background, as described previously (Ling et al., 2002; Bereczky et al., 2003). 35s1 and 35s2 plants were complemented by FER overexpression and grew similar to wild type (Ling et al., 2002).
Gene Expression Analysis
Total RNA was extracted using Invisorb Spin Plant RNA mini kit (Invitek, Berlin) according to the manufacturer's instructions. One microgram of DNase I-treated RNA was used for cDNA synthesis using the RevertAid First Strand cDNA synthesis kit (MBI Fermentas, Burlington, Canada). Semiquantitative RT-PCR was performed as described in Bereczky et al. (2003). The reactions were analyzed by agarose gel electrophoresis and Southern-blot hybridization according to standard procedures. The FER expression signals (5′-tttcggagcgcaaaaggagag-3′ and 5′-cttgattgctggataataggttgtgaaat-3′, amplified in 20 cycles) were normalized according to the constitutive control product of the elongation factor gene LeEF-1a (5′-actggtggttttgaagctggtatctcc-3′ and 5′-cctcttgggctcgttaatctggtc-3′, amplified in 15 cycles).
Recombinant Protein and Antibody Production and Purification
The entire coding region of the FER cDNA (5′-aatggagagtggtaatgcatcaatgg-3′ and 5′-ttagaccaacggagatgtctcgaagt-3′), the region between the first ATG and the helix-loop-helix domain (N-FER; 5′-aatggagagtggtaatgcatcaatgg-3′ and 5′-ttaggctttatccatctttgtgatattaggaact-3′), and the region between the helix-loop-helix domain and the stop codon (C-FER; 5′-aatgaatttcacaacctattatccagcaat-3′ and 5′-ttagaccaacggagatgtctcgaagt-3′) were amplified by PCR and cloned into pCR II plasmid via TA cloning (Invitrogen, Carlsbad, CA). After sequence verification, the fragments were subcloned into the expression vector pET-29a (Novagen, Madison, WI) by using the EcoRI restriction site. Protein expression was performed in the Escherichia coli strain HMS174 with 0.5 mm isopropylthio-β-galactoside induction at OD600 of 0.6 for 3 h at 30°C. The expressed proteins were purified using S-protein Agarose (Novagen) columns according to the manufacturer's instructions. The purified N-FER protein was used to obtain a rabbit polyclonal antiserum by a service facility of the Institute of Plant Genetics and Crop Plant Research. NHS-activated agarose (Amersham, Buckinghamshire, UK) was covalently linked to purified FER protein and used to affinity purify the anti-N-FER antiserum. The purified anti-N-FER antiserum recognized a 37-kD full-length FER protein by western-blot analysis using E. coli-expressed FER protein as well as plant samples. No protein of the correct size was detected in fer mutant protein extracts. Throughout the following sections, the affinity-purified anti-N-FER antiserum is named anti-N-FER antiserum. For western-blot experiments conducted on leaf protein extracts, an additional antiserum was generated, anti-FER antiserum. Anti-FER antiserum was directed against the full-length FER protein and affinity purified against C-FER, following the above procedure. The anti-FER antiserum recognized a single band in leaf and root protein extracts that was of the expected size of FER at 37 kD.
Western-Blot Analysis on Plant Protein Extracts
Total plant protein extracts were obtained as follows: Leaves and roots were harvested and weighed after grinding. The plant material was extracted in 2× Laemmli loading buffer and subsequently centrifuged for 5 min at 10,000g. The amounts of 2× Laemmli buffer added were adjusted according to the weights of ground material. Crude nuclear protein fractions were isolated according to Escobar et al. (2001). Protein concentrations were measured using the 2D-Quant kit (Amersham Biosciences, Uppsala). Equal amounts of the supernatants containing the total protein extracts (9 μg) were denatured at 95°C for 5 min and loaded onto a 12% SDS-polyacrylamide gel for separation. Samples were transferred to nitrocellulose, stained with Ponceau S, and photographed. Subsequently, the membranes were probed with anti-N-FER antiserum or anti-FER antiserum (1:2,000) followed by goat anti-rabbit horseradish peroxidase secondary antibody (Pierce Chemical, Rockford, IL; 1:4,000). Western blots were developed using ECL chemiluminescence detection reagents (Amersham Biosciences) according to the manufacturer's instructions. The accuracy of loading was further controlled by Coomassie Blue staining of protein gels loaded with the same amounts of protein samples (9 μg) as used for western blots.
Immunolocalization on Single Nuclei
Single nuclei were obtained from paraformaldehyde-fixed root tips after cellulase/pectinase enzyme treatment and subsequent cell disruption (Houben et al., 1999). The isolated nuclei were probed with anti-N-FER antiserum (1:200) followed by rhodamine red-conjugated goat anti-rabbit secondary antibody (Dianova, Hamburg, Germany; 1:100) and counterstained with DAPI (Molecular Probes, Eugene, OR). The fluorescent signals were detected by a confocal laser scanning microscope (CLSM 510 Meta; Zeiss, Jena, Germany). For DAPI, the 364-nm line of an argon laser was used for excitation and the emission was measured at 450 to 490 nm. For rhodamine red fluorescence, the excitation used was 543 nm (helium-neon laser) with a band-pass filter at 560 to 600 nm. The numbers of rhodamine red fluorescent signal peaks per nucleus were counted using Zeiss LSM image examiner software after generation of six-step diagrams where the intensity of the signal was plotted over the area of the confocal image. Different levels of relative pixel intensities were presented as RGU: 1 to 50, 51 to 100, 101 to 150, and 151 to 200.
Immunolocalization on Root Cross-Sections
Tomato roots from plants grown under different iron supply conditions were formaldehyde fixed, eosin counterstained, and embedded in Paraplast Plus (Sherwood Medical, St. Louis). Immunolocalization was performed on 10-μm transverse sections using anti-N-FER antiserum (1: 200) followed by anti-rabbit alkaline phosphatase-conjugated secondary antibody (Sigma, St. Louis) according to Smith et al. (1992). The signals were visualized by a nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate toluidine salt color reaction (violet staining) according to Roche Diagnostics (Mannheim, Germany). Images were recorded using an Axioplan 2 imaging microscope (Zeiss).
GFP Localization
Three different FER C-terminal GFP fusion constructs were generated by first amplifying cDNA fragments: 35S∷FER∷GFP, the whole coding sequence of FER (5′-aatggagagtggtaatgcatcaatgg-3′ and 5′-ttagaccaacggagatgtctcgaagt-3′); 35S∷N-FER∷GFP, the N-terminal coding sequence in front of the helix-loop-helix domain (5′-aatggagagtggtaatgcatcaatgg-3′ and 5′-ttaggctttatccatctttgtgatattaggaact-3′); 35S∷C-FER∷GFP, the C-terminal coding sequence behind the bHLH domain (5′-aatgaatttcacaacctattatccagcaat-3′ and 5′-ttagaccaacggagatgtctcgaagt-3′). PCR fragments contained a KpnI and SalI restriction site at the 5′ and 3′ termini, respectively, and were cloned behind the 35S promoter into a modified pFF19 vector that contained mGFP5 (Hofius et al., 2004). The verified constructs were transformed into Arabidopsis (Arabidopsis thaliana) protoplasts according to Reidt et al. (2000). GFP fluorescent signals were detected using a laser scanning microscope (CLSM 510 Meta; Zeiss) by 488-nm argon laser excitation and a band-pass filter at 505 to 525 nm.
Yeast One-Hybrid Assay
A FER∷GAL4 DNA-binding domain fusion construct was created by cloning the EcoRI restriction fragment from the respective pET-29a full-length FER construct (see above) into pGBKT7 (CLONTECH Laboratories, Palo Alto, CA). The verified construct was transformed into the yeast (Saccharomyces Cerevisiae) strain AH109 (CLONTECH) according to Gietz et al. (1992) and grown on synthetic dextrose/Trp medium. The obtained colonies were assayed for lacZ reporter gene activation according to the manufacturer's instructions. Yeast cells transformed with the empty pGBKT7 vector were assayed in parallel and used as a negative control.
Sequence data from this article are available at the EMBL/GenBank data libraries under accession numbers AF437878 (LeFER) and XI4449 (LeEF-1a).
Acknowledgments
We thank Renate Manteuffel for polyclonal antiserum generation and Bernhard Claus for confocal laser scanning microscopy. The help of Andreas Houben with single-nuclei immunolocalization is gratefully acknowledged. Daniel Hofius kindly provided plasmid pFF19G, and Annegret Tewes kindly provided Arabidopsis protoplasts. We are also grateful to Frederik Börnke for assistance with the yeast one-hybrid assay and for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grants in the Emmy Noether program to P.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.054270.
References
- Andrews NC, Fleming MD, Gunshin H (1999) Iron transport across biologic membranes. Nutr Rev 57: 114–123 [DOI] [PubMed] [Google Scholar]
- Bagg A, Neilands JB (1987) Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26: 5471–5477 [DOI] [PubMed] [Google Scholar]
- Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I (2004) Analysis of sequence, map position and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol 136: 4169–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704 [DOI] [PubMed] [Google Scholar]
- Brown JC, Ambler JE (1974) Iron-stress response in tomato (Lycopersicon esculentum) 1. Sites of Fe reduction, absorption and transport. Physiol Plant 31: 221–224 [Google Scholar]
- Brown JC, Chaney RL, Ambler JE (1971) A new mutant inefficient in the transport of iron. Physiol Plant 25: 48–53 [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML (2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH (1997) Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev 61: 319–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF (2003) Iron transport and signalling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar C, Aristizábal F, Navas A, del Campo FF, Fenoll C (2001) Isolation of active DNA-binding nuclear proteins from tomato galls induced by root-knot nematodes. Plant Mol Biol Rep 19: 375a–375h [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181: 6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Hajirezaei MR, Geiger M, Tschiersch H, Melzer M, Sonnewald U (2004) RNAi-mediated tocopherol deficiency impairs photoassimilate export in transgenic potato plants. Plant Physiol 135: 1256–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Wako T, Furushima-Shimogawara R, Presting G, Kunzel G, Schubert I, Fukui K (1999) Short communication: the cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J 18: 675–679 [DOI] [PubMed] [Google Scholar]
- Kaplan J (2002) Strategy and tactics in the evolution of iron acquisition. Semin Hematol 39: 219–226 [DOI] [PubMed] [Google Scholar]
- Li L, Cheng X, Ling HQ (2004) Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol Biol 54: 125–136 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaieb RE, Saneoka H, Fujita K (2004) Shoot regeneration from GUS-transformed tomato (Lycopersicon esculentum) hairy root. Cell Mol Biol Lett 9: 439–449 [PubMed] [Google Scholar]
- Pantopoulos K (2004) Iron metabolism and the IRIE/IRP regulatory system: an update. Ann N Y Acad Sci 1012: 1–13 [DOI] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerstrom M, Czihal A, Tewes A, Ezcurra I, Rask L, Baumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21: 401–408 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Scholz G, Becker R, Pich A, Stephan UW (1992) Nicotianamine: a common constituent of strategy-I and strategy-II of iron acquisition by plants: a review. J Plant Nutr 15: 1647–1665 [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Stephan UW, Prochazka Z (1989) Physiological disorders of the nicotianamine-auxotroph tomato mutant chloronerva at different levels of iron nutrition. I. Growth characteristics and physiological abnormalities related to iron and nicotianamine supply. Acta Bot Neerl 38: 147–153 [Google Scholar]
- Van Ho A, Ward DM, Kaplan J (2002) Transition metal transport in yeast. Annu Rev Microbiol 56: 237–261 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Dancis A, Klausner RD (1995) AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 14: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD (1996) Iron-regulated DNA binding by the AFT1 protein controls the iron regulation in yeast. EMBO J 15: 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Ueta R, Fukunaka A, Sasaki R (2002) Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J Biol Chem 277: 18914–18918 [DOI] [PubMed] [Google Scholar]